Abstract
During infection in the gastrointestinal tract, enterohemorrhagic Escherichia coli (EHEC) O157:H7 is exposed to a wide range of signaling molecules, including the eukaryotic hormones epinephrine and norepinephrine, and bacterial signal molecules such as indole. Since these signaling molecules have been shown to be involved in the regulation of phenotypes such as motility and virulence that are crucial for EHEC infections, we hypothesized that these molecules also govern the initial recognition of the large intestine environment and attachment to the host cell surface. Here, we report that, compared to indole, epinephrine and norepinephrine exert divergent effects on EHEC chemotaxis, motility, biofilm formation, gene expression, and colonization of HeLa cells. Using a novel two-fluorophore chemotaxis assay, it was found that EHEC is attracted to epinephrine and norepinephrine while it is repelled by indole. In addition, epinephrine and norepinephrine also increased EHEC motility and biofilm formation while indole attenuated these phenotypes. DNA microarray analysis of surface-associated EHEC indicated that epinephrine/norepinephrine up-regulated the expression of genes involved in surface colonization and virulence while exposure to indole decreased their expression. The gene expression data also suggested that autoinducer 2 uptake was repressed upon exposure to epinephrine/norepinephrine but not indole. In vitro adherence experiments confirmed that epinephrine and norepinephrine increased attachment to epithelial cells while indole decreased adherence. Taken together, these results suggest that epinephrine and norepinephrine increase EHEC infection while indole attenuates the process.
The intestinal tract is colonized by approximately 1012 commensal bacteria consisting of hundreds of bacterial species, including the genus Escherichia (9, 10, 19). The introduction of pathogenic bacteria such as enterohemorrhagic Escherichia coli (EHEC) O157:H7 into the human gastrointestinal (GI) tract results in colonization of host cells and leads to the onset of bloody diarrhea and hemolytic uremic syndrome (24, 25). EHEC infections progress through a three-step mechanism, the first of which involves adhesion of bacteria to host cells and the formation of microcolonies (23, 46). EHEC infections pose a serious clinical problem as they are often associated with complications and permanent disabilities, including neurological defects, hypertension, and renal insufficiency (35). Understanding the mechanisms underlying EHEC pathogenicity could lead to better approaches for attenuating the deleterious consequences associated with GI tract infections (52).
E. coli O157:H7 is exposed to a wide range of signaling molecules in the GI tract. These include bacterial quorum-sensing molecules such as autoinducer 2 and 3 (AI-2 and AI-3, respectively) that are involved in the regulation of phenotypes crucial for virulence and infection (44, 49). For example, Sperandio et al. (43, 44) have shown that the adhesion of pathogenic E. coli to host cells, which is an important initial step in the infection process, is regulated by these soluble signals. An EHEC luxS mutant that is deficient in the synthesis of AI-2 and AI-3 demonstrated markedly decreased expression of flagella and motility genes required for adherence to epithelial cells (17), and we have found that the direct addition of AI-2 stimulates E. coli biofilm formation (18). The concentration of these bacterium-derived soluble signals is expected to be high in the GI tract, as both pathogenic as well as nonpathogenic commensal bacteria that reside in the GI tract (9) produce AI-2 and AI-3. Other bacterial signaling molecules, such as the stationary phase signal indole (50), that are produced by commensal E. coli (13) are also expected to be present at high concentrations in the GI tract. Indole decreases motility and biofilm formation of nonpathogenic E. coli (13) through SdiA (28) and could be especially important in EHEC infections since it has been shown to regulate virulence genes in enteropathogenic E. coli (4).
Recent studies have also shown that eukaryotic signals in the GI tract also play an important role in EHEC adhesion and infections. Catecholamines such as epinephrine (EPI) and norepinephrine (NE) have been shown to enhance the growth of pathogenic bacteria (15, 16, 33) and production of virulence factors (30, 32). NE has also been shown to increase adhesion of EHEC to cecal mucosa (7), colonic mucosa (19), and ileum (48). Sperandio et al. (44) initially demonstrated that EPI and NE function similarly to AI-3 by activating the virulence genes (e.g., LEE genes) in EHEC (44). Recent studies by Clarke et al. (8) and Walters and Sperandio (49) have also shown that AI-3 and EPI utilize the same receptor (QseC) and suggest a synergistic relationship between these two molecules in the expression of virulence genes.
The goal of this work was to investigate the effect of eukaryotic and prokaryotic extracellular molecules on phenotypes important in EHEC infections. We hypothesized that EHEC infection is influenced to different degrees by the different eukaryotic and prokaryotic molecules present in the GI tract. Therefore, we investigated the effect of three molecules—the eukaryotic hormones EPI and NE and the E. coli stationary phase signal indole—on EHEC chemotaxis, biofilm formation, and adherence to epithelial cells. The molecular basis of the alterations in EHEC physiology upon exposure to EPI, NE, and indole was also investigated using DNA microarrays. Our results suggest molecular-level interactions between prokaryotic and eukaryotic signaling molecules in EHEC colonization. To our knowledge, this is the first report of EHEC chemotaxis, motility, and biofilm formation in response to eukaryotic signaling molecules.
MATERIALS AND METHODS
Bacterial strains, materials, and growth.
E. coli O157:H7 (CDC EDL933) and E. coli TG1 were obtained from ATCC (Manassas, VA) and Stratagene (La Jolla, CA), respectively. Plasmids pCM18 (20) and pDS-RedExpress (Clontech, California) were used to constitutively express the green fluorescent protein (GFP) and the red fluorescent protein (RFP), respectively. Luria-Bertani medium (LB) was used for overnight cultures of E. coli O157:H7 (ATCC, Manassas, VA). LB supplemented with 0.2% (wt/vol) glucose (LB-Glu) was used in all biofilm experiments. EPI and NE were purchased from MP Biomedicals (Irvine, CA). Indole was purchased from Sigma Chemical Co. (St. Louis, MO).
Agarose plug chemotaxis assay.
Chemotaxis experiments in agarose plugs were performed by adapting the procedure of Yu and Alam (55) to include live cells tagged with GFP and dead cells tagged with RFP. Briefly, an overnight culture of E. coli O157:H7 (pCM18) was used to inoculate 10 ml of growth medium (10 g/liter tryptone, 5 g/liter yeast extract, 2 mM MgSO4, 2 mM CaCl2) to a turbidity of ∼0.05 at 600 nm and grown to mid-exponential phase (turbidity at 600 nm of ∼0.7). The chemotaxis compound was also added to the medium during growth. Cells were harvested, washed twice, and resuspended in 1 ml of chemotaxis buffer (1× phosphate-buffered saline [PBS], pH 7.4, 0.1 mM EDTA, 10 mM sodium succinate). In addition, kanamycin-killed E. coli TG1 (pDS-RedExpress) was also added to the cell suspension such that the ratio of live cells to dead cells was 2:1. In parallel, the chemotaxis compound was mixed with a 2% low-melting-point agarose in chemotaxis buffer containing trace amounts of bromophenol blue (to help visualization of the plug) at 55°C. A 10-μl aliquot of the agarose solution containing the chemotaxis compound was added between a glass slide and a raised glass coverslip. Two hundred microliters of the cell suspension was immediately added to the slide, and the migration of bacteria toward or away from the agarose plug was imaged on a Zeiss Axiovert 200 fluorescence microscope (Thornwood, NY). Green and red fluorescence images were obtained every 5 min for 30 min. Images were processed and analyzed using the Zeiss Axiovision software, version 4.3.
Motility and biofilm assays.
Motility assays were performed as described by Sperandio et al. (42). Briefly, overnight cultures of EHEC were subcultured 1:100 in LB medium and grown to a turbidity of ∼1.0 at 37°C. EPI (50 μM), NE (50 μM), or indole (500 μM) was added to the motility agar plates (1% tryptone, 0.25% NaCl, and 0.3% agar), and the sizes of the motility halos were measured after 8 h. Six motility plates were used for each molecule, and the experiment was repeated with three independent cultures (a total of 18 motility plates per molecule).
Biofilm assays were performed in 96-well polystyrene plates according to the protocol described by Pratt and Kolter (37). Cells were grown for 24 h without shaking in the presence of EPI (50 μM), NE (50 μM), or indole (500 μM). Each data point was averaged from 18 replicate wells (six wells from three independent cultures).
Biofilms for microarray experiments were formed on glass wool for RNA extraction and DNA microarray analysis (21). Briefly, an overnight culture of E. coli O157:H7 was used to inoculate 10 g of glass wool in 250 ml of LB-Glu medium to a starting turbidity of ∼0.03. Biofilms (n = 3) were developed at 37°C for 7 h with shaking in the presence of EPI (50 μM), NE (50 μM), or indole (500 μM). Based on our unpublished data showing that extracellular indole levels increase after 4 h of culturing (due to depletion of glucose) and that indole biosynthesis is subject to catabolite repression (22), the glucose in the medium was replenished after 3.5 h for all samples so as to minimize the effects of intracellularly produced indole on the observed gene expression patterns. The glass wool was removed from the culture quickly and gently washed two times in 100 ml of 0.85% NaCl buffer at 0°C (less than 30 s). Biofilm cells were removed from glass wool by sonication for 2 min in 200 ml of 0.85% NaCl buffer at 0°C, cell pellets were stored at −80°C, and RNA was isolated as described previously (21).
RNA isolation and DNA microarrays.
Total RNA was isolated from EHEC biofilms in triplicate, and RNA quality was assessed using gel electrophoresis (39). E. coli Genome 2.0 arrays (Affymetrix, California) containing approximately 10,000 probe sets for all 20,366 genes present in four strains of E. coli, including E. coli O157:H7 EDL933, were used to profile changes in gene expression using one of the triplicate RNA samples for each treatment. Hybridization was performed for 16 h, and the total cell intensity was scaled automatically in the software to an average value of 500. The data were inspected for quality and analyzed according to the procedures described by the manufacturer (1) which include using premixed polyadenylated transcripts of the B. subtilis genes (lys, phe, thr, and dap) at different concentrations. Genes were identified as differentially expressed if the expression ratio (between biofilms treated with EPI, NE, or indole and untreated control biofilm cells) was greater than 2.0 (based on the standard deviation between values measuring relative changes in expression) (38) and the change in the P value was less than 0.05. The differentially expressed genes were annotated using gene ontology definitions available in the Affymetrix NetAffx Analysis Center (http://www.affymetrix.com/analysis/index.affx).
Quantitative RT-PCR.
Quantitative reverse transcription-PCR (RT-PCR) was performed using an iCycler (Bio-Rad, California) real-time PCR unit (28). Approximately 50 ng of total RNA from control or EPI-treated EHEC was used for RT. Gene sequences were downloaded from the ASAP (for a systematic annotation package for community analysis of genomes) website (https://asap.ahabs.wisc.edu/asap/home.php?formSubmitReturn=1), and gene-specific primers were used for lsrA, lsrB, lsrC, and rpoA (housekeeping control) (Table 1). Threshold cycle numbers were calculated using MyiQ software (Bio-Rad), and PCR products were verified using agarose electrophoresis. RT-PCR experiments were initially performed using the same sample used for microarray analysis, followed by verification with replicate samples.
TABLE 1.
Primers used for quantitative RT-PCR
| Gene | ASAP identifier | Forward primer | Reverse primer |
|---|---|---|---|
| lsrA | ABH-0025965 | 5′-AACATCCTGTTTGGGCTGGCAA-3′ | 5′-AAACAAGCGTTCGGTTTCCGCA-3′ |
| lsrB | ABH-0025962 | 5′-AGCATCCTGGCTGGGAAATTGT-3′ | 5′-AAATTCTTTCACCGTGCCGCGT-3′ |
| lsrC | ABH-0025964 | 5′-ACAGCGTTTGGACGCAGTTT-3′ | 5′-ACGCAGGCTGCAATTGCTTT-3′ |
| rpoA | ABH-0028324 | 5′-CGCGGTCGTGGTTATGTG-3′ | 5′-GCGCTCATCTTCTTCCGAAT-3′ |
In vitro adhesion assays.
Adhesion of EHEC to HeLa S3 cells was performed using a previously described protocol (46). HeLa S3 cells (ATCC, Manassas, VA) were routinely cultured and propagated in Dulbecco's modified Eagle's medium with 10% heat-inactivated fetal bovine serum according to standard protocols (ATCC). Low-passage-number HeLa cells were cultured in standard 24-well tissue culture plates and grown at 37°C in a 5% CO2 humidified environment until ∼80% confluence. HeLa cell monolayers were washed three times with sterile PBS to remove unattached cells, and the growth medium was replaced with antibiotic-free Dulbecco's modified Eagle's medium with 10% fetal bovine serum. Approximately 5 × 106 cells of a freshly grown E. coli O157:H7 culture (turbidity of ∼1.0) were added to each well and incubated for 3 h at 37°C in a 5% CO2 environment. Loosely attached cells were removed by washing the wells three times with sterile PBS, and the HeLa cells were lysed in the wells using 0.1% Triton X-100 in PBS. The cell suspension in each well was vigorously vortexed, and serial dilutions of the bacteria were plated on LB plates. Colonies were counted after a 24-h incubation at 37°C.
Microarray data accession number.
The expression data have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE5552.
RESULTS
Effect of EPI, NE, and indole on E. coli O157:H7 chemotaxis, motility, and biofilm formation.
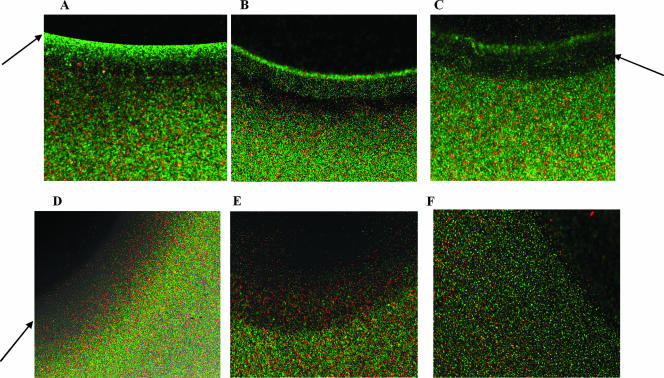
A standard agarose plug assay (55) was modified to include a live and a dead strain, each tagged with a different fluorophore, to create a new assay for studying chemoattraction or repulsion exhibited by EHEC toward EPI, NE, and indole. EHEC expressing GFP and kanamycin-killed E. coli TG1 with RFP were mixed together and introduced into the vicinity of an agarose plug containing the chemotaxis molecule. Diffusion of EPI and NE from the agarose plug resulted in attraction of EHEC and accumulation near the agarose plug while the dead cells were evenly distributed (Fig. 1A and B). The concentration at the edge of the plug was not inhibitory to EHEC as cells were observed attached to the agarose plug after 30 min of exposure. A distinct clearance zone was also detected between the attracted cells and cells in the bulk fluid (i.e., away from the agarose plug), which also suggested a concentration-dependent migration of cells toward EPI and NE. This chemoattractive response was similar to that observed with the positive control Casamino Acids (Fig. 1C).
FIG. 1.
Agarose plug chemotaxis. Attraction or repulsion of E. coli O157:H7 to different chemical agents was determined using a modified agarose plug assay. Fluorescence images from bacteria exposed to 500 μM EPI (A), 500 μM NE (B), 2% Casamino Acids (C), 5,000 μM indole (D), 1 × 10−4 M glycerol (E), and 1× M9 salts solution (F) are shown. Green cells are E. coli O157:H7 expressing GFP, and red cells are kanamycin-killed E. coli TG1 expressing RFP. Fluorescence images were obtained on a Zeiss Axiovert 200 microscope after 30 min using a 10× objective. Data shown are representative images from three independent experiments. Arrows indicate chemoattractant ring (A), clearance zone (C), and chemorepellant band (D).
In contrast, exposure to indole resulted in migration of EHEC away from the agarose plug, and a clearance zone was formed closer to the agarose plug (Fig. 1D); this repulsion was similar to that observed with the negative control glycerol (Fig. 1E). Exposure to a neutral chemical medium (M9 salts) did not result in any attraction or repulsion (Fig. 1F). These results clearly show that EHEC demonstrates chemotactic attraction toward EPI and NE but migrates away from indole.
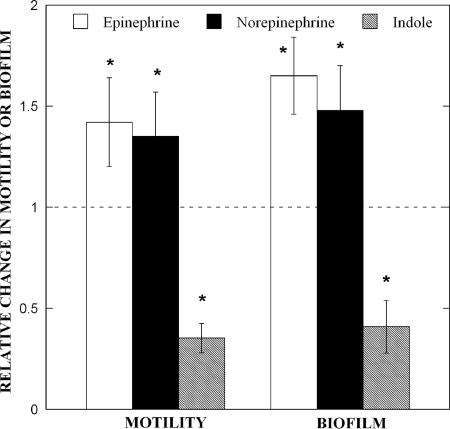
Based on the significant changes in chemotaxis and its relation to cell motility, the effect of EPI, NE, and indole on EHEC motility was also investigated. EPI and NE were used at concentrations of 50 μM based on published reports (44), while indole was used at a higher concentration (500 μM) based on the extracellular concentration of indole (∼400 μM) in stationary phase cultures of E. coli O157:H7 (27). At these concentrations, no significant alteration in the growth rate of EHEC was observed (1.35 ± 0.02 h−1, 1.35 ± 0.02 h−1, and 1.33 ± 0.07 h−1 for EHEC, EHEC with 50 μM EPI, and EHEC with 50 μM NE, respectively), indicating that the observed results are not due to changes in growth rate. Addition of EPI and NE increased motility by 1.4-fold, compared to untreated controls, while indole decreased EHEC motility by 2.8-fold (Fig. 2).
FIG. 2.
EPI, NE, and indole affect E. coli O157:H7 motility and biofilm formation. The relative change in EHEC motility and biofilm upon exposure to EPI (50 μM), NE (50 μM), and indole (500 μM) was determined. Motility data are shown as means ± 1 standard deviation from 18 motility agar plates from three independent experiments. Biofilm data are means ± 1 standard deviation from 36 wells and three independent experiments. The asterisk indicates statistical significance determined using a Student t test (P < 0.01).
Given the changes in motility and their effect on biofilm (53), the effect of these molecules on EHEC biofilm formation was also determined. Figure 2 also shows that EPI and NE resulted in a 1.7- and 1.5-fold increase, respectively (statistically significant at a P value of <0.01 using a Student t test), in EHEC biofilm formation on polystyrene after 24 h. On the other hand, the addition of indole decreased biofilm formation by 2.4-fold after 24 h. Together, our data indicate that EPI, NE, and indole affect EHEC motility and biofilm formation in a manner consistent with their effect on chemotaxis.
Gene expression in EPI-, NE-, and indole-treated E. coli O157:H7 biofilms.
The molecular basis of the effect of EPI, NE, and indole on chemotaxis, motility, and biofilm formation was investigated using DNA microarrays. EHEC biofilms were formed in LB-Glu medium for 7 h on glass wool in the presence of EPI, NE, or indole. The glass wool biofilm model was used to mimic surface-associated short-term growth of EHEC, which is similar to the localized adherence required for infection (41). Genes whose expression was altered by at least twofold were selected and sorted into functional categories (e.g., metabolism or transport) (Table 2), and the expression trends with the different stimuli were analyzed.
TABLE 2.
Summary of changes in E. coli O157:H7 biofilm gene expression
| Gene type or function and name | Relative change in expression (n-fold) after exposure to:a
|
Description | ||
|---|---|---|---|---|
| EPI | NE | IND | ||
| AI-2 group | ||||
| c1993 | 1.4 | 1.4 | 1.6 | Putative integral membrane transport protein |
| lsrA | −9.8 | −5.3 | 1.0 | Fused AI-2 transporter subunits of ABC superfamily; ATP-binding components |
| lsrB | −7.0 | −5.3 | −1.4 | Putative LACI-type transcriptional regulator |
| lsrC | −4.3 | −2.3 | 1.2 | AI-2 transporter |
| lsrD | −3.2 | −1.4 | 1.5 | AI-2 transporter |
| lsrG | −3.2 | −3.0 | −1.1 | AI-2 modifying protein LsrG |
| lsrK | −2.8 | −2.3 | 1.2 | AI-2 kinase |
| lsrR | −3.7 | −3.7 | 1.1 | lsr operon transcriptional repressor |
| Phosphate related | ||||
| phnD | −2.1 | 1.2 | 2.1 | Phosphonates-binding periplasmic protein precursor |
| phnG | −1.1 | 1.5 | 2.3 | PhnG protein |
| phnP | −1.2 | 1.3 | 2.1 | PhnP protein |
| phoB | 2.1 | 3.0 | −2.6 | Phosphate regulon transcriptional regulatory protein phoB |
| phoH | −4.9 | −4.6 | −1.9 | PhoH protein |
| phoR | 2.6 | 3.5 | −1.6 | Phosphate regulon sensor protein phoR |
| phoU | 1.9 | 2.1 | −3.7 | Phosphate transport system protein phoU |
| pstA | 2.5 | 2.6 | −3.2 | Phosphate transport system permease protein pstA |
| pstB | 1.3 | 1.5 | −3.7 | Phosphate transport ATP-binding protein pstB |
| pstC | 2.6 | 3.0 | −2.3 | Phosphate transport system permease protein pstC |
| pstS | 1.1 | 1.1 | −4.6 | Phosphate-binding periplasmic protein precursor |
| purE | 3.0 | 3.7 | 1.4 | Phosphoribosylaminoimidazole carboxylase catalytic subunit |
| purK | 1.2 | 2.6 | 1.6 | Phosphoribosylaminoimidazole carboxylase ATPase subunit |
| purT | −1.4 | 2.5 | 2.1 | Phosphoribosylglycinamide formyltransferase 2 |
| Hydrognease | ||||
| hyaA | −3.0 | −3.2 | 1.1 | Hydrogenase-1 large chain |
| hyaB | −4.0 | −3.5 | 1.2 | Hydrogenase-1 operon protein hyaE |
| hyaC | −3.7 | −3.0 | 1.1 | Hydrogenase-1 operon protein hyaF |
| hyaD | −3.2 | −2.8 | 1.2 | Hydrogenase-1 small chain precursor |
| hyaE | −3.5 | −2.5 | 1.3 | Hydrogenase-1 large chain |
| hyaF | −3.0 | −2.6 | 1.1 | Hydrogenase-1 operon protein hyaE |
| PTS | ||||
| mtlA | −13.9 | −11.3 | 1.0 | PTS system; mannitol-specific IIABC component |
| manX | −4.6 | −3.5 | 1.1 | PTS system; mannose-specific IIAB component |
| manY | −2.8 | −2.3 | 1.1 | PTS system; mannose-specific IIC component |
| manZ | −2.8 | −2.1 | 1.2 | PTS system; mannose-specific IID component |
| fruB | −1.7 | −2.1 | 1.2 | PTS system; fructose-specific IIA/FPr component |
| Glycerol related | ||||
| glgC | −2.5 | −3.0 | −1.3 | Glucose-1-phosphate adenylyltransferase |
| glpA | −4.9 | −1.7 | 1.7 | Anaerobic glycerol-3-phosphate dehydrogenase subunit A |
| glpD | −9.2 | −18.4 | 1.3 | sn-Glycerol-3-phosphate dehydrogenase (aerobic) |
| glpF | −5.7 | −5.7 | 1.3 | Glycerol uptake facilitator protein |
| glpK | −7.5 | −6.1 | 1.1 | Glycerol kinase |
| glpQ | −2.1 | −2.5 | −1.1 | Glycerophosphoryl diester phosphodiesterase; periplasmic precursor |
| glpR | −1.6 | −2.1 | −1.1 | Glycerol-3-phosphate regulon repressor |
| ugpB | −7.0 | −6.1 | −1.4 | Glycerol-3-phosphate-binding periplasmic protein precursor |
| Histidine biosynthesis | ||||
| hisA | −2.1 | −2.8 | −1.6 | 1-(5-Phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase |
| hisB | −2.3 | −2.6 | −2.1 | Bifunctional: histidinol-phosphatase (N terminus) and imidazoleglycerol-phosphate dehydratase (C terminus) |
| hisC | −4.0 | −4.6 | −2.8 | Histidinol-phosphate aminotransferase |
| hisD | −4.3 | −4.3 | −2.6 | Histidine biosynthesis |
| hisF | −3.0 | −3.0 | −2.0 | Imidazole glycerol phosphate synthase subunit hisF |
| hisG | 1.1 | −1.3 | −2.8 | ATP phosphoribosyltransferase |
| hisH | −1.6 | −2.1 | −2.0 | Imidazole glycerol phosphate synthase subunit hisH |
| hisI | −2.1 | −2.0 | −1.9 | Histidine biosynthesis |
| hisL | 3.0 | 1.1 | −3.7 | His operon leader peptide |
| hisLEDL | 1.7 | −1.2 | −3.5 | Histidine biosynthesis; hisL gene from strain EDL933 |
| hisP | −2.5 | −1.4 | −1.1 | Nucleotide binding |
| Two-component signal transduction | ||||
| dos | 1.3 | 2.1 | 1.4 | Putative phosphodiesterase, oxygen-sensing protein |
| evgS | −3.2 | −1.5 | −1.2 | Two-component signal transduction system (phosphorelay) |
| fimZ | 1.1 | 2.1 | 3.0 | Fimbiral protein Z; putative transcriptional regulator of fimbrial expression (LuxR/UhpA family) |
| glnL | 3.2 | 2.5 | −1.9 | Two-component signal transduction system (phosphorelay) |
| narL | −2.6 | −2.0 | −1.4 | Nitrate/nitrite response regulator protein narL |
| prpR | −3.5 | −3.7 | 1.1 | Regulator for prp operon |
| rstA | 2.5 | 1.9 | −1.3 | Transcriptional Regulatory protein rstA |
| yedV | 1.4 | 2.6 | 2.0 | Putative two-component sensor protein |
| ygeV | −2.1 | −1.4 | −1.5 | Two-component signal transduction system (phosphorelay) |
| Sulfate | ||||
| cysD | −3.0 | −3.0 | −1.6 | Sulfate adenylyltransferase (ATP) activity |
| cysI | −3.5 | −3.0 | −1.2 | Sulfite reductase [NADPH] hemoprotein beta-component |
| cysJ | −4.3 | −2.8 | 1.0 | Sulfite reductase [NADPH] flavoprotein alpha-component |
| sseA | −2.1 | −1.9 | 1.4 | 3-Mercaptopyruvate sulfurtransferase |
| Motility/chemotaxis | ||||
| acs | −8.0 | −4.3 | 1.1 | Acetyl-coenzyme A synthetase |
| c2129 | 3.0 | 2.5 | 1.5 | Cell division activator cedA |
| c4004 | 1.3 | 2.1 | 1.1 | Hypothetical protein |
| csgA | −4.3 | −2.8 | 1.5 | Cell adhesion |
| csgB | −5.7 | −4.6 | −1.1 | Minor curlin subunit precursor |
| csgF | −3.7 | −2.5 | −1.4 | Curli production assembly/transport component csgF precursor |
| fimZ | 1.1 | 2.1 | 3.0 | Fimbiral protein Z; putative transcriptional regulator of fimbrial expression (LuxR/UhpA family) |
| fliD | 2.0 | 2.1 | 1.4 | Cell motility |
| fliR | −1.1 | 1.9 | 2.3 | Flagellar biosynthetic protein fliR |
| gidB | 1.4 | 2.1 | 1.5 | Methyltransferase gidB |
| motB | −2.3 | −1.2 | 1.3 | Chemotaxis motB protein |
| mreC | 1.5 | 2.1 | 1.2 | Rod shape-determining protein mreC |
| sfmA | −1.2 | 1.5 | 2.1 | Putative fimbrial-like protein |
| sfmF | 1.0 | 1.5 | 2.3 | Putative fimbrial-like protein |
| sfmH | −2.0 | 1.0 | 2.5 | Fimbrial assembly protein |
| yhhP | 1.7 | 2.5 | 1.5 | SirA protein |
| Z0020 | 1.5 | 2.5 | 2.1 | Cell adhesion |
| Z4971 | −1.9 | 3.7 | 1.9 | Cell adhesion |
| Cytochrome | ||||
| appB | −3.0 | −2.3 | 1.2 | Cytochrome bd II oxidase subunit II |
| appC | −2.5 | −2.1 | 1.1 | Cytochrome bd II oxidase subunit I |
| cyoC | −2.1 | −3.0 | 1.0 | Cytochrome o ubiquinol oxidase subunit III |
| nrfA | −3.2 | −1.5 | 1.5 | Cytochrome c-552 precursor |
| nrfB | −2.8 | 1.0 | 1.5 | Cytochrome c-type protein nrfB precursor |
| Cold shock protein | ||||
| cspA | 1.5 | 1.5 | −3.2 | Cold shock protein cspA |
| cspE | 2.3 | 2.5 | −1.1 | Cold shock-like protein cspE |
| cspG | 8.0 | 6.1 | −4.0 | Cold shock-like protein cspG |
| cspH | 22.6 | 21.1 | −2.5 | Cold shock-like protein cspH |
| deaD | 2.1 | 2.6 | −1.2 | Cold shock DEAD-box protein A |
| Antisense RNA | ||||
| dsrA | 3.5 | 4.9 | 1.3 | Antisense RNA; silencer of rcsA gene, interacts with rpoS translation |
| rdlD | 4.0 | 4.0 | −2.5 | Antisense RNA; trans-acting regulator of ldrD translation |
| sokB | 9.2 | 7.0 | −1.1 | Antisense RNA blocking mokB and hokB translation |
| sokC | 2.0 | 2.6 | −1.6 | Antisense RNA blocking mokC (orf69) and hokC (gef) translation |
| Glutamine/glutamate | ||||
| c0018 | 24.3 | 24.3 | 5.7 | Putative glutamate dehydrogenase |
| glnA | 1.6 | 1.3 | −2.3 | Glutamine synthetase |
| glnH | −1.2 | 1.0 | −2.1 | Glutamine-binding periplasmic protein precursor |
| glnP | 2.5 | 3.0 | −1.9 | Glutamine transport system permease protein glnP |
| gltJ | 3.7 | 4.3 | −1.6 | Glutamate/aspartate transport system permease protein gltJ |
| gltK | 2.6 | 2.6 | −1.6 | Glutamate/aspartate transport system permease protein gltK |
| gltL | 1.4 | 1.7 | −1.9 | Glutamate/aspartate transport ATP-binding protein gltL |
| ybeJ | −1.3 | −1.1 | −2.5 | Glutamate/aspartate periplasmic binding protein precursor |
| Iron related | ||||
| c3773 | 2.0 | 2.1 | 1.6 | Putative iron compound permease protein of ABC transporter family |
| cydA | 2.0 | 2.8 | −1.1 | Cytochrome d ubiquinol oxidase subunit I |
| cydB | 1.9 | 2.8 | −1.2 | Cytochrome d ubiquinol oxidase subunit II |
| feoA | 2.5 | 2.6 | −1.3 | Ferrous iron transport protein A |
| feoB | 2.0 | 3.2 | 1.0 | Ferrous iron transport protein B |
| fhuB | 1.3 | 2.5 | 1.1 | Ferrichrome transport system permease protein fhuB |
| fhuC | 1.6 | 2.5 | 1.0 | Ferrichrome transport ATP-binding protein fhuC |
| fhuD | 2.5 | 4.6 | 1.1 | Ferrichrome-binding periplasmic protein precursor |
| yodB | 2.5 | 2.6 | −1.3 | Cytochrome b561 homolog 1 |
| NADH dehydrogenase | ||||
| nuoA | −1.3 | −2.5 | 1.1 | NADH dehydrogenase I chain A |
| nuoB | −2.0 | −3.5 | 1.0 | NADH dehydrogenase I chain B |
| nuoC | −1.7 | −4.0 | −1.1 | NADH dehydrogenase I chain C/D |
| nuoE | −2.0 | −4.3 | −1.1 | NADH dehydrogenase I chain E |
| nuoI | −1.1 | −2.5 | 1.1 | NADH dehydrogenase I chain I |
| nuoJ | −1.3 | −2.5 | 1.3 | NADH dehydrogenase I chain J |
| nuoK | −1.3 | −2.3 | 1.4 | NADH dehydrogenase I chain K |
| nuoL | −1.2 | −2.3 | 1.4 | NADH dehydrogenase I chain L |
| Tricarboxylic acid cycle | ||||
| sdhD | −13.0 | −21.1 | 1.1 | Succinate dehydrogenase hydrophobic membrane anchor protein |
| sdhC | −12.1 | −18.4 | 1.1 | Succinate dehydrogenase cytochrome b-556 subunit |
| sdhA | −11.3 | −16.0 | 1.3 | Succinate dehydrogenase, catalytic and NAD/flavoprotein subunit |
| sdhB | −10.6 | −12.1 | 1.0 | Succinate dehydrogenase iron-sulfur protein |
| sucA | −7.5 | −11.3 | 1.1 | 2-Oxoglutarate dehydrogenase E1 component |
| sucB | −7.0 | −9.2 | −1.1 | Dihydrolipoamide succinyltransferase component of 2-oxoglutarate dehydrogenase complex |
| sucD | −6.5 | −8.6 | 1.0 | Succinyl-coenzyme A synthetase alpha chain |
| sucC | −6.5 | −8.6 | 1.0 | Succinyl-coenzyme A synthetase beta chain |
| acnB | −2.0 | −4.3 | 1.0 | Aconitate hydratase activity |
| icdA | −1.9 | −2.6 | 1.1 | Isocitrate dehydrogenase [NADP] |
| yojH | −1.2 | −2.3 | −1.5 | Malate:quinone oxidoreductase |
| mdh | −2.0 | −2.1 | −1.1 | Malate dehydrogenase |
Important changes are shown in boldface. IND, indole.
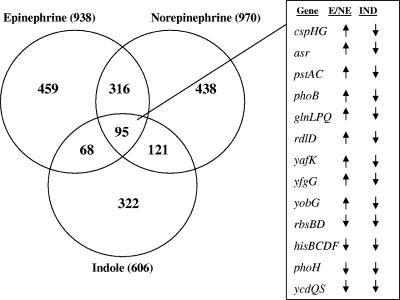
The number of genes differentially expressed in response to EPI, NE, and indole treatments were initially compared (Fig. 3). A total of 938 and 970 genes were differentially expressed by EPI and NE, respectively, with 411 differentially expressed genes common between the two treatments (Fig. 3, 316 plus 95). In comparison, 163 and 216 E. coli O157:H7 genes were commonly expressed between EPI and indole or NE and indole, respectively. Nearly equal numbers of common differentially expressed genes were either similarly (86 out of 163 genes up-regulated by both EPI and indole) or divergently (77 out of 163 genes down-regulated by both EPI and indole) expressed upon exposure of E. coli O157:H7 to either EPI or indole. On the other hand, a majority (197 out of 216) of the common genes differentially regulated after exposure to NE and indole were similarly expressed (i.e., genes up-regulated by NE were also up-regulated upon exposure to indole and vice-versa). These results indicate that EPI and NE, although similar in structure, exert different effects on E. coli O157:H7 biofilm gene expression.
FIG. 3.
Gene expression in E. coli O157:H7 biofilms upon exposure to EPI, NE, or indole. The effects of EPI (50 μM), NE (50 μM), and indole (500 μM) on gene expression in EHEC biofilms on glass wool were determined. The number of differentially expressed genes for each molecule, as well as the number of genes common to other molecules, is indicated in the Venn diagram. Genes common to all three molecules are not included in any of the other categories to facilitate interpretation. Annotated genes common to EPI, NE, and indole treatments (i.e., divergently expressed between EPI/NE and indole) are shown at right along with arrows indicating an increase or decrease in expression. E, epinephrine; IND, indole.
The common EHEC genes significantly expressed after exposure to EPI, NE, and indole were further analyzed (Fig. 3, intersection between EPI, NE, and indole). Since EPI and NE exhibit divergent effects on chemotaxis, motility, and biofilm formation compared to indole, we sought to classify genes which were either similarly or divergently altered in expression by EPI/NE and indole. The addition of EPI and NE significantly altered several genes, including those that function in AI-2 uptake, phosphate metabolism, glutamate/glutamine metabolism, cold shock, antisense RNA, hydrogenase activity, phosphotransferase system (PTS), tricarboxylic acid cycle, iron uptake and transport, and two-component signal transduction systems (Table 2).
The expression levels of genes involved in the transport and uptake of AI-2 were decreased in EHEC biofilms upon exposure to EPI and NE. The entire lsr operon (lsrACDBFG) that encodes an AI-2 uptake and modification system (51, 54), as well as regulators of the lsr operon (lsrK and lsrR), demonstrated decreases in expression ranging from 1.4- to 9.8-fold upon exposure to EPI and NE (Table 3). However, the AI-2 exporter gene, ydgG (21), was not significantly altered in expression. The trends observed with the microarrays were further validated using real-time RT-PCR, and comparable changes in expression were observed for lsrA (−9.8-fold in microarrays versus −4.8-fold with RT-PCR), lsrB (−7-fold in microarrays versus −2.5-fold with RT-PCR), and lsrC (−4.3-fold in microarrays versus −2.4-fold with RT-PCR). Together, these results suggest that the uptake of AI-2, but not export, was down-regulated by EPI and NE in EHEC biofilms.
TABLE 3.
Changes in the expression of AI-2 uptake genes in E. coli O157:H7 biofilms after exposure to EPI and NE
| Gene | Biological process description | Relative change in expression (n-fold) determined by the indicated methoda
|
||
|---|---|---|---|---|
| Microarray
|
RT-PCR (EPI) | |||
| EPI | NE | |||
| lsrB | AI-2 transporter | −7.0 | −5.3 | −2.5 |
| lsrG | Antibiotic biosynthesis | −3.2 | −3.0 | |
| lsrD | Transport | −3.2 | −1.4 | |
| lsrA | Transport | −9.8 | −5.3 | −4.8 |
| lsrC | Transport | −4.3 | −2.3 | −2.4 |
| lsrK | Carbohydrate metabolism | −2.8 | −2.3 | |
| lsrR | Regulation of transcription; DNA dependent | −3.7 | −3.7 | |
Values are based on an exposure of 7 h. Significant changes are in boldface.
Genes belonging to three functional groups—cold shock response, phosphate metabolism, and glutamate/glutamine metabolism—were also divergently regulated by indole compared to EPI and NE in E. coli O157:H7 biofilms. For example, the cspH gene was up-regulated 22.6-fold by EPI and 21.1-fold by NE but was down-regulated by 2.5-fold upon indole treatment. Changes in the expression of these divergently regulated genes are discussed below.
Cold shock genes.
Stress-related cold shock genes were differentially altered in E. coli O157:H7 biofilms by EPI, NE, and indole. The data show that cold shock regulator genes (cspGH) increased in expression in EPI- and NE-treated biofilms by 6- to 23-fold, while indole decreased the expression of these genes by 2.5- to 4-fold. Earlier work from our laboratory (13) has shown that cold shock genes are up-regulated in developing (7 h) but not in mature E. coli K-12 biofilms. The changes in the cold shock response genes, along with the alterations in biofilm formation (Fig. 2A), further support the hypothesis that EPI and NE increase E. coli O157:H7 colonization while indole decreases biofilm formation.
Phosphate-related genes.
The expression levels of phosphate-related genes in E. coli O157:H7 biofilms were also divergently regulated upon exposure to EPI and NE or indole. Several belonging to the pho (phoBRU) and pst (pstAC) operons were increased in expression upon exposure to EPI or NE whereas the expression of the pstSCAB genes and phoBU genes was significantly down-regulated in indole-treated biofilms. The phosphate transport system (encoded by the pstSCAB-phoU operon) has been previously shown to play an important role in E. coli CFT073 virulence and pathogenicity (12). Recently, Buckles et al. (5) have also identified PhoU as a virulence factor in urinary tract colonization. The changes in pst and pho gene expression observed with EPI and NE suggest that these signal molecules increase E. coli O157:H7 virulence while indole inhibits expression of genes involved in virulence.
Glutamate/glutamine genes.
The expression of genes involved in glutamate/glutamine biosynthesis and transport were increased in EPI- and NE-treated E. coli O157:H7 biofilms. These included genes involved in glutamate and transport (e.g., gltJ and glnP) that function in importing glutamate into the cell (29, 36). A decrease in the expression of these genes was observed with indole; however, these changes in expression were not significant. Genes involved in nitrogen metabolism are known to be involved in virulence of gram-positive and gram-negative bacteria (26). Recently, Walters et al. (49) reported that glutamate and oxaloacetate are utilized to generate l-aspartate that is converted to homocysteine and used for AI-3 production. The increase in the expression of genes involved in glutamate intake suggests that EPI and NE might stimulate AI-3 production in E. coli O157:H7 during biofilm formation, whereas indole might inhibit the synthesis of AI-3 (although the changes in the expression of glnP and gltJ are slightly below the twofold significance threshold).
Adherence to HeLa cells in the presence of EPI, NE, and indole.
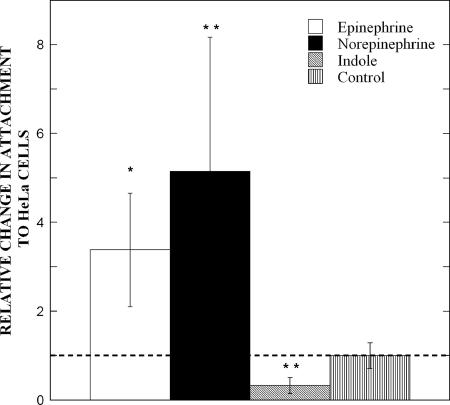
Based on the chemoattraction of E. coli O157:H7 to EPI and NE and the repulsion by indole, we hypothesized that these molecules would also divergently impact the EHEC adhesion that leads to virulence and infection. The human cervical carcinoma epithelial cell line, HeLa S3, was used for determining the effect of EPI, NE, and indole on E. coli O157:H7 adherence to epithelial cells. Although not of intestinal origin, HeLa cells have been extensively used for studying E. coli O157:H7 infections as they exhibit several classical features of infection including the formation of attaching and effacing lesions (46, 47). Figure 4 shows that exposure to a 50 μM concentration of EPI or NE for 3 h increased the attachment of EHEC to HeLa cells by 3.4- and 5.2-fold, respectively. On the other hand, adherence to HeLa cells in the presence of a 500 μM concentration of indole was reduced by 3.1-fold compared to untreated controls. These results are consistent with the effect that these molecules have on E. coli O157:H7 chemotaxis, motility, biofilm formation, and gene expression.
FIG. 4.
EPI, NE, and indole affect E. coli O157:H7 attachment to HeLa cells. The relative change in EHEC attachment to HeLa cells after 3 h of exposure to EPI (50 μM), NE (50 μM), or indole (500 μM) was determined. Cell counts (means ± 1 standard deviation) are from duplicate LB agar plates and were generated from five HeLa cell culture wells. Control values are based on EHEC cell counts obtained without the addition of any molecule. Statistical significance was determined using a Student t test. *, P < 0.01; **, P < 0.005.
DISCUSSION
The data presented in this report clearly demonstrate that the eukaryotic signaling molecules EPI and NE impact E. coli O157:H7 chemotaxis, motility, biofilm formation, gene expression, and attachment to epithelial cells in a manner different from that of the prokaryotic signal indole. The mechanisms utilized by this enteric pathogen to “recognize” its environs and initiate colonization are not fully understood. Kaper and Sperandio (24) have proposed that pathogenic E. coli detect quorum-sensing molecules from commensal bacteria, as well as other eukaryotic signals in the GI tract, to initiate colonization and infection. Our data provide evidence that EHEC can utilize eukaryotic and prokaryotic signals to “sense” and detect the appropriate location (i.e., large intestine) prior to colonization.
Sensing of EPI, NE, and indole by E. coli O157:H7 could be the first step in a sequence of events leading to infection, as phenotypes such as adherence and colonization, which are downstream of chemotactic recognition and important for infection, are also up-regulated by EPI and NE. Indeed, catecholamines have been shown to increase the adherence of E. coli O157:H7 to intestinal mucosa in different in vivo models of infection (7, 19). Our observations on the recognition of EPI and NE by EHEC are also in agreement with other reports on the regulation by EPI and NE of the expression of virulence genes and infection (8, 44, 49) and, together, suggest an important role for these molecules in EHEC infections. It should be noted that the addition of 50 μM EPI or NE did not increase the growth rate of EHEC, which is in contrast to prior reports describing an NE-mediated increase in growth of EHEC and other bacteria (14, 16). However, the similar growth rates observed between control and EPI/NE-treated cultures further emphasize the fact that the changes observed in chemotaxis, motility, biofilm formation, and HeLa cell adherence are not an artifact of increased cell density arising from increased growth rate upon exposure to EPI or NE.
Even though the presence and concentration of EPI and NE in the intestinal lumen has not been directly measured, it is reasonable to assume that EHEC encounters these molecules during infection. Very high local concentrations of catecholamines exist in the GI tract, which can result in NE leakage into the lumen through diffusion driven by the concentration gradient (31). Evidence for this has been presented by Chen et al. (6), who demonstrated that the addition of 10 μM NE to the contraluminal side of mucosal explants from cecum and colon increases EHEC adherence on the luminal side. The likelihood of NE's also being present in the lumen is further supported by the work of Ahlman et al. (2, 3), who detected a different neuroendocrine molecule, serotonin, in the intestinal lumen. However, it should be noted that EPI is less likely to be present in the intestinal lumen, as there have been no prior reports on the generation of EPI through the enteric nervous system or by the mesenteric organs (11).
We restricted our investigation to gene expression changes in a surface-associated (biofilm) model as opposed to planktonic cultures because the localized adherence of microcolonies to epithelial cells is more relevant for infections. Our data clearly show that 15 genes (encoding for functions related to virulence and biofilm formation) are divergently regulated between EPI/NE and indole. It should also be noted that these signals also exhibited similar effects on the expression of nearly 80 other genes (Fig. 3). However, a majority of these similarly regulated genes are hypothetical proteins, and of the annotated genes, none is related thus far to surface attachment or virulence. Therefore, based on the available annotation information, we propose that EPI/NE and indole play antagonistic roles during E. coli O157:H7 colonization and infection of epithelial cells. Identification and characterization of the 80 similarly regulated genes is needed to generate a comprehensive picture of the role that these molecules play during E. coli O157:H7 infection in the GI tract. Our unpublished data indicate that some of the genes (e.g., pstA) identified in this study are important in the EHEC response to NE and in infection, as an EHEC pstA mutant does not demonstrate a significant increase in attachment to HeLa cells upon exposure to NE (data not shown).
The changes in the expression of glutamate transport genes suggest an intriguing possibility that EPI and NE are involved in up-regulating AI-3 production. Since glutamate is involved in AI-3 synthesis (49), it is possible that this increase in glutamate uptake underlies an increase in AI-3 synthesis and, thereby, virulence. While some genes related to virulence (e.g., phoU) were altered in expression, none of the virulence genes (e.g., eae) per se demonstrated any significant changes in expression upon treatment with EPI and NE. These results are in contrast to other studies (19, 31) that have reported increased production of Shiga toxins by E. coli O157:H7 upon exposure to catecholamines. However, the experimental conditions between the two studies are sufficiently different (rich LB medium in our study versus serum-based basal medium; gene expression measurement at 7 h in our study versus Shiga toxin measurement at 12, 24, and 48 h) and possibly underlie the observed differences. It is also possible that colonization of abiotic surfaces (such as glass wool in our study) does not lead to expression of virulence genes as cues from epithelial cells that may be necessary for induction of the virulence genes may be missing (27). Current work in our laboratory focuses on comparing virulence gene expression between EHEC colonization of abiotic and biotic surfaces.
The entire lsr operon was down-regulated upon exposure to EPI and NE but not indole, which suggests that the uptake of AI-2 was down-regulated upon exposure to EPI and NE and does not play a significant role in biofilm formation. However, unpublished data from our laboratory show that the addition of AI-2 increases E. coli O157:H7 biofilm formation by 9.9-fold after 24 h. The increase in E. coli O157:H7 biofilm upon addition of AI-2 is also consistent with our prior work (18, 21) showing that an AI-2 export-deficient K-12 mutant (ΔydgG) demonstrates a 7,000-fold increase in biofilm formation (21). Together, these findings suggest the possibility that a second AI-2 transporter is present in E. coli O157:H7 (that is not impacted by the addition of EPI or NE), as our data (not shown) also show no difference in intracellular AI-2 levels between control and EPI/NE-treated E. coli O157:H7. The notion of a second AI-2 transporter is also consistent with explanations previously proposed by Wang et al. (51) for nonpathogenic E. coli and Taga et al. (45) for Salmonella enterica serovar Typhimurium.
It is interesting that some of the genes up-regulated by NE in EHEC biofilms are also similarly altered by other bacterial molecules. Recent work from our laboratory (27) shows that the indole derivative isatin (1-H-indole-2,3-dione), which induces biofilm formation, up-regulated the expression of cold shock response genes (cspGH and deaD) by 4.6- to 17.1-fold and down-regulated the expression of the AI-2 uptake genes (lsrACDBFG) by 10.6- to 64-fold, both of which are consistent with the data presented in this study for EPI and NE (Table 2). Similarly, both NE and isatin down-regulated the expression of genes involved in sulfur metabolism (cys genes), which are important in biofilm formation (40). Together, these results suggest commonalities between the effect of NE and isatin on EHEC physiology, especially with respect to AI-2 signaling and biofilm formation.
The consistency with which EPI, NE, and indole impact five different responses—chemotaxis, motility, biofilm formation, gene expression, and adherence to HeLa cells—strongly indicates that these molecules are important determinants of E. coli O157:H7 colonization in the GI tract. However, the differences in the response to EPI/NE versus indole suggest that interactions between these abundant molecules could be important in GI tract infections. While the current study establishes the response of E. coli O157:H7 to a single concentration of these molecules, the colonizing pathogen is likely to encounter diffusion-driven gradients of different signals in the intestinal lumen. Current work in our laboratory is focusing on presenting different concentration gradients of signaling molecules and their combinations to E. coli O157:H7 and investigating their effect on chemotaxis and colonization.
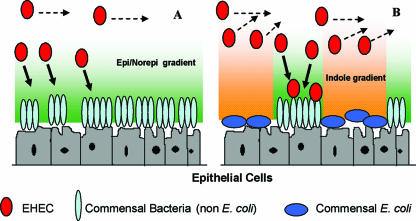
Based on the divergent regulation of E. coli O157:H7 chemotaxis, motility, and biofilm formation by EPI/NE and indole, we propose a signal-dependent model for E. coli O157:H7 colonization in the GI tract (Fig. 5). In this model, it is assumed that neuroendocrine signals such as EPI and NE are present in the intestinal lumen throughout the large intestine while spatial (positional) heterogeneities in the concentration of indole exist in the large intestine due to heterogeneities in the distribution of commensal bacteria (especially that of indole-producing nonpathogenic E. coli) (34). We propose that the migration of EHEC to the epithelial cell surface is primarily driven by sensing of EPI and NE but occurs only in regions of the biofilm where indole concentration is low or below a critical threshold value. It follows that EHEC colonization occurs to a large extent in regions of the GI tract that are not colonized by nonpathogenic E. coli (i.e., regions with low levels of indole). Furthermore, since E. coli O157:H7 itself secretes indole (∼400 μM) (27), subsequent colonization will occur only in places that are not already colonized by the pathogen, thereby contributing to further colonization and the spread of infection. Thus, the extent of EHEC colonization is likely to be strongly influenced by the spatial distribution of both eukaryotic signals and indole-producing commensal bacteria in the GI tract.
FIG. 5.
Hypothetical model for E. coli O157:H7 colonization in the GI tract. Gradients of EPI and NE influence the chemotactic migration of E. coli O157:H7 to epithelial cell surfaces. Cells that do not encounter high concentrations of EPI and NE will continue to move parallel to, and not toward, the epithelial cell surface. (A) In the absence of indole-secreting commensal E. coli, colonization of the nonpathogenic biofilm occurs throughout the epithelial cell surface. (B) In the presence of commensal E. coli, the pathogen is exposed to gradients of indole which repel it from the host cells. In this scenario, colonization occurs only in regions where nonpathogenic, non-E. coli bacteria are present. The direction of migration is indicated by arrows.
Acknowledgments
This work was supported by funds from the Texas Engineering Experiment Station (to A.J.) and the NIH (EB003872-01) (to T.K.W.).
We thank R. Jayaraman for critical reading of the manuscript.
Editor: D. L. Burns
Footnotes
Published ahead of print on 25 June 2007.
REFERENCES
- 1.Affymetrix. 2004. GeneChip expression: data analysis fundamentals. Affymetrix, Inc., Santa Clara, CA.
- 2.Ahlman, H., H. N. Bhargava, A. Dahlström, I. Larsson, B. Newson, and G. Pettersson. 1981. On the presence of serotonin in the gut lumen and possible release mechanisms. Acta Physiol. Scand. 112:263-269. [DOI] [PubMed] [Google Scholar]
- 3.Ahlman, H., L. DeMagistris, M. Zinner, and B. M. Jaffe. 1981. Release of immunoreactive serotonin into the lumen of the feline gut in response to vagal nerve stimulation. Science 213:1254-1255. [DOI] [PubMed] [Google Scholar]
- 4.Anyanful, A., J. M. Dolan-Livengood, T. Lewis, S. Sheth, M. N. Dezalia, M. A. Sherman, L. V. Kalman, G. M. Benian, and D. Kalman. 2005. Paralysis and killing of Caenorhabditis elegans by enteropathogenic Escherichia coli requires the bacterial tryptophanase gene. Mol. Microbiol. 57:988-1007. [DOI] [PubMed] [Google Scholar]
- 5.Buckles, E. L., X. Wang, C. V. Lockatell, D. E. Johnson, and M. S. Donnenberg. 2006. PhoU enhances the ability of extraintestinal pathogenic Escherichia coli strain CFT073 to colonize the murine urinary tract. Microbiology 152:153-160. [DOI] [PubMed] [Google Scholar]
- 6.Chen, C., M. Lyte, M. P. Stevens, L. Vulchanova, and D. R. Brown. 2006. Mucosally directed adrenergic nerves and sympathomimetic drugs enhance non-intimate adherence of Escherichia coli O157:H7 to porcine cecum and colon. Eur. J. Pharmacol. 539:116-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, C., D. R. Brown, Y. Xie, B. T. Green, and M. Lyte. 2003. Catecholamines modulate Escherichia coli O157:H7 adherence to murine cecal mucosa. Shock 20:183-188. [DOI] [PubMed] [Google Scholar]
- 8.Clarke, M. B., D. T. Hughes, C. Zhu, E. C. Boedeker, and V. Sperandio. 2006. The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. USA 103:10420-10425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke, M. B., and V. Sperandio. 2005. Events at the host-microbial interface of the gastrointestinal tract. III. Cell-to-cell signaling among microbial flora, host, and pathogens: there is a whole lot of talking going on. Am. J. Physiol. Gastrointest Liver. 288:G1105-1109. [DOI] [PubMed] [Google Scholar]
- 10.Collier-Hyams, L. S., and A. S. Neish. 2005. Innate immune relationship between commensal flora and the mammalian intestine. Cell Mol. Life Sci. 62:1339-1348. [DOI] [PubMed] [Google Scholar]
- 11.Costa, M., S. J. Brookes, and G. W. Hennig. 2000. Anatomy and physiology of the enteric nervous system. Gut 47:15-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daigle, F., J. M. Fairbrother, and J. Harel. 1995. Identification of a mutation in the pst-phoU operon that reduces pathogenicity of an Escherichia coli strain causing septicemia in pigs. Infect. Immun. 63:4924-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domka, J., J. Lee, and T. K. Wood. 2006. YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl. Environ Microbiol. 72:2449-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freestone, P. P., R. D. Haigh, and M. Lyte. 2007. Specificity of catecholamine-induced growth in Escherichia coli O157:H7, Salmonella enterica, and Yersinia enterocolitica. FEMS Microbiol. Lett. 269:221-228. [DOI] [PubMed] [Google Scholar]
- 15.Freestone, P. P., R. D. Haigh, P. H. Williams, and M. Lyte. 1999. Stimulation of bacterial growth by heat-stable, norepinephrine-induced autoinducers. FEMS Microbiol. Lett. 172:53-60. [DOI] [PubMed] [Google Scholar]
- 16.Freestone, P. P., P. H. Williams, R. D. Haigh, A. F. Maggs, C. P. Neal, and M. Lyte. 2002. Growth stimulation of intestinal commensal Escherichia coli by catecholamines: a possible contributory factor in trauma-induced sepsis. Shock 18:465-470. [DOI] [PubMed] [Google Scholar]
- 17.Girón, J. A., A. G. Torres, E. Freer, and J. B. Kaper. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44:361-379. [DOI] [PubMed] [Google Scholar]
- 18.González Barrios, A. F., R. Zuo, Y. Hashimoto, L. Yang, W. E. Bentley, and T. K. Wood. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol. 188:305-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green, B. T., M. Lyte, C. Chen, Y. Xie, M. A. Casey, A. Kulkarni-Narla, L. Vulchanova, and D. R. Brown. 2004. Adrenergic modulation of Escherichia coli O157:H7 adherence to the colonic mucosa. Am. J. Physiol. Gastrointest. Liver Physiol. 287:G1238-1246. [DOI] [PubMed] [Google Scholar]
- 20.Hansen, M. C., R. J. J. Palmer, C. Udsen, D. C. White, and S. Molin. 2001. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 147:1383-1391. [DOI] [PubMed] [Google Scholar]
- 21.Herzberg, M., I. K. Kaye, W. Peti, and T. K. Wood. 2006. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J. Bacteriol. 188:587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isaacs, H., D. Chao, C. Yanofsky, and M. H. Saier. 1994. Mechanism of catabolite repression of tryptophanase synthesis in Escherichia coli. Microbiology 140:2125-2134. [DOI] [PubMed] [Google Scholar]
- 23.Kaper, J. B., J. P. Nataro, and H. L. T. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-139. [DOI] [PubMed] [Google Scholar]
- 24.Kaper, J. B., and V. Sperandio. 2005. Bacterial cell-to-cell signaling in the gastrointestinal tract. Infect. Immun. 73:3197-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karch, H., P. I. Tarr, and M. Bielaszewska. 2005. Enterohaemorrhagic Escherichia coli in human medicine. Int. J. Med. Microbiol. 295:405-418. [DOI] [PubMed] [Google Scholar]
- 26.Kloosterman, T. G., W. T. Hendriksen, J. J. Bijlsma, H. J. Bootsma, S. A. van Hijum, J. Kok, P. W. Hermans, and O. P. Kuipers. 2006. Regulation of glutamine and glutamate metabolism by GlnR and GlnA in Streptococcus pneumoniae. J. Biol. Chem. 281:25097-25109. [DOI] [PubMed] [Google Scholar]
- 27.Lee, J., T. Bansal, A. Jayaraman, W. Bentley, and T. K. Wood. 2007. Enterohemorrhagic Escherichia coli biofilms are inhibited by 7-hydroxyindole and stimulated by isatin. Appl. Environ Microbiol. 73:4100-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, J., A. Jayaraman, and T. K. Wood. 2007. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 7:42. doi: 10.1186/1471-2180-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linton, K. J., and C. F. Higgins. 1998. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 28:5-13. [DOI] [PubMed] [Google Scholar]
- 30.Lyte, M., B. Arulanandam, K. Nguyen, C. Frank, A. Erickson, and D. Francis. 1997. Norepinephrine induced growth and expression of virulence associated factors in enterotoxigenic and enterohemorrhagic strains of Escherichia coli. Adv. Exp. Med. Biol. 412:331-339. [DOI] [PubMed] [Google Scholar]
- 31.Lyte, M., and M. T. Bailey. 1997. Neuroendocrine-bacterial interactions in a neurotoxin-induced model of trauma. J. Surg. Res. 70:195-201. [DOI] [PubMed] [Google Scholar]
- 32.Lyte, M., A. K. Erickson, B. Arulanandam, C. Frank, M. A. Crawford, and D. Francis. 1997. Norepinephrine-induced expression of the K99 pilus adhesin of enterotoxigenic Escherichia coli. Biochem. Biophys. Res. Commun. 232:682-686. [DOI] [PubMed] [Google Scholar]
- 33.Lyte, M., and S. Ernst. 1992. Catecholamine induced growth of Gram-negative bacteria. Life Sci. 50:203-212. [DOI] [PubMed] [Google Scholar]
- 34.Macfarlane, S., and J. F. Dillon. 2007. Microbial biofilms in the human gastrointestinal tract. J. Appl. Microbiol. 102:1187-1196. [DOI] [PubMed] [Google Scholar]
- 35.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nohno, T., T. Saito, and J. S. Hong. 1986. Cloning and complete nucleotide sequence of the Escherichia coli glutamine permease operon (glnHPQ). Mol. Gen. Genet. 205:260-269. [DOI] [PubMed] [Google Scholar]
- 37.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 38.Ren, D., L. A. Bedzyk, R. W. Ye, S. M. Thomas, and T. K. Wood. 2004. Differential gene expression shows natural brominated furanones interfere with the autoinducer-2 bacterial signaling system of Escherichia coli. Biotechnol. Bioeng. 88:630-642. [DOI] [PubMed] [Google Scholar]
- 39.Ren, D., L. A. Bedzyk, R. W. Ye, S. M. Thomas, and T. K. Wood. 2004. Stationary-phase quorum-sensing signals affect autoinducer-2 and gene expression in Escherichia coli. Appl. Environ Microbiol. 70:2038-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren, D., R. Zuo, A. F. González Barrios, L. A. Bedzyk, G. R. Eldridge, M. E. Pasmore, and T. K. Wood. 2005. Differential gene expression for investigation of Escherichia coli biofilm inhibition by plant extract ursolic acid. Appl. Environ. Microbiol. 71:4022-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sircili, M. P., M. Walters, L. R. Trabulsi, and V. Sperandio. 2004. Modulation of enteropathogenic Escherichia coli virulence by quorum sensing. Infect. Immun. 72:2329-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sperandio, V., C. C. Li, and J. B. Kaper. 2002. Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect. Immun. 70:3085-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sperandio, V., A. G. Torres, J. A. Girón, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sperandio, V., A. G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. USA 100:8951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taga, M. E., S. T. Miller, and B. L. Bassler. 2003. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol. Microbiol. 50:1411-1427. [DOI] [PubMed] [Google Scholar]
- 46.Torres, A. G., and J. B. Kaper. 2003. Multiple elements controlling adherence of enterohemorrhagic Escherichia coli O157:H7 to HeLa cells. Infect. Immun. 71:4985-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torres, A. G., X. Zhou, and J. B. Kaper. 2005. Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect. Immun. 73:18-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vlisidou, I., M. Lyte, P. M. van Diemen, P. Hawes, P. Monaghan, T. S. Wallis, and M. P. Stevens. 2004. The neuroendocrine stress hormone norepinephrine augments Escherichia coli O157:H7-induced enteritis and adherence in a bovine ligated ileal loop model of infection. Infect. Immun. 72:5446-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walters, M., and V. Sperandio. 2006. Autoinducer 3 and epinephrine signaling in the kinetics of locus of enterocyte effacement gene expression in enterohemorrhagic Escherichia coli. Infect. Immun. 74:5445-5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, D., X. Ding, and P. N. Rather. 2001. Indole can act as an extracellular signal in Escherichia coli. J. Bacteriol. 183:4210-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, L., Y. Hashimoto, C. Y. Tsao, J. J. Valdes, and W. E. Bentley. 2005. Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli. J. Bacteriol. 187:2066-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong, C. S., S. Jelacic, R. L. Habeeb, S. L. Watkins, and P. I. Tarr. 2000. The risk of the hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N. Engl. J. Med. 342:1930-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wood, T. K., A. F. González Barrios, M. Herzberg, and J. Lee. 2006. Motility influences biofilm architecture in Escherichia coli. Appl. Microbiol. Biotechnol. 72:361-367. [DOI] [PubMed] [Google Scholar]
- 54.Xavier, K. B., and B. L. Bassler. 2005. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 187:238-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu, H. S., and M. Alam. 1997. An agarose-in-plug bridge method to study chemotaxis in the Archaeon Halobacterium salinarum. FEMS Microbiol. Lett. 156:265-269. [DOI] [PubMed] [Google Scholar]