Abstract
Enterococcus faecalis is an important nosocomial pathogen associated with high morbidity and mortality for patients who are immunocompromised or who have severe underlying diseases. The E. faecalis genome encodes numerous surface-exposed proteins that may be involved in virulence. This work describes the characterization of the first internalin-like protein in E. faecalis, ElrA, belonging to the recently identified WxL family of surface proteins. ElrA contains an N-terminal signal peptide for export, a leucine-rich repeat domain that may interact with host cells, and a C-terminal WxL domain that interacts with the peptidoglycan. Disruption of the elrA gene significantly attenuates bacterial virulence in a mouse peritonitis model. The elrA deletion mutant also displays a defect in infection of host macrophages and a decreased interleukin-6 response in vivo. Finally, elrA expression is induced in vivo. Altogether, these results demonstrate a role for ElrA in the E. faecalis infectious process in vivo and suggest that this surface protein may contribute to E. faecalis virulence by stimulating the host inflammatory response.
Enterococci belong to the normal gastrointestinal tract microflora of hosts ranging from mammals to insects. Even if considered opportunistic pathogens, enterococci are a major cause of nosocomial infections in North America and in Europe (7, 33, 52), with Enterococcus faecalis being the most common enterococcal species. This bacterium is mainly responsible for bloodstream, urinary tract, and surgical site infections and endocarditis (32). Notably, E. faecalis seems to modulate the host systemic inflammatory response in mono- or polymicrobial infections, leading sometimes to severe sepsis or septic shock in immunocompromised patients (3, 39). Several enterococcal virulence factors, such as lipoteichoic acid (6, 64), aggregation substance, and enterococcal binding substance (56), have been implicated in the stimulation of the inflammatory response in vitro. Like the capsular polysaccharides Epa and Cps (29, 31, 62) and transcriptional regulators HypR and Ers (24, 67, 68), the aggregation substance is involved in the resistance to macrophages and/or to phagocytosis (50, 61, 65).
Bacteria have evolved a plethora of sophisticated molecular mechanisms to adhere to and invade host cells and tissues and to resist host defenses (49). Interestingly, several proteins with well-known roles in host-pathogen interactions, such as SspH of Salmonella enterica serovar Typhimurium (44, 63), YopM of Yersinia enterocolitica (26), IpaH of Shigella flexneri (20), and internalins of Listeria spp. (41), share a common leucine-rich repeat (LRR) domain. This module is present in a number of eukaryotic and prokaryotic proteins with different functions and appears to play a direct role in protein-protein interactions (10).
The best-characterized bacterial LRR proteins, InlA and InlB of Listeria monocytogenes, are the prototypes of an internalin multigene family of Listeria spp. Both proteins impact on L. monocytogenes virulence by interacting with host cell receptors in a species-specific manner and inducing endocytic pathways for invasion of and dissemination in tissues (28). Internalin-like proteins have been recently predicted in genomes of several gram-positive bacteria (8, 48, 54, 69). All characterized or predicted internalins are assigned to cell surface or secreted locations (8). Recently, we and others identified a new family of surface proteins in low-GC-percent gram-positive bacteria including E. faecalis and Lactobacillus plantarum, which are characterized by a WxL domain that mediates a novel type of C-terminal cell wall association (9, 59). Two E. faecalis WxL proteins, EF2686 and EF2250, were predicted to be internalin-like proteins (9). The present study reports the first characterization of an E. faecalis WxL internalin-like protein, named ElrA for enterococcal leucine-rich protein A. Inactivation of the elrA gene significantly reduced virulence in a mouse peritonitis model and dissemination to the spleen and liver. Our results thus identify ElrA as a virulence factor and further indicate that ElrA has a role in eliciting the inflammatory interleukin-6 (IL-6) response.
MATERIALS AND METHODS
Bacterial strains, macrophages, and culture conditions.
E. faecalis OG1RF (15) and Escherichia coli TG1 (25) were grown in brain heart infusion (BHI) and Luria-Bertani medium (Difco Laboratories), respectively, at 37°C. Antibiotics used were ampicillin (80 μg/ml) and erythromycin (150 μg/ml for E. coli and 30 μg/ml for E. faecalis). Murine monocytic-macrophage cell line J774 was propagated in Dulbecco's modified Eagle's medium (DMEM; Gibco Invitrogen, Cergy Pontoise, France) supplemented with 10% decomplemented fetal calf serum, 2 mM glutamine, and 1% nonessential amino acids (referred to as supplemented DMEM). Cells were incubated at 37°C under 10% CO2.
A total of 147 E. faecalis strains including 83, 51, and 13 isolates of clinical, food, and commensal origins, respectively, from different locations (Argentina, Egypt, England, France, and the United States) were also included to investigate the distribution of EF_2686 and its operon in different isolates. The clinical isolates were obtained from the Centre Hospitalier de Versailles (France), the Centre Hospitalier Universitaire of Lyon (France) (22), or the Health Sciences Center at the University of Oklahoma. Food isolates were provided by the CNRZ collection at Jouy-en-Josas (France) and the INRA collection at Aurillac (France). Commensal isolates were obtained from the Health Sciences Center at the University of Oklahoma or isolated from fecal specimens of healthy volunteers.
General DNA techniques.
General molecular biology techniques were performed according to standard protocols (55). Total DNA was extracted from 4 ml of late-exponential-phase bacterial cultures as described previously (21). PCR amplifications were carried out in a Perkin-Elmer (Courtaboeuf, France) or an Eppendorf apparatus using Taq DNA polymerase (Qbiogene, Illkirch, France) or Expand Taq (Roche Diagnostics, Meylan, France) according to the manufacturers' recommendations. The primers used in this study are listed in Table 1. The standard program was as follows: 95°C for 1 min; 30 cycles of 95°C for 1 min, 48 to 52°C for 1 min, and 72°C for 1 min; and then a final extension step of 72°C for 7 min. PCR products and DNA restriction fragments were purified with QIAquick kits (QIAGEN S.A., Courtaboeuf, France). Plasmids were purified using QIAprep kits (QIAGEN). E. coli and E. faecalis strains were transformed by electroporation using a Gene Pulser apparatus (Bio-Rad Laboratories, Marnes-la-Coquette, France) as described previously (13, 16). Sequence inserts were determined with Amersham dye terminator chemistry using a MegaBACE DNA sequencer (Amersham Biosciences, Freiburg, Germany).
TABLE 1.
Primers used in this study
| Name | Sequence (5′-3′)a | Target |
|---|---|---|
| OEF2 | CTATTTTGCACTGCCATCAGG | Reverse elrA |
| OEF8 | AAACGACCGAAACAATCGC | Forward elrA |
| OEF10 | CATTCTGGTTGTGTTTAGCG | Forward elrA |
| OEF14 | AATCAGGTGCTACGTTGCC | Forward elrA |
| OEF15 | TATTCGATGTTGGCGTTGG | Forward EF_2687 |
| OEF16 | TATCTGGATTCACTGGATCG | Reverse EF_2684 |
| OEF18 | GGAGGATGCGATTGTTTCG | Reverse elrA |
| OEF55 | GTTTCACCGAACTTTGTCCG | Reverse EF_2681 |
| OEF62 | TTAGATGAGGGACTTCGTG | Forward rpoB |
| OEF63 | TCCATTTCTAACCATGCACC | Reverse rpoB |
| OEF96 | CGCGGATCCCAACTGTTGGGAAGGGC | Forward Pusp45b |
| OEF97 | AACTGTTCTTTTTTAATTTTTCC | Reverse Pusp45 |
| OEF98 | GGAAAAATTAAAAAAGAACAGTTATGAAAAAAATGTGCATCTCC | Forward Pusp45::elrA |
| OEF99 | CAACAGTCGTTAACTCTTCTG | Reverse elrA |
| OEF210 | CTCTTCTGATTGGGGAATCTCC | Reverse elrA |
| OEF212 | CTCTTCTGCCGATGAAGTTTCTGG | Reverse elrA |
| OEF214 | CGCTATGCCTATACAGTAGC | Forward EF_2682 |
| OEF217 | CGATAGGTGTCACAGGATC | Reverse EF_2683 |
| OEF218 | GGTCCTGATTTGACTAATCC | EF_2684 |
| OEF219 | CAGACGTAAAATGATGAACAGG | Between EF_2682 and EF_2681 |
| OEF220 | CACTCCGTTGCTGTTTGTTG | EF_2682 |
| OEF221 | GTGCTGGGAAATGGTCATAC | EF_2683 |
| OEF246 | TGGCTAATAGTTTGCTACTTCCTG | Forward elrA |
| OEF247 | TACTGGTTGCTGAATTTTGCTCTT | Reverse elrA |
| OEF248 | FAM-CGCCGAAACGACCGAAACAATCGC-TAMRA | elrA TaqMan probe |
| OEF249 | GTTTATTACACGCAATCTTCGGG | Forward rpoB |
| OEF250 | CCAGCTTCACGAGTAAAGATTTTC | Reverse rpoB |
| OEF251 | Texas Red-CGATACCGCCGCCACCGTGAGG-BHQ2 | rpoB TaqMan probe |
Underlined nucleotides specify the restriction enzyme site used for cloning. FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine; BHQ2, black hole quencher 2.
usp45 promoter region.
Construction of an in-frame elrA deletion mutant in E. faecalis strain OG1RF.
A markerless in-frame ΔelrA deletion mutant of E. faecalis OG1RF was constructed as described for E. faecalis strain JH2-2 by using the same primers (9). Deletion of elrA was confirmed by PCR amplification and Southern hybridization with a probe corresponding to a fragment that overlapped the deletion in elrA.
Complementation of the E. faecalis ΔelrA strain.
To complement the ΔelrA strain in trans, the elrA gene was cloned under the control of the Lactococcus lactis usp45 promoter, which was previously shown to be expressed in E. faecalis (9). The elrA gene was amplified using OEF2 and OEF10 primers (Table 1) and cloned in pCRII-TOPO (Invitrogen) to obtain pVE14001. A 226-bp elrA fragment starting from the elrA start codon was then amplified from pVE14001 with primers OEF98 and OEF99. A 384-bp fragment encompassing the Pusp45 promoter was amplified from pVE3618 (38) with primers OEF96 and OEF97. The two PCR products were fused by PCR using the external primers, and the resulting DNA segment was cloned into pCRII-TOPO to obtain pVE14129. The 664-bp BamHI/HpaI fragment of pVE14129 was fused to the 5,888-bp BamHI/HpaI fragment of pVE14001. The resulting plasmid, pVE14131, was fused to L. lactis plasmid pIL253 at the BamHI site, to obtain pVE14132, which was electroporated into the ΔelrA strain.
RNA extraction, RT-PCR, and RACE-PCR.
Total RNA of E. faecalis strains was isolated using the High Pure RNA extraction kit (Roche Diagnostics). Briefly, cultures of E. faecalis at an optical density at 600 nm of 0.6 were centrifuged at 10,000 × g for 5 min at 4°C. Cell pellets were suspended in 500 μl sterilized H2O that was DNase and RNase free and transferred in a 1.5-ml screw-cap microtube containing 500 mg of glass beads (150 to 200 μm) and 500 μl of phenol (pH 4.7)-chloroform (5:1; Sigma, Saint Quentin Fallavier, France). To this was added 30 μl of 10% sodium dodecyl sulfate and 30 μl of 3 M sodium acetate (NaAc), pH 5.2. Bacteria were disrupted using a Fast Prep FP120 apparatus (Bio101 Savant Ozyme, Saint-Quentin-en-Yvelines, France) with four pulses of 45 s at a speed of 6.5 m/s and centrifuged for 15 min at 15,000 × g at 4°C. The supernatant was transferred into a new Eppendorf tube and extracted once with phenol (pH 4.7)-chloroform and once with chloroform. RNA was further purified using the High Pure RNA isolation kit (Roche Diagnostics). Purification steps were performed according to the manufacturer's instructions, except that 1 volume of lysis buffer was added before RNA was loaded on the column. Removal of DNA contamination was ensured by treating eluted RNA with 50 U of RNase-free DNase I (Roche Diagnostics) for 30 min at 25°C. DNase I was eliminated with phenol-chloroform and chloroform extractions. Total RNA was precipitated with 1/10 volume of 3 M NaAc, pH 5.2, and 2 volumes of 100% ethanol for 1 h at −80°C. After centrifugation at 13,000 × g for 45 min at 4°C, the pellet was washed with 200 μl of 75% ethanol and dissolved in 50 μl of RNase- and DNase-free water. The absence of DNA contamination was verified by PCR amplification of an internal rpoB gene fragment using OEF62 and OEF63 primers on total RNA (Table 1). Reverse transcription (RT) was performed on 2 μg of total RNA using random nonamers (Amersham Biosciences) and Power Script reverse transcriptase (Clontech, Saint-Quentin-Yvelines, France) according to the manufacturer's instructions. After heat inactivation of the enzyme for 15 min at 70°C, PCR was performed to amplify cDNA with primers listed in Table 1. The transcriptional start point of elrA was determined by using the rapid amplification of cDNA ends (RACE) 5′/3′ kit (Roche Diagnostics) according to the manufacturer's recommendations with primers OEF210 for cDNA synthesis and OEF212 for nested PCR and sequencing of the PCR product (Table 1).
In vitro macrophage survival assay using immunofluorescence.
J774 macrophages were diluted in 24-well tissue culture plates on a glass slide, at approximately 1 × 105 cells per ml. Monolayers produced after 24 h of incubation were used for infection studies. Exponential-phase cultures of the E. faecalis OG1RF and ΔelrA strains were prepared in BHI medium. Cells were then pelleted by centrifugation, washed three times with phosphate-buffered saline (PBS), and adjusted to a concentration of 1 × 106 bacteria per ml in supplemented DMEM. Macrophages were washed twice in supplemented DMEM and infected at a multiplicity of infection of 5:1 (bacterium/cell ratio) for 1 h at 37°C with 10% CO2. Infected macrophages were washed twice in supplemented DMEM and then fixed with 4% (wt/vol) paraformaldehyde in PBS for 20 min at room temperature. After three washes in PBS, cells were incubated with Streptococcus group D antiserum (BD Diagnostics, Le Pont de Claix, France; dilution, 1/2,000) diluted in PBS containing 0.5% (wt/vol) bovine serum albumin for 45 min to label extracellular bacteria. Cells were rinsed three times with PBS and incubated with secondary goat anti-rabbit Cy3-conjugated antibody (Amersham Biosciences; dilution, 1/300) for 45 min. Cells were then permeabilized with 0.4% Triton X-100 in PBS for 4 min and washed three times with PBS before incubation with Streptococcus group D antiserum (as described above) for 45 min. After three washes in PBS, cells were incubated with secondary goat anti-rabbit Alexa 488-conjugated antibody (Molecular Probes; dilution, 1/300) and DAPI (4′,6′-diamidino-2-phenylindole; dilution, 1/200) for 45 min to label intracellular bacteria. In all experiments, washes and antibody incubation steps were performed at room temperature. Preparations were observed with a Zeiss Axiovert 135 microscope. Image acquisition from the Zeiss microscope was carried out with a cooled charge-coupled device camera (Princeton), and images were processed with Metamorph software (Universal Imaging Corporation, Downingtown, PA).
In vivo-in vitro macrophage survival assays.
E. faecalis survival in mouse peritoneal macrophages was tested using an in vivo-in vitro infection model as described previously (23, 68). Experiments were performed five times, and results were subjected to statistical analysis using Student's t test. P values of <0.05 were considered statistically significant.
Mouse peritonitis model.
Testing of the OG1RF, ΔelrA, and complemented mutant strains was performed as described previously (60). Briefly, bacterial cells were harvested by centrifugation, washed twice with ice-cold 0.85% saline solution, and suspended in the same solution to a density of ∼1.5 × 1010 cells per ml. The inoculum size was confirmed by determining the number of CFU on BHI agar. According to previous studies (60), each inoculum was 10-fold diluted in 25% sterile rat fecal extract prepared from a single batch (46). Groups of 4- to 6-week-old (22 to 25 g) outbred (ICR) female mice (Harlan Italy S.r.l., San Pietro al Natisone, Udine, Italy) were challenged intraperitoneally with 1 ml of each bacterial inoculum, housed five per cage, and fed ad libitum. A control group of mice was injected with 25% sterile rat fecal extract only. Survival was monitored every 3 to 6 h. Survival estimates were constructed by the Kaplan-Meier method and compared by log-rank analysis. All strains were tested more than once. In another set of experiments, groups of mice were killed 20 h postinfection, and livers and spleens were removed, weighed, homogenized, and serially diluted in saline solution for colony counts and RT-PCR analyses (see below). The experiments were made in duplicate, and comparison among the organ counts was performed by use of an unpaired t test.
All statistical analyses were performed using GraphPad Prism version 4.03 for Windows (GraphPad Software, San Diego, CA), and comparisons with P values of <0.05 were considered to be significant.
Quantitative real-time RT-PCR.
Total RNA was extracted from exponential-phase enterococcal cultures or mouse peritoneal fluids or organ homogenates with an RNeasy Protect Mini kit (QIAGEN), after cells were suspended in RNAlater solution (QIAGEN), as described previously (11, 12). Expression of the elrA gene was quantitatively assessed with real-time RT-PCR in an iCycler iQ system (Bio-Rad Laboratories, Hercules, CA), using rpoB as the normalization gene. Comparative studies of in vitro/in vivo expression were performed in order to assess the validity of rpoB as a reference gene. Thus, a standard curve was constructed by plotting serial dilutions of a cloned rpoB gene fragment (range, 106 to 101 copies/reaction) and used to quantify rpoB transcripts in samples of RNA extracted from cultured enterococcal cells or from infected fluids or tissue homogenates. The number of CFU was determined in each sample, and the results were expressed as the amount of rpoB transcript per CFU. Similar amounts of rpoB transcript were found in the cells grown in vitro and in those recovered from the infected peritoneal fluids or organs (data not shown), indicating that this gene is expressed at nearly identical levels in E. faecalis OG1RF cells grown in vivo and in vitro.
For each gene, a set of primer pairs and a TaqMan probe were designed with Beacon Designer 2 (version 2.06) software (Premier Biosoft International, Palo Alto, CA) and synthesized by MWG Biotech. elrA primers and probe were OEF246, OEF247, and OEF248. rpoB primers and probe were OEF249, OEF250, and OEF251 (Table 1). PCRs were performed in a 50-μl volume containing the following reagents: 25 μl of the Platinum Quantitative RT-PCR ThermoScript reaction mix (Invitrogen Inc., Milan, Italy), 1.5 U of ThermoScript Plus/Platinum Taq mix (Invitrogen), each primer pair and Taqman probe at a concentration of 0.5 μM, 5 μl of total RNA sample, and distilled water up to final volume. Samples were subjected to an initial step at 52°C for 45 min for RT, 94°C for 5 min to inactivate the ThermoScript Plus reverse transcriptase and to activate the Platinum Taq polymerase, and 50 cycles each consisting of 15 s at 94°C and 1 min at 59°C. Fluorescence data were collected during the 59°C annealing/extension step and analyzed with the iCycler iQ software. Each reaction was run in quadruplicate, and amplification efficiencies for the target gene were determined. The relative mRNA expression level of the target gene in each sample was calculated using the comparative cycle threshold as described previously (43).
Multiplex cytokine assay.
Mouse inflammatory cytokines were quantified in triplicate using a Q-Plex array (Ozyme) according to the manufacturer's instructions. Samples tested were from peritoneal fluid and serum of mice infected with E. faecalis OG1RF and the ΔelrA strain 20 h postinfection. Chemiluminescence was quantified using a Fluorchem 8000 apparatus (Alpha Innotech Corporation, San Leandro, CA). Data were analyzed using the Quansys program (Quansys Biosciences, San Diego, CA). Data were expressed as mean values ± standard variations of three mice.
RESULTS
Analysis of the E. faecalis EF_2686 locus.
Gene EF_2686 of the E. faecalis V583 clinical isolate encodes a surface protein that we recently identified as a prototype for proteins associated with the peptidoglycan by a WxL motif (9). Gene EF_2686 is immediately followed by EF_2685, predicted to encode a small protein with an LPXTG anchor motif (48), with only an in-frame stop codon (TAG) between the two genes (http://cmr.tigr.org/tigr-scripts/CMR/shared/GenePage.cgi?locus=EF2686). To examine the distribution of EF_2686 in different E. faecalis isolates, a total of 147 strains including 83, 51, and 13 isolates of clinical, food, and fecal origins, respectively, were analyzed by PCR using internal EF_2686-specific primers OEF2 and OEF8. A 2.1-kb fragment was obtained for all strains, except for seven food (14%), three clinical (3.6%), and three fecal (23%) isolates, indicating that EF_2686 is widespread among the isolates. To confirm that the stop codon that separate EF_2686 from EF_2685 was conserved in the different strains, we amplified and sequenced a 784-bp region spanning the two open reading frames (ORFs) from 96 E. faecalis isolates using primers OEF14 and OEF16 (Table 1). All isolates had the in-frame stop codon (data not shown), indicating that the EF_2686 product is not a pseudoprotein, in contrast to what was originally proposed in the E. faecalis V583 annotation (48). In addition, the possibility of an in vivo read-through product was examined by fusing a region spanning EF_2686 and EF_2685 in frame with the staphylococcal nuclease reporter gene as described previously (9). Under laboratory growth conditions, expression of the resulting fusion protein in E. faecalis failed to detect read-through, whereas the protein corresponding to the Nuc-EF2686 fusion was detected by Western blotting (data not shown). These results indicate that EF_2686 is a full-length gene that encodes a WxL protein.
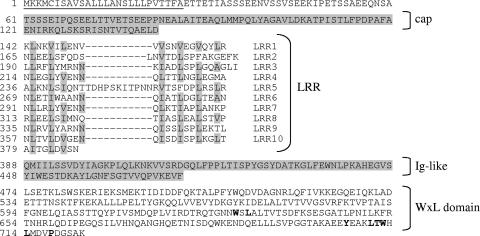
The EF_2686 gene is predicted to encode a 723-amino-acid protein with a molecular mass of 80 kDa and a pI of 4.7. Primary sequence analysis using NCBI blastP, PFAM, and SIGNALP 3.0 revealed domains characteristic of internalin family members that are well characterized in Listeria spp. (Fig. 1). EF2686 comprises an N-terminal signal peptide (amino acids 1 to 26) followed by a short conserved cap (amino acids 61 to 141), an LRR domain, an immunoglobulin-like domain conserved in several internalin family members (14, 41, 57), and a C-terminal WxL domain. The best hit (35% identity and 59% homology) was observed with InlA of Listeria monocytogenes between residues 102 and 395 of EF2686 and between residues 36 and 338 of InlA, and the overall predicted structure of EF2686 resembles that of InlA. Eight full-length LRRs contain 22 residues, whereas LRR2 and LRR5 contain 25 and 33 residues, respectively. They all contain a conserved 11-residue segment (LxxLxLxxNxL) giving a predicted horseshoe-shaped protein. They also contain the characteristic residues of the consensus sequence for LRR internalin proteins (36, 41). Interestingly, LRR5 of ElrA is atypical as it contains an insertion of nine residues (TDHPSKITP). The C-terminal part of EF2686 differs from InlA in that it contains a WxL domain, which we demonstrated confers cell surface localization on the protein (9). Altogether these observations indicate that EF_2686 encodes a member of the internalin family in E. faecalis. Based on the leucine-rich character of Ef2686, we named it ElrA (encoded by elrA) for enterococcal leucine-rich protein.
FIG. 1.
Principal characteristics of the amino acid sequence of ElrA. The predicted signal peptide is underlined. N-terminal cap and immunoglobulin (Ig)-like domains are indicated on a gray background. Conserved residues of the WxL C-terminal domain are in bold. LRR residues corresponding to LRR consensus sequences are highlighted in gray. Amino acid positions of residues are indicated on the left.
elrA is clustered in an operon.
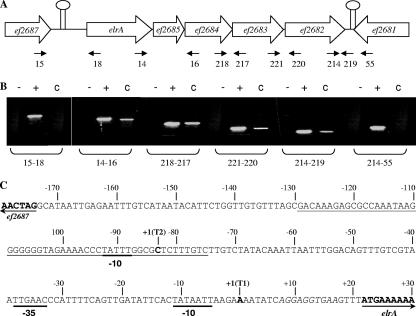
elrA is the first gene of a putative operon comprising five ORFs (Fig. 2A). They encode, in order, ElrA; EF2685, a small protein with an LPXTG surface anchoring motif; EF2684 and EF2683, two WxL proteins of unknown function (9); and EF2682, a putative transmembrane protein. Putative rho-independent transcription terminators flank the EF_2682 and elrA genes. To investigate whether the five genes were cotranscribed as an operon, total RNA was extracted from exponential-phase OG1RF cultures, and RT-PCR analysis was performed using intragenic specific primers flanking all the predicted junctions between genes elrA and EF_2681 (Fig. 2B). Amplification profiles indicated that transcription started downstream of the putative terminator located upstream of elrA and ended at the terminator located downstream of EF_2682. Moreover, cDNA was amplified from all junctions between each of the five genes. These results indicate that the five ORFs from elrA to EF_2682 constitute an operon.
FIG. 2.
Expression analysis of the putative elrA operon. (A) Genetic organization of the putative elrA operon. Each ORF is named according to its TIGR gene identification. Solid black spacers indicate predicted intergenic regions. Putative terminators are indicated as lollipops. (B) RT-PCR of junctions in the putative operon comprising ORFs elrA and EF_2682. Each sample was run in three reactions with three different templates: RNA (−), genomic DNA (+), and cDNA (C). (C) Schematic representation of elrA promoter region organization. Arrows indicate the start of EF_2687 and elrA coding sequences. The ribosome binding site is in italics. Transcriptional initiation nucleotides (T1 and T2) are indicated as +1 in bold. The −35 and −10 boxes are underlined in bold. The 55-bp inverted repeat is underlined.
The transcriptional start site of the operon comprising elrA was mapped by 5′-RACE PCR. The nucleotide sequence of the 5′-RACE PCR products revealed two transcriptional start sites, T1 and T2, at the A and C nucleotides located 21 and 42 bases, respectively, upstream of the elrA translational start codon (data not shown). Perfect −10 (TATAAT) and −35 (TTGAAC) σ70 recognition sequences, separated by 21 bp, were identified upstream of T1 (Fig. 2C). Transcriptional start T2 is located in a predicted inverted repeat of 55 bp. A possible −10 sequence (TATTT) was identified upstream of T2, whereas no clear −35 box was identified.
elrA inactivation attenuates E. faecalis virulence in a mouse peritonitis model.
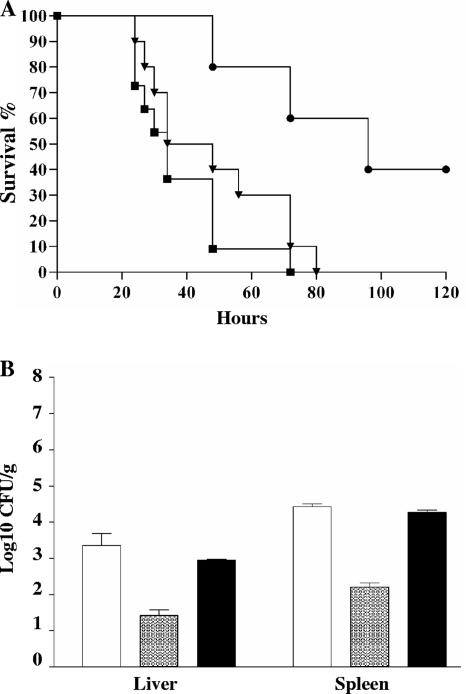
To study the potential role of ElrA in enterococcal virulence, we generated an in-frame elrA deletion mutant by allelic exchange in E. faecalis strain OG1RF, as previously described (9). In preliminary experiments, we showed that deletion of elrA did not affect the growth of the ΔelrA strain in BHI at 37°C, compared with the wild-type parental strain (data not shown). The virulence was examined using the mouse peritonitis model (60). Growth of OG1RF and mutant strains in the peritoneal cavity was first tested by plating peritoneal lavage fluids from infected mice on BHI agar. Bacterial counts were not statistically different, indicating that the two strains had similar abilities to grow in the mouse peritoneal cavity. Mice were then infected intraperitoneally with 8.5 × 108 and 9 × 108 CFU of OG1RF and the ΔelrA strain, respectively, and mortality rates were compared. Mortality was significantly reduced for mice injected with the ΔelrA strain; 40% of the mice were still alive after 120 h, whereas 100% of mice died after 70 h when injected with the OG1RF strain (Fig. 3A). To further confirm that attenuated virulence was due to the loss of a functional elrA gene, we constructed a complemented ΔelrA strain, which carried a pIL253 plasmid harboring the elrA gene under the control of Pusp45, known to be active in E. faecalis (9). This construct restored virulence to nearly OG1RF levels in the mouse peritonitis model (Fig. 3A), thereby indicating that ElrA plays a role in E. faecalis virulence.
FIG. 3.
Effect of elrA inactivation on virulence. (A) Kaplan-Meier survival analysis in a mouse peritonitis model with OG1RF (squares), isogenic ΔelrA mutant (circles), and the complemented mutant strain (inverted triangles). Mice were infected intraperitoneally with 8.5 × 108, 9.0 × 108, and 9.3 × 108 CFU of OG1RF, the ΔelrA strain, and the complemented mutant strain, respectively. (B) Dissemination levels of OG1RF (white bars), the ΔelrA strain (gray bars), and the complemented mutant (black bars) were compared in mouse spleen and liver. Mice were infected with ∼108 CFU of each strain. The results represent the means ± standard deviations of the number of bacteria able to colonize the spleen and liver at 20 h postinfection.
We also compared the virulence phenotypes of the ΔelrA, complemented, and OG1RF strains by determining bacterial counts in organs of mice at 20 h postinfection (Fig. 3B). A 2-log10 decrease in the bacterial counts in the liver and spleen was observed for the ΔelrA strain compared to the OG1RF and complemented strains, when mice were challenged with inocula of ∼108 CFU. Similar decreases in the organ counts of the ΔelrA strain were observed when inocula of ∼109 and ∼107 CFU were used (data not shown). These results further support the observation that ElrA is implicated in E. faecalis virulence.
elrA gene inactivation has no effect on in vitro phagocytosis.
Phagocytic cells constitute a first host defense barrier to bacterial infection. To examine whether the ΔelrA strain was impaired at a particular stage of E. faecalis interaction with phagocytic cells, entry efficiencies of ΔelrA and OG1RF strains were compared in J774 murine macrophages, using a double immunofluorescence assay. In J774 macrophages, entry efficiency of the ΔelrA bacteria detected after 1 h of infection at an initial multiplicity of infection of 10 bacteria per cell (81% ± 3%) was similar to that observed for the OG1RF strain (78.5% ± 2.5%). These results indicated that ElrA does not directly affect phagocytosis in vitro, at least in the cell line tested.
elrA inactivation affects bacterial infection of peritoneal macrophages.
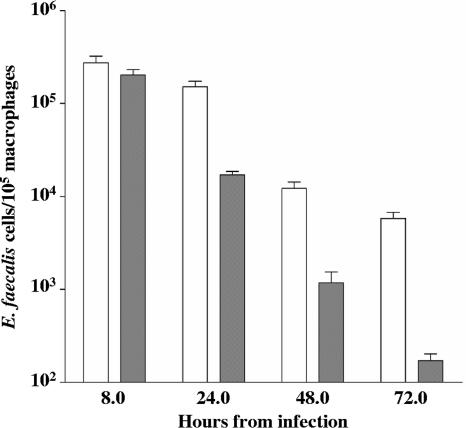
The reduced colonization by ΔelrA of the spleen and liver of infected mice suggested that this strain might be attenuated for infection of host macrophages, which can serve as vehicles for dissemination of E. faecalis (23). OG1RF and the ΔelrA mutant were tested in an in vivo-in vitro macrophage infection model, which consists of intraperitoneal infection of mice with 4.5 × 107 and 4.9 × 107 CFU, respectively; recovery of peritoneal macrophages 6 h postinfection; and their in vitro culture to monitor intracellular survival of bacteria over a 72-h period (Fig. 4). Whereas no significant difference was observed at 8 h postinfection, ΔelrA strain bacterial counts recovered from macrophages were significantly decreased at 24 h postinfection (more than 1 log10; P < 0.05) compared to those of OG1RF. This difference was maintained at the same proportion over the 48- and 72-h periods, indicating that long-term survival rates of the two strains are comparable. Together, these results suggested that the ΔelrA strain was defective for infection of peritoneal macrophages, likely due to increased early killing by phagocytes or to decreased initial bacterial multiplication.
FIG. 4.
Time course of intracellular survival of OG1RF and ΔelrA strains within murine peritoneal macrophages. Results correspond to the means ± standard variations of viable intracellular bacteria per 105 macrophages of four independent experiments. White bars, OG1RF; gray bars, ΔelrA strain.
elrA expression is induced in vivo.
Since elrA plays a role in E. faecalis virulence, we examined whether this gene was overexpressed during a bacterial infection in vivo. Quantitative RT-PCR analyses were performed to compare the abundance of elrA mRNAs within total RNA extracts obtained from the OG1RF strain grown in BHI medium or isolated from the peritoneal cavities and spleens of mice 8 h and 20 h postinfection, respectively. Levels of elrA transcripts were 3.5- ± 0.06- and 3.6- ± 0.06-fold higher in the bacteria recovered from peritoneal cavity and spleen, respectively, than in those grown in vitro, indicating that elrA may be specifically upregulated in vivo. This finding was consistent with the lack of difference between the OG1RF and ΔelrA strains during in vitro J774 macrophage infection tests, a laboratory growth condition where ElrA may be insufficiently produced to be fully functional, and with the reduced infection of peritoneal macrophages and virulence of the ΔelrA strain displayed in vivo.
The elrA gene contributes to IL-6 cytokine induction.
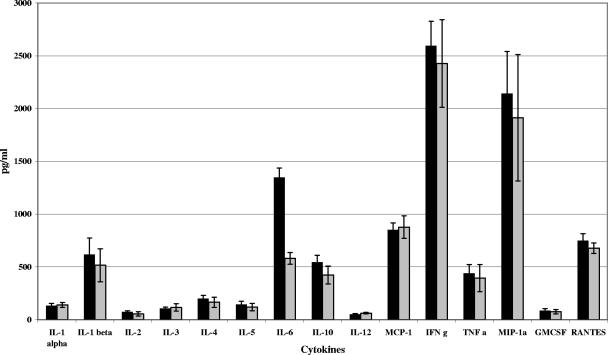
We also examined the contribution of elrA to the inflammatory response to E. faecalis. Using a Q-Plex mouse cytokine array (see Materials and Methods), we semiquantified 16 mouse inflammatory cytokines in peritoneal fluid of mice infected with the ΔelrA or OG1RF strain (Fig. 5). The two strains induced similar cytokine expression profiles, with one exception: levels of IL-6 were at least twofold lower in mice infected with the ΔelrA strain (579 ± 55 pg/ml) than in mice infected with the OG1RF strain (1,348 ± 87 pg/ml), indicating that ElrA is involved in the E. faecalis-mediated induction of IL-6.
FIG. 5.
Cytokine expression profile of peritoneal fluid of mice infected with OG1RF and ΔelrA strains. Peritoneal fluid was recovered from mice infected intraperitoneally with ∼108 CFU of the OG1RF (gray bars) or ΔelrA (black bars) strain. Results show the average cytokine concentration ± standard deviation in pg/ml for three mice. IFN g, gamma interferon; GMCSF, granulocyte-macrophage colony-stimulating factor.
DISCUSSION
In this study, we identified a new E. faecalis virulence factor, ElrA, of the internalin-like protein family. We showed that the elrA gene, which is widespread among E. faecalis isolates, is upregulated and that its product is required during E. faecalis infection in a mouse peritonitis model. Moreover, this protein may be involved in the inflammatory response by stimulating production of IL-6 cytokine, which plays a role in sepsis (27).
ElrA is a LRR-containing protein structurally related to the L. monocytogenes internalin family. Its LRRs contain the 13 amino acid positions implicated in the structure of the internalin LRR domain (41). However, ElrA displays an unusual characteristic compared to canonical internalins, with LRR5 of ElrA containing an insertion of nine residues. Such an insertion has never been observed for other bacterial LRR proteins. Interestingly, seven of the human Toll-like receptors contain insertions at the same position among the LRRs of their ectodomains (5). These insertions are thought to be involved in ligand recognition. In particular, an insertion in LRR8 of TLR9 contains a CpG DNA binding motif that may play a role in binding its ligand (5). Taking into account the similarities of the LRR domain between ElrA, InlA, and InlB and the features of the LRR region, we propose that ElrA may be involved in the interaction with the host and that LRR5 of ElrA may be involved in recognition of a potential host cell ligand.
Most L. monocytogenes internalins are surface proteins predicted to be anchored to the cell wall by an LPXTG motif (14, 54). Some exceptions are InlB, which is loosely associated with bacterial cell wall lipoteichoic acid (34), and four proteins, like InlC, thought to be secreted (17). Just one L. monocytogenes internalin, Lmo0549, has a WxL domain at present uncharacterized (8). In contrast, the only two identified internalin-like proteins in E. faecalis, ElrA and EF2250, both contain a WxL C-terminal region that promotes binding to peptidoglycan (9). They each belong to a gene cluster that was proposed to encode a cell surface complex (59). We show here that the genes in the putative elrA cluster actually comprise an operon. The elrA operon is representative of the organization of other WxL protein clusters, which include a small LPXTG protein and a predicted membrane protein with a DU916 domain (4) of as-yet-unknown function.
Bacterial LRR proteins reportedly contribute to the virulence of gram-negative and gram-positive bacteria (19, 20, 26, 44, 51). In the case of ElrA, its role in virulence was established in the peritonitis murine model, where elrA inactivation resulted in strong attenuation in both infectivity and lethality and correlated with reduced infection of host macrophages in vivo. In contrast, no effect of elrA inactivation on virulence was observed in a Caenorhabditis elegans oral infection (data not shown). These results may be explained in part by the lack of bacterial phagocytosis in C. elegans (18). Alternatively, ElrA interaction with the host may be species specific, as was demonstrated for L. monocytogenes InlA and InlB proteins (35, 37).
Inactivation of elrA did not affect in vitro phagocytosis in J774 murine macrophage cultured cell lines nor in vivo invasion of peritoneal macrophages, indicating that ElrA is not critical for initial entry within macrophages. However, the intracellular survival of the ΔelrA strain in in vivo-infected macrophages was significantly reduced at 24 h postinfection, suggesting that ElrA is involved in early intracellular survival in macrophages, during which wild-type bacteria are still within the phagocytic vacuole (23). Pathogenic bacteria utilize a variety of strategies to avoid macrophage killing (1, 53). For E. faecalis, escape mechanisms have not been elucidated, although oxidative stress response and extracellular polysaccharide synthesis appear to be important (2, 62, 66, 68). This study reveals that ElrA may contribute to survival by evading early bactericidal activities of macrophages.
Expression studies revealed an almost fourfold induction of elrA expression in bacteria extracted from the mouse peritoneal cavity. Low expression of virulence factor ElrA under laboratory conditions may obscure the ΔelrA phenotype. The elrA gene of E. faecalis strain MMH594 was reportedly induced to similar levels during logarithmic growth in serum and in urine and strongly induced during stationary growth in urine compared to a medium devoid of mammal-derived components (2YT) (58). We failed to detect transcriptional induction of elrA of OG1RF in urine and serum using lacZ and luciferase reporter genes (data not shown), suggesting that elrA may be differentially regulated in the two strains. While we consider it likely that ElrA protein expression is increased with the increased mRNA levels, direct detection of ElrA protein is hampered by its cell wall localization (9), as also seen for surface virulence factors in other gram-positive bacteria (8). These observations raise the possibility that in addition to ElrA protein abundance, its accessibility at the bacterial surface may be modulated during the infectious process in vivo, as proposed for InlB of Listeria and Blr and Slr of streptococci (8, 69).
Proinflammatory cytokines play a key role in the inflammatory response and in activation of phagocytes during sepsis (27). Induction of proinflammatory cytokines such as tumor necrosis factor alpha (TNF-α) and IL-6 is mediated through pathogen-associated molecular patterns including surface-associated proteins from various bacteria (30). E. faecalis is known to play a role in the inflammatory response in monomicrobial and polymicrobial intra-abdominal infections by inducing a rapid and robust localized and systemic IL-6 response (45, 47). The mouse peritonitis model that we used mimics sepsis (42). Interestingly, the reduced lethality of the elrA mutant was correlated with a significant reduction in IL-6 proinflammatory cytokine level in the peritoneal cavity. To our knowledge, ElrA protein is the first enterococcal protein that appears to have a significant role in IL-6 induction. Reductions in IL-6 response in in vivo phagocytosis upon infection with the elrA mutant lead us to propose that ElrA may be involved in stimulating the proinflammatory response by favoring E. faecalis infection of macrophages or of other immune cells. However, ElrA is only partially responsible for IL-6 induction, since residual activity remains with the ΔelrA strain, suggesting that IL-6 induction by E. faecalis is multifactorial. Indeed, lipoteichoic acid from E. faecalis stimulates IL-6 production from human monocytes in vitro (6). Interestingly, whereas InlB of L. monocytogenes induces IL-6 and TNF-α through NF-κB (40), ElrA does not seem to induce TNF-α, suggesting that ElrA may induce IL-6 through another regulatory pathway.
By revealing a role of ElrA in the proinflammatory response in a sepsis model, the present study should contribute to elucidating the mechanisms involved in E. faecalis infection. Further investigations will focus on characterizing ElrA expression in vivo and identifying its potential host cell ligand.
Acknowledgments
We are very grateful to A. Gruss and L. Rigottier-Gois for careful reading of the manuscript. We thank P. Y. Allouch, M. Gilmore, M. C. Montel, J. Anba, D. B. Clewell, J. J. Gratadoux, and E. Vandenesch for providing E. faecalis isolates. We thank P. Cossard, I. Schwartz, S. Riffault, X. Roux, S. Chedin, and J. Grosclaude for valuable discussions and for providing access to cell culture facilities and E. Pradel for help with the C. elegans infection model.
This work was supported by the Institut National de la Recherche Agronomique. S.B. was supported by fellowships from the Région Ile-de-France and the Fondation de la Recherche Médicale.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 9 July 2007.
REFERENCES
- 1.Amer, A. O., and M. S. Swanson. 2002. A phagosome of one's own: a microbial guide to life in the macrophage. Curr. Opin. Microbiol. 5:56-61. [DOI] [PubMed] [Google Scholar]
- 2.Baldassarri, L., L. Bertuccini, R. Creti, P. Filippini, M. G. Ammendolia, S. Koch, J. Huebner, and G. Orefici. 2005. Glycosaminoglycans mediate invasion and survival of Enterococcus faecalis into macrophages. J. Infect. Dis. 191:1253-1262. [DOI] [PubMed] [Google Scholar]
- 3.Bar, K., H. Wisplinghoff, R. P. Wenzel, G. M. Bearman, and M. B. Edmond. 2006. Systemic inflammatory response syndrome in adult patients with nosocomial bloodstream infections due to enterococci. BMC Infect. Dis. 6:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell, J. K., G. E. Mullen, C. A. Leifer, A. Mazzoni, D. R. Davies, and D. M. Segal. 2003. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 24:528-533. [DOI] [PubMed] [Google Scholar]
- 6.Bhakdi, S., T. Klonisch, P. Nuber, and W. Fischer. 1991. Stimulation of monokine production by lipoteichoic acids. Infect. Immun. 59:4614-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biedenbach, D. J., G. J. Moet, and R. N. Jones. 2004. Occurrence and antimicrobial resistance pattern comparisons among bloodstream infection isolates from the SENTRY Antimicrobial Surveillance Program (1997-2002). Diagn. Microbiol. Infect. Dis. 50:59-69. [DOI] [PubMed] [Google Scholar]
- 8.Bierne, H., C. Sabet, N. Personnic, and P. Cossart. 2007. Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect. doi: 10.1016/j.micinf.2007.05.003. [DOI] [PubMed]
- 9.Brinster, S., S. Furlan, and P. Serror. 2007. C-terminal WxL domain mediates cell wall binding in Enterococcus faecalis and other gram-positive bacteria. J. Bacteriol. 189:1244-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchanan, S. G., and N. J. Gay. 1996. Structural and functional diversity in the leucine-rich repeat family of proteins. Prog. Biophys. Mol. Biol. 65:1-44. [DOI] [PubMed] [Google Scholar]
- 11.Delogu, G., M. Sanguinetti, B. Posteraro, S. Rocca, S. Zanetti, and G. Fadda. 2006. The hbhA gene of Mycobacterium tuberculosis is specifically upregulated in the lungs but not in the spleens of aerogenically infected mice. Infect. Immun. 74:3006-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delogu, G., M. Sanguinetti, C. Pusceddu, A. Bua, M. J. Brennan, S. Zanetti, and G. Fadda. 2006. PE_PGRS proteins are differentially expressed by Mycobacterium tuberculosis in host tissues. Microbes Infect. 8:2061-2067. [DOI] [PubMed] [Google Scholar]
- 13.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dramsi, S., P. Dehoux, M. Lebrun, P. L. Goossens, and P. Cossart. 1997. Identification of four new members of the internalin multigene family of Listeria monocytogenes EGD. Infect. Immun. 65:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. USA 75:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunny, G. M., L. N. Lee, and D. J. LeBlanc. 1991. Improved electroporation and cloning vector system for gram-positive bacteria. Appl. Environ. Microbiol. 57:1194-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelbrecht, F., S. K. Chun, C. Ochs, J. Hess, F. Lottspeich, W. Goebel, and Z. Sokolovic. 1996. A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol. Microbiol. 21:823-837. [DOI] [PubMed] [Google Scholar]
- 18.Fares, H., and I. Greenwald. 2001. Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics 159:133-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fedhila, S., N. Daou, D. Lereclus, and C. Nielsen-LeRoux. 2006. Identification of Bacillus cereus internalin and other candidate virulence genes specifically induced during oral infection in insects. Mol. Microbiol. 62:339-355. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez-Prada, C. M., D. L. Hoover, B. D. Tall, A. B. Hartman, J. Kopelowitz, and M. M. Venkatesan. 2000. Shigella flexneri IpaH7.8 facilitates escape of virulent bacteria from the endocytic vacuoles of mouse and human macrophages. Infect. Immun. 68:3608-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fouet, A., and A. L. Sonenshein. 1990. A target for carbon source-dependent negative regulation of the citB promoter of Bacillus subtilis. J. Bacteriol. 172:835-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gauduchon, V., L. Chalabreysse, J. Etienne, M. Celard, Y. Benito, H. Lepidi, F. Thivolet-Bejui, and F. Vandenesch. 2003. Molecular diagnosis of infective endocarditis by PCR amplification and direct sequencing of DNA from valve tissue. J. Clin. Microbiol. 41:763-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gentry-Weeks, C. R., R. Karkhoff-Schweizer, A. Pikis, M. Estay, and J. M. Keith. 1999. Survival of Enterococcus faecalis in mouse peritoneal macrophages. Infect. Immun. 67:2160-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giard, J.-C., E. Riboulet, N. Verneuil, M. Sanguinetti, Y. Auffray, and A. Hartke. 2006. Characterization of Ers, a PrfA-like regulator of Enterococcus faecalis. FEMS Immunol. Med. Microbiol. 46:410-418. [DOI] [PubMed] [Google Scholar]
- 25.Gibson, T. J. 1984. Studies on the Epstein-Barr virus genome. Ph.D. thesis. University of Cambridge, Cambridge, United Kingdom.
- 26.Grosdent, N., I. Maridonneau-Parini, M. P. Sory, and G. R. Cornelis. 2002. Role of Yops and adhesins in resistance of Yersinia enterocolitica to phagocytosis. Infect. Immun. 70:4165-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hack, C. E., L. A. Aarden, and L. G. Thijs. 1997. Role of cytokines in sepsis. Adv. Immunol. 66:101-195. [DOI] [PubMed] [Google Scholar]
- 28.Hamon, M., H. Bierne, and P. Cossart. 2006. Listeria monocytogenes: a multifaceted model. Nat. Rev. Microbiol. 4:423-434. [DOI] [PubMed] [Google Scholar]
- 29.Hancock, L. E., and M. S. Gilmore. 2002. The capsular polysaccharide of Enterococcus faecalis and its relationship to other polysaccharides in the cell wall. Proc. Natl. Acad. Sci. USA 99:1574-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henderson, B., S. Poole, and M. Wilson. 1996. Bacterial modulins: a novel class of virulence factors which cause host tissue pathology by inducing cytokine synthesis. Microbiol. Rev. 60:316-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huebner, J., Y. Wang, W. A. Krueger, L. C. Madoff, G. Martirosian, S. Boisot, D. A. Goldmann, D. L. Kasper, A. O. Tzianabos, and G. B. Pier. 1999. Isolation and chemical characterization of a capsular polysaccharide antigen shared by clinical isolates of Enterococcus faecalis and vancomycin-resistant Enterococcus faecium. Infect. Immun. 67:1213-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jett, B. D., M. M. Huycke, and M. S. Gilmore. 1994. Virulence of enterococci. Clin. Microbiol. Rev. 7:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones, M. E., D. C. Draghi, C. Thornsberry, J. A. Karlowsky, D. F. Sahm, and R. P. Wenzel. 2004. Emerging resistance among bacterial pathogens in the intensive care unit—a European and North American surveillance study (2000-2002). Ann. Clin. Microbiol. Antimicrob. 3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jonquieres, R., H. Bierne, F. Fiedler, P. Gounon, and P. Cossart. 1999. Interaction between the protein InlB of Listeria monocytogenes and lipoteichoic acid: a novel mechanism of protein association at the surface of Gram-positive bacteria. Mol. Microbiol. 34:902-914. [DOI] [PubMed] [Google Scholar]
- 35.Khelef, N., M. Lecuit, H. Bierne, and P. Cossart. 2006. Species specificity of the Listeria monocytogenes InlB protein. Cell. Microbiol. 8:457-470. [DOI] [PubMed] [Google Scholar]
- 36.Kobe, B., and A. V. Kajava. 2001. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11:725-732. [DOI] [PubMed] [Google Scholar]
- 37.Lecuit, M., S. Dramsi, C. Gottardi, M. Fedor-Chaiken, B. Gumbiner, and P. Cossart. 1999. A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 18:3956-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Loir, Y., S. Nouaille, J. Commissaire, L. Bretigny, A. Gruss, and P. Langella. 2001. Signal peptide and propeptide optimization for heterologous protein secretion in Lactococcus lactis. Appl. Environ. Microbiol. 67:4119-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linden, P. 2003. Can enterococcal infections initiate sepsis syndrome? Curr. Infect. Dis. Rep. 5:372-378. [DOI] [PubMed] [Google Scholar]
- 40.Mansell, A., L. Braun, P. Cossart, and L. A. O'Neill. 2000. A novel function of InIB from Listeria monocytogenes: activation of NF-kappaB in J774 macrophages. Cell. Microbiol. 2:127-136. [DOI] [PubMed] [Google Scholar]
- 41.Marino, M., L. Braun, P. Cossart, and P. Ghosh. 2000. A framework for interpreting the leucine-rich repeats of the Listeria internalins. Proc. Natl. Acad. Sci. USA 97:8784-8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matlow, A. G., J. M. Bohnen, C. Nohr, N. Christou, and J. Meakins. 1989. Pathogenicity of enterococci in a rat model of fecal peritonitis. J. Infect. Dis. 160:142-145. [DOI] [PubMed] [Google Scholar]
- 43.Meijerink, J., C. Mandigers, L. van de Locht, E. Tonnissen, F. Goodsaid, and J. Raemaekers. 2001. A novel method to compensate for different amplification efficiencies between patient DNA samples in quantitative real-time PCR. J. Mol. Diagn. 3:55-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miao, E. A., C. A. Scherer, R. M. Tsolis, R. A. Kingsley, L. G. Adams, A. J. Baumler, and S. I. Miller. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34:850-864. [DOI] [PubMed] [Google Scholar]
- 45.Montravers, P., J. Mohler, L. Saint Julien, and C. Carbon. 1997. Evidence of the proinflammatory role of Enterococcus faecalis in polymicrobial peritonitis in rats. Infect. Immun. 65:144-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pai, S. R., K. V. Singh, and B. E. Murray. 2003. In vivo efficacy of the ketolide ABT-773 (cethromycin) against enterococci in a mouse peritonitis model. Antimicrob. Agents Chemother. 47:2706-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papasian, C. J., R. Silverstein, J. J. Gao, D. M. Bamberger, and D. C. Morrison. 2002. Anomalous role of tumor necrosis factor alpha in experimental enterococcal infection. Infect. Immun. 70:6628-6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paulsen, I. T., L. Banerjei, G. S. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 49.Pizarro-Cerda, J., and P. Cossart. 2006. Bacterial adhesion and entry into host cells. Cell 124:715-727. [DOI] [PubMed] [Google Scholar]
- 50.Rakita, R. M., N. N. Vanek, K. Jacques-Palaz, M. Mee, M. M. Mariscalco, G. M. Dunny, M. Snuggs, W. B. Van Winkle, and S. I. Simon. 1999. Enterococcus faecalis bearing aggregation substance is resistant to killing by human neutrophils despite phagocytosis and neutrophil activation. Infect. Immun. 67:6067-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reid, S. D., A. G. Montgomery, J. M. Voyich, F. R. DeLeo, B. Lei, R. M. Ireland, N. M. Green, M. Liu, S. Lukomski, and J. M. Musser. 2003. Characterization of an extracellular virulence factor made by group A Streptococcus with homology to the Listeria monocytogenes internalin family of proteins. Infect. Immun. 71:7043-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richards, M. J., J. R. Edwards, D. H. Culver, and R. P. Gaynes. 2000. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect. Control Hosp. Epidemiol. 21:510-515. [DOI] [PubMed] [Google Scholar]
- 53.Rosenberger, C. M., and B. B. Finlay. 2003. Phagocyte sabotage: disruption of macrophage signalling by bacterial pathogens. Nat. Rev. Mol. Cell Biol. 4:385-396. [DOI] [PubMed] [Google Scholar]
- 54.Sabet, C., M. Lecuit, D. Cabanes, P. Cossart, and H. Bierne. 2005. LPXTG protein InlJ, a newly identified internalin involved in Listeria monocytogenes virulence. Infect. Immun. 73:6912-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 56.Schlievert, P. M., P. J. Gahr, A. P. Assimacopoulos, M. M. Dinges, J. A. Stoehr, J. W. Harmala, H. Hirt, and G. M. Dunny. 1998. Aggregation and binding substances enhance pathogenicity in rabbit models of Enterococcus faecalis endocarditis. Infect. Immun. 66:218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schubert, W. D., G. Gobel, M. Diepholz, A. Darji, D. Kloer, T. Hain, T. Chakraborty, J. Wehland, E. Domann, and D. W. Heinz. 2001. Internalins from the human pathogen Listeria monocytogenes combine three distinct folds into a contiguous internalin domain. J. Mol. Biol. 312:783-794. [DOI] [PubMed] [Google Scholar]
- 58.Shepard, B. D., and M. S. Gilmore. 2002. Differential expression of virulence-related genes in Enterococcus faecalis in response to biological cues in serum and urine. Infect. Immun. 70:4344-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siezen, R., J. Boekhorst, L. Muscariello, D. Molenaar, B. Renckens, and M. Kleerebezem. 2006. Lactobacillus plantarum gene clusters encoding putative cell-surface protein complexes for carbohydrate utilization are conserved in specific gram-positive bacteria. BMC Genomics 7:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh, K. V., X. Qin, G. M. Weinstock, and B. E. Murray. 1998. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J. Infect. Dis. 178:1416-1420. [DOI] [PubMed] [Google Scholar]
- 61.Sussmuth, S. D., A. Muscholl-Silberhorn, R. Wirth, M. Susa, R. Marre, and E. Rozdzinski. 2000. Aggregation substance promotes adherence, phagocytosis, and intracellular survival of Enterococcus faecalis within human macrophages and suppresses respiratory burst. Infect. Immun. 68:4900-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teng, F., K. D. Jacques-Palaz, G. M. Weinstock, and B. E. Murray. 2002. Evidence that the enterococcal polysaccharide antigen gene (epa) cluster is widespread in Enterococcus faecalis and influences resistance to phagocytic killing of E. faecalis. Infect. Immun. 70:2010-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsolis, R. M., S. M. Townsend, E. A. Miao, S. I. Miller, T. A. Ficht, L. G. Adams, and A. J. Bäumler. 1999. Identification of a putative Salmonella enterica serotype Typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect. Immun. 67:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsutsui, O., S. Kokeguchi, T. Matsumura, and K. Kato. 1991. Relationship of the chemical structure and immunobiological activities of lipoteichoic acid from Streptococcus faecalis (Enterococcus hirae) ATCC 9790. FEMS Microbiol. Immunol. 3:211-218. [DOI] [PubMed] [Google Scholar]
- 65.Vanek, N. N., S. I. Simon, K. Jacques-Palaz, M. M. Mariscalco, G. M. Dunny, and R. M. Rakita. 1999. Enterococcus faecalis aggregation substance promotes opsonin-independent binding to human neutrophils via a complement receptor type 3-mediated mechanism. FEMS Immunol. Med. Microbiol. 26:49-60. [DOI] [PubMed] [Google Scholar]
- 66.Verneuil, N., A. Maze, M. Sanguinetti, J. M. Laplace, A. Benachour, Y. Auffray, J. C. Giard, and A. Hartke. 2006. Implication of (Mn)superoxide dismutase of Enterococcus faecalis in oxidative stress responses and survival inside macrophages. Microbiology 152:2579-2589. [DOI] [PubMed] [Google Scholar]
- 67.Verneuil, N., A. Rince, M. Sanguinetti, Y. Auffray, A. Hartke, and J. C. Giard. 2005. Implication of hypR in the virulence and oxidative stress response of Enterococcus faecalis. FEMS Microbiol. Lett. 252:137-141. [DOI] [PubMed] [Google Scholar]
- 68.Verneuil, N., M. Sanguinetti, Y. Le Breton, B. Posteraro, G. Fadda, Y. Auffray, A. Hartke, and J. C. Giard. 2004. Effects of the Enterococcus faecalis hypR gene encoding a new transcriptional regulator on oxidative stress response and intracellular survival within macrophages. Infect. Immun. 72:4424-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waldemarsson, J., T. Areschoug, G. Lindahl, and E. Johnsson. 2006. The streptococcal Blr and Slr proteins define a family of surface proteins with leucine-rich repeats: camouflaging by other surface structures. J. Bacteriol. 188:378-388. [DOI] [PMC free article] [PubMed] [Google Scholar]