Abstract
Concerted morphological and sequencing-based strategies revealed the identity of a nonsporulating clinical isolate as Petromyces alliaceus (anamorph Aspergillus alliaceus). This rare Aspergillus sp. was recovered as the etiological agent of invasive pulmonary aspergillosis and had reduced in vitro susceptibilities to amphotericin B and caspofungin, which correlated with clinical failure of therapy.
A 54-year-old woman underwent sibling donor allogeneic hematopoietic cell transplantation for refractory pre-B acute lymphoblastic leukemia. On day 2 posttransplantation, amphotericin B lipid complex (ABLC) therapy (1 mg/kg daily) was begun as antifungal prophylaxis. Glucocorticosteroids were begun immediately posttransplantation for graft-versus-host disease prophylaxis and continued through her hospital course. On day 10 itraconazole (ITZ) was substituted for the ABLC because of nephrotoxicity; on day 26 the patient developed cardiopulmonary failure and required intubation. A chest radiograph at that time revealed pulmonary infiltrates. Bronchoscopy revealed serosanguinous fluid, and cytologic examination of the bronchoalveolar lavage fluid showed branching septate hyphae; however, a KOH smear did not reveal any hyphae, nor did the culture grow any fungus at this time. A diagnosis of probable invasive pulmonary fungal infection was made based upon the EORTC/MSG criteria (i.e., the European Organization for Research and Treatment of Cancer/Mycoses Study Group [1]) since the first bronchoscopy demonstrated fungal hyphae on cytologic examination, and the second bronchoscopy grew out the mold. On day 27, ITZ treatment was discontinued, and ABLC (5 mg/kg daily) and caspofungin (CAS; 70 mg loading dose, followed by 50 mg daily) treatment were initiated. A serum Aspergillus galactomannan enzyme immunoassay on day 28 was negative; this could be, as previously shown (9), because the patient was on prophylactic antifungal therapy prior to the onset of infection. On day 38 a second bronchoscopy was performed; the bronchoalveolar lavage fluid was negative for fungal elements as determined by direct microscopy. However, the fluid grew a “white, fuzzy mold” that was reported as “nonsporulating mold.” A chest computed tomography scan obtained on day 43 revealed three cavitary lesions with patchy areas of consolidation and diffuse interstitial infiltrates. The patient continued to deteriorate and died on day 48. An autopsy was not performed. The patient had been on high doses of corticosteroids throughout her posttransplantation period.
No further attempt was made to identify the fungal isolate, and the organism was sent to the Fungal Reference Unit, Centers for Disease Control and Prevention, for detailed investigation as part of a surveillance study for invasive fungal infections in transplant recipients (10). The isolate was subjected to both morphological and molecular identification. Genomic DNA was extracted from the fungus, and the internal transcribed spacer regions (ITS) and 28S ribosomal regions were PCR amplified and sequenced as described previously (7, 8). Primer sequences used for both PCR and sequencing were as follows: forward, 5′-TCCTCCGCTTATTGATATGC and GGAAGTAAAAGTGGTAACAAGG (ITS4 and ITS5 [8]) for the ITS regions; and forward, 5′-GCATATCAATAAGCGGAGGAAAAG, and reverse, 5′-GGTCCGTGTTTCAAGACGG, for the D1 and D2 regions of the 28S rRNA (7). The resultant sequences (amplicon sizes of 594 and 577 bp for the ITS and 28S regions, respectively) were then compared to the available sequences in the GenBank database using the program BLASTn. The results revealed that the sequence was 100% homologous to Petromyces alliaceus (GenBank accession nos. AF149755 and AF149754) and 98% homologous to P. albertensis (GenBank accession no. AJ005673) and A. lanosus (GenBank accession no. AF203810) in the ITS region. When the more conserved 28S region was analyzed, the queried sequence was 100% homologous to P. alliaceus (GenBank accession no. PAU15496 and PAU28913), P. albertensis (GenBank accession no. PAU28910), and A. lanosus (GenBank accession no. ALU28914).
The sequences generated in this study have been submitted to GenBank under accession numbers EF447422 and EF447423.
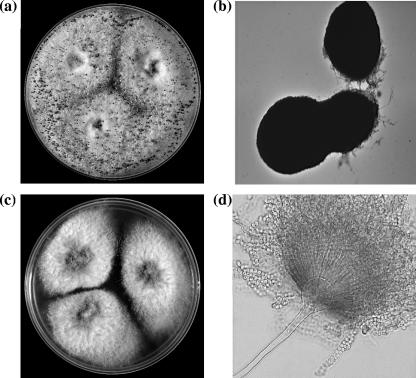
Subsequently, the fungus was subcultured on Sabouraud dextrose agar (SDA; Becton Dickinson, Sparks, MD) supplemented with chloramphenicol and gentamicin at 25°C. The isolate grew predominantly as sterile hyphae interspersed with large ellipsoidal to cylindrical dark brown ascomata produced directly from the vegetative hyphae (Fig. 1a and b). Microscopic examinations of sections of these structures revealed that they were composed of sclerenchymatous tissue and were devoid of spores. Asexual sporulation was induced by culturing the isolate on SDA (without antibiotics) at 25°C in the presence of a constant supply of light. Under these conditions, the isolate produced orange yellow conidial heads indicative of the genus Aspergillus. Conidiophores were smooth with globose vesicles that gave rise to radiating, biseriate conidial heads producing smooth-walled, globose conidia (Fig. 1d). Figure 1c shows colonies growing from three-point inoculation that is used typically for Penicillium and Aspergillus species identification. The isolate grew well at 30, 35, and 37°C and sporulated abundantly in the presence of light.
FIG. 1.
(a) Plate showing 10-day-old colonies on SDA with sterile hyphae interspersed with black ascomata produced in concentric circles after incubation at 30°C. (b) Ellipsoidal, thick-walled, dark ascomata produced by P. alliaceus. Magnification, ×100. (c) Colonies of P. alliaceus growing predominantly in the asexual stage producing orange yellow conidia after growth on SDA for 10 days at 37°C. (d) Smooth conidiophores with radiating, biseriate conidial heads. Magnification, ×400.
The hallmark of P. alliaceus is the production of ascomata, whereas A. lanosus does not produce ascomata; furthermore, P. albertensis is now considered synonymous with P. alliaceus (14). Although not observed in this study, P. alliaceus upon prolonged incubation (several months) is known to produce ascocarps within the stromata (6). Thus, with the combined phenotypic and genotypic evidence, the unknown fungal isolate was identified as P. alliaceus. The teleomorphic species P. alliaceus is one of only three members assigned to the genus Petromyces. Although Petromyces spp. have been isolated from surfaces of plants and soil, the P. alliaceus is rarely recovered from an invasive fungal infection and has been previously isolated from a case of chronic otorrhea (drainage from the auditory canal). Petromyces alliaceus produces a variety of secondary metabolites, including ochratoxin A and B. Although the anamorph of the teleomorphic genus Petromyces was originally assigned to the Aspergillus section Circumdati, recent chemotaxonomic and genotypic evidence places this genus in Aspergillus section Flavi, whose members often produce sclerotia (13, 14).
The in vitro susceptibilities of P. alliaceus to the antifungal drugs amphotericin B (AmB), natamycin (NAT), voriconazole (VRZ), ITZ, and CAS were determined by the broth microdilution method as outlined in CLSI document M38A (5) and for all drugs except NAT by the Etest methodologies (12). The results of the broth microdilution method showed that the antifungal drug MICs were high (AmB, 8 μg/ml; NAT, >32 μg/ml; ITZ, 0.125 μg/ml; and 1 μg/ml, VRZ) for the isolate and that the minimum effective concentration (defined as the minimum concentration of CAS that produced morphological alterations as viewed under a light microscope) was >8 μg/ml for CAS. These MICs were reproducible (the analyses were repeated twice with similar results). The Etest MICs were comparable with an MIC of >32 μg/ml for both AmB and CAS. The patient failed therapy with both ABLC and CAS, and this correlated well with the high in vitro MICs for both of these antifungals. Higher MICs for the cell wall active drugs (AmB, NAT, and CAS) may indicate differences in cell wall architecture in this previously unrecognized Aspergillus isolate.
It is being increasingly regarded that traditional approaches to fungal identification will gradually be replaced by DNA-based identification methods (11). While P. alliaceus isolated in the present study was at first refractory to classical identification (because of the absence of sporulation), morphologically identifiable features were eventually induced by growth in different conditions, media, and temperatures. Although molecular methods directed the identification, delineation between the two closely related species A. lanosus and P. alliaceus was secured by the observation of ascomata produced by P. alliaceus.
Genotyping methods are more rapid and easier to perform than phenotypic methods, and yet the latter approach has and will continue to be used in the clinical microbiology laboratory due to its simplicity, economy, and reliable identification most of the time. Thus, a prudent approach to fungal identification would be to augment, rather than to replace, classical methods with molecular methods. Another advantage of pursuing morphology-based methods for identifying aspergilli is that antifungal susceptibility profiles can be generated with the induction of a sporulating phenotype; as shown in this study, this information may be important in guiding treatment strategies given the low susceptibility of this isolate to several antifungal agents.
Although isolation of Petromyces as a cause of invasive infection is rare, there appear to be increasing reports of such previously unrecognized aspergilli with low susceptibilities to antifungal agents in clinical settings (2-4). The present study highlights two important, but often overlooked factors: (i) classical methods, if applied appropriately, have a sustained and significant role in the clinical microbiology laboratory, and (ii) the identification of “nonsporulating” filamentous aspergilli from ultra-high-risk patients may be important, given the reduced susceptibility of the P. alliaceus strain isolated here.
Acknowledgments
We thank the members of the TransNet (surveillance for invasive fungal infections in transplant recipients) Study Group from the University of Alabama at Birmingham, the Centers for Disease Control and Prevention, and the participating sites.
The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the CDC.
Footnotes
Published ahead of print on 30 May 2007.
REFERENCES
- 1.Ascioglu, S., J. H. Rex, B. de Pauw, J. E. Bennett, J. Bille, F. Crokaert, D. W. Denning, J. P. Donnelly, J. E. Edwards, Z. Erjavec, D. Fiere, O. Lortholary, J. Maertens, J. F. Meis, T. F. Patterson, J. Ritter, D. Selleslag, P. M. Shah, D. A. Stevens, and T. J. Walsh. 2002. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clin. Infect. Dis. 34:7-14. [DOI] [PubMed] [Google Scholar]
- 2.Balajee, S. A., J. Gribskov, M. Brandt, J. Ito, A. Fothergill, and K. A. Marr. 2005. Mistaken identity: Neosartorya pseudofischeri and its anamorph masquerading as Aspergillus fumigatus. J. Clin. Microbiol. 43:5996-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balajee, S. A., J. L. Gribskov, E. Hanley, D. Nickle, and K. A. Marr. 2005. Aspergillus lentulus sp. nov., a new sibling species of A. fumigatus. Eukaryot. Cell 4:625-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balajee, S. A., D. Nickle, J. Varga, and K. A. Marr. 2006. Molecular studies reveal frequent misidentification of Aspergillus fumigatus by morphotyping. Eukaryot. Cell 5:1705-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical Laboratory Standards Institute. 2002. Reference method for broth dilution antifungal susceptibility testing of conidium-forming filamentous fungi: approved standard M38-A. CLSI, Wayne, PA.
- 6.Klich, M. A. 2002. Identification of common Aspergillus species, vol. 1. Centralbureau voor Schimmelcultures, Utrecht, The Netherlands.
- 7.Kurtzman, C. P., and C. J. Robnett. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 35:1216-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larena, I., O. Salazar, V. Gonzalez, M. C. Julian, and V. Rubio. 1999. Design of a primer for ribosomal DNA internal transcribed spacer with enhanced specificity for ascomycetes. J. Biotechnol. 75:187-194. [DOI] [PubMed] [Google Scholar]
- 9.Marr, K. A., and W. Leisenring. 2005. Design issues in studies evaluating diagnostic tests for aspergillosis. Clin. Infect. Dis. 41(Suppl. 6):S381-S386. [DOI] [PubMed] [Google Scholar]
- 10.Morgan, J., K. A. Wannemuehler, K. A. Marr, S. Hadley, D. P. Kontoyiannis, T. J. Walsh, S. K. Fridkin, P. G. Pappas, and D. W. Warnock. 2005. Incidence of invasive aspergillosis following hematopoietic stem cell and solid organ transplantation: interim results of a prospective multicenter surveillance program. Med. Mycol. 43(Suppl. 1):S49-S58. [DOI] [PubMed] [Google Scholar]
- 11.Odds, F. C. 2003. Reflections on the question: what does molecular mycology have to do with the clinician treating the patient? Med. Mycol. 41:1-6. [PubMed] [Google Scholar]
- 12.Pfaller, M. A., S. A. Messer, L. Boyken, R. J. Hollis, and D. J. Diekema. 2003. In vitro susceptibility testing of filamentous fungi: comparison of Etest and reference M38-A microdilution methods for determining posaconazole MICs. Diagn. Microbiol. Infect. Dis. 45:241-244. [DOI] [PubMed] [Google Scholar]
- 13.Rigo, K., J. Varga, B. Toth, J. Teren, A. Mesterhazy, and Z. Kozakiewicz. 2002. Evolutionary relationships within Aspergillus section Flavi based on sequences of the intergenic transcribed spacer regions and the 5.8S rRNA gene. J. Gen. Appl. Microbiol. 48:9-16. [DOI] [PubMed] [Google Scholar]
- 14.Varga, J., B. Toth, E. Kevei, A. Palagyi, and Z. Kozakiewicz. 2000. Analysis of genetic variability within the genus Petromyces. Antonie Leeuwenhoek 77:83-89. [DOI] [PubMed] [Google Scholar]