Abstract
Yersinia enterocolitica bioserogroup 4/O3 is the predominant causative agent of yersiniosis in Europe and North America. Multiple-locus variable-number tandem-repeat analysis (MLVA) was developed to improve the resolution power of classical genotyping methods. MLVA based on six loci was able to distinguish 76 genotypes among 91 Y. enterocolitica isolates of worldwide origin and 41 genotypes among 51 nonepidemiologically linked bioserogroup 4/O3 isolates, proving that it has a high resolution power. However, only a slight correlation of the MLVA genotypes and the geographic distribution of the isolates was observed. Although MLVA was also capable of distinguishing strains of Y. enterocolitica subsp. palearctica O9 and O5,27, there was only a minor correlation between the MLVA genotypes and serogroups. MLVA may be a helpful tool for epidemiological investigations of Y. enterocolitica subsp. palearctica outbreaks.
Yersinia enterocolitica is a pathogen that is found worldwide and that is associated with yersiniosis in humans and animals. A large variety of clinical and immunologic manifestations, ranging from mild diarrhea, pseudoappendicular syndrome, and transfusion-related septicemia to postinfectious complications like erythema nodosum and reactive arthritis, have been described (7). Y. enterocolitica is a highly heterogeneous species (5, 7, 15, 22, 30). The species has recently been subdivided into two subspecies: Y. enterocolitica subsp. palearctica and Y. enterocolitica subsp. enterocolitica, which comprise all European and American pathogenic bioserovars, respectively (22). Since then, Howard and coworkers, using a comparative phylogenomic analysis of Y. enterocolitica strains, obtained data that argue toward the establishment of a third subspecies (15). The virulent strains of Y. enterocolitica subsp. palearctica serogroups O3, O5,27, and O9 are the main causative agents of yersiniosis in Europe. Moreover, despite the initial predominance of Y. enterocolitica subsp. enterocolitica serogroup O8 in the United States in the middle to late 1980s, serogroup O3 has emerged in that part of the world (5). Thus, Y. enterocolitica O3 strains are now considered the most frequent causative agent of yersiniosis worldwide.
Pathogenic strains belonging to the serotypes mentioned above have been proved to possess a relatively high degree of genetic homogeneity compared to that for the nonpathogenic strains of bioserogroup 1A/O5 isolated from environmental samples (30). Therefore, several typing techniques based on both phenotypic and genetic markers have been applied to discriminate pathogenic Y. enterocolitica isolates. The most discriminatory methods indicated that isolates of serotype O3 are the most clonal among other pathogenic serotypes of Y. enterocolitica subsp. palearctica (10, 16).
Biochemical differentiation (biotyping), the most simple and widely available method, is able to separate Y. enterocolitica O3 into biotypes 2, 3, and 4, with biotype 4 (BT4) causing the majority of clinical cases (11, 12, 20). Most Y. enterocolitica 4/O3 isolates belong to phage type VIII (21). The low discriminatory power of the phenotype-related methods prompted the application of genotyping. However, the basic genotyping techniques, i.e., restriction endonuclease analysis of chromosomal DNA and ribotyping, offered only a low resolution (6). A more advanced PCR-ribotyping method generated only seven profiles for more than 60 strains of Y. enterocolitica O3, with the majority of BT4 strains grouped in PCR-ribotype SR1 (20). Application of enterobacterial repetitive intergenic consensus and repetitive extragenic palindromic sequences resulted in only a minor improvement of the resolution (31).
A higher degree of heterogeneity of Y. enterocolitica O3 isolates was reported by use of the current genotyping standard, pulsed-field gel electrophoresis (PFGE). Eleven NotI pulsotypes were distinguished among 20 strains of bioserogroup 4/O3 of worldwide origin (21). Application of an additional enzyme was reported to improve the discriminatory power of PFGE (1, 11). The recently developed fluorescent amplified fragment length polymorphism method demonstrated a relatively high discriminatory power by distinguishing 13 genotypes among 17 strains of Y. enterocolitica 4/O3 (10). However, this method required the use of an automated DNA sequencer and sophisticated software.
One of the most recent developments in molecular typing involves the analysis of variable-number tandem-repeat (VNTR) sequences (9, 17, 19, 23, 25, 29). VNTRs are repeated sequences and range in number from a pair to hundreds of nucleotides found in tandem. Various copy numbers are generated through a number of mechanisms, including slippage and mispairing during DNA replication, creating a different allele (9). These mutations can be detected by PCR with primers complementary to regions flanking the VNTR sequences. Analysis of multiple VNTRs enables high-resolution genotyping (29). Multiple-locus VNTR analysis (MLVA) may use two up to dozens of VNTR loci (19, 29). In general, the use of five to eight loci provides satisfactory resolution, and the workload that results from the use of this number of loci is acceptable (17, 22, 27).
MLVA has already been used for the efficient genotyping of many bacterial pathogens (9, 22, 27, 28), including Yersinia pestis (14, 18, 25) and Bacillus anthracis (17, 18, 19), the most homogeneous species known. However, recent examination of mutational events in VNTR loci in Escherichia coli O157:H7 showed that the use of hypervariable loci may limit the validity of MLVA for epidemiological investigations (24).
In 2003 the VNTR of the virginiamycin acetyltransferase gene vat (orf528) of Y. enterocolitica was used to differentiate 14 genotypes (3), but the resolution power from the use of a single locus proved to be insufficient for epidemiological investigations.
The present study describes MLVA, a novel, high-resolution genotyping tool designed for Y. enterocolitica subsp. palearctica subtyping based on six VNTR markers. This technique requires only basic molecular biology equipment and conventional cluster analysis software. It is therefore easily implemented in the routine laboratory.
MATERIALS AND METHODS
Bacterial strains.
A total of 99 Y. enterocolitica strains of worldwide origin were tested, including strains of serogroups O3 (n = 80), O5,27 (n = 4), O8 (n = 6), and O9 (n = 5). Among the Y. enterocolitica O3 strains, there were isolates from the same town and the same clinic. Strains of bioserogroup 4/O3 (n = 62) were mainly obtained from clinical cases in Europe. Three isolates (isolates 45A/06, 45B/06, and 46/06) were from blood transfusion-related cases of yersiniosis in Poland. Two groups of epidemiologically linked Y. enterocolitica 4/O3 isolates of porcine origin (groups hut1 [n = 3] and hut2 [n = 9]) were included. All strains tested are characterized in Table 1. Six colonies of two laboratory Y. enterocolitica O3 strains (strains P2 and P9) were subjected to 20 serial passages at 27°C on solid medium (LB; Difco, Germany) in order to test stabilities of the VNTR genotypes. This process yielded an in vitro population representing about 2,400 generations for each strain.
TABLE 1.
Y. enterocolitica strains grouped by serogroup, biotype, phage type, and geographic location and source
| No. of isolatesa | Strain(s)b | Serogroup | Biotype | Phage type | Country | Source |
|---|---|---|---|---|---|---|
| 1 | 305/96 | O3 | 4 | —c | Poland | Human |
| 20 | P2, P5 to P23 | O3 | 4 | — | Poland | Human |
| 3 | UG1 to UG3d | O3 | 4 | — | Poland (hut1) | Pig |
| 9 | UG10 to UG18d | O3 | 4 | — | Poland (hut2) | Pig |
| 3 | 45A/06, 45B/06, 46/06d | O3 | 4 | — | Poland | Blood |
| 1 | Y11 | O3 | 4 | — | Germany | Human |
| 1 | 108 | O3 | 4 | — | Germany | Human |
| 2 | M1, M2d | O3 | — | — | Germany | Human |
| 9 | M5, M7 to M12 | O3 | — | — | Germany | Human |
| 1 | 131 | O3 | — | — | Germany | — |
| 2 | Y745, Y746 | O3 | 3 | II | Japan | Human |
| 1 | Y747 (IP134) | O3 | 4 | — | Sweden | Human |
| 1 | 556 (8265) | O3 | 4 | — | France | Human |
| 1 | Y748 (IP21981) | O3 | 4 | — | France | Human |
| 1 | Y749 (IP1601) | O3 | 4 | — | Japan | Human |
| 1 | Y750 (IP18718) | O3 | 4 | VIII | China | Human |
| 1 | Y751 (IP23222) | O3 | 4 | VIII | Great Britain | Human |
| 1 | Y752 (IP23357) | O3 | 4 | VIII | Brazil | Human |
| 2 | Y753, Y754 | O3 | 4 | VIII | New Caledonia | Human |
| 2 | Y755, Y756 | O3 | 4 | IXa | South Africa | Human |
| 2 | Y757, Y758 | O3 | 4 | IXa | Hungary | Human |
| 3 | Y759, Y763, Y764 | O3 | 4 | IXb | Canada | Human |
| 4 | Y765 to Y768 | O3 | 4 | IXb | Australia | Human |
| 3 | Y769 to Y771 | O3 | 4 | IXb | New Zealand | Human |
| 1 | 560 (SW13123) | O3 | 4 | — | Japan | Pig |
| 1 | 531 (SW13711) | O3 | — | — | Japan | Pig |
| 1 | 123 | O3 | — | — | Belgium | Human |
| 2 | 141, 143 | O3 | — | — | Belgium | Pig |
| 1 | 148 (Y485) | O3 | — | — | — | Pig |
| 1 | 150 (Y486) | O3 | — | — | — | Calve |
| 1 | 568 (Ye873) | O5,27 | 2 | — | Canada | Pig |
| 1 | 586 (H567/90) | O5,27 | 3 | — | Germany | Human |
| 1 | 534 (D113) | O5,27 | 2 | — | Japan | Dog |
| 1 | O527 | O5,27 | — | — | — | — |
| 1 | Y738 (IP22393) | O9 | 2 | X3 | France | Human |
| 1 | 96 | O9 | — | — | Germany | Human |
| 1 | 564 (7Oulua) | O9 | 2 | — | Finland | Human |
| 1 | Y736 (IP636) | O8 | 1B | Xz | United States | Human |
| 1 | P24 | O8 | 1B | — | Poland | Human |
| 1 | WA-314 | O8 | 1B | — | United States | Human |
| 1 | 589 (900/90) | O8 | 1B | — | Japan | Human |
| 1 | 575 (893/87) | O8 | 1B | — | Italy | Human |
| 1 | 8081 | O8 | 1B | — | — | Human |
| 1 | Y775 (IP124) | O5 | 1A | Xz | France | Pony |
| 1 | Y744 | O1,2a,3 | — | II | Holland | — |
| 1 | Y772 (IP1) | O2a,2b,3 | 5 | I | France | Hare |
| 1 | Y773 (IP178) | O2a,2b,3 | 5 | II | France | Hare |
A total of 99 strains were tested.
The alternative names of strains described elsewhere are included in parentheses.
—, no data available.
Epidemiologically linked strains.
Computer-aided searches for VNTRs.
The draft of the Y. enterocolitica subsp. palearctica O3 type strain Y11 genome (data not published) was scanned for tandem repeats by using the Tandem Repeats Finder program (version 2.02), by Gary Benson, by using standard scanning features (4). The Y11 draft genome was obtained in cooperation with Integrated Genomics (Jena, Germany) and represents 4× coverage of the genome.
DNA purification.
Highly purified DNA was obtained from 8 ml of an overnight culture incubated at 27°C by using a Nucleo-Bond AXG 100 kit (Macherey-Nagel, Germany), in accordance with the protocol of the manufacturer.
PCR analysis.
The VNTR markers were amplified by using the primers listed in Table 2 at a concentration of 0.2 μM and AmpliTaq Gold Hot-Start DNA polymerase (Perkin-Elmer, Germany) in the reaction buffer with 1.5 mM MgCl2 supplied by the manufacturer in a 20-μl reaction volume. A general program consisting of 35 cycles for 30 s each of denaturation at 94°C, annealing at 58°C, and elongation at 72°C was used for amplification. Finally, DNA synthesis was completed at 72°C for 3 min. Prior to cycling, a 10-min denaturation step at 94°C was included.
TABLE 2.
Nucleotide sequences of tandem repeats and primers used for amplification of VNTR markers
| Marker locus | Motif | Period (bp) | Primer sequence
|
Total no. of variants | |
|---|---|---|---|---|---|
| Forward | Reverse | ||||
| V2A | TCTCAC | 6 | CAGCGCTTCTTTATTTGCTGC | GCGTTATCTACCTGATGGTGC | 14 |
| V4 | CGGCAAC | 7 | GTCACATTGGCCTTAATCACC | TCGTACTCAATTTCCTGATGC | 8 |
| V5 | GGTGCA | 6 | ACAGTTATTGCAAGAGATGGG | AACTGGTTGAACTAGAACACC | 15 |
| V6 | GACTCA | 6 | ATTGCTCTGCGGTGTATTACG | CTTCTCGGCGATCCAGAAGCC | 12 |
| V7 | GTGCTG | 6 | CCATAATCTAGACCTCTTTGG | AGAATTCGTTGGCCTGTTTGG | 14 |
| V9 | ATGTCGGTAGAA | 12 | AGGGTATTCATGCACAGAAGC | ATGGCTAAAATACGTTCAGCC | 7 |
| V10 | GTTCTGGT | 8 | TTATCTAAGTGCAGGACGGAG | TTGGTTCATCGGAGGTTAAGC | 13 |
Electrophoretic analysis was performed by using an Amplisize 2.5% agarose gel (Bio-Rad, Germany) in Tris-acetate electrophoresis buffer (pH 8.0) (26). Gels supporting a separation distance of 17 cm were run in a horizontal chamber at a constant voltage of 100 V for 7 h, stained with ethidium bromide (2 μg/ml) for 10 min, and photographed (Gel-Doc image station; Bio-Rad).
DGE.
Denaturing gel electrophoresis (DGE) was performed to determine the amplicon size. A Sequi-Gen GT chamber (Bio-Rad) with 50-cm-long plates and 0.4-mm-thick spacers was used to support the long migration distance and the homogenous thermal conditions through the entire gel plate. Electrophoresis was conducted in 0.75× Tris-borate EDTA (26) at 55 W, in accordance with the manufacturer's recommendations. Two microliters of the PCR product was mixed with 18 μl of denaturing buffer (60% [vol/vol] deionized formamide, 10% [vol/vol] saturated urea, 19.6% [vol/vol] glycerol, 0.02% [wt/vol] xylene cyanol FF, 0.02% [wt/vol] bromophenol blue, 10 mM EDTA), heated at 95°C for 3 min, and then immediately cooled by placing the mixture onto a frozen aluminum rack. Three microliters of the denatured sample was loaded onto an 8% polyacrylamide gel (K4 acrylamide and K4 bisacrylamide mixture [29:1]; Applichem, Germany) containing 7 M urea. The amplicons of V4, V9, and V10 and the amplicons of V2a, V5, V6, and V7 were separated for 4 h and 5 h, respectively. The DNA bands were visualized by silver staining and were analyzed as described previously (13). The size of each MLVA marker was determined by using a 20-bp step DNA ladder (Sigma-Aldrich Co., Germany) as the main standard for calculation. A 50-bp step ladder (Sigma-Aldrich Co.) was applied as a reference for the V2A marker.
DNA sequencing.
To determine the exact correspondence between the allele size measured and the number of tandem repeats of the MLVA markers, representative amplicons of each size group for all seven markers used in this study were subjected to sequence analysis with automated fluorescent DNA sequencer 377 and the BigDye Terminator version 3.1 kit (Applied Biosystems), in accordance with the manufacturer's instructions. Since VNTR motifs are generally well conserved (17, 25), in most cases only a single strand of the analyzed amplicon was sequenced with the forward primer (Table 2) to determine the number of repeats. In the cases of V9 and V10, additional primers 9seq (GTGACTGTACGGCATTTATTGC) and 10seq (CTGAATTGAAAAGAGCGACAG), located several nucleotides upstream of the forward primers of the MLVA, were used.
Cluster analysis.
Only strains which were positive for the six MLVA markers tested (V2a, V4, V5, V6, V7, and V9) were subjected to cluster analysis. Prior to analysis, the allele sizes determined by DGE were converted to the sizes determined by DNA sequencing (Table 3). Cluster analysis was performed by the average linkage (unweighted pair group method with arithmetic mean) agglomeration method of WinSTAT software (version 2001.1).
TABLE 3.
Comparison of tandem repeat numbers and marker sizes determined by nucleotide sequence analysis and DGE
| TRNa | Marker size (bp) detected by the indicated method for the following MLVA marker locus:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V2A
|
V4
|
V5
|
V6
|
V7
|
V9
|
V10
|
||||||||
| SEQb | DGE | SEQ | DGE | SEQ | DGE | SEQ | DGE | SEQ | DGE | SEQ | DGE | SEQ | DGE | |
| 2 | 268 | 263 | 117 | 117 | —c | — | — | — | — | — | 103 | 101 | — | — |
| 3 | — | — | — | — | — | — | — | — | 183 | 179 | 115 | 113 | 108 | 106 |
| 4 | — | — | 131 | 131 | 194 | 191 | — | — | 189 | 185 | 127 | 125 | — | — |
| 5 | 286 | 281 | 138 | 137 | 200 | 197 | 188 | 184 | 195 | 192 | 139 | 137 | — | — |
| 6 | 292 | 288 | 145 | 144 | 206 | 203 | 194 | 190 | 201 | 198 | 151 | 148 | 132 | 130 |
| 7 | 298 | 294 | 152 | 151 | 212 | 210 | 200 | 196 | 207 | 204 | — | — | 140 | 138 |
| 8 | 304 | 299 | 159 | 158 | 218 | 216 | 206 | 202 | 213 | 210 | 175 | 171 | 148 | 146 |
| 9 | 310 | 305 | 166 | 165 | 224 | 222 | 212 | 208 | 219 | 216 | 187 | 182 | 156 | 154 |
| 10 | 316 | 312 | 173 | 172 | 230 | 227 | 218 | 214 | 225 | 222 | — | — | 164 | 162 |
| 11 | 322 | 317 | — | — | 236 | 234 | 224 | 220 | 231 | 228 | — | — | 172 | 170 |
| 12 | 328 | 323 | — | — | 242 | 239 | 230 | 226 | 237 | 234 | — | — | 180 | 178 |
| 13 | 334 | 328 | — | — | 248 | 245 | — | — | 243 | 240 | — | — | 188 | 186 |
| 14 | 340 | 333 | — | — | 254 | 251 | 242 | 238 | 249 | 246 | — | — | 196 | 195 |
| 15 | 348 | 338 | — | — | — | — | 248 | 244 | 255 | 252 | — | — | 204 | 202 |
| 16 | 352 | 343 | — | — | 266 | 263 | 254 | 249 | — | — | — | — | — | — |
| 17 | 358 | 350 | — | — | 272 | 268 | 260 | 255 | 267 | 264 | — | — | 220 | 217 |
| 18 | — | — | — | — | 278 | 274 | — | — | — | — | — | — | — | — |
| 19 | — | — | — | — | 284 | 279 | — | — | — | — | — | — | — | — |
| 20 | — | — | — | — | — | — | — | — | — | — | — | — | 244 | 241 |
TNR, tandem repeat number.
SEQ, nucleotide sequence analysis.
—, tandem repeat numbers not observed in this study.
RESULTS
Tracking potential VNTRs.
The search for tandem repeats in the draft sequence of the Y. enterocolitica Y11 genome resulted in the detection of 97 sequences containing VNTRs. Among these, only 14 met the criteria for easy handling, i.e., a repeat length greater than 5 nucleotides and a copy number of at least 4 with a highly conserved repeated sequence (no less than 90% matches). Two of these 14 VNTR markers selected (V8 and V11) were located on the pYV virulence plasmid. Since pYV may easily be lost during manipulation in the laboratory, these markers were excluded. Furthermore, five markers were eliminated due to their low rate of occurrence (in only 36% of 25 strains selected for a pilot study) in Yersinia strains. Thus, only 7 of the 14 VNTR loci detected were selected for further analysis (Table 2).
VNTR marker size determination.
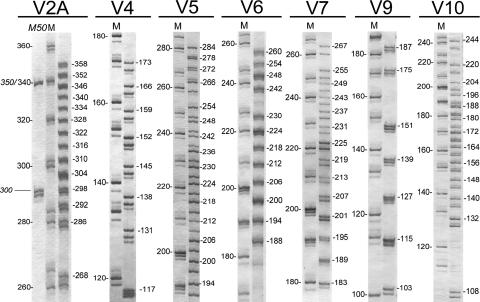
Electrophoresis under native conditions with a special grade of agarose was successfully applied for the detection of the VNTR polymorphisms. This method was capable of detecting differences of 6 nucleotides in length if the amplicons were run in adjacent lanes. However, calculation of the size of the same amplicon on different gels was not possible. In contrast, gel-to-gel variations in the sizes of the amplicons determined by electrophoresis under denaturing conditions (DGE) did not exceed 2 nucleotides. Figure 1 shows all size variants detected for MLVA markers. Single-strand mass asymmetry has been observed by DGE (13, 17, 19). The single-strand DNA asymmetry between the DNA size ladder and the amplicons tested may lead to an inaccurate size determination by DGE compared with that determined by sequence analysis. Table 3 shows the discrepancies observed between the sizes of the VNTR variants determined by DGE and those determined by sequence analysis. As a reference, the size determined by sequence analysis is indicated near the representative band of each VNTR variant shown in Fig. 1.
FIG. 1.
DGE separation of the VNTR types for each of the seven MLVA markers investigated. The representative amplicons of each size group were amplified separately and mixed prior to electrophoresis. This mixture was loaded close to the 20-bp step ladder (lanes M). For V2A, an additional 50-bp step ladder was used (lane M50). The amplicon length of only the representative band of each size variant determined by DNA sequence analysis is marked. The real dimensions of the gel fragments shown were transformed proportionally to support the highest printout resolution.
Repeat variability.
The repeat sequences of the VNTRs and their maximal numbers are shown in Table 2. The corresponding numbers of tandem repeats determined by sequence analysis are shown in Table 3. Generally, in Y. enterocolitica subsp. palearctica, the tandem repeat number varied from 2 to 20 copies and the sequences neighboring the repeats showed homology.
Genetic stability of MLVA genotypes.
Comparative MLVA of primary colonies of strains P2 and P9 and their clones obtained after 20 serial passages revealed no single repeat shift for any of the seven VNTR marker sequences, demonstrating the in vitro stability of the method and suitability for use for routine diagnostics. Additional evidence of the stability of the marker loci in vivo was provided by two Y. enterocolitica O3 strains (strains M1 and M2) isolated from a single patient at an interval of 45 days. The two strains were indistinguishable by MLVA. The identity of the MLVA genotype was also observed for three isolates from cases of blood transfusion-related septicemia. Strains 45A/06 and 45B/06 were isolated from a patient, while strain 46/06 was obtained from the blood administered to the patient. The three strains were indistinguishable, even though strains 45A/06 and 45B/06 were isolated from separate blood samples obtained within an interval of a few hours.
In the light of recent studies by Noller and colleagues (23, 24), it is strongly recommended that a group of epidemiologically linked strains be tested to determine whether the VNTR markers selected are stable in clonal isolates obtained from different hosts of the same population (19). We analyzed the stability of the MLVA markers in strains of Y. enterocolitica 4/O3 from two groups of pigs from two farms. Strains from the same hut were indistinguishable by PFGE with the NotI and XbaI enzymes (data not shown). All isolates were calcium dependent; exhibited affinity for Congo red; and possessed the main virulence genes virF, ail, ystA, and myfA, as determined by previously described methods (12). The three strains from group hut1 were indistinguishable. Three MLVA genotypes were detected for nine strains from group hut2. The differences were noticed for VNTR loci V2a and V10. Minor diversity was observed in locus V2a. Eight strains revealed type 334, while type 340 was detected only in strain UG15. A higher diversity was observed in locus V10, where five types were found. Predominant type 220 was detected in four strains, and type 188 was detected in two strains, while types 196, 204, and 228 were each observed in single isolates (data not shown). Since such diversity suggested that the V10 locus has a hypervariable nature, it was excluded from MLVA.
MLVA genotype variability.
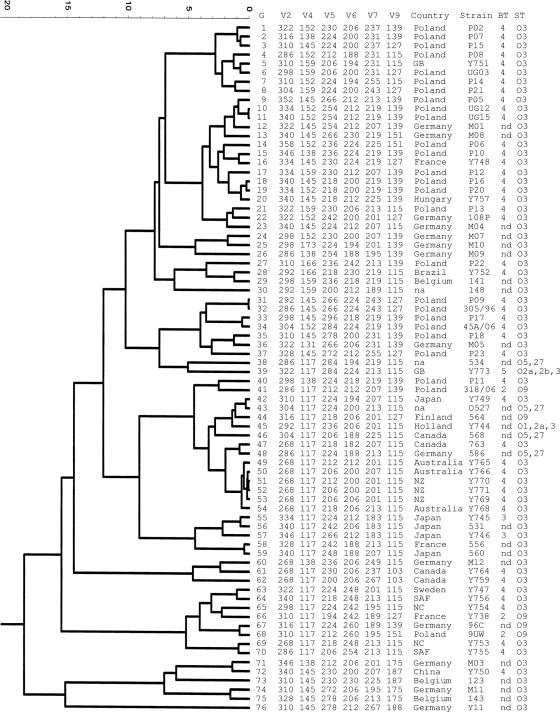
Ninety-one (92%) of the strains tested yielded PCR products with primers for all six VNTR loci (V2a, V4, V5, V6, V7, and V9) finally selected for use for MLVA. These strains were analyzed by cluster analysis. Among eight strains that could not be genotyped by MLVA, six belonged to bioserogroup 1B/O8. The remaining two nontypeable strains (strains Y772 and Y775) belonged to serotypes O2a2b3 and O5, respectively. Finally, 76 MLVA genotypes were distinguished by cluster analysis (Fig. 2).
FIG. 2.
Cluster analysis of 76 MLVA genotypes of Y. enterocolitica subsp. palearctica compared to their geographic origin and bioserogroup. The tree was generated by the average linkage (unweighted pair group method with arithmetic mean) agglomeration method. Abbreviations: G, genotype; ST, serogroup; BT, biotype; SAF, South Africa; NC, New Caledonia; NZ, New Zealand; GB, Great Britain; na, not available; nd, not determined.
Excluding epidemiologically linked isolates, the majority of strains tested belonged to unique genotypes. Few strains with no known epidemiological link belonged to the same MLVA genotype. Such strains were isolated within the same country (Hungary, Poland, and Germany) as well as in different ones (Germany and Sweden). Forty-five genotypes were detected among 62 isolates of bioserogroup 4/O3. Among the isolates in this group, 36 strains from Poland were separated into 24 MLVA genotypes. As expected, the discriminatory capacity of the MLVA developed in this study was higher when epidemiologically linked strains were excluded. Among the strains with no known epidemiological link (n = 21), 20 genotypes were detected. Figure 2 illustrates the genotype relatedness determined by cluster analysis. The dendrogram reflects the genetic diversity of the MLVA genotypes of Y. enterocolitica subsp. palearctica. Strains of serogroup O3 displayed high levels of genotype heterogeneity and were present in all main branches. Generally, no relatedness of the geographic distribution and the MLVA genotype of a strain was found, but strains from New Zealand and Australia were grouped in a single cluster, close to isolates from Japan. No correlation between phage type and MLVA genotype was observed, but a few type IXb strains originating from the same geographic region (Australia and New Zealand) were assigned to a single cluster (data not shown). Despite the presence of multiple isolates from Poland and Germany, no country-specific genotype pattern was observed.
DISCUSSION
In this study a draft genome sequence of Y. enterocolitica subsp. palearctica 4/O3 type strain Y11 was used as a template for the search for VNTRs. Twelve VNTR marker loci were initially selected for genotyping. However, only six loci met the criteria for use for MLVA. It is noteworthy that the same number of marker loci allowed outbreak and sporadic Escherichia coli O157:H7 isolates to be distinguished (23). This low number of loci is usually sufficient for outbreak investigations and strain clustering (29), while an analysis based on multiple loci is recommended for phylogeny studies (18, 19).
Any method able to estimate the PCR product size or to distinguish the allele length with a sufficient resolution with respect to the repeat unit size can be applied for MLVA. The currently used techniques are agarose gel electrophoresis, DGE, and DNA sequencing (13, 29). Although the first method is cheap and easy to perform, it was found to be unreliable. The automated sequencing technology is highly reproducible and effective, but it requires expensive equipment and reagents. For this reason, DGE (13), which has a high resolution, which is reproducible, and for which the cost of reagents is low, was used. However, our MLVA can also be used in laboratories equipped with an automated DNA sequencer.
Although DGE is considered a method that allows the determination of DNA length with the accuracy of a single nucleotide, a high degree of reliability may be achieved only when the concentrations of acrylamide and bisacrylamide as well as the migration distance are optimized (26). In this study, the use of a gel concentration of 8% and a migration distance of 40 to 50 cm was proposed for the optimal separation of all MLVA marker amplicons in a single gel. The only disadvantage of applying uniform electrophoresis conditions that was observed was a minor inaccuracy of the DNA size compared to the amplicon size determined by DNA sequencing. Interestingly, repeated DGE analyses showed that this inaccuracy was reproducible. Lista and colleagues (19) observed the same phenomenon using an automated DNA sequencer for the separation of VNTR locus amplicons for B. anthracis MLVA genotyping. Those authors recommended the use of a tandem repeat copy number or a size calculated from DNA sequencing for cluster analysis and genotype determination. Therefore, in order to support conformity with the results obtained in laboratories equipped with DNA sequencers, the VNTR marker size estimated by DGE was converted to the size determined by sequencing (Table 3).
Still, the majority of various genotyping techniques applied to Y. enterocolitica require the use of reference strains to ascertain the consistency of interlaboratory results. In contrast, MLVA is determinative and no reference strain is needed for calibration. A local reference strain from which amplicons have been sequenced is sufficient for gel-to-gel comparison. Since the results of MLVA are integers, the genotypes determined in different laboratories may be easily compared and joint databases can be produced, as shown in various studies (18, 29). MLVA demonstrated a high discriminatory power not only for Y. enterocolitica bioserogroup 4/O3 strains of worldwide origin but also for those strains isolated within a country or even within a hospital. In this study, 20 MLVA genotypes were distinguished in 21 nonepidemiologically linked strains of Y. enterocolitica 4/O3 from Poland. Buchrieser and colleagues, who tested 17 bioserogroup 4/O3 isolates from Austria, distinguished nine genotypes when the PFGE results for NotI and XbaI were combined (8). Among 17 of the 4/O3 strains from England and Wales, fluorescent amplified fragment length polymorphism analysis distinguished 13 genotypes (10). The best resolution for bioserogroup 4/O3 reported so far was obtained by PFGE with three endonucleases. This method distinguished 30 genotypes among 128 strains isolated worldwide (11). Overall, the data showed that on a countrywide scale, MLVA would offer a higher discriminatory power for Y. enterocolitica 4/O3 than the methods mentioned above. On the other hand, Noller and colleagues (24) concluded that the high resolution of MLVA may be in part a result of a genetic noise produced by the VNTR loci which are hypervariable. Three of the seven VNTR loci used by those authors for discrimination of sporadic and epidemic E. coli O157:H7 isolates mutated after multiple serial passages under laboratory conditions. One of them, TR2, indicated hypermutability. Similar phenomena were observed in our study. Although the seven VNTR loci selected at the beginning were stable during multiple serial passages in the laboratory, a comparison of the MLVA genotypes of epidemiologically linked isolates showed that they exhibited a single variation in the V2a locus and hypermutability of the V10 locus. Therefore, the latter locus was excluded from the MLVA scheme that was developed, while the six remaining loci were finally considered reliable because of their genotype stability, which is required for routine manipulations.
Although this study was conducted with a limited number of epidemiologically linked isolates, it is noteworthy that our results support the generalizations made by Noller and colleagues (24) about the interpretation of MLVA data during outbreak investigations. Briefly, among a group of epidemiologically linked strains, isolates indistinguishable by MLVA originated from the same source, while isolates indicating diversity in a single or a double tandem repeat in a single VNTR locus came from a point source. If an isolate differs at two VNTR loci, a degree of variation in these loci needs to be considered. Taken together, the data presented in our study and those previously reported by Noller and colleagues (24) demonstrate that each MLVA scheme addressed for epidemiological investigations must be tested with a group of epidemiologically linked strains.
It is noteworthy that the VNTR markers designed for Y. enterocolitica subsp. palearctica O3 were also present in strains of other pathogenic serogroups, i.e., O5,27 and O9. Despite the low number of strains tested, a high degree of diversity of genotypes was observed, as each strain tested had its own genotype. In addition, two VNTR markers, V5 and V7, were found to be present in the American Y. enterocolitica subsp. enterocolitica serogroup O8 strains (data not shown), indicating their potential usefulness for the genotyping of isolates belonging to this highly pathogenic group.
The use of different genotyping techniques, including the method considered to be the “gold standard” for the subtyping of Y. enterocolitica O3 (PFGE), showed that isolates originating from an outbreak or from a given geographical area were often so homogeneous that they could not be differentiated (11, 16). Considering the wide spectrum of loci analyzed by a variety of genotyping methods, strains of Y. enterocolitica O3 were supposed to be highly clonal (1, 2, 3, 15, 20, 21). Correspondingly, the relatively high number of MLVA genotypes observed in this study could argue for support of the assumption that the Y. enterocolitica O3 VNTR loci evolved at a faster speed than that which occurs on the evolutionary scale (29) and therefore does not argue against the clonal nature of Y. enterocolitica O3. As a result of the high-speed evolution of VNTR loci, a convergence of single VNTR markers may appear even in strains of distant geographic origins. To reduce the influence of such convergence, more than one VNTR marker must be used for genotyping (18, 19, 24).
Due to its high discriminatory power, MLVA appears to be a helpful tool for distinguishing Y. enterocolitica subsp. palearctica isolates which are difficult to differentiate by PFGE but which are suspected of being epidemiologically unrelated. In addition, as was shown for the blood transfusion-related strains used in our study, the MLVA genotype may serve as a fingerprint for epidemiological investigations.
In summary, the MLVA genotyping tool developed in this study was capable of distinguishing 45 genotypes among 62 isolates of Y. enterocolitica 4/O3 of worldwide origin tested. Our results showed that the clonal structure of Y. enterocolitica O3 was highly diverse in the VNTR loci. The MLVA genotyping technique described in the present study appears to be a promising tool for epidemiological investigations of Y. enterocolitica O3 outbreaks.
Acknowledgments
We thank Christina Nolting (Max von Pettenkoffer-Institut für Hygiene und Medizinische Mikrobiologie, Ludwig Maximilians Universität München, Germany) for assistance with DNA sequence analysis. We are thankful to Antoni Jakubczak (Department of Microbiology, Faculty of Biology, Podlaska Academy, Siedlce, Poland) and Jolanta Szych (National Institute of Hygiene, Warsaw, Poland) for providing epidemiologically linked isolates and all the colleagues from routine laboratories of the three institutes for providing clinical isolates of Y. enterocolitica. We are grateful to Gilles Vergnaud (Institute of Genetics and Microbiology, University Paris XI, and Division d'Analyses Biologiques, Centre d'Etudes du Bouchet) for critically reading the manuscript and helpful suggestions.
Since the greatest part of this study was done in the Max von Pettenkoffer-Institut für Hygiene und Medizinische Mikrobiologie, Rafał Gierczyński is grateful to FEMS for a fellowship grant supporting his stay at MPI, Munich.
Footnotes
Published ahead of print on 6 June 2007.
REFERENCES
- 1.Asplund, K., T. Johansson, and A. Siitonen. 1998. Evaluation of pulsed-field gel electrophoresis of genomic restriction fragments in the discrimination of Yersinia enterocolitica O3. Epidemiol. Infect. 121:579-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, P. M., and J. J. Farmer III. 1982. New bacteriophage typing system for Yersinia enterocolitica, Yersinia kristensenii, Yersinia frederiksenii, and Yersinia intermedia: correlation with serotyping, biotyping and antibiotic susceptibility. J. Clin. Microbiol. 15:491-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benito, I., M. E. Cano, J. Aguero, and J. M. G. Lobo. 2004. A polymorphic tandem repeat potentially useful for typing in the chromosome of Yersinia enterocolitica. Microbiology 150:199-204. [DOI] [PubMed] [Google Scholar]
- 4.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bissett, M. L., C. Powers, L. S. Abbott, and J. M. Janda. 1990. Epidemiologic investigations of Yersinia enterocolitica and related species: sources, frequency and serogroup distribution. J. Clin. Microbiol. 28:910-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blumberg, H. M., J. A. Kiehlbauch, and I. K. Wachsmuth. 1991. Molecular epidemiology of Yersinia enterocolitica O:3 infections: use of chromosomal DNA restriction fragment length polymorphisms of rRNA genes. J. Clin. Microbiol. 29:2368-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bottone, E. J. 1997. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10:257-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchrieser, C., O. Buchrieser, A. Kristl, and C. W. Kaspar. 1994. Clamped homogeneous electric field (CHEF) gel-electrophoresis of DNA restriction fragments for comparing genomic variations among strains of Yersinia enterocolitica and Yersinia spp. Zentbl. Bakteriol. Parasitenkd. Infektkrankh. Hyg. Abt. 1 Orig. 281:457-470. [DOI] [PubMed] [Google Scholar]
- 9.Farlow, J., K. L. Smith, J. Wong, M. Abrams, M. Lytle, and P. Keim. 2001. Francisciella tularenis strain typing using multiple-locus variable-number tandem repeat analysis. J. Clin. Microbiol. 39:3186-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fearnley, C., S. L. W. On, B. Kokotovic, G. Manning, T. Cheasty, and D. G. Newell. 2005. Application of fluorescent amplified fragment length polymorphism for comparison of human and animal isolates of Yersinia enterocolitica. Appl. Environ. Microbiol. 71:4960-4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frederiksson-Ahomaa, M., T. Autio, and H. Korkeala. 1999. Efficient subtyping of Yersinia enterocolitica O:3 bioserotype 4/O:3 with pulsed-field gel electrophoresis. Lett. Appl. Microbiol. 29:308-312. [DOI] [PubMed] [Google Scholar]
- 12.Gierczyński, R., M. Jagielski, and W. Rastawicki. 2002. Molecular virulence attributes and occurrence of pYV-bearing strains among human clinical isolates of Yersinia enterocolitica in Poland. Eur. J. Clin. Microbiol. Infect. Dis. 21:158-159. [DOI] [PubMed] [Google Scholar]
- 13.Gierczyński, R., S. Kałużewski, A. Rakin, M. Jagielski, A. Zasada, A. Jakubczak, B. Borkowska-Opacka, and W. Rastawicki. 2004. Intriguing diversity of Bacillus anthracis in eastern Poland—the molecular echoes of the past outbreaks. FEMS Microbiol. Lett. 239:235-240. [DOI] [PubMed] [Google Scholar]
- 14.Girard, J. M., D. M. Wagner, A. M. Vogler, C. Keys, C. J. Allender, L. C. Drickamer, and P. Keim. 2004. Differential plague-transmission dynamics determine Yersinia pestis population genetic structure on local, regional, and global scales. Proc. Natl. Acad. Sci. USA 101:8408-8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howard, S. L., M. W. Gaunt, J. Hinds, A. A. Witney, R. Stabler, and B. W. Wren. 2006. Application of comparative phylogenomics to study the evolution of Yersinia enterocolitica and to identify genetic differences relating to pathogenicity. J. Bacteriol. 188:3645-3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iteman, I., A. Guiyoule, and E. Carniel. 1996. Comparison of three molecular methods for typing and subtyping pathogenic Yersinia enterocolitica strains. J. Med. Microbiol. 45:48-56. [DOI] [PubMed] [Google Scholar]
- 17.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple locus variable number tandem repeats analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Flèche, P., Y. Hauck, L. Onteniente, A. Prieur, F. Denoeud, V. Ramisse, P. Sylvestre, G. Benson, F. Ramisse, and G. Vergnaud. 2001. A tandem repeats database for bacterial genomes: application to the genotyping of Yersinia pestis and Bacillus anthracis. BMC Microbiol. 1:2. (http://www.biomedcentral.com/1471-2180/1/2.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lista, F., G. Faggioni, S. Valjevac, A. Ciammaruconi, J. Vaissaire, C. le Doujet, O. Gorge, R. De Santis, A. Carattoli, A. Ciervo, A. Fasanella, F. Orsini, R. D'Amelio, C. Pourcel, A. Cassone, and G. Vergnaud. 2006. Genotyping of Bacillus anthracis strains based on automated capillary 25-loci multiple locus variable-number tandem repeats analysis. BMC Microbiol. 6:33. (http://www.biomedcentral.com/1471-2180/6/33.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lobato, M. J., E. Landeras, M. A. Gonzalez-Hevia, and M. C. Mendoza. 1998. Genetic heterogeneity of clinical strains of Yersinia enterocolitica traced by ribotyping and relationships between ribotypes, serotypes, and biotypes. J. Clin. Microbiol. 36:3297-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Najdenski, H., I. Iteman, and E. Carniel. 1994. Efficient subtyping of pathogenic Yersinia enterocolitica strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 32:2913-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neubauer, H., S. Aleksic, A. Hensel, E. J. Finke, and H. Meyer. 2000. Yersinia enterocolitica 16S rRNA gene types belong to the same genospecies but form three homology groups. Int. J. Med. Microbiol. 290:61-64. [DOI] [PubMed] [Google Scholar]
- 23.Noller, A. C., M. C. McEllistrem, A. G. Pacheco, D. J. Boxrud, and L. H. Harrison. 2003. Multilocus variable-tandem repeat analysis distinguishes outbreak and sporadic Escherichia coli O157:H7 isolates. J. Clin. Microbiol. 41:5389-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noller, A. C., M. C. McEllistrem, K. A. Shutt, and L. H. Harrison. 2006. Locus specific mutational events in a multilocus variable-number tandem repeat analysis of Escherichia coli O157:H7. J. Clin. Microbiol. 44:374-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pourcel, C., F. Andre-Mazeaud, H. Neubauer, F. Ramisse, and G. Vergnaud. 2004. Tandem repeats analysis for the high resolution phylogenetic analysis of Yersinia pestis. BMC Microbiol. 4:22. (http://www.biomedcentral.com/1471-2180/4/22.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Schouls, L. M., A. van der Ende, M. Damen, and I. van de Pol. 2006. Multiple-locus variable-number tandem repeat analysis of Neisseria meningitidis yields groupings similar to those obtained by multilocus sequence typing. J. Clin. Microbiol. 44:1509-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Belkum, A., S. Scherer, W. van Leeuwen, D. Willemse, L. van Alphen, and H. Verbrugh. 1997. Variable number of tandem repeats in clinical strains of Haemophilus influenzae. Infect. Immun. 65:5017-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vergnaud, G., and C. Pourcel. 2006. Multiple locus VNTR (variable number of tandem repeat) analysis (MLVA), pp. 83-104. In E. Stackebrandt (ed.), Molecular identification, systematics and population structure of prokaryotes. Springer-Verlag, Berlin, Germany.
- 30.Virdi, J. S., and P. Sachdeva. 2005. Molecular heterogeneity in Yersinia enterocolitica and “Y. enterocolitica-like” species—implications for epidemiology, typing and taxonomy. FEMS Immun. Microbiol. 45:1-10. [DOI] [PubMed] [Google Scholar]
- 31.Wojciech, Ł., Z. Staroniewicz, A. Jakubczak, and M. Ugorski. 2004. Typing of Yersinia enterocolitica isolates by ITS profiling, REP- and ERIC-PCR. J. Vet. Med. 51:238-244. [DOI] [PubMed] [Google Scholar]