Abstract
Reliable and rapid identification of staphylococcal strains continues to be a problem faced by many microbiology laboratories. This study evaluates a simplified method that uses a flowchart to assist in the identification of 12 clinical species of Staphylococcus, including eight subspecies. A total of 198 isolates and 11 control strains were identified by the reference method, which employed 22 tests. The results were compared with those obtained by two other methods: an automated system (MicroScan WalkAway) and a simplified method composed of nine tests. The simplified scheme showed an accuracy of 98.5%, while the automated method showed an accuracy of 79.3% (P < 0.001), in identifying staphylococcal species. Atypical phenotypic profiles were detected by both the reference (55.6%) and the simplified (19.7%) methods. The simplified method proposed here was shown to be reliable, with the advantage of being more practical and economic than the reference method.
Staphylococci are major human pathogens, causing a large variety of infections worldwide (3). Staphylococcus aureus is frequently isolated from community and hospital infections, including septicemia, lower respiratory, urinary tract, and skin infections (4), whereas coagulase-negative staphylococci (CoNS) most frequently are isolated from blood cultures, accounting for one-third of nosocomial bacteremias (5, 21, 26). At present, more than 80% of the CoNS strains isolated from this infection type are of the species S. epidermidis, S. haemolyticus, and S. hominis (22, 24). The remarkable ability of both S. aureus and CoNS to acquire antibiotic resistance limits therapy options and consequently may increase patient morbidity and mortality (8).
To shed light on the clinical significance of each Staphylococcus species in infections and to provide data for control and epidemiological measures, the reliable identification of these organisms is crucial (14). The reference Staphylococcus identification method, composed of 36 tests (3), is reliable but is relatively cumbersome for use in routine laboratories. Moreover, in general, this technique requires 5 to 7 days for identification. In the last decade, several phenotypic (11, 20, 23, 24) and genotypic (10, 11, 17, 27) systems for staphylococcal identification have been developed and tested as alternatives to the reference method. However, the majority of these methods present obstacles, such as cost, the need for trained personnel, prolonged incubation time, and/or poor accuracy. Thus, many routine laboratories, mainly in developing countries, including Brazil, continue to identify staphylococci by using a limited scheme that involves a rapid screening test for S. aureus, while non-S. aureus isolates still are reported as CoNS (8, 12).
Simple phenotypic schemes composed of a few tests to identify staphylococcal species have been evaluated (6, 7, 13, 18, 19) in order to help clinical routine laboratories, especially in situations where automated systems are not available. Nevertheless, these methods continue to use a large number of tests for the identification of a few species of Staphylococcus and, in general, require a long incubation period. In this study, we present a novel scheme, involving nine phenotypic tests chosen from the reference method, to identify 12 Staphylococcus species isolated from infections within 72 h. The results obtained were compared to those found by using an automated system and the reference method.
MATERIALS AND METHODS
Clinical bacterial strains.
In the present study, we evaluated 198 staphylococcal strains isolated from different clinical sites (72.2% from blood, 7.1% from urine, 6% from nares, 3.5% from surgical sites, and 11.1% from other sites) from patients at five Brazilian hospitals between 2001 and 2006. The isolates were grown to confluence on blood agar base plates (Oxoid, Basingstoke, Hampshire, England) supplemented with 5% sheep blood, scraped off the plates, resuspended in trypticase soy broth (Oxoid) containing 20% glycerol, and kept at −20°C as a heavy suspension. For most tests, the isolates were grown on blood agar base plates at 35°C for 48 h. The organisms initially were identified by Gram staining, the catalase test, acid production from glucose in Hugh and Leifson's OF base medium (15), and susceptibility to 0.04 U bacitracin (CECON, São Paulo, Brazil) to characterize the genus Staphylococcus. The inhibition zone of resistance for bacitracin was ≤10 mm (1).
Control strains.
A total of 10 type strains were used in the present study as a control: S. epidermidis (ATCC 14990), S. haemolyticus (ATCC 29970), S. aureus (ATCC 12600), S. hominis subsp. hominis (ATCC 27844), S. capitis subsp. capitis (ATCC 27840), S. saprophyticus subsp. saprophyticus (ATCC 15305), S. cohnii subsp. cohnii (ATCC 29974), S. xylosus (ATCC 29971), S. lugdunensis (DSMZ 4804), and S. schleiferi subsp. schleiferi (DSMZ 4807). The reference strain S. warneri (ATCC 10209) also was used as a control.
Staphylococcal identification methods. (i) Reference method.
All 198 clinical strains and 11 control staphylococcal strains were identified to the species level by the reference method, according to the methods of Bannerman (3) and MacFaddin (15). Twenty-two tests were used: coagulase, hemolysis (using sheep blood), clumping factor (Slidex Staph Plus; bioMérieux S/A, Inc., Durham, NC), pyrrolidonyl arylamidase (PYR), urease, alkaline phosphatase, ornithine and arginine decarboxylase, nitrate reduction, acetoin production, susceptibility to 5 μg novobiocin (CECON), and acid production from d-trehalose, sucrose, d-ribose, d-cellobiose, d-xylose, α-lactose, d-mannitol, maltose, and d-mannose. Susceptibilities to 100 μg desferrioxamine (Sigma Chemical Co., St. Louis, MO) and 300 IU polymyxin B (CECON) were determined according to the methods of Monsen and coworkers (18).
The strains initially were evaluated with hemolysis, PYR, clumping factor, and coagulase tests. Subsequently, these organisms were used for inoculation in the remaining tests, including tube and disk assays. To obtain the staphylococcal identification, the disk tests were incubated for 24 h, while the other tests were read after up to 72 h of incubation. A dense bacterial suspension (0.25 ml; equivalent to a 2 McFarland standard) was inoculated into each tube, except for those of the acetoin and amino acid tests, which required 0.05-ml suspensions (15). For the novobiocin (3), polymyxin B, and desferrioxamine (18) susceptibility tests, the bacterial inoculum was obtained from a 0.5 McFarland standard. Inhibition zones of susceptibility for novobiocin and polymyxin B were ≥16 mm, and for desferrioxamine they were ≥7 mm.
(ii) MicroScan WalkAway automation method.
MicroScan WalkAway automated system (Dade Behring, Inc., West Sacramento, CA) results were provided along with the samples by the hospitals where the patients were treated. The method is composed of 20 tests: nitrate reduction, production of two glucosidases, indoxyl phosphatase, acetoin production, alkaline phosphatase, PYR, arginine dihydrolase, urease, and acid production from raffinose, α-lactose, d-trehalose, d-mannose, l-sorbose, l-arabinose, d-ribose, d-mannitol, inulin, and pyruvate. Susceptibility to novobiocin also was determined by this methodology. The bacterial inoculum and the incubation time were in accordance with the manufacturers' recommendations.
(iii) Simplified method.
The simplified identification method proposed in this study was composed of nine tests selected from the reference method. The scheme combined two susceptibility tests, using 5 μg novobiocin and 100 μg desferrioxamine disks, with tests that detect the production of clumping factor, PYR, urease, and alkaline phosphatase. Acid production from d-mannose, d-trehalose, and d-xylose also was included. The tests were carried out as described above.
Interpretation of results.
Results of staphylococcal identifications obtained by the automated system and the simplified method were compared to those obtained by the reference method. In order to ensure the accuracy of results and to exclude technical errors, the isolates with initial ambiguous results compared to the results of the reference method were analyzed at least twice. Final ambiguous results were considered misidentifications.
Statistical test.
All comparisons were performed using the chi-square method.
RESULTS
Staphylococcus species identification.
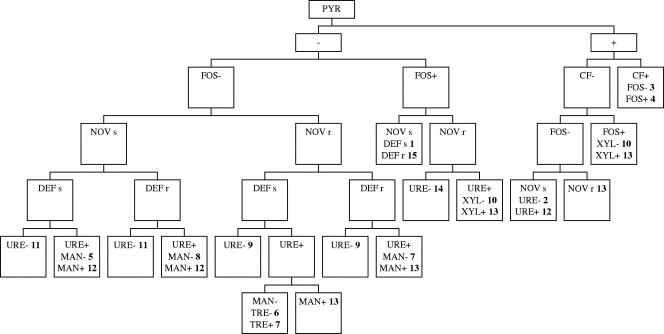
Table 1 shows the results of the scheme composed of nine phenotypic tests that were used in the simplified method to identify 12 species and eight subspecies of staphylococci found in clinical specimens. S. schleiferi subsp. schleiferi, which was not present among the clinical isolates evaluated, was included in this classification, because it was analyzed as a control organism and was easily differentiated from other staphylococcal species. The flowchart used to help with the identification is presented in Fig. 1. Staphylococcus identification by the reference, simplified, and automated methods is shown in Table 2. Among 198 staphylococcal isolates evaluated by the reference method, we found 69 S. epidermidis (34.8%) isolates, 44 S. haemolyticus (22.2%) isolates, 25 S. hominis (12.6%) isolates, and 17 S. aureus (8.6%) isolates. The other species identified were S. saprophyticus subsp. saprophyticus (eight strains), S. warneri (seven strains), S. lugdunensis (six strains), S. capitis subsp. capitis (six strains), S. cohnii subsp. urealyticus (four strains), S. sciuri subsp. sciuri (four strains), S. xylosus (four strains), S. capitis subsp. urealyticus (three strains), and S. cohnii subsp. cohnii (one strain). The simplified method correctly identified 195 (98.5%) isolates. Moreover, from the 198 isolates evaluated, 45.4% were identified in 48 h, and the remaining isolates were identified within 72 h. Three strains were not identified by this method (isolates of the species S. epidermidis, S. haemolyticus, and S. lugdunensis). The automated system identified 157 (79.3%) strains correctly; however, 10 (5%) strains were not identified, and 31 (15.6%) strains were misidentified. Among the misidentified isolates, clinically prevalent species, such as S. haemolyticus, S. epidermidis, S. hominis, and S. aureus, were found. A significant decrease in accuracy was observed for the automation method compared to the accuracy of the simplified method (P < 0.001; chi-square method). All control strains were correctly identified by the reference and simplified methods.
TABLE 1.
Simplified method using nine tests for the staphylococci identification
| Species | Result with the biochemical testa:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| CF | PYR | URE | FOS | MAN | TRE | XYL | NOV | DEF | |
| S. aureus | + | − | v | + | + | + | − | s | r |
| S. epidermidis | − | − | + | + | + | − | − | s | s |
| S. haemolyticus | − | + | − | − | − | + | − | s | r |
| S. lugdunensis | + | + | v | − | + | + | − | s | ND |
| S schleiferi subsp. schleiferi | + | + | − | + | + | v | − | s | ND |
| S. warneri | − | − | + | − | − | + | − | s | r |
| S. capitis subsp. capitis | − | − | − | − | + | − | − | s | ND |
| S. capitis subsp. urealyticus | − | v | + | − | + | − | − | s | ND |
| S. hominis subsp. hominis | − | − | + | − | − | v | − | s | s |
| S. hominis subsp. novobiosepticus | − | − | + | − | − | − | − | r | s |
| S. saprophyticus subsp. saprophyticus | − | − | + | − | − | + | − | r | ND |
| S. cohnii subsp. cohnii | − | − | − | − | v | + | − | r | ND |
| S. cohnii subsp. urealyticus | − | v | + | + | + | + | − | r | ND |
| S. sciuri | − | − | − | + | v | + | v | r | ND |
| S. xylosus | − | v | + | v | + | + | + | r | ND |
CF, clumping factor; URE, urease; FOS, alkaline phosphatase; MAN, d-mannose; TRE, d-trehalose; XYL, d-xylose; NOV, novobiocin susceptibility; DEF, desferrioxamine susceptibility; +, 90% or more strains positive; −, 90% or more strains negative; s, susceptible; r, resistant; v, 11 to 89% of strains positive; ND, not determined.
FIG. 1.
Flowchart of the simplified method for identification of Staphylococcus species. Symbols and abbreviations: FOS, alkaline phosphatase; CF, clumping factor; NOV, novobiocin susceptibility; DEF, desferrioxamine susceptibility; URE, urease; MAN, d-mannose; TRE, d-trehalose; XYL, d-xylose; +, positive test result; -, negative test result; s, susceptible; r, resistant. Staphylococcal species are numbered in boldface as follows: 1, S. epidermidis; 2, S. haemolyticus; 3, S. lugdunensis; 4, S. schleiferi subsp. schleiferi; 5, S. hominis subsp. hominis; 6, S. hominis subsp. novobiosepticus; 7, S. saprophyticus; 8, S. warneri; 9, S. cohnii subsp. cohnii; 10, S. cohnii subsp. urealyticus; 11, S. capitis subsp. capitis; 12, S. capitis subsp. urealyticus; 13, S. xylosus; 14, S. sciuri; and 15, S. aureus.
TABLE 2.
Identification of 198 staphylococcal isolates by the reference, simplified, and automated methods
| Species identified by the reference method (no. of strains/% of total no. of strains) | Identity of strain (no.) by the simplified method | Identity of strain (no.) by the automated method |
|---|---|---|
| S. epidermidis (69/34.8) | S. epidermidis (68) | S. epidermidis (59) |
| Staphylococcus spp. (1) | S. haemolyticus (3) | |
| Staphylococcus spp. (2) | ||
| S. auricularis (2) | ||
| S. simulans (1) | ||
| S. capitis subsp. urealyticus (1) | ||
| Not identified (1) | ||
| S. haemolyticus (44/22.2) | S. haemolyticus (43) | S. haemolyticus (32) |
| S. epidermidis (4) | ||
| Staphylococcus spp. (1) | Staphylococcus spp. (2) | |
| S. warneri (1) | ||
| S. simulans (1) | ||
| S. hominis subsp. hominis (1) | ||
| S. aureus (1) | ||
| S. auricularis (1) | ||
| S. saprophyticus (1) | ||
| S. aureus (17/8.6) | S. aureus (17) | S. aureus (14) |
| S. capitis subsp. urealyticus (1) | ||
| S. lugdunensis (1) | ||
| Staphylococcus spp. (1) | ||
| S. hominis subsp. hominis (16/8.1) | S. hominis subsp. hominis (16) | S. hominis subsp. hominis (10) |
| S. epidermidis (3) | ||
| Staphylococcus spp. (2) | ||
| S. warneri (1) | ||
| S. hominis subsp. novobiosepticus (9/4.5) | S. hominis subsp. novobiosepticus (9) | S. hominis subsp. novobiosepticus (8) |
| S. hominis subsp. hominis (1) | ||
| S. saprophyticus subsp. saprophyticus (8/4) | S. saprophyticus subsp. saprophyticus (8) | S. saprophyticus subsp. saprophyticus (8) |
| S. warneri (7/3.5) | S. warneri (7) | S. warneri (7) |
| S. lugdunensis (6/3) | S. lugdunensis (5) | S. lugdunensis (5) |
| Staphylococcus spp. (1) | S. haemolyticus (1) | |
| S. capitis subsp. capitis (6/3) | S. capitis subsp. capitis (6) | S. capitis subsp. capitis (4) |
| S. epidermidis (1) | ||
| S. xylosus (1) | ||
| S. cohnii subsp. urealyticus (4/2) | S. cohnii subsp. urealyticus (4) | S. cohnii subsp. urealyticus (1) |
| S. saprophyticus (1) | ||
| S. aureus (1) | ||
| Staphylococcus spp. (1) | ||
| S. sciuri (4/2) | S. sciuri (4) | S. sciuri (4) |
| S. xylosus (4/2) | S. xylosus (4) | S. xylosus (2) |
| S. cohnii subsp. urealyticus (2) | ||
| S. capitis subsp. urealyticus (3/1.5) | S. capitis subsp. urealyticus (3) | S. capitis subsp. urealyticus (2) |
| Staphylococcus spp. (1) | ||
| S. cohnii subsp. cohnii (1/0.5) | S. cohnii subsp. cohnii (1) | S. cohnii subsp. cohnii (1) |
| Total 198 (100% identified) | 195 (98.5%) | 157 (79.3%) |
Atypical biochemical profiles presented by the staphylococcal species.
Atypical strains were defined as those that showed contrary results for tests that are considered not variable by the reference method (23). Variable test results are represented as “v,” and positive results are achieved with 11 to 89% of strains (3). Among the 198 staphylococcal isolates evaluated, 110 (55.6%) were found to be atypical by at least one test of the reference method, while 39 (19.7%) strains showed atypical profiles in the simplified scheme. A large number of atypical strains was detected for the species S. haemolyticus (28/44; 63.6%), S. epidermidis (33/69; 47.8%), and S. hominis (10/25; 40%) by the reference method. The frequency of atypical staphylococcal strains in relation to each one of the 22 phenotypic tests is shown in Table 3. The majority of the strains were atypical for susceptibility to polymyxin B (37 strains; 18.7%) and for acetoin production (21; 10.6%). Coagulase, PYR, and ornithine decarboxylase tests did not present any strain with an atypical biochemical profile, while novobiocin susceptibility and clumping factor tests showed only one isolate with this profile type. The other tests presented <7% atypical isolates.
TABLE 3.
Frequency of atypical strains among 198 staphylococcal isolates as determined by 22 tests of the reference method
| Species (total no. of strains) | No. (%) of atypical strains identified with the biochemical testa:
|
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CG | HEM | CF | PYR | URE | FOS | MAN | MAT | TRE | LAC | MAL | SUC | RIB | CEL | XYL | ORN | ARG | NIT | ACT | NOV | DEF | POL | |
| S. epidermidis (69) | 0 | − | 0 | 0 | 6 (8.7) | 2 (2.9) | 5 (7.2) | 5 (7.2) | 3 (4.3) | 1 (1.4) | 0 | 1 (1.4) | − | 4 (5.8) | 4 (5.8) | − | − | 7 (10.1) | 4 (5.8) | 1 (1.4) | 0 | 5 (7.2) |
| S. haemolyticus (44) | 0 | 0 | 0 | 0 | 0 | 0 | 3 (6.8) | − | 2 (4.5) | − | 0 | 1 (2.3) | − | 0 | 1 (2.3) | 0 | 0 | 0 | 12 (27.3) | 0 | 2 (4.5) | 13 (29.5) |
| S. aureus (17) | 0 | 2 (11.8) | 0 | 0 | − | 0 | 0 | 1 (5.9) | 1 (5.9) | 1 (5.9) | 0 | 1 (5.9) | 1 (5.9) | 1 (5.9) | 0 | 0 | 0 | 0 | 1 (5.9) | 0 | 1 (5.9) | 0 |
| S. hominis subsp. hominis (16) | 0 | 1 (6.3) | 0 | 0 | 0 | 2 (12.5) | 3 (18.8) | 2 (12.5) | − | − | 1 (6.3) | 0 | 2 (12.5) | 1 (6.3) | 0 | 0 | − | − | − | 0 | 0 | 2 (12.5) |
| S. hominis subsp. novobiosepticus (9) | 0 | 1 (11.1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | − | 0 | 0 | − | 0 | 0 | 0 | 0 | − | − | 0 | 0 | − |
| S. saprophyticus subsp. saprophyticus (8) | 0 | 1 (12.5) | 0 | 0 | 0 | 0 | 0 | − | 0 | − | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 (25) | 0 | − | 4 (50) |
| S. warneri (7) | 0 | − | 0 | 0 | 0 | 0 | 1 (14.3) | − | 0 | − | 0 | 0 | − | 0 | 0 | 0 | − | − | 2 (28.6) | 0 | 0 | 6 (85.7) |
| S. lugdunensis (6) | 0 | 0 | 1 (16.7) | 0 | − | 0 | 0 | 0 | 0 | 1 (16.7) | 1 (16.7) | 0 | 1 (16.7) | 0 | 0 | 0 | 1 (16.7) | 0 | 0 | 0 | − | − |
| S. capitis subsp. capitis (6) | 0 | − | 0 | 0 | 0 | 0 | 0 | 1 (16.7) | 0 | 0 | 3 (50) | − | 0 | 0 | 0 | 0 | − | − | − | 0 | − | 2 (33.3) |
| S. cohnii subsp. urealyticus (4) | 0 | − | 0 | − | 0 | 1 (25) | 0 | 0 | 0 | 0 | 0 | 3 (75) | 2 (50) | 0 | 0 | 0 | 1 (25) | 1 (25) | − | 0 | − | 2 (50) |
| S. sciuri (4) | 0 | − | 0 | 0 | 0 | 0 | − | 0 | 0 | − | − | 0 | 0 | − | − | 0 | 1 (25) | 0 | 0 | 0 | − | 3 (75) |
| S. xylosus (4) | 0 | 2 (50) | 0 | − | 0 | − | 0 | 0 | 0 | − | − | − | − | 0 | 0 | 0 | 0 | − | − | 0 | − | 0 |
| S. capitis subsp. urealyticus (3) | 0 | − | 0 | − | 0 | 0 | 0 | 0 | 3 (100) | − | 0 | 1 (33.3) | − | 0 | 0 | 0 | 0 | 0 | − | 0 | 0 | 0 |
| S. cohnii subsp. cohnii (1) | 0 | − | 0 | 0 | 0 | 0 | − | − | 0 | 0 | − | 0 | 0 | 0 | 0 | 0 | 0 | 0 | − | 0 | − | 0 |
| Total (198) | 0 (0.0) | 7 (3.5) | 1 (0.5) | 0 (0.0) | 6 (3) | 5 (2.5) | 12 (6.1) | 9 (4.5) | 9 (4.5) | 3 (1.5) | 5 (2.5) | 7 (3.5) | 6 (3) | 6 (3) | 5 (2.5) | 0 (0.0) | 3 (1.5) | 8 (4) | 21 (10.6) | 1 (0.5) | 3 (1.5) | 37 (18.7) |
CG, coagulase; HEM, hemolysin; CF, clumping factor; URE, urease; FOS, alkaline phosphatase; MAN, d-mannose, MAT, d-mannitol; TRE, d-trehalose; LAC, α-lactose; MAL, maltose; SUC, sucrose; RIB, d-ribose; CEL, d-cellobiose; XYL, d-xylose; ORN, ornithine decarboxylase; ARG, arginine decarboxylase; NIT, nitrate reduction; ACT, acetoin production; NOV, novobiocin susceptibility; DEF, desferrioxamine susceptibility; POL, polymyxin B susceptibility; −, variable biochemical profile for the test (11 to 89% of positive strains according to Bannerman [3]).
Molecular methods for staphylococcal identification, such as the PCR technique, have been reported as accurate methods (17, 27). We selected all atypical isolates of the S. epidermidis species as well as unidentified isolates and used the PCR method, according to the methods of Martineau and coworkers (17), to confirm the results found by the simplified and reference methods. After analysis by PCR, all 32 isolates were confirmed to be S. epidermidis (data not shown), showing the high accuracy of the phenotypic scheme proposed.
DISCUSSION
In this study, we used a simplified method composed of nine tests to identify 198 staphylococcal isolates, and we obtained an accuracy level of 98.5% with this analysis. Conversely, when we evaluated the isolates by an automated system, the level of accuracy of identification was significantly lower (79.3%; P < 0.001). Other authors have found similar results with the automated method. Weinstein and coworkers (27) verified that the MicroScan method correctly identified only 76.5% of the staphylococci isolated from blood that were identified by a reference method similar to the one used in our study. Other authors also have found low accuracy values, ranging from 61.7% (11) to 77% (16). Methods that use a large number of tests, such as the reference method, have a tendency to be accurate. Automated systems also present various and numerous tests; however, the short incubation time used for staphylococcal identification in these systems seems to be unsuitable, specially for CoNS analysis, thus making accurate identification difficult.
On the other hand, the rates of accuracy that have been observed by some authors with the automated systems are higher than results obtained with miniaturized methods, such as the ID 32 Staph identification system (9, 20, 24, 25). However, these data may have been masked, since the accuracy of the staphylococcal identification by miniaturized tests also has been found to be lower than that of the reference method, ranging from 82% (23) to 85% accuracy (2, 7, 11).
In the last decade, simplified schemes showing good levels of accuracy for identifying staphylococci have been developed. However, in general, these methods identify a few species of Staphylococcus and/or continue to use a large number of tests, making its use in routine laboratories difficult. Ieven and coworkers (13) developed a phenotypic scheme showing 97.7% accuracy and composed of seven tests, but they were able to identify only eight species and one subspecies of staphylococci. A simple scheme has been designed for CoNS identification (7) that provided results for 11 species and seven subspecies of Staphylococcus. However, this method used 13 phenotypic tests and had a long incubation time, varying from 3 to 6 days. The authors obtained an accuracy of 97.5%. Cunha and colleagues (6) proposed a method based on eight tests in a first step and on five tests in a second step, for a total of 13 different tests. This method also required a long period of incubation (3 to 6 days), and it identified 13 species and eight subspecies of Staphylococcus. The simplified method developed in the present study was able to identify 12 species of Staphylococcus, including eight subspecies, by using nine phenotypic tests with incubation times of up to 72 h. Although the species S. schleiferi subsp. schleiferi was not one of the clinical isolates evaluated, this organism was analyzed as a control strain during this study, and it was easily characterized and distinguished from the other species by the simplified method. Therefore, the novel scheme proposed here could be used to identify a total of 12 species and eight subspecies of staphylococci, as shown in Table 1.
To form the simplified method, the majority of tests giving atypical results were not included in the scheme, such as acetoin production and polymyxin B susceptibility, which found 10.6% and 18.7% atypical isolates, respectively. More appropriate tests were selected, in particular, those that were practical and fast and that showed few atypical strains. PYR and clumping factor tests, followed by the alkaline phosphatase test and novobiocin and desferrioxamine disk susceptibility tests, were some of the tests that showed these characteristics and also provided results within 24 h. To our knowledge, this is the first study to date that reports and discusses the use of atypical phenotypic characteristics in the identification of identify staphylococci isolates.
To confirm the presence of the S. lugdunensis species, the ornithine decarboxylase test could have been used, as reported by other authors (6, 7). However, the clumping factor test was preferred for identifying S. lugdunensis isolates, because the results were provided in a few seconds, making the test faster than the ornithine decarboxylase test. Moreover, we have worked with this species in the laboratory, and we observed that few S. lugdunensis isolates have presented atypical results for the clumping factor test, showing that it is reliable.
Some tests selected for inclusion in the simplified method were very important for distinguishing certain species. Novobiocin and desferrioxamine disk susceptibility tests were effective in identifying the novobiocin-resistant species S. hominis subsp. novobiosepticus, S. saprophyticus, S. cohnii, S. sciuri, and S. xylosus and in distinguishing the species S. hominis and S. warneri, respectively. The urease test was used to distinguish S. cohnii subsp. cohnii from S. cohnii subsp. urealyticus, and the same test was used to separate the subspecies S. capitis subsp. capitis from S. capitis subsp. urealyticus. Although the urease test is not depicted as a key test to identify the S. epidermidis species in the flowchart (Fig. 1), it helped to distinguish S. epidermidis isolates that presented atypical results for the other tests, and it is included in Table 1. Only a few staphylococcal species present variable results for the mannose and trehalose tests (3). Thus, in this study these tests also were employed and were important for staphylococcal identification.
S. xylosus and S. cohnii subsp. urealyticus are species that present very similar phenotypic characteristics. In these specific cases, acid production from d-xylose was the test used to discriminate these organisms, as shown in the flowchart in Fig. 1.
In the present study, we tested different bacterial inocula at McFarland standards of 0.5, 1.0, and 2.0 with the simplified and conventional methods to define what inoculum would provide better identification of the different species of staphylococci. Some tests from the conventional method have been assessed by using dense bacterial suspensions (3, 15). However, the authors do not specify the exact bacterial suspension used. The inoculum equivalent to a 2.0 McFarland standard used in our study was the suspension that showed better results for staphylococcal identification; only for the susceptibility tests was a 0.5 McFarland standard used.
The flowchart shown in Fig. 1 was composed of identification tests that showed no or a low percentage (<7%) of atypical isolates, and it was useful in the identification of clinical isolates of staphylococci. Therefore, the identification of typical and atypical staphylococcal isolates could be performed with the simultaneous use of Table 1 and the flowchart (Fig. 1).
In conclusion, the simplified scheme proposed here was shown to be a reliable method, with the advantages of being more practical, more economic, and faster than the reference method. Thus, this scheme is a good candidate for use as a routine method in microbiology laboratories.
Acknowledgments
We thank Walter Oelemann for critical reading of the manuscript.
This study was supported by grants from Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento Pessoal de Nível Superior (CAPES), Fundação Universitária José Bonifácio (FUJB), and Programa de Núcleos de Excelência (PRONEX).
Footnotes
Published ahead of print on 30 May 2007.
REFERENCES
- 1.Baker, J. S., M. F. Hackett, and D. J. Simard. 1986. Variations in bacitracin susceptibility observed in Staphylococcus and Micrococcus species. J. Clin. Microbiol. 23:963-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannerman, T. L., K. T. Kleeman, and W. E. Kloos. 1993. Evaluation of the Vitek Systems Gram-Positive Identification card for species identification of coagulase-negative staphylococci. J. Clin. Microbiol. 31:1322-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannerman, T. L. 2003. Staphylococcus, Micrococcus, and other catalase-positive cocci that grow aerobically, p. 384-404. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, DC.
- 4.Chambers, H. F. 2001. The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 7:178-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costa, S. F., M. H. Miceli, and E. J. Anaissie. 2004. Mucosa or skin as source of coagulase-negative staphylococcal bacteraemia? Lancet Infect. Dis. 4:278-286. [DOI] [PubMed] [Google Scholar]
- 6.Cunha, M. L. R. S., Y. K. Sinzato, and L. V. A. Silveira. 2004. Comparison of methods for the identification of coagulase-negative staphylococci. Mem. Inst. Oswaldo Cruz. 99:855-860. [DOI] [PubMed] [Google Scholar]
- 7.De Paulis, A. N., S. C. Predari, C. D. Chazarreta, and J. E. Santoianni. 2003. Five-test simple scheme for species-level identification of clinically significant coagulase-negative staphylococci. J. Clin. Microbiol. 41:1219-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diekema, D. J., M. A. Pfaller, F. J. Schmitz, J. Smayevsky, J. Bell, R. N. Jones, M. Beach, et al. 2001. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin. Infect. Dis. 32(Suppl. 2):S114-S132. [DOI] [PubMed] [Google Scholar]
- 9.Fahr, A. M., U. Eigner, M. Armbrust, A. Caganic, G. Dettori, C. Chezzi, L. Bertoncini, M. Benecchi, and M. G. Menozzi. 2003. Two-center collaborative evaluation of the performance of the BD Phoenix automated microbiology system for identification and antimicrobial susceptibility testing of Enterococcus spp. and Staphylococcus spp. J. Clin. Microbiol. 41:1135-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujita, S. I., Y. Senda, T. Iwagami, and T. Hashimoto. 2005. Rapid identification of staphylococcal strains from positive-testing blood culture bottles by internal transcribed spacer PCR followed by microchip gel electrophoresis. J. Clin. Microbiol. 43:1149-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heikens, E., A. Fleer, A. Paauw, A. Florijn, and A. C. Fluit. 2005. Comparison of genotypic and phenotypic methods for species-level identification of clinical isolates of coagulase-negative staphylococci. J. Clin. Microbiol. 43:2286-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huebner, J., and D. A. Goldmann. 1999. Coagulase-negative staphylococci: role as pathogens. Annu. Rev. Med. 50:223-236. [DOI] [PubMed] [Google Scholar]
- 13.Ieven, M., J. Verhoeven, S. R. Pattyn, and H. Goossens. 1995. Rapid and economical method for species identification of clinically significant coagulase-negative staphylococci. J. Clin. Microbiol. 33:1060-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarlov, J. O. 1999. Phenotypic characteristics of coagulase-negative staphylococci: typing and antibiotic susceptibility. APMIS Suppl. 91:1-42. [PubMed] [Google Scholar]
- 15.MacFaddin, J. F. 1977. Biochemical tests for identification of medical bacteria. Waverly Press, Baltimore, MD.
- 16.Marsou, R., M. Bes, Y. Brun, M. Boudouma, L. Idrissi, H. Meugnier, J. Freney, and J. Etienne. 2001. Molecular techniques open up new vistas for typing of coagulase-negative staphylococci. Pathol. Biol. 49:205-215. [DOI] [PubMed] [Google Scholar]
- 17.Martineau, F., F. J. Picard, P. H. Roy, M. Ouellette, and M. G. Bergeron. 1996. Species-specific and ubiquitous DNA-based assays for rapid identification of Staphylococcus epidermidis. J. Clin. Microbiol. 34:2888-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monsen, T., M. Rönnmark, C. Olofsson, and J. Wiström. 1998. An inexpensive and reliable method for routine identification of staphylococcal species. Eur. J. Clin. Microbiol. Infect. Dis. 17:327-335. [DOI] [PubMed] [Google Scholar]
- 19.Mulder, J. G. 1995. A simple and inexpensive method for the identification of Staphylococcus epidermidis and Staphylococcus hominis. Eur. J. Clin. Microbiol. Infect. Dis. 14:1052-1056. [DOI] [PubMed] [Google Scholar]
- 20.Nonhoff, C., S. Rottiers, and M. J. Struelens. 2005. Evaluation of the Vitek 2 system for identification and antimicrobial susceptibility testing of Staphylococcus spp. Clin. Microbiol. Infect. 11:150-153. [DOI] [PubMed] [Google Scholar]
- 21.Otto, M. 2004. Virulence factors of the coagulase-negative staphylococci. Front. Biosci. 9:841-863. [DOI] [PubMed] [Google Scholar]
- 22.Pfaller, M. A., R. N. Jones, G. V. Doern, H. S. Sader, K. C. Kugler, M. L. Beach, et al. 1999. Survey of blood stream infections attributable to gram-positive cocci: frequency of occurrence and antimicrobial susceptibility of isolates collected in 1997 in the United States, Canada, and Latin America from the SENTRY Antimicrobial Surveillance Program. Diagn. Microbiol. Infect. Dis. 33:283-297. [DOI] [PubMed] [Google Scholar]
- 23.Renneberg, J., K. Rieneck, and E. Gutschik. 1995. Evaluation of Staph ID 32 system and Staph-Zym system for identification of coagulase-negative staphylococci. J. Clin. Microbiol. 33:1150-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spanu, T., M. Sanguinetti, D. Ciccaglione, T. D'Inzeo, L. Romano, F. Leone, and G. Fadda. 2003. Use of the VITEK 2 system for rapid identification of clinical isolates of staphylococci from bloodstream infections. J. Clin. Microbiol. 41:4259-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spanu, T., M. Sanguinetti, T. D'Inzeo, D. Ciccaglione, L. Romano, F. Leone, P. Mazzella, and G. Fadda. 2004. Identification of methicillin-resistant isolates of Staphylococcus aureus and coagulase-negative staphylococci responsible for bloodstream infections with the Phoenix system. Diagn. Microbiol. Infect. Dis. 48:221-227. [DOI] [PubMed] [Google Scholar]
- 26.von Eiff, C., G. Peters, and C. Heilmann. 2002. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect. Dis. 2:677-685. [DOI] [PubMed] [Google Scholar]
- 27.Weinstein, M. P., S. Mirrett, L. V. Pelt, M. McKinnon, B. L. Zimmer, W. Kloos, and L. B. Reller. 1998. Clinical importance of identifying coagulase-negative staphylococci isolated from blood cultures: evaluation of MicroScan Rapid and Dried Overnight Gram-Positive panels versus a conventional reference method. J. Clin. Microbiol. 36:2089-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yugueros, J., A. Temprano, M. Sánchez, J. M. Luengo, and G. Naharro. 2001. Identification of Staphylococcus spp. by PCR-restriction fragment length polymorphism of gap gene. J. Clin. Microbiol. 39:3693-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]