Abstract
We investigated in vitro whether storage of blood samples influences the time to positivity used for the calculation of the differential time to positivity (DTP) and the results of the Gram stain-acridine orange leukocyte Cytospin (AOLC) test. A 24-hour storage of blood samples at room temperature may lead to false-negative DTP and false-positive Gram stain-AOLC test results, whereas storage at 4°C does not.
The most common pathogen isolated from catheter-related bloodstream infections (CRBSI) is Staphylococcus epidermidis, followed by gram-negative rods like Escherichia coli (9). Two methods for the diagnosis of CRBSI that do not require removal of the central venous catheters (CVC) have been described recently. Both the differential time to positivity (DTP) method and the Gram stain-acridine orange leukocyte Cytospin (AOLC) test are highly specific and sensitive for the detection of CRBSI (2, 6, 7). The DTP method is based on the difference in the microbial loads between blood drawn from the CVC and blood drawn peripherally. The DTP is considered positive and hence suggestive of CRBSI if the culture from blood drawn through the CVC becomes positive at least 120 min earlier than a culture from a blood sample drawn simultaneously from a peripheral vein (2). In contrast, the Gram stain-AOLC test provides results very quickly, making it an attractive tool for the diagnosis of CRBSI. The Gram stain-AOLC test has a suggested absolute threshold of 103 to 104 CFU/ml blood (6; P. Kite, Leeds, United Kingdom, personal communication), which is consistent with the microbiological cutoff of various diagnostic methods for CRBSI (1, 3-5, 8).
Blood samples for the DTP method and the Gram stain-AOLC test have to be processed without major delay to maintain the microbial load present at the time when the blood is drawn. It has been speculated that delayed processing may lead to false DTP and Gram stain-AOLC test results (9). We investigated whether delayed processing of blood samples and different storage conditions influence the DTP and Gram stain-AOLC test results.
To accurately simulate the time to positivity (TTP; used for the calculation of the DTP) and Gram stain-AOLC tests in vitro, we used human whole blood for the experiments. Confirmed sterile whole-blood bags were used for further tests. S. epidermidis ATCC 35984 and E. coli ATCC 35218 were cultured overnight in brain heart infusion broth. Two milliliters of each culture was diluted with whole human blood to achieve an inoculum of 107 CFU/ml blood and then serially diluted with blood.
Five milliliters of each dilution (101 to 107 CFU/ml) described above was added to three aerobic BACTEC blood culture bottles (Becton Dickinson, Heidelberg, Germany) and subsequently processed as follows. Blood culture bottles in group 1 were immediately placed in the BACTEC 9240 automatic blood culture detection system (Becton Dickinson, Heidelberg, Germany). Group 2 was stored at room temperature for 24 h and then processed in the same manner as group 1. Group 3 was stored at 4°C for 24 h and subsequently processed in the same manner as group 1. The TTP for each blood culture bottle was recorded. Blood culture bottles remained in the automatic blood culture detection system until the system recorded positivity, up to 7 days. After removal of the bottles from the system, 100 μl from each blood culture bottle was cultured on blood agar and strains were identified by routine microbiological methods to confirm the presence of the inoculated strain. All tests were performed five times.
To ensure that storage at 4°C for 24 h does not inhibit detection of certain bacteria or fungi, we also inoculated Pseudomonas aeruginosa ATCC 27853, Acinetobacter baumannii cultured from patient 1, Staphylococcus aureus ATCC 29213, Enterococcus faecalis ATCC 29212, Stenotrophomonas maltophilia ATCC 51331, and Candida albicans C7:CAF2-1 (kindly provided by B. Hube, Robert Koch-Institut, Berlin) into blood culture bottles. All of these control strains were detected by the BACTEC system after blood culture bottles were stored at 4°C for 24 h, and each control strain could subsequently be recultured on blood agar.
Serial dilutions of S. epidermidis ATCC 35984 and E. coli ATCC 35218 (as described above) were used for preparation of Gram stain-AOLC tests and were divided into three groups. Serial dilutions of group 1 were immediately used for preparation of Gram stain-AOLC test slides as previously described (6). Serial dilutions of group 2 were stored at room temperature for 24 h, and serial dilutions of group 3 were stored at 4°C for 24 h; both groups were subsequently processed in the same manner as group 1. Examination of slides was performed as described previously (6). All tests were performed five times.
One-way analysis of variance with Bonferroni's multiple-comparison test was used to compare the TTP results of the three groups. To compare two groups, an unpaired t test was used. Throughout the study, a P of <0.05 was considered significant.
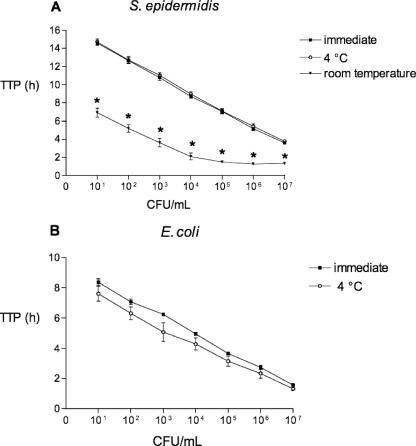
The TTP results from the serial dilution of S. epidermidis and E. coli are shown in Fig. 1. Blood culture bottles with S. epidermidis (dilutions, 101 to 107 CFU/ml) stored at room temperature for 24 h were recognized as positive by the BACTEC system significantly earlier than the blood culture bottles with S. epidermidis processed immediately (P < 0.05) and those stored at 4°C for 24 h (P < 0.05). There was no difference in the TTPs between blood culture bottles with S. epidermidis (dilutions, 101 to 107 CFU/ml) stored at 4°C for 24 h and those that were processed immediately.
FIG. 1.
Influence of storage temperature of blood samples on TTP. (A) Storage of blood samples (dilutions, 101 to 107 CFU/ml of S. epidermidis) at room temperature significantly reduces the TTP compared to that after storage at 4°C and to that after immediate processing (P < 0.05). (B) There is no difference in TTP results (dilutions, 101 to 107 CFU/ml of E. coli) between blood samples that were processed immediately and those that were stored at 4°C. As the blood culture detection system did not recognize blood cultures inoculated with E. coli at 102 to 107 CFU/ml blood as positive, the data are not shown. The detection of E. coli at these dilutions was inhibited due to exhausted CO2 production, on which the BACTEC detection system is based. Asterisks indicate values that are significantly different at a P of <0.05 compared to the values obtained for samples processed immediately and those stored at 4°C (one-way analysis of variance).
Similarly, there was no difference in the TTPs between blood culture bottles with E. coli (dilutions, 101 to 107 CFU/ml) stored at 4°C for 24 h and those that were processed immediately. In the group of blood culture bottles with E. coli stored for 24 h at room temperature, the dilution of 101 CFU/ml became positive after 0.92 ± 0.22 h (mean ± standard deviation) compared to 8.37 ± 0.37 h for those processed immediately and 7.62 ± 0.85 h for those stored at 4°C for 24 h (P < 0.05). Blood culture bottles containing E. coli (dilutions, 102 to 107 CFU/ml) stored at room temperature for 24 h were not detected as positive by the BACTEC 9240 system within 7 days. Subsequent Gram-stained slides and cultures on blood agar showed gram-negative rods and growth of E. coli, respectively.
In the serial dilutions of S. epidermidis and E. coli cultures from samples that were processed immediately, the detection limit of the Gram stain-AOLC tests was a dilution of 104 CFU/ml. In the serial dilutions stored at room temperature for 24 h, the detection limit of the Gram stain-AOLC test decreased to 101 CFU/ml for both microorganisms. Gram stain-AOLC test results for serial dilutions stored at 4°C for 24 h resembled those for the immediately processed samples (Fig. 2).
FIG. 2.
Influence of storage temperature of blood cultures (S. epidermidis and E. coli) on Gram stain-AOLC test results. The detection limit was a dilution of 104 CFU/ml when blood samples were processed immediately or stored at 4°C for 24 h. Due to bacterial growth during the storage period, the detection limit decreased to a dilution of 101 CFU/ml when blood samples were stored at room temperature for 24 h.
Both the DTP and Gram stain-AOLC tests are based on the microbial loads of blood samples. It is therefore crucial to maintain the bacterial inoculum (Gram stain-AOLC test) or the ratio of microbes in central and peripheral blood cultures (DTP test) for correct results. A recent paper demonstrated that for the DTP test, the ratio of microbes was maintained if the blood cultures were kept at room temperature for up to 8 h, but detailed data were not shown (9).
We demonstrate that 24-hour storage of blood culture bottles at room temperature markedly influenced the TTP. Furthermore, 24-hour storage at room temperature of blood culture bottles with fast-growing strains (e.g., E. coli) and high bacterial loads inhibited microbial detection due to exhausted CO2 production, on which the BACTEC detection system is based. In clinical practice, this could lead to false-negative DTP results. In addition, 24-hour storage at room temperature can lead to false-positive Gram stain-AOLC test results due to an increase in microbial load. Bacterial growth during the storage period at room temperature accounts for the positive Gram stain-AOLC test results for the dilutions of 103 to 101 CFU/ml.
As a consequence, laboratory personnel should be available 24 h a day to process blood samples drawn for DTP or Gram stain-AOLC test purposes. However, a 24-hour microbiological laboratory service is not feasible in most hospitals. We therefore evaluated a simple and clinically practical storage condition to maintain the initial microbial inoculum. We demonstrate that after blood sample storage at 4°C for 24 h, both TTP and Gram stain-AOLC test results resembled the results for samples that were processed immediately.
We conclude that 24-hour storage at room temperature of blood cultures and blood samples may lead to false-negative DTP and false-positive Gram stain-AOLC test results. In daily clinical practice, blood samples drawn for the DTP test and/or the Gram stain-AOLC test that cannot be processed within 8 h should be stored at 4°C to obtain correct results in suspected cases of CRBSI.
Acknowledgments
We gratefully acknowledge the excellent technical assistance of Christina Strempfl and Waltraut Eberharth.
Footnotes
Published ahead of print on 30 May 2007.
REFERENCES
- 1.Bjornson, H. S., R. Colley, R. H. Bower, V. P. Duty, J. T. Schwartz-Fulton, and J. E. Fischer. 1982. Association between microorganism growth at the catheter insertion site and colonization of the catheter in patients receiving total parenteral nutrition. Surgery 92:720-727. [PubMed] [Google Scholar]
- 2.Blot, F., G. Nitenberg, E. Chachaty, B. Raynard, N. Germann, S. Antoun, A. Laplanche, C. Brun-Buisson, and C. Tancrede. 1999. Diagnosis of catheter-related bacteraemia: a prospective comparison of the time to positivity of hub-blood versus peripheral-blood cultures. Lancet 354:1071-1077. [DOI] [PubMed] [Google Scholar]
- 3.Brun-Buisson, C., F. Abrouk, P. Legrand, Y. Huet, S. Larabi, and M. Rapin. 1987. Diagnosis of central venous catheter-related sepsis. Critical level of quantitative tip cultures. Arch. Intern. Med. 147:873-877. [PubMed] [Google Scholar]
- 4.Cleri, D. J., M. L. Corrado, and S. J. Seligman. 1980. Quantitative culture of intravenous catheters and other intravascular inserts. J. Infect. Dis. 141:781-786. [DOI] [PubMed] [Google Scholar]
- 5.Crump, J. A., and P. J. Collignon. 2000. Intravascular catheter-associated infections. Eur. J. Clin. Microbiol. Infect. Dis. 19:1-8. [DOI] [PubMed] [Google Scholar]
- 6.Kite, P., B. M. Dobbins, M. H. Wilcox, and M. J. McMahon. 1999. Rapid diagnosis of central-venous-catheter-related bloodstream infection without catheter removal. Lancet 354:1504-1507. [DOI] [PubMed] [Google Scholar]
- 7.Raad, I., H. A. Hanna, B. Alakech, I. Chatzinikolaou, M. M. Johnson, and J. Tarrand. 2004. Differential time to positivity: a useful method for diagnosing catheter-related bloodstream infections. Ann. Intern. Med. 140:18-25. [DOI] [PubMed] [Google Scholar]
- 8.Raad, I. I., M. F. Sabbagh, K. H. Rand, and R. J. Sherertz. 1992. Quantitative tip culture methods and the diagnosis of central venous catheter-related infections. Diagn. Microbiol. Infect. Dis. 15:13-20. [DOI] [PubMed] [Google Scholar]
- 9.Seifert, H., O. Cornely, K. Seggewiss, M. Decker, D. Stefanik, H. Wisplinghoff, and G. Fätkenheuer. 2003. Bloodstream infection in neutropenic cancer patients related to short-term nontunnelled catheters determined by quantitative blood cultures, differential time to positivity, and molecular epidemiological typing with pulsed-field gel electrophoresis. J. Clin. Microbiol. 41:118-123. [DOI] [PMC free article] [PubMed] [Google Scholar]