Abstract
Salmonella enterica serovar Virchow is highly prevalent in humans and farm animals in Israel. In addition to high rates of resistance to multiple antibiotics, this serovar exhibits a high incidence of resistance to nalidixic acid. More than 90% of Salmonella serovar Virchow isolates of human and poultry origin obtained from 1997 to 2004 were resistant to nalidixic acid (MIC ≥ 128 μg/ml), with reduced susceptibility to ciprofloxacin (MIC between 0.125 and 0.250 μg/ml). Most isolates belonged to two predominant, closely related pulsed-field gel electrophoresis image types. Investigation of the mechanisms of quinolone resistance revealed that this pathogen probably emerged from a parental clone that overproduced the AcrAB efflux pump and had a single point mutation in gyrA leading to the Asp87Tyr substitution. The close resemblance between human and poultry isolates points to poultry as a likely source of Salmonella serovar Virchow in the food chain.
The global increase in the prevalence of Salmonella strains with a reduced susceptibility to quinolones constitutes a major concern, since these pathogens have been associated with a significant burden of hospitalization and mortality (18, 19, 30) and with clinical failures of therapy (6-8, 11, 23, 30, 36, 38, 39). Quinolone resistance in gram-negative pathogens is usually acquired by chromosomal mutations, primarily in the genes that encode DNA gyrase and topoisomerase IV (22). Additionally, mutations affecting the uptake or efflux of the quinolones result in decreased accumulation of the antibiotics in the bacterial cell (32). Plasmids that harbor qnr genes that encode flouroquinolone-inactivating enzymes have also been discovered (15, 37).
Salmonella enterica serovar Virchow, a serovar that is rare in the United States and uncommon in Europe, has emerged and spread among humans and farm animals in Israel to become one of the three ranking Salmonella serovars. The higher proportion of Salmonella serovar Virchow infections among blood isolates reflected its increased invasiveness in young children (43, 44). In addition to high rates of resistance to multiple antibiotics, this serovar displays notable rates of nalidixic acid resistance (about 90%) (44). The unique epidemiology in Israel, where Salmonella serovar Virchow is both highly prevalent among humans and poultry and unusually highly resistant to nalidixic acid, presents an opportunity to investigate the existence and evolution of the various mechanisms of quinolone resistance.
MATERIALS AND METHODS
Bacterial strains.
A total of 336 isolates of Salmonella serovar Virchow were available for our studies. Human stool and blood isolates (n = 243) collected between 1997 and 2004 were obtained from the Government Central Laboratories (44), and poultry isolates (n = 93) collected between 2001 and 2003 from the Veterinary Services (random samples).
Typing by PFGE.
Typing of the isolates was carried out by pulsed-field gel electrophoresis and analyzed as previously described (44).
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing was performed by the disk diffusion technique according to the disk manufacturer's (Oxoid) guidelines, which, except for guidelines pertaining to colistin and polymyxin B, are based on the Clinical and Laboratory Standards Institute (CLSI) guidelines (31). Guidelines for colistin and polymycin B are based on the approval of the U.S. Food and Drug Administration.
MICs of nalidixic acid and ciprofloxacin were determined by the broth dilution method according to the CLSI guidelines (10).
Cyclohexane resistance.
Cyclohexane resistance was determined by the method of Asako et al. (1). Escherichia coli K-12 AG100, AG102, (resistant strain) and AG100-A (sensitive strain) (16, 45) were used for controls.
Determination of mutations within gyrA.
Mutations in gyrA were determined by using pyrosequencing and/or sequencing of the gyrA amplicon. For sequence analysis, the gyrA gene was amplified by PCR from the extracted genomic DNA of the isolates using the primers CACCCGAATAAAGCATTGTCTGG/ACGGACGCGAAATCAGCG. Primers were designed based on the gyrA sequence of Salmonella serovar Typhimurium (accession no. X78977). Pyrosequencing of the locus encoding the codons of amino acids 83 to 88 of GyrA was performed on an additional 87 isolates to expand the sampling. The experiment was performed and data were analyzed as previously described (21) with the PCR primers 5′-AAAATCTGCCCGTGTCGT-3′ and 5′Biotine-C-TGCGCCATACGAACGAT-3′ and the sequencing primer 5′-ATCCCCACGGCGATT-3′. Controls were serovar Virchow isolates that had been sequenced and clinical isolates of serovars Agona and Typhimurium with known mutations.
PCR screening for the qnr genes.
The presence of qnrA, qnrB, and qnrS genes was studied by PCR as previously described (15). Positive controls were E. coli strains with the plasmids pMG252, pMG298, and pMG306 (15).
Determination of transcription levels of acrAB, marA, and soxS.
Transcription analysis of acrAB, marA, and soxS was carried out using the green fluorescent protein (GFP) signals expressed from the gfp gene that was fused to the promoter/operator regions of these genes. Construction of the plasmids was previously described (E. Hartog, L. Ben-Shalom, D. Shachar, K. R. Matthews, and S. Yaron, presented at the 2nd ASM Conference on Salmonella: From Pathogenesis to Therapeutics, Victoria, Canada, 9 to 13 September 2006). E. coli K-12 AG100 and AG102 and Salmonella serovar Typhimurium ATCC 14028 were used for comparison. Each isolate was also transformed with a promoterless plasmid for control. Growth, fluorescence detection, and data analysis were carried out as described previously (3).
Statistical methods.
Proportions were compared using the chi-square test, and two-tailed P values were reported. Analyses were performed using SPSS version 13.
RESULTS AND DISCUSSION
A high rate of resistance was detected when we determined the susceptibility of the Salmonella serovar Virchow isolates to 12 antibiotics (Table 1). Overall, the resistance rates were comparable between the two sources, poultry and humans, except for the excessive frequency of resistance to colistin and polymyxin B among the poultry isolates. The highest resistance rate was associated with nalidixic acid-more than 90%. However, all the tested isolates were susceptible to ciprofloxacin according to the CLSI criteria (31). There was no change in the resistance pattern to nalidixic acid during the study period.
TABLE 1.
Frequency of resistance to antimicrobial agents in Salmonella serovar Virchow isolates from human and poultry sources
| Resistance to:a | Human isolates (n = 243)
|
Chicken isolates (n = 93)
|
P valueb | ||
|---|---|---|---|---|---|
| No. | % | No. | % | ||
| No antibiotic agents | 13 | 5.3 | 2 | 2.2 | NS |
| Colistin | 4 | 1.6 | 15 | 16.1 | <0.001 |
| Polymyxin B | 0 | 0 | 14 | 15.1 | <0.001 |
| Ampicillin | 31 | 12.8 | 19 | 20.7 | NS |
| Neomycin | 1 | 0.4 | 4 | 4.3 | NS |
| Gentamicin | 1 | 0.4 | 0 | 0.0 | NS |
| Nalidixic acid | 221 | 90.9 | 86 | 92.5 | NS |
| Trimethoprim-sulfamethoxazole | 113 | 46.5 | 52 | 55.9 | NS |
| Tetracycline | 124 | 51.0 | 59 | 63.4 | 0.05 |
| Chloramphenicol | 85 | 35.0 | 37 | 39.8 | NS |
| Streptomycin | 125 | 51.4 | 48 | 51.6 | NS |
| Ciprofloxacin | 0 | 0 | 0 | 0 | NS |
| Ceftriaxone | 0 | 0 | 0 | 0 | NS |
| ≥4 Antibiotic agents | 113 | 46.5 | 52 | 55.9 | NS |
Antibiotics and the amounts in disks were as follows: chloramphenicol, nalidixic acid, neomycin, ceftriaxone, tetracycline, and polymyxin B, 30 μg each; ampicillin, streptomycin, and colistin, 10 μg each; ciprofloxacin and gentamicin, 5 μg each; trimethoprim-sulfamethoxazole 1:19, 25 μg. The quality control strains that were used for the susceptibility testing are Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922, and Pseudomonas aeruginosa ATCC 27853.
NS, not significant. For all NS cell entries, the difference between human isolates and chicken isolates is not statistically significant (P > 0.05).
PFGE analysis showed 20 pulsotypes. Most isolates from human and poultry sources belonged to two predominant pulsotypes, A1 (43%) and B1 (36%), which differ by two bands only (44). Other pulsotypes (see Table 3) were also closely related. Based on published PFGE images, the predominant pulsotypes in Belgium, France, and Australia were similar to the less common A4 pulsotype in Israel (4, 5). Resistance to nalidixic acid was not associated with a particular PFGE profile.
TABLE 3.
Mutation in gyrA gene in Salmonella serovar Virchow isolates
| Nalidixic acid MIC (μg/ml) | Ciprofloxacin MIC (μg/ml) | No. of isolates | PFGE result | GyrA mutation |
|---|---|---|---|---|
| Resistant isolates | ||||
| 128-512 | 0.125-0.25 | 78 | A1 (43%), B1 (30%), A4 (8%), A3 (5%), B2 (4%), A2 (4%), B6, B7, A6, A9 (1.3%) | Asp87-Tyr |
| 0.125 | 7 | A1 (29%), B1 (42%), A4 (29%) | No mutation | |
| 128 | ≤0.03 | 2 | A1 (100%) | No mutation |
| Resistanta | Susceptiblea | 26 | B1 (58%), A1 (23%), B4 (12%), A10 (4%), A4 (4%) | Asp87-Tyr |
| Sensitive isolates | ||||
| ≤8 | ≤0.03 | 3 | B1 (33%), A1 (33%), A3 (33%) | No mutation |
| 4 | ≤0.03 | 1 | A1 | Asp87-Tyr |
MICs were not determined; disk diffusion was performed.
Approximately one-third of the nalidixic acid-resistant isolates from both sources (105/307) and four susceptible isolates were randomly selected for further testing of their resistance to nalidixic acid and ciprofloxacin by the broth dilution method. Resistance to ciprofloxacin was not detected, but the ciprofloxacin MIC for the nalidixic acid-resistant isolates was usually higher than that for the sensitive isolates, and a significant correlation between nalidixic acid resistance and an elevated ciprofloxacin MIC was observed (P < 0.001) (Table 2). To the best of our knowledge, a stable frequency of more than 90% nalidixic acid resistance with MIC ≥ 128 μg/ml and with elevated ciprofloxacin MIC in human and veterinary isolates over an 8-year study period had not been described with Salmonella (22). This rate of quinolone resistance was by far higher than the rates reported for Salmonella serovar Virchow in Europe (40). However, nalidixic acid resistance among isolates of Salmonella serovar Virchow in Belgium doubled during the period of 2000 to 2003 to 84% (5). Resistance to nalidixic acid, among other prevalent Salmonella serovars in Israel, was also less common: 37% in serovar Typhimurium, 17% in serovar Hadar, and 1% in serovar Enteritidis (29).
TABLE 2.
Correlation of MICs between nalidixic acid and ciprofloxacin for Salmonella serovar Virchow isolatesa
Numbers in table cells represent numbers of isolates.
NAL, nalidixic acid.
CIP, ciprofloxacin.
It was previously shown that mutations in gyrA can be sufficient to cause high-level resistance to nalidixic acid in Salmonella (17, 22). We screened for mutations in gyrA to target the reason for the high incidence of resistance among the isolates. Initially we sequenced the gyrA gene of 27 nalidixic acid-resistant isolates and 3 susceptible isolates. The same single-point mutation was observed in all resistant isolates: GAC was switched to TAC (Asp87Tyr). The same mutation was also found in a susceptible isolate, while the gyrA gene of the other susceptible isolates was identical to the gyrA of Salmonella serovar Typhimurium (accession no. X78977). Pyrosequencing was performed on an additional 87 isolates to expand the sampling. The only substitution that was detected in the serovar Virchow isolates was Asp87Tyr. Table 3 summarizes the sequence results of all isolates with regard to their resistance and PFGE typing. Overall, there was a significant correlation between the presence of the Asp87Tyr substitution and resistance to nalidixic acid (P = 0.013, chi-square test), as well as an elevated ciprofloxacin MIC (P < 0.001). No correlation was observed between the presence/absence of the Asp87Tyr substitution and any PFGE profile.
The Asp87Tyr substitution was reported with Salmonella serovar Virchow as well as with other serovars. Similar to the results of this study, for the different serovars with the Asp87Tyr substitution, the nalidixic acid MICs were between 64 and 512 μg/ml and ciprofloxacin MICs were between 0.125 and 1 μg/ml (12, 20, 24, 27, 42), lower MICs than for strains with other mutations in the codons of Asp87 or Ser83 (26).
A high incidence of nalidixic acid resistance could arise from either rare mutation events with subsequent clonal expansion and dissemination or from frequent mutation and selection events. Clonal expansion would result in relatively homogeneous resistant organisms, whereas the frequent mutation hypothesis should result in greater heterogeneity (24). The observation that most of the resistant isolates from 1997 to 2004 shared the same mutation is indicative of the spread of a single resistant clone of Salmonella serovar Virchow. This is also supported by the observations that all isolates from human and poultry sources had closely related PFGE profiles. Evidence for the spread of a resistant clone with the Asp87Tyr switch was also described for Salmonella serovar Enteritidis (24). Similarly, a single substitution (Ser83Phe) was found in a clone of extended-spectrum beta-lactamase-producing Salmonella serovar Virchow with an elevated MIC of ciprofloxacin (5).
To examine whether quinolone resistance of Salmonella serovar Virchow is also plasmid encoded, we screened the isolates for the presence of qnrA, qnrB, and qnrS. All PCRs were negative, in contrast to those of the E. coli strains which served as positive controls.
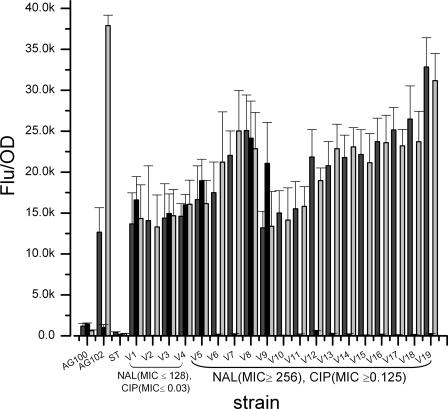
Mechanisms resulting in a decreased quinolone accumulation may also contribute to the elevated resistance. The major mechanism identified in Salmonella is active efflux mediated by AcrAB-TolC (2, 14). This can be achieved by overexpression of AcrAB or the activators MarA and SoxS (25, 28, 35). Using the GFP reporting system, we compared the transcription levels of soxS, marA, and acrA in 19 isolates with different resistance properties. A high correlation was observed between the transcription of acrA and that of marA (P < 0.0001); however, the transcription of neither acrA nor marA correlated with resistance to nalidixic acid or ciprofloxacin (Fig. 1). The transcription of acrA and marA was significantly higher in all tested Salmonella serovar Virchow isolates than in Salmonella serovar Typhimurium or E. coli AG100 isolates. E. coli AG102 is a mutant that overexpresses MarA and AcrAB due to mutation in the repressor MarR (16). The transcription of acrA in AG102 was in the range of the Salmonella serovar Virchow isolates. A notable variability was observed in the transcription of soxS, and a high proportion (3/6) of isolates that overexpressed soxS were susceptible to nalidixic acid. This agrees with the previous reports that flouroquinolone-resistant isolates of Salmonella displayed increased expression of marA and acrA but not soxS (13, 41). The reason for the high transcription levels of acrA and marA in the Virchow isolates has not been elucidated, as no mutation was detected in the sequence of these genes or their promoters. All these loci were found to be identical to Salmonella serovar Typhimurium (GenBank accession no. NC_003277). The high transcription levels of marA and acrA in all isolates (including the sensitive ones) could suggest that this overproduction appeared prior to the mutation in gyrA.
FIG. 1.
Transcription of acrAB, marA, and soxS in E. coli K-12 strains AG100 and AG102, Salmonella serovar Typhimurium ATCC 14028 (ST), and representative isolates of Salmonella serovar Virchow (V1-V19). Numbers represent the means of normalized GFP fluorescence intensities of cultures with the following promoter-gfp fusions: acrAp::gfp (dark gray), soxSp::gfp (black), and marAp::gfp (light gray). The bars represent the standard errors of the means of the results of three experiments; each experiment was conducted in triplicate. SV10 and SV11 are cyclohexane-resistant isolates.
Recent reports pointed to a possible correlation between antibiotic resistance and cyclohexane resistance (9, 33, 34). Since the isolates produced high levels of acrA and marA transcripts, we hypothesized that they would be resistant to cyclohexane. All but two tested isolates were found to be sensitive to cyclohexane. The two isolates (one from humans and one from poultry) that were resistant to cyclohexane and to nalidixic acid had the Asp87Tyr exchange in gyrA (Fig. 1, SV10 and SV11). The infrequent resistance to cyclohexane indicates a lack of correlation between resistance to cyclohexane and antibiotic resistance or the transcription levels of acrA and marA. Similar observations were also reported for other Salmonella serovars (13).
Ciprofloxacin is routinely used in Israel for the treatment of severe gastrointestinal infections in adults. Other flouroquinolones like enrofloxacin, norfloxacin, danofloxacin, and ofloxacin are widely used with farm animals. Despite the persistent antibiotic pressure, we observed a high stability of a single type of mutation over the 8-year period studied without the development of ciprofloxacin resistance. Our data suggest that the high prevalence of nalidixic acid resistance with increased ciprofloxacin MICs among Salmonella serovar Virchow isolates in Israel probably emerged from the spread of a parental clone that overproduced the AcrAB efflux pump and had a single mutation in the gyrA gene. The same clone emerged in both humans and poultry, indicating that poultry was probably the main source for Salmonella serovar Virchow in the food chain. The veterinary and health authorities should target this clone for the application of more intensive and efficacious control measures.
Acknowledgments
We thank A. Reisfeld and R. Yishai from the Israeli Government Central Laboratories and E. Berman from the Veterinary Services, Bet Dagan for donation of the Salmonella serovar Virchow strains, S. B. Levy for the E. coli AG100, AG102, and AG100-A strains, and G. Jacoby for the qnr-positive E. coli strains.
This work was funded by grant 1275/04 from the Israel Science Foundation (ISF).
Footnotes
Published ahead of print on 27 June 2007.
REFERENCES
- 1.Asako, H., H. Nakajima, K. Kobayashi, M. Kobayashi, and R. Aono. 1997. Organic solvent tolerance and antibiotic resistance increased by overexpression of marA in Escherichia coli. Appl. Environ. Microbiol. 63:1428-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baucheron, S., H. Imberechts, E. Chaslus-Dancla, and A. Cloeckaert. 2002. The AcrB multidrug transporter plays a major role in high-level fluoroquinolone resistance in Salmonella enterica serovar Typhimurium phage type DT204. Microb. Drug Resist. 8:281-289. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Barak, Z., W. Streckel, S. Yaron, S. Cohen, R. Prager, and H. Tschape. 2006. The expression of the virulence-associated effector protein gene avrA is dependent on a Salmonella enterica-specific regulatory function. Int. J. Med. Microbiol. 296:25-38. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, C. M., C. Dalton, M. Beers-Deeble, A. Milazzo, E. Kraa, D. Davos, M. Puech, A. Tan, and M. W. Heuzenroeder. 2003. Fresh garlic: a possible vehicle for Salmonella Virchow. Epidemiol. Infect. 131:1041-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertrand, S., F. X. Weill, A. Cloeckaert, M. Vrints, E. Mairiaux, K. Praud, K. Dierick, C. Wildemauve, C. Godard, P. Butaye, H. Imberechts, P. A. Grimont, and J. M. Collard. 2006. Clonal emergence of extended-spectrum beta-lactamase (CTX-M-2)-producing Salmonella enterica serovar Virchow isolates with reduced susceptibilities to ciprofloxacin among poultry and humans in Belgium and France (2000 to 2003). J. Clin. Microbiol. 44:2897-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casin, I., J. Breuil, J. P. Darchis, C. Guelpa, and E. Collatz. 2003. Fluoroquinolone resistance linked to GyrA, GyrB, and ParC mutations in Salmonella enterica typhimurium isolates in humans. Emerg. Infect. Dis. 9:1455-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiu, C. H., T. L. Wu, L. H. Su, C. Chu, J. H. Chia, A. J. Kuo, M. S. Chien, and T. Y. Lin. 2002. The emergence in Taiwan of fluoroquinolone resistance in Salmonella enterica serotype Choleraesuis. N. Engl. J. Med. 346:413-419. [DOI] [PubMed] [Google Scholar]
- 8.Chiu, C. H., T. L. Wu, L. H. Su, J. W. Liu, and C. Chu. 2004. Fluoroquinolone resistance in Salmonella enterica serotype Choleraesuis, Taiwan, 2000-2003. Emerg. Infect. Dis. 10:1674-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, S. H., J. H. Woo, J. E. Lee, S. J. Park, E. J. Choo, Y. G. Kwak, M. N. Kim, M. S. Choi, N. Y. Lee, B. K. Lee, N. J. Kim, J. Y. Jeong, J. Ryu, and Y. S. Kim. 2005. Increasing incidence of quinolone resistance in human non-typhoid Salmonella enterica isolates in Korea and mechanisms involved in quinolone resistance. J. Antimicrob. Chemother. 56:1111-1114. [DOI] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 11.Crump, J. A., T. J. Barrett, J. T. Nelson, and F. J. Angulo. 2003. Reevaluating fluoroquinolone breakpoints for Salmonella enterica serotype Typhi and for non-Typhi salmonellae. Clin. Infect. Dis. 37:75-81. [DOI] [PubMed] [Google Scholar]
- 12.Eaves, D. J., E. Liebana, M. J. Woodward, and L. J. Piddock. 2002. Detection of gyrA mutations in quinolone-resistant Salmonella enterica by denaturing high-performance liquid chromatography. J. Clin. Microbiol. 40:4121-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eaves, D. J., V. Ricci, and L. J. Piddock. 2004. Expression of acrB, acrF, acrD, marA, and soxS in Salmonella enterica serovar Typhimurium: role in multiple antibiotic resistance. Antimicrob. Agents Chemother. 48:1145-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escribano, I., J. C. Rodriguez, L. Cebrian, and G. Royo. 2004. The importance of active efflux systems in the quinolone resistance of clinical isolates of Salmonella spp. Int. J. Antimicrob. Agents 24:428-432. [DOI] [PubMed] [Google Scholar]
- 15.Gay, K., A. Robicsek, J. Strahilevitz, C. H. Park, G. Jacoby, T. J. Barrett, F. Medalla, T. M. Chiller, and D. C. Hooper. 2006. Plasmid-mediated quinolone resistance in non-Typhi serotypes of Salmonella enterica. Clin. Infect. Dis. 43:297-304. [DOI] [PubMed] [Google Scholar]
- 16.George, A. M., and S. B. Levy. 1983. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J. Bacteriol. 155:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giraud, E., S. Baucheron, and A. Cloeckaert. 2006. Resistance to fluoroquinolones in Salmonella: emerging mechanisms and resistance prevention strategies. Microbes Infect. 8:1937-1944. [DOI] [PubMed] [Google Scholar]
- 18.Helms, M., J. Simonsen, and K. Molbak. 2004. Quinolone resistance is associated with increased risk of invasive illness or death during infection with Salmonella serotype Typhimurium. J. Infect. Dis. 190:1652-1654. [DOI] [PubMed] [Google Scholar]
- 19.Helms, M., P. Vastrup, P. Gerner-Smidt, and K. Molbak. 2002. Excess mortality associated with antimicrobial drug-resistant Salmonella typhimurium. Emerg. Infect. Dis. 8:490-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirose, K., A. Hashimoto, K. Tamura, Y. Kawamura, T. Ezaki, H. Sagara, and H. Watanabe. 2002. DNA sequence analysis of DNA gyrase and DNA topoisomerase IV quinolone resistance-determining regions of Salmonella enterica serovar Typhi and serovar Paratyphi A. Antimicrob. Agents Chemother. 46:3249-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopkins, K. L., C. Arnold, and E. J. Threlfall. 2007. Rapid detection of gyrA and parC mutations in quinolone-resistant Salmonella enterica using pyrosequencing technology. J. Microbiol. Methods 68:163-171. [DOI] [PubMed] [Google Scholar]
- 22.Hopkins, K. L., R. H. Davies, and E. J. Threlfall. 2005. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int. J. Antimicrob. Agents 25:358-373. [DOI] [PubMed] [Google Scholar]
- 23.Kadhiravan, T., N. Wig, A. Kapil, S. K. Kabra, K. Renuka, and A. Misra. 2005. Clinical outcomes in typhoid fever: adverse impact of infection with nalidixic acid-resistant Salmonella typhi. BMC Infect. Dis. 5:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kilmartin, D., D. Morris, C. O'Hare, G. Corbett-Feeney, and M. Cormican. 2005. Clonal expansion may account for high levels of quinolone resistance in Salmonella enterica serovar Enteritidis. Appl. Environ. Microbiol. 71:2587-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levy, S. B. 2002. Active efflux, a common mechanism for biocide and antibiotic resistance. J. Appl. Microbiol. 92(Suppl.):65S-71S. [PubMed]
- 26.Liebana, E., C. Clouting, C. A. Cassar, L. P. Randall, R. A. Walker, E. J. Threlfall, F. A. Clifton-Hadley, A. M. Ridley, and R. H. Davies. 2002. Comparison of gyrA mutations, cyclohexane resistance, and the presence of class I integrons in Salmonella enterica from farm animals in England and Wales. J. Clin. Microbiol. 40:1481-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ling, J. M., E. W. Chan, A. W. Lam, and A. F. Cheng. 2003. Mutations in topoisomerase genes of fluoroquinolone-resistant salmonellae in Hong Kong. Antimicrob. Agents Chemother. 47:3567-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin, R. G., and J. L. Rosner. 2004. Transcriptional and translational regulation of the marRAB multiple antibiotic resistance operon in Escherichia coli. Mol. Microbiol. 53:183-191. [DOI] [PubMed] [Google Scholar]
- 29.Mates, A., V. Agmon, R. Yishai, N. Andorn, and A. Reisfeld. 2002. Salmonella 1997-2000. A report of the National Salmonella Center. State of Israel Ministry of Health, Jerusalem, Israel.
- 30.Molbak, K. 2005. Human health consequences of antimicrobial drug-resistant Salmonella and other foodborne pathogens. Clin. Infect. Dis. 41:1613-1620. [DOI] [PubMed] [Google Scholar]
- 31.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial disk susceptibility tests for bacteria that grow aerobically. Approved standard M2-A7. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 32.Paulsen, I. T. 2003. Multidrug efflux pumps and resistance: regulation and evolution. Curr. Opin. Microbiol. 6:446-451. [DOI] [PubMed] [Google Scholar]
- 33.Randall, L. P., S. W. Cooles, L. J. Piddock, and M. J. Woodward. 2004. Effect of triclosan or a phenolic farm disinfectant on the selection of antibiotic-resistant Salmonella enterica. J. Antimicrob. Chemother. 54:621-627. [DOI] [PubMed] [Google Scholar]
- 34.Randall, L. P., S. W. Cooles, A. R. Sayers, and M. J. Woodward. 2001. Association between cyclohexane resistance in Salmonella of different serovars and increased resistance to multiple antibiotics, disinfectants and dyes. J. Med. Microbiol. 50:919-924. [DOI] [PubMed] [Google Scholar]
- 35.Randall, L. P., and M. J. Woodward. 2002. The multiple antibiotic resistance (mar) locus and its significance. Res. Vet. Sci. 72:87-93. [DOI] [PubMed] [Google Scholar]
- 36.Renuka, K., S. Sood, B. K. Das, and A. Kapil. 2005. High-level ciprofloxacin resistance in Salmonella enterica serotype Typhi in India. J. Med. Microbiol. 54:999-1000. [DOI] [PubMed] [Google Scholar]
- 37.Robicsek, A., G. A. Jacoby, and D. C. Hooper. 2006. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6:629-640. [DOI] [PubMed] [Google Scholar]
- 38.Rupali, P., O. C. Abraham, M. V. Jesudason, T. J. John, A. Zachariah, S. Sivaram, and D. Mathai. 2004. Treatment failure in typhoid fever with ciprofloxacin susceptible Salmonella enterica serotype Typhi. Diagn. Microbiol. Infect. Dis. 49:1-3. [DOI] [PubMed] [Google Scholar]
- 39.Slinger, R., M. Desjardins, A. E. McCarthy, K. Ramotar, P. Jessamine, C. Guibord, and B. Toye. 2004. Suboptimal clinical response to ciprofloxacin in patients with enteric fever due to Salmonella spp. with reduced fluoroquinolone susceptibility: a case series. BMC Infect. Dis. 4:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Threlfall, E. J., I. S. Fisher, C. Berghold, P. Gerner-Smidt, H. Tschape, M. Cormican, I. Luzzi, F. Schnieder, W. Wannet, J. Machado, and G. Edwards. 2003. Antimicrobial drug resistance in isolates of Salmonella enterica from cases of salmonellosis in humans in Europe in 2000: results of international multi-centre surveillance. Eur. Surveill. 8:41-45. [DOI] [PubMed] [Google Scholar]
- 41.Tibbetts, R. J., T. L. Lin, and C. C. Wu. 2003. Phenotypic evidence for inducible multiple antimicrobial resistance in Salmonella choleraesuis. FEMS Microbiol. Lett. 218:333-338. [DOI] [PubMed] [Google Scholar]
- 42.Walker, R. A., N. Saunders, A. J. Lawson, E. A. Lindsay, M. Dassama, L. R. Ward, M. J. Woodward, R. H. Davies, E. Liebana, and E. J. Threlfall. 2001. Use of a LightCycler gyrA mutation assay for rapid identification of mutations conferring decreased susceptibility to ciprofloxacin in multiresistant Salmonella enterica serotype Typhimurium DT104 isolates. J. Clin. Microbiol. 39:1443-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinberger, M., and N. Keller. 2005. Recent trends in the epidemiology of non-typhoid Salmonella and antimicrobial resistance: the Israeli experience and worldwide review. Curr. Opin. Infect. Dis. 18:513-521. [DOI] [PubMed] [Google Scholar]
- 44.Weinberger, M., H. Solnik-Isaac, D. Shachar, A. Reisfeld, L. Valinsky, N. Andorn, V. Agmon, R. Yishai, R. Bassal, A. Fraser, S. Yaron, and D. Cohen. 2006. Salmonella enterica serotype Virchow: epidemiology, resistance patterns and molecular characterisation of an invasive Salmonella serotype in Israel. Clin. Microbiol. Infect. 12:999-1005. [DOI] [PubMed] [Google Scholar]
- 45.White, D. G., K. Maneewannakul, E. von Hofe, M. Zillman, W. Eisenberg, A. K. Field, and S. B. Levy. 1997. Inhibition of the multiple antibiotic resistance (mar) operon in Escherichia coli by antisense DNA analogs. Antimicrob. Agents Chemother. 41:2699-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]