Abstract
We compared ChromID VRE medium with Enterococcosel containing vancomycin for the detection of vancomycin-resistant Enterococcus in 1,007 specimens. ChromID VRE in combination with Gram straining provided a higher specificity than Enterococcosel, irrespective of the incubation time and enrichment.
Glycopeptide-acquired resistance has emerged in Enterococcus faecalis and Enterococcus faecium, which are designated vancomycin-resistant Enterococcus species (VRE). The resistance phenotype VanA is the most common and features high-level resistance to both vancomycin and teicoplanin. hospital outbreaks of VRE have been reported extensively in the United States, with a prevalence as high as 47% in some studies (2). In Europe, a high prevalence has also been observed in the United Kingdom (10.4%) and Italy (19.6%) (9). In France, the prevalence remains low (<2%) (12, 17). However, an outbreak of VRE was observed in 2004 in the teaching hospital of Clermont-Ferrand (14). Since then, at least five others have been reported in French hospitals (8, 15). Early detection of VRE in fecal specimens is important for nosocomial prevention measures, epidemiologic infectious disease follow-up, and also prevention of vancomycin-resistant Staphylococcus aureus emergence (10, 19). Several agar and broth medium formulations containing vancomycin have been developed for this purpose (1, 3, 5, 11, 13, 18, 20). However, no consensus has been established for medium base, vancomycin concentration, or method of use. ChromID VRE is a new selective chromogenic medium developed for the detection and identification of vancomycin-resistant E. faecium and E. faecalis. The aims of this study were to assess the performance of chromID VRE medium in a low-prevalence context and to determine the best method of use. ChromID VRE was compared with a modified bile esculin agar, which is one of the best media for the isolation of VRE (3).
A total of 1,007 fecal specimens (861 rectal swabs and 146 stool samples) were plated directly or after overnight enrichment in brain heart infusion broth (bioMérieux) with 3 μg/ml vancomycin onto chromID VRE (bioMérieux, Marcy l'Étoile, France) and Enterococcosel agar (Becton Dickinson, Cockeysville, MD). These media contained 8 μg/ml vancomycin. The plates were incubated aerobically at 37°C and examined after 24 h and 48 h of incubation. Identification by chromID VRE was based on the detection of enzymatic activity. E. faecium was stained purple, and E. faecalis was stained blue or blue-green. Enterococci had brownish black to black halos on Enterococcosel with vancomycin (BEAv). Biochemical identification and antibiotic testing were performed with Vitek 2 GP and Vitek 2 AST-P532 cards (bioMérieux). Vancomycin and teicoplanin MICs were determined by E-test for resistant strains. The identification of VRE was confirmed by molecular investigation with the GenoType Enterococcus test (Hain Lifescience, Nehren, Germany) (6).
In 1,007 specimens, 22 VRE were detected with the chromID VRE and BEAv media. These VRE were E. faecium carrying the vanA gene. The large number of samples investigated during this study and the low prevalence of VRE provided the opportunity to explore the false positives. The sensitivities of the chromID VRE and BEAv media were similar under various conditions of use (Table 1, P > 0.05). The rate of VRE detection was significantly increased by the enrichment step, as previously demonstrated (7, 11, 16). However, fecal carriage of these VRE was transitory in patients and this increased sensitivity may not be essential for the management of most outbreaks (4). The comparison of our results with those of other studies is difficult since in most of the latter direct plating was used without enrichment methods. In our study, when enrichment was not performed, the sensitivity of chromID VRE was 92%. The same sensitivity was found in our laboratory for VCA3 agar (5). The bile-esculin-based media supplemented with vancomycin had sensitivities of 80 to 100% (1, 3, 20). Other agar formulations have been tested, but their sensitivities were low (40 to 80%) (3, 13, 20).
TABLE 1.
Sensitivity, specificity, PPV, and NPV for chromID VRE and BEAv media with 24h and 48h of incubation with or without an enrichment step and without or with Gram staininga
| Enrichment (incubation time [h]) and medium used | TP | FP | TN | FN | Se | Sp | PPV | NPV |
|---|---|---|---|---|---|---|---|---|
| Yes (24) | ||||||||
| chromID VRE | 22 | 48 | 937 | 0 | 100 | 95.13 | 31.43 | 100.00 |
| 22 | 1 | 984 | 0 | 100 | 99.90 | 95.65 | 100.00 | |
| BEAv | 22 | 89 | 896 | 0 | 100 | 90.96 | 19.82 | 100.00 |
| 22 | 52 | 933 | 0 | 100 | 94.72 | 29.73 | 100.00 | |
| No (24) | ||||||||
| chromID VRE | 12 | 17 | 968 | 10 | 54.55 | 98.27 | 41.38 | 98.98 |
| 12 | 0 | 985 | 10 | 54.55 | 100.00 | 100.00 | 98.99 | |
| BEAv | 9 | 25 | 960 | 13 | 40.91 | 97.46 | 26.47 | 98.66 |
| 9 | 24 | 961 | 13 | 40.91 | 97.56 | 27.27 | 98.67 | |
| Yes (48) | ||||||||
| chromID VRE | 22 | 108 | 877 | 0 | 100 | 89.04 | 16.92 | 100.00 |
| 22 | 1 | 984 | 0 | 100 | 99.90 | 95.65 | 100.00 | |
| BEAv | 22 | 137 | 848 | 0 | 100 | 86.09 | 13.84 | 100.00 |
| 22 | 75 | 910 | 0 | 100 | 92.39 | 22.68 | 100.00 | |
| No (48) | ||||||||
| chromID VRE | 12 | 147 | 838 | 10 | 54.55 | 85.08 | 7.55 | 98.82 |
| 12 | 23 | 962 | 10 | 54.55 | 97.66 | 34.29 | 98.97 | |
| BEAv | 13 | 116 | 869 | 9 | 59.09 | 88.22 | 10.08 | 98.97 |
| 13 | 79 | 906 | 9 | 59.09 | 91.98 | 14.13 | 99.02 |
Results obtained with Gram staining are in bold. TP, true positive; FN, false negative; TN, true negative; FP, false positive; Se, sensitivity of detection; Sp, specificity of coloration.
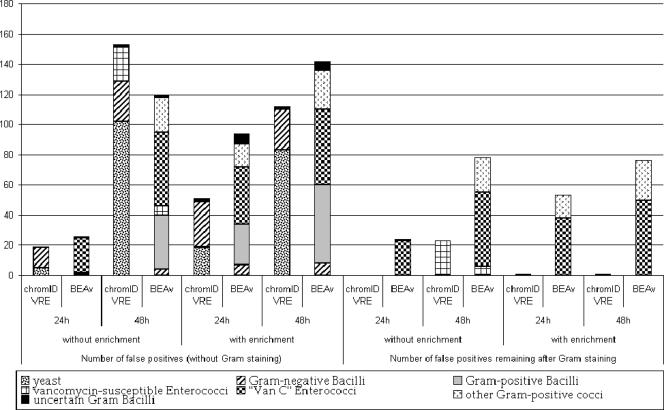
Unlike BEAv, ChromID VRE conferred characteristic colors of E. faecalis and E. faecium that were different from those of Enterococcus gallinarum, Enterococcus casseliflavus, and gram-positive bacilli (n = 114). However, colonies of yeast (n = 124) and gram-negative bacilli (n = 53) could be confused with VRE. Yeasts isolated on chromID VRE were mainly detected after 48 h of incubation (>80%) (Fig. 1). Additionally, 23 Enterococcus strains susceptible to vancomycin grew after 48 h of incubation on chromID VRE, compared to 6 on BEAv. Most gram-positive bacteria were isolated on BEAv after 48 h of incubation (Fig. 1). Thus, the best specificities and positive predictive values (PPV) were obtained for both media at 24 h of incubation and were not significantly different (Table 1). After enrichment, the chromID VRE medium was more specific than BEAv medium (P < 0.05). Gram staining significantly improved the specificity and PPV of chromID VRE since rods and yeasts were excluded (Table 1). Thus, chromID VRE has a higher specificity and PPV than does BEAv (P < 10−6). After 24 h of incubation, Gram staining of typically colored strains allowed the direct identification of VRE without complementary tests (specificity and PPV, 100%; negative predictive value [NPV], 99%). Moreover, prolongation of the incubation period to 48 h was not useful and affected specificity (P < 0.05). For the same response delay, reading of chromID VRE at 24 h of incubation with enrichment provided better performance than reading at 48 h of incubation after direct plating.
FIG. 1.
Distribution of false positives on chromID VRE and BEAv media and contribution of Gram staining to the isolation of VRE.
The costs of chromID VRE and BEAv when used with broth enrichment, 24 h of incubation, and Gram staining were compared. These conditions allowed the best compromise between sensitivity and specificity. The cost of chromID VRE was greater than that of BEAv (additional cost for 1,007 samples, 201€). However, with BEAv, VRE detection required numerous subcultures, supplementary identifications, and susceptibility tests (materials, 730€, exclusive of tax; technician time, 95€) because of the false positives. Therefore, the use of chromID VRE would allow a saving of at least 0.62€ per sample for an annual VRE prevalence of 2.2%.
In conclusion, chromID VRE agar is a useful tool for the easy, economical, and efficient detection of vanA-harboring VRE. The recommendations from this study are to incubate in chromID VRE for 24 h and perform Gram staining of typically colored strains.
Footnotes
Published ahead of print on 6 June 2007.
REFERENCES
- 1.Brown, D. F. J., and E. Walpole. 2003. Evaluation of selective and enrichment media for isolation of glycopeptide-resistant enterococci from faecal specimens. J. Antimicrob. Chemother. 51:289-296. [DOI] [PubMed] [Google Scholar]
- 2.Cetinkaya, Y., P. Falk, and C. G. Mayhall. 2000. Vancomycin-resistant enterococci. Clin. Microbiol. Rev. 13:686-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chadwick, P. R., D. F. J. Brown, M. H. Wilcox, T. A. Collyns, E. Walpole, J. Dillon, R. Smith, G. Gopal Rao, and B. A. Oppenheim. 1997. Comparison of agar-based media for the primary isolation of glycopeptide-resistant enterococci. Clin. Microbiol. Infect. 3:559-563. [DOI] [PubMed] [Google Scholar]
- 4.Cookson, B. D., M. B. Macrae, S. P. Barrett, D. F. J. Brown, C. Chadwick, G. L. French, P. Hateley, I. K. Hosein, and J. J. Wade. 2006. Guidelines for the control of glycopeptide-resistant enterococci in hospitals. J. Hosp. Infect. 62:6-21. [DOI] [PubMed] [Google Scholar]
- 5.Delmas, J., F. Robin, J. P. Romaszko, R. Baraduc, O. Lesens, J. Sirot, and R. Bonnet. 2005. VCA agar (bioMérieux) for selective isolation of vancomycin-resistant enterococci (VRE) from fecal specimens. Pathol. Biol. 53:8-9. [DOI] [PubMed] [Google Scholar]
- 6.Eigner, U., A. Fahr, M. Weizenegger, and W. Witte. 2005. Evaluation of a new molecular system for simultaneous identification of four Enterococcus species and their glycopeptide resistance genotypes. J. Clin. Microbiol. 43:2920-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ford, M., J. D. Perry, F. K. Gould, and K. E. Orr. 1996. Neomycin blood agar as a selective medium for vancomycin resistant Enterococcus faecium. J. Clin. Pathol. 49:437-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fortineau, N., R. Leclercq, S. Maugat, J. Robert, and the members of Réseaux de l'ONERBA. 2006. Le portage des enterocoques resistants aux glycopeptides (ERV): les enquêtes de l'ONERBA Réunion Interdisciplinaire de Chimiothérapie Anti-Infectieuse. ONERBA, Paris, France. http://www.onerba.org/download/ERV_portage_RICAI06.pdf.
- 9.Goossens, H., D. Jabes, R. Rossi, C. Lammens, G. Privitera, and P. Courvalin. 2003. European survey of vancomycin-resistant enterococci in at-risk hospital wards and in vitro susceptibility testing of ramoplanin against these isolates. J. Antimicrob. Chemother. 51:iii5-iii12. [DOI] [PubMed] [Google Scholar]
- 10.Hospital Infection Control Practices Advisory Committee. 1995. Recommendations for preventing the spread of vancomycin resistance. Recommendations of the Hospital Infection Control Practices Advisory Committee. Morbid. Mortal. Wkly. Rep. 44(RR12):1-13. [Google Scholar]
- 11.Ieven, M., E. Vercauteren, P. Descheemaeker, F. van Laer, and H. Goossens. 1999. Comparison of direct plating and broth enrichment culture for the detection of intestinal colonization by glycopeptide-resistant enterococci among hospitalized patients. J. Clin. Microbiol. 37:1436-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones, M. E., D. C. Draghi, C. Thornsberry, J. A. Karlowsky, D. F. Sahm, and R. P. Wenzel. 2004. Emerging resistance among bacterial pathogens in the intensive care unit: a European and North American surveillance study (2000-2002). Ann. Clin. Microbiol. Antimicrob. 3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landman, D., J. M. Quale, E. Oydna, B. Willey, V. Ditore, M. Zaman, K. Patel, G. Saurina, and W. Huang. 1996. Comparison of five selective media for identifying fecal carriage of vancomycin-resistant enterococci. J. Clin. Microbiol. 34:751-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lesens, O., L. Mihaila, F. Robin, O. Baud, J. P. Romaszko, O. Tourniac, J. M. Constantin, B. Souweine, R. Bonnet, A. Bouvet, J. Beytout, O. Traore, and H. Laurichese. 2006. Outbreak of colonization and infection with vancomycin-resistant Enterococcus faecium in a French university hospital. Infect. Control Hosp. Epidemiol. 27:984-986. [DOI] [PubMed] [Google Scholar]
- 15.Naas, T., N. Fortineau, R. Snanoudj, C. Spicq, A. Durrbach, and P. Nordmann. 2005. First nosocomial outbreak of vancomycin-resistant Enterococcus faecium expressing a VanD-like phenotype associated with a vanA genotype. J. Clin. Microbiol. 43:3642-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novicki, T. J., J. M. Schapiro, B. K. Ulness, A. Sebeste, L. Busse-Johnston, K. M. Swanson, S. R. Swanzy, W. Leisenring, and A. P. Limaye. 2004. Convenient selective differential broth for isolation of vancomycin-resistant Enterococcus from fecal material. J. Clin. Microbiol. 42:1637-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schouten, M. A., J. A. Hoogkamp-Korstanje, J. F. Meis, A. Voss, and the European VRE Study Group. 2000. Prevalence of vancomycin-resistant enterococci in Europe. Eur. J. Clin. Microbiol. Infect. Dis. 19:816-822. [DOI] [PubMed] [Google Scholar]
- 18.Swenson, J. M., N. C. Clark, M. J. Ferraro, D. F. Sahm, G. Doern, M. A. Pfaller, L. B. Reller, M. P. Weinstein, R. J. Zabransky, and F. C. Tenover. 1994. Development of a standardized screening method for detection of vancomycin-resistant enterococci. J. Clin. Microbiol. 32:1700-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tenover, F. C., L. M. Weigel, P. C. Appelbaum, L. K. McDougal, J. Chaitram, S. McAllister, N. Clark, G. Killgore, C. M. O'Hara, L. Jevitt, J. B. Patel, and B. Bozdogan. 2004. Vancomycin-resistant Staphylococcus aureus isolate from a patient in Pennsylvania. Antimicrob. Agents Chemother. 48:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Horn, K. G., C. A. Gedris, and K. M. Rodney. 1996. Selective isolation of vancomycin-resistant enterococci. J. Clin. Microbiol. 34:924-927. [DOI] [PMC free article] [PubMed] [Google Scholar]