Abstract
We have developed and validated a nested real-time PCR (NRT-PCR) for the genotyping of Chlamydia trachomatis and used it specifically for the typing of either eight genovars from D to K or three genovars of lymphogranuloma venereum (LGV). The 11 probes used in the NRT-PCR correctly identified the DNA from D to K and LGV reference strains and did not cross-react with the DNA from 26 strains representing the bacterial pathogens and commensals of the oropharynx, genital tract, and rectum. The NRT-PCR had a 95% probability of detection at four genome copies (confidence interval, three to six copies) of C. trachomatis per reaction. One hundred cervical and urethral swab specimens containing C. trachomatis DNA from 63 women and 37 men were used to validate the method. The results from the NRT-PCR and the DNA sequencing of amplicons generated from the omp1 gene showed 100% correlation for these samples. The assay also identified the LGV-II genotype in 24 of 48 rectal swab specimens containing C. trachomatis DNA that were obtained from men having sex with men. The Sexually Transmitted Bacteria Reference Laboratory, London, independently confirmed these results using group-specific LGV real-time PCR and restriction fragment length polymorphism analysis. Compared with the NRT-PCR, non-NRT-PCR was found to be less sensitive: it typed C. trachomatis DNA in only 80% of the genital samples and 90% of the rectal swab samples. This is the first successful demonstration of the use of real-time PCR for the genotype-specific typing of C. trachomatis strains that cause sexually transmitted diseases.
Chlamydia trachomatis infection is a significant global health problem. C. trachomatis is the causative agent of trachoma and sexually transmitted diseases (STDs), i.e., lymphogranuloma venereum (LGV) and other genital manifestations. Trachoma is the second major cause of blindness (cataract is the first) and affects 6 million people worldwide (17), while the estimated global incidence of STDs caused by C. trachomatis, is about 92 million (19). This organism is an intracellular bacterium, and its different genovars produce specific clinical manifestations; i.e., types A, B, Ba, and C cause trachoma, types LGV-I, -II, and -III cause LGV, and types D to K cause oculogenital diseases (14). Trachoma is hyperendemic in many of the poorest areas of Africa, Asia, Central and South America, Australia, and the Middle East (17). The prevalence of LGV is highest in the tropics and subtropics (14). A number of LGV outbreaks among men who have sex with men have been reported recently in different western countries (3, 11, 18). Types D to K are prevalent worldwide (19). Due to extensive genetic diversity between different genovars, together with their distinct geographical distribution and clinical manifestations, a better appreciation of the molecular epidemiology of C. trachomatis infection is crucial to control this infection. The genotyping of C. trachomatis is also important for understanding the pathogenesis of infection and sequelae of the infection, for monitoring therapy and contact tracing, and for the development of a vaccine.
The genotyping of C. trachomatis is based mainly on nested PCR targeting of the omp1 gene. The amplicons are being used for typing by restriction fragment length polymorphism (RFLP) analysis (2, 7, 9, 12), DNA sequencing (1, 8, 15, 21), reverse line blot hybridization (10, 16, 20), and oligonucleotide arrays (22). These studies have resulted in more than 60 reports since 1991 according to PubMed. However, due to the complexity and laborious nature of the above-mentioned methods, all of these publications have described only small-scale studies. For large national and international molecular epidemiology projects, a simpler technique with potential for automation is urgently needed. Real-time PCR fulfils these criteria. This is the first report describing the development of a nested real-time PCR (NRT-PCR) for the genotyping of C. trachomatis genovars that cause STDs. We have also compared the sensitivity of NRT-PCR with that of non-NRT-PCR (NNRT-PCR).
MATERIALS AND METHODS
Bacterial strains.
Fourteen isolates representing all the genovars of C. trachomatis and 26 bacterial isolates representing the pathogenic bacteria and commensals of the oropharynx, genital tract, and rectum were used to investigate the specificities of primers and probes. The identities of these organisms are shown in Tables 1 and 2 in a previous publication (5), while their culture and the extraction of DNA from bacterial suspensions were performed as described in the same paper (5). The ability of the NRT-PCR to generate correct results from mixed infections was investigated by testing 11 pools of DNA from different strains of C. trachomatis. The pools of 100-μl volumes were prepared by mixing 1 ng of DNA from one strain and 100 ng of DNA from each of the other two or three strains according to the format of each assay (Fig. 1). The composition of each pool is described below. Pool 1 contained 1 ng DNA from genotype D and 100 ng DNA from each of genotypes E, H, and I. Pool 2 contained 1 ng DNA from genotype E and 100 ng DNA from each of genotypes D, H, and I. Pool 3 contained 1 ng DNA from genotype H and 100 ng DNA from each of genotypes D, E, and I. Pool 4 contained 1 ng DNA from genotype I and 100 ng DNA from each of genotypes D, E, and H. Pool 5 contained 1 ng DNA from genotype F and 100 ng DNA from each of genotypes G, J, and K. Pool 6 contained 1 ng DNA from genotype G and 100 ng DNA from each of genotypes F, J, and K. Pool 7 contained 1 ng DNA from genotype J and 100 ng DNA from each of genotypes F, G, and K. Pool 8 contained 1 ng DNA from genotype K and 100 ng DNA from each of genotypes F, G, and J. Pool 9 contained 1 ng DNA from genotype LGV-I and 100 ng DNA from each of genotypes LGV-II and LGV-III. Pool 10 contained 1 ng DNA from genotype LGVII and 100 ng DNA from each of genotypes LGV-I and LGV-III. Pool 11 contained 1 ng DNA from genotype LGV-III and 100 ng DNA from each of genotypes LGV-I and LGV-II.
TABLE 1.
Sequence of primers and probes
| Primer or probe | Sequence (5′ to 3′) | Locationa | Fluorophore/quencherb |
|---|---|---|---|
| Primers | |||
| P1 | GACTTTGTTTTCGACCGTGTT | 199 | |
| P2 | AGCRTATTGGAAAGAAGCBCCTAA | 657 | |
| P3 | AAACWGATGTGAATAAAGARTT | 224 | |
| P4 | TCCCASARAGCTGCDCGAGC | 617 | |
| P5 | ACARAATACATCAAARCGATCCCA | 417 | |
| P6 | TGGGATCGYTTTGATGTATTYTGT | 394 | |
| Probes | |||
| D | CAACTGATACAGGCAATAGTG | 201 | JOE/BHQ-1 |
| E | CAAGTACTACAGGCAATGCTG | 201 | ROX/BHQ-2 |
| F | CGAAACCTGCTGCAGATAGT | 503 | ROX/BHQ-2 |
| G | CGCAGCCTGCTGCAACAAGT | 503 | Cy5/BHQ-2 |
| H | CCAACGATGCAGCAGACTTA | 204 | Cy5/BHQ-2 |
| I | CTACCAAGGATGTAG-MGB | 201 | FAM/NFQ |
| J | CACAAGCTTCTAGCT-MGB | 494 | FAM/NFQ |
| K | CACAATATTCTAAGTTTA-MGB | 494 | VIC/NFQ |
| LGV-I | GTCAAAAAGGATGC-MGB | 508 | VIC/NFQ |
| LGV-II | TGCTACAGTTTCAG-MGB | 501 | FAM/NFQ |
| LGV-III | GATACAGCGGGCTTATCAAA | 209 | ROX/BHQ-2 |
Nucleotide positions of primers and probes are shown relative to those of GenBank accession no. AF304857.
FAM, 6-carboxyfluorescein.
TABLE 2.
Comparison of NRT-PCR and NNRT-PCR assays for genotyping of C. trachomatis
| Genotype | No. of samples correctly typed by indicated technique
|
||||
|---|---|---|---|---|---|
| Validation panel
|
Rectal swabs
|
||||
| DNA sequencing | NRT-PCR | NNRT-PCR | NRT-PCR | NNRT-PCR | |
| D | 19 | 19 | 15 | 10 | 9 |
| E | 38 | 38 | 26 | 0 | 0 |
| F | 24 | 24 | 24 | 0 | 0 |
| G | 3 | 3 | 1 | 7 | 6 |
| H | 2 | 2 | 0 | 0 | 0 |
| I | 0 | 0 | 0 | 0 | 0 |
| J | 4 | 4 | 4 | 7 | 6 |
| K | 10 | 10 | 10 | 0 | 0 |
| LGV-I | 0 | 0 | 0 | 0 | 0 |
| LGV-II | 0 | 0 | 0 | 24 | 22 |
| LGV-III | 0 | 0 | 0 | 0 | 0 |
| Total | 100 | 100 | 80 | 48 | 43 |
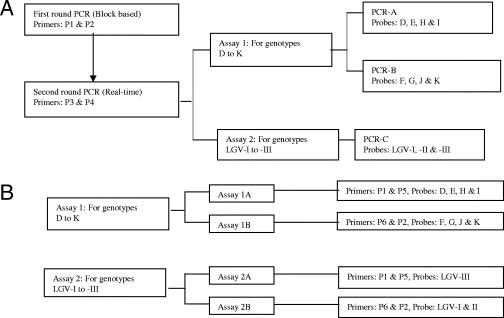
FIG. 1.
Schematic representations of the formats of the NRT-PCR (A) and NNRT-PCR (B) assays. The sequences and nucleotide positions of primers and probes are shown in Table 1.
Validation panel.
The validation panel included DNA from 100 anonymous samples, consisting of urethral swab specimens from 37 men and both urethral and cervical swab specimens in a single tube from 63 women. These samples were collected as described previously (4). DNA was extracted using the MagNA Pure LC total nucleic acid isolation kit and MagNA Pure LC Robot, according to the protocol of the manufacturer, Roche Molecular Systems, Inc., CA (4). C. trachomatis genotyping was performed by DNA sequencing of amplicons generated by nested PCR from the omp1 gene (6). All samples were coded, and genotyping results were masked before validation experiments. The samples were tested by both NRT-PCR and NNRT-PCR.
Rectal swab specimens.
The Sexually Transmitted Bacteria Reference Laboratory (STBRL), London, United Kingdom, provided an aliquot from each of 48 anonymous rectal swab samples from men who have sex with men. These samples were collected by different regional laboratories in the United Kingdom and sent to the STBRL for epidemiological investigations for LGV. The STBRL detected C. trachomatis DNA in rectal swab samples using an in-house real-time PCR assay which targeted the cryptic plasmid. LGV DNA was detected in 24 of 48 samples using a previously described LGV group-specific real-time PCR assay (LGV-PCR) (13). In accordance with their policy, the STBRL performed RFLP analysis only on LGV-PCR-positive samples, as described previously (9). RFLP analysis identified LGV-II DNA in 21 of 24 samples but failed for the remaining 3. The STBRL performed all of the above-described experiments as part of a routine diagnostic service. The aliquots of samples were coded before being sent to our laboratory. DNA from rectal swab samples was extracted using an XTR1-1 kit (Sigma-Aldrich, Steinheim, Germany) and the CAS-1820 X-Tractor gene automated DNA/RNA extraction system (Corbett Robotics, Australia), according to the manufacturer's protocol. All samples were tested with both NRT-PCR and NNRT-PCR.
Nested PCR. (i) First round of block-based PCR.
Block-based PCR was performed with a 50-μl volume containing 5 μl of DNA from a clinical sample, a bacterial isolate, or a pool of DNA from different strains of C. trachomatis; a 200 μM concentration of each deoxynucleoside triphosphate; 25 pmol of each primer (P1 and P2); 4 mM MgCl2; 5 μl of 10× PCR buffer; and 1.5 units of recombinant Taq DNA polymerase (Invitrogen Life Technologies, Paisley, United Kingdom). DNAs from different strains of C. trachomatis were used as positive controls, according to the format of the assay, and nuclease-free water was used as a negative control for amplification. Samples and controls were denatured at 95°C for 3 min, followed by 20 cycles of amplification in a PTC-200 Peltier thermal cycler (Genetic Research Instrumentation Ltd., Essex, United Kingdom). Each cycle consisted of denaturation at 95°C for 10 seconds, annealing of primers at 50°C for 10 seconds, and extension at 72°C for 1 min, followed by a final extension at 72°C for 7 min.
(ii) Second round of real-time PCR.
Real-time PCR was performed using a Rotorgene 3000 apparatus (Corbett Robotics, Australia) essentially as described previously but with a few modifications (5). A pair of primers, P3 and P4, and three to four labeled probes in TaqMan format were used for each component of the assay (Fig. 1A). PCR was performed in a 25-μl volume containing 1 μl of amplicons generated by blocked-base PCR, 12.5 μl of Platinum Quantitative PCR SuperMix-UDG (Invitrogen Life Technologies, Paisley, United Kingdom), 6.25 pmol of each primer, and 2.5 pmol of each probe. The amplification reaction profile included heating at 50°C for 2 min and at 95°C for 2 min followed by 30 cycles of 95°C for 1 second and 55°C for 90 seconds. The signal was acquired at 55°C during each cycle. All primers and probes were synthesized by Metabion International AGi, Germany, except the MGB-labeled probes, which were synthesized by Applied Biosystems, Cheshire, United Kingdom (Table 1). The two-step nested PCR, as used in this study and in previously published studies (1, 7, 8, 9, 10, 15, 20, 21, 22), carries an inherent risk of contaminating equipment and laboratory environment. Stringent physical separation of different steps in the assays, namely, the use of dedicated equipment for each step, a one-way flow of work from extraction to amplification, meticulous techniques to prevent the production of aerosols from tubes containing amplicons, and filtered tips avoided this problem.
NNRT-PCR.
NNRT-PCR was performed as the second round of the NRT-PCR, with two changes. The reaction mixture contained 5 μl of DNA from either a clinical sample or a bacterial isolate instead of 1-μl amplicons, and the number of amplification cycles was increased from 30 to 50. Different combinations of primers and probes were used for NNRT-PCR assays. These combinations are shown in Fig. 1B. The sequences of all primers and probes used in this study are shown in Table 1.
RESULTS
Analytic sensitivity.
The lower limit of detection for NNRT-PCR was determined by testing a dilution series using C. trachomatis genotype E (ATCC code, VR-348B) at the following concentrations: 32, 16, 8, 4, 2, and 1 genome copies per reaction mixture. Each concentration was tested in 10 replicates, and Probit analysis of the result was carried out using StatsDirect version 2.4.1. According to Probit analysis, the assay has a 95% probability of detection with four genome copies (confidence interval, three to six copies) of C. trachomatis per reaction. For DNA from cultures of genovar E, the sensitivity of NNRT-PCR was comparable with that of NRT-PCR. However, the cycle threshold value for each of the 10-fold serial dilutions, ranging from 5,000 to 5 genome copies per reaction mixture, as determined by NNRT-PCR was 17 to 18 cycles higher than that of NRT-PCR.
Analytic specificity.
The primers and probes used in this study did not cross-react with the DNA from any of the pathogenic bacteria or commensals tested from the oropharynx, genital tract, and rectum. The primers amplified DNA from all strains of C. trachomatis, and the probes correctly identified their respective genovars without any cross-reactivity. Examples of the results for NRT-PCR are shown in Fig. 2 and 3. All genotypes were also correctly identified within pools containing various concentrations of DNA from different strains of C. trachomatis.
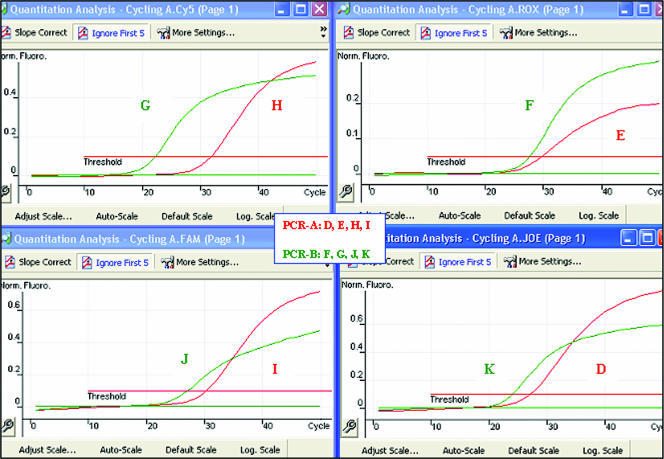
FIG. 2.
Specificities of NRT-PCR assay 1, PCR-A (for genotypes D, E, H, and I), and PCR-B (for genotypes F, G, J, and K). The results for eight amplification reactions are shown. Each reaction mixture contained four probes and DNA from a single reference strain of C. trachomatis. Red and green peaks represent PCR-A and -B, respectively. Norm. Fluoro., normal fluorescence; FAM, 6-carboxyfluorescein.
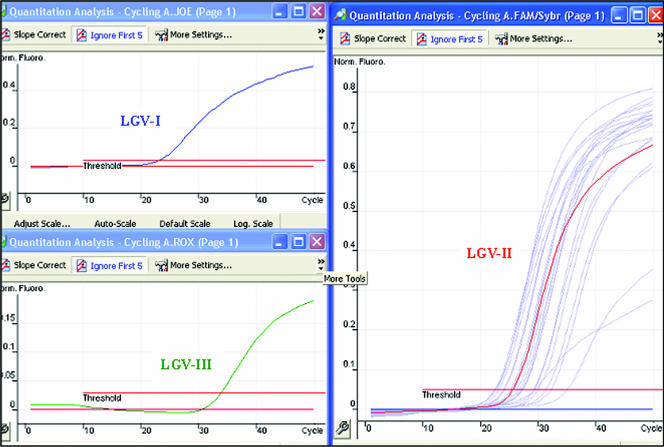
FIG. 3.
Specificities of NRT-PCR assay 2 and PCR-C (for genotypes LGV-I, -II, and -III). Blue, red, and green peaks represent results from three amplification reactions. Each reaction mixture contained three probes and DNA from a single reference strain of LGV. Light-blue peaks in the 6-carboxyfluorescein (FAM) channel represent clinical samples. Norm. Fluoro., normal fluorescence.
Validation panel.
Using DNA sequencing as a gold standard, NRT-PCR typed all of 100 samples in the validation panel correctly. NNRT-PCR was unable to genotype C. trachomatis DNA in 20 of the same 100 samples. However, the typing results generated by this assay from the remaining 80 samples were correct. The results are summarized in Table 2.
Rectal swab samples.
NRT-PCR typed C. trachomatis DNA in all 48 rectal swab samples. NNRT-PCR was able to do it in only 43 of these samples. Of those that contained LGV DNA, 22 were correctly identified by NNRT-PCR, and all 24 were identified by NRT-PCR. The results are summarized in Table 2.
DISCUSSION
The omp1 gene is the most variable genetic marker of C. trachomatis for epidemiological investigations. It has regions of highly conserved and highly variable nucleotide sequences, and this characteristic has been exploited by various molecular techniques for the genotyping of C. trachomatis (1, 2, 16, 22). We designed primers from the conserved regions and probes from the variable regions of the omp1 gene. The primers amplified DNA from all genovars of C. trachomatis, including genovars for trachoma. The probes were designed only for genovars D to K and LGV. Our primers and probes did not cross-react with DNA from the pathogens or commensals of the oropharynx, genital tract, and rectum samples. After extensive trials and errors in combining probes for different types, we describe here a highly sensitive and specific NRT-PCR for the genotyping of C. trachomatis.
In recent publications, 9% (10) and 13% (21) of clinical samples were reported to contain DNA from more than one genovar of C. trachomatis. RFLP analysis and sequencing are unable to identify genotypes in such samples without cloning of amplicons. Such cloning is a laborious, time-consuming, and cumbersome technique unsuitable for epidemiological studies. Reverse line blot hybridization (10, 16, 21) and our NRT-PCR assays, due to the use of a specific probe for each genovar, are ideal techniques for typing C. trachomatis in clinical samples containing DNA from multiple genovars. Unfortunately, such samples were not available for this study. However, experiments performed on artificially mixed DNA from different genotypes demonstrated the ability of NRT-PCR to type multiple genotypes accurately in an amplification reaction.
Regardless of the natures of the techniques, most published methods use two-step nested PCR for the genotyping of C. trachomatis (1, 7, 8, 9, 10, 15, 20, 21, 22). In line with these methods, we have now developed for the same purpose a two-step NRT-PCR. Its sensitivity for chlamydia in culture was the same as that of NNRT-PCR, while its sensitivity with both genital and rectal swabs was superior to that of NNRT-PCR, which typed for 80% of the genital samples and 90% of the rectal samples. NRT-PCR produced peaks which were easy to interpret even from samples with low levels of DNA. The peaks generated by NNRT-PCR from such samples were very close to the threshold of detection and often difficult to interpret.
Real-time PCR, without two steps for nesting, could be fully automated by the use of robotic liquid handling, but two-step PCR makes automation difficult. Further work is needed to convert NRT-PCR to a single-tube nested amplification. In conclusion, the present study has demonstrated the feasibility of using real-time PCR for genotyping the strains of C. trachomatis that cause STDs. Such assays could be adapted to the needs of different epidemiological projects, i.e., the genotyping of C. trachomatis strains that cause LGV or the identification of genotypes D to K in clinical samples. A similar assay could be developed to type the strains of C. trachomatis that cause trachoma.
Acknowledgments
We are grateful to Ian Silver and Maria Erecinska for their suggestions to improve the presentation of this article. We are also grateful to the STBRL for providing rectal swab specimens for this study.
Footnotes
Published ahead of print on 13 June 2007.
REFERENCES
- 1.Bandea, C. I., K. Kubota, T. M. Brown, P. H. Kilmarx, V. Bhullar, S. Yanpaisarn, P. Chaisilwattana, W. Siriwasin, and C. M. Black. 2001. Typing of Chlamydia trachomatis strains from urine samples by amplification and sequencing the major outer membrane protein gene (omp1). Sex. Transm. Infect. 77:419-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frost, E. H., S. Deslandes, S. Veilleux, and D. Bourgaux-Ramoisy. 1991. Typing Chlamydia trachomatis by detection of restriction fragment length polymorphism in the gene encoding the major outer membrane protein. J. Infect. Dis. 163:1103-1107. [DOI] [PubMed] [Google Scholar]
- 3.Halse, T. A., K. A. Musser, and R. J. Limberger. 2006. A multiplexed real-time PCR assay for rapid detection of Chlamydia trachomatis and identification of serovar L-2, the major cause of lymphogranuloma venereum in New York. Mol. Cell. Probes 20:290-297. [DOI] [PubMed] [Google Scholar]
- 4.Jalal, H., H. Stephen, A. Al-Suwaine, C. Sonnex, and C. Carne. 2006. The superiority of polymerase chain reaction over an amplified enzyme immunoassay for the detection of genital chlamydial infections. Sex. Transm. Infect. 82:37-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jalal, H., H. Stephen, M. D. Curran, J. Burton, M. Bradley, and C. Carne. 2006. Development and validation of a Rotor-Gene real-time PCR assay for detection, identification and quantification of Chlamydia trachomatis in a single reaction. J. Clin. Microbiol. 44:206-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jalal, H., H. Stephen, C. Sonnex, and C. Carne. Molecular epidemiology of genital HPV and Chlamydia trachomatis among patients attending the GU medicine clinic in Cambridge—will vaccine protect? Int. J. STD AIDS, in press. [DOI] [PubMed]
- 7.Kaltenboeck, B., K. G. Kousoulas, and J. Storz. 1992. Two-step polymerase chain reactions and restriction endonuclease analyses detect and differentiate ompA DNA of Chlamydia spp. J. Clin. Microbiol. 30:1098-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klint, M., M. Löfdahl, C. Ek, Å. Airell, T. Berglund, and B. Herrmann. 2006. Lymphogranuloma venereum prevalence in Sweden among men who have sex with men and characterization of Chlamydia trachomatis ompA genotypes. J. Clin. Microbiol. 44:4066-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan, J., J. M. M. Walboomers, R. Roosendaal, G. J. J. van Doornum, D. M. MacLaren, C. J. L. M. Meijer, and A. J. C. van den Brule. 1993. Direct detection and genotyping of Chlamydia trachomatis in cervical scrapes by using polymerase chain reaction and restriction fragment length polymorphism analysis. J. Clin. Microbiol. 31:1060-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molano, M., C. J. L. M. Meijer, S. A. Morré, R. Pol, and A. J. C. van den Brule. 2004. Combination of PCR targeting the VD2 of the omp1 and reverse line blot analysis for typing of urogenital Chlamydia trachomatis serovars in cervical scrape specimens. J. Clin. Microbiol. 42:2935-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nieuwenhuis, R. F., J. M. Ossewaarde, H. M. Gotz, J. Dees, H. B. Thio, M. G. Thomeer, J. C. den Hollander, M. H. Neumann, and W. I. van der Meijden. 2004. Resurgence of lymphogranuloma venereum in Western Europe: an outbreak of Chlamydia trachomatis serovar L2 proctitis in The Netherlands among men who have sex with men. Clin. Infect. Dis. 39:996-1003. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez, P., A. Vekris, B. de Barbeyrac, B. Dutilh, J. Bonnet, and C. Bebear. 1991. Typing of Chlamydia trachomatis by restriction endonuclease analysis of the amplified major outer membrane protein gene. J. Clin. Microbiol. 29:1132-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Servaas, A. M., J. Spaargaren, J. S. A. Fennema, H. J. C. de Vries, R. A. Coutinho, and A. S. Pena. 2005. Real-time polymerase chain reaction to diagnose lymphogranuloma venereum. Emerg. Infect. Dis. 11:1311-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stamm, W. E., R. B. Jones, and B. E. Batteiger. 2005. Chlamydia trachomatis (trachoma, perinatal infections, lymphogranuloma venereum, and other genital infections), p. 2239-2255. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, 6th ed. Elsevier Inc., Philadelphia, PA.
- 15.Stothard, D. R., G. Boguslawski, and R. B. Jones. 1998. Phylogenetic analysis of the Chlamydia trachomatis major outer membrane protein and examination of potential pathogenic determinants. Infect. Immun. 68:3618-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stothard, D. R. 2001. Use of reverse blot procedure to identify the presence of multiple serovars in Chlamydia trachomatis urogenital infection. J. Clin. Microbiol. 39:2655-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thylerfors, B., A. D. F. Negrel, R. Pararajasegaram, and K. Y. Dadzie. 1995. Global data on blindness. Bull. W. H. O. 73:115-121. [PMC free article] [PubMed] [Google Scholar]
- 18.Ward, H., I. Martin, N. Macdonald, S. Alexander, I. Simms, K. Fenton, P. French, G. Dean, and C. Ison. 2007. Lymphogranuloma venereum in the United Kingdom. Clin. Infect. Dis. 44:26-32. [DOI] [PubMed] [Google Scholar]
- 19.WHO. 2001. Global prevalence and incidence of selected curable sexually transmitted infections: overview and estimates. World Health Organization, Geneva, Switzerland.
- 20.Xiong, L., F. Kong, H. Zhou, and G. L. Gilbert. 2006. Use of PCR and reverse line blot hybridization assay for rapid simultaneous detection and serovar identification of Chlamydia trachomatis. J. Clin. Microbiol. 44:1413-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang, C. L., I. Maclean, and R. C. Brunham. 1993. DNA sequence polymorphism of the Chlamydia trachomatis omp1 gene. J. Infect. Dis. 168:1225-1230. [DOI] [PubMed] [Google Scholar]
- 22.Zheng, H., L. Jiang, D. Fang, Y. Xue, Y. Wu, J. Huang, and Z. Ou. 2007. Application of an oligonucleotide array assay for rapid detecting and genotyping of Chlamydia trachomatis from urogenital specimens. Diagn. Microbiol. Infect. Dis. 57:1-6. [DOI] [PubMed] [Google Scholar]