Abstract
Streptococcus mutans is the major pathogen of dental caries, a biofilm-dependent infectious disease, and occasionally causes infective endocarditis. S. mutans strains have been classified into four serotypes (c, e, f, and k). However, little is known about the S. mutans population, including the clonal relationships among strains of S. mutans, in relation to the particular clones that cause systemic diseases. To address this issue, we have developed a multilocus sequence typing (MLST) scheme for S. mutans. Eight housekeeping gene fragments were sequenced from each of 102 S. mutans isolates collected from the four serotypes in Japan and Finland. Between 14 and 23 alleles per locus were identified, allowing us theoretically to distinguish more than 1.2 × 1010 sequence types. We identified 92 sequence types in these 102 isolates, indicating that S. mutans contains a diverse population. Whereas serotype c strains were widely distributed in the dendrogram, serotype e, f, and k strains were differentiated into clonal complexes. Therefore, we conclude that the ancestral strain of S. mutans was serotype c. No geographic specificity was identified. However, the distribution of the collagen-binding protein gene (cnm) and direct evidence of mother-to-child transmission were clearly evident. In conclusion, the superior discriminatory capacity of this MLST scheme for S. mutans may have important practical implications.
Streptococcus mutans is the major pathogen of dental caries, a biofilm-dependent infectious disease. These organisms prevail in the complex microcommunity of the oral biofilm in the presence of sucrose, under the extremely low pHs responsible for tooth demineralization. This organism is also a possible causative agent of infective endocarditis (9). S. mutans has been classified into four serotypes (c, e, f, and k) based on the chemical composition of its cell surface rhamnose-glucose polymers. The genes involved in the synthesis of serotype-specific polymers have been cloned and sequenced. Four rml genes (rmlA-rmlD) are related to the synthesis of dTDP-l-rhamnose (37, 38), and gluA is involved in the production of the immediate precursor of the glucose side chain (42). The six-gene operon (rgpA-rgpF) and rgpG, which are required for the synthesis of rhamnose-glucose polymers, have also been cloned and sequenced (41, 43). Serotype-specific genes just downstream from the rgpA-rgpF operon have also been sequenced, and this region is highly diverse among these serotypes (33). Therefore, the strains of each serotype of S. mutans are thought to have evolved and spread independently.
In a previous study, we showed that some blood isolates of S. mutans that cannot be classified into the c, e, or f serotype have negligible amounts of glucose side chains, despite the presence of the rhamnose backbone in serotype-specific polysaccharides (21). We designated the novel serotype as serotype “k” and showed that serotype k strains are less susceptible to phagocytosis by human polymorphonuclear leukocytes. Most oral isolates are of serotype c (approximately 70% to 80%), followed by serotype e (approximately 20%) and serotype f (less than 5%) isolates. Serotype k was defined in 2004, and its distribution frequency in the oral cavity is estimated to be 2% to 5% (21, 22). S. mutans strains of serotype k were isolated from the blood of patients in the early 1990s. However, no serotype k strains were identified from 1,326 stock strains isolated between 1982 and 1990. These results suggest that serotype k strains have acquired or lost some genetic elements in recent years. However, no information is available about the evolution of this unique serotype.
Although serotyping has been widely used to differentiate S. mutans strains, this method has limited power to discriminate the genetic relationships of strains within the same serotype. Several genotypic methodologies have been used to subtype S. mutans, including multilocus enzyme electrophoresis, ribotyping, and random amplification of polymorphic DNA (8, 16, 28). Another discriminatory method for the subtyping of S. mutans is pulsed-field gel electrophoresis (19). However, these methods differ in their discriminatory powers and reproducibilities. In general, DNA-based typing approaches display better discriminatory power than do phenotypic approaches in investigating S. mutans infections. In this study, we designed a multilocus sequence typing (MLST) method, using housekeeping loci, to evaluate the evolution of the three classical serotypes and the newly identified serotype k from epidemiological samples.
MATERIALS AND METHODS
Bacterial isolates.
A total of 102 S. mutans strains were examined in this study (Table 1). Strains MT8148 (serotype c) (26) and UA159 (c) (1) were used as the standard laboratory strains. Eighty-one clinical isolates of S. mutans were obtained from the Pedodontics Clinic of Osaka University Dental Hospital, Suita, Osaka, Japan, with the informed consent of patients obtained according to the protocol approved by the Ethics Committee of Osaka University Dental Hospital. Four isolates from the blood of patients with bacteremia after tooth extraction (TW295 [k]) or with infective endocarditis (TW871 [k], TW964 [f], and TW1378 [e]) were included (6). Blood and oral isolates from a patient with infective endocarditis (V1 [c] and P1 [c], respectively) (24) and another oral isolate collected in 2005 from a patient with an aortic aneurysm (strain OR22P1 [k]) were also included. We also examined seven blood isolates from Finnish patients with bacteremia or infective endocarditis and five oral strains isolated from Finnish subjects. All the strains were confirmed to be S. mutans by conventional physiological tests, including rough colony morphology on mitis-salivarius agar (Difco Laboratories, Detroit, MI), bacitracin resistance, and fermentation of sorbitol, mannitol, raffinose, or melibiose (1% each) in a phenol red broth base (Difco).
TABLE 1.
S. mutans strains used in this study
| Strain | No. of isolates by indicated serotype
|
Total no. of isolates | Yr(s) of isolation | No. from indicated source type
|
Origin | Description (reference or source) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| c | e | f | k | Oral | Blood | |||||
| UA159 | 1 | 0 | 0 | 0 | 1 | 1980s | 1 | 0 | United States | Reference strain (1) |
| MT8148 | 1 | 0 | 0 | 0 | 1 | 1980s | 1 | 0 | Japan | Reference strain (25) |
| MT4000 series | 10 | 10 | 3 | 0 | 23 | 1982-1990 | 23 | 0 | Japan | Clinical isolates from children (this study) |
| NN2000 series | 8 | 8 | 7 | 5 | 28 | 2002 | 28 | 0 | Japan | Clinical isolates from 25 children and 3 adults (20) |
| LJ series | 16 | 10 | 3 | 1 | 30 | 2006 | 30 | 0 | Japan | Clinical isolates from 18 children and 6 mother-child pairs (this study)a |
| TW series | 0 | 1 | 1 | 2 | 4 | 1990-1993 | 0 | 4 | Japan | Clinical isolates from 1 bacteremia patient and 3 IE patientsb (5) |
| V1/P1 | 2 | 0 | 0 | 0 | 2 | 2005 | 1 | 1 | Japan | Clinical isolates from IE patientsb (23) |
| OR22P1 | 0 | 0 | 0 | 1 | 1 | 2005 | 1 | 0 | Japan | Clinical isolate from an aortic aneurysm patient (this study) |
| SA series | 5 | 2 | 2 | 3 | 12 | 1990s | 5 | 7 | Finland | Clinical isolates from 5 children and 7 IE patientsb (this study) |
Clinical isolates LJ3 and LJ4, LJ11 and LJ12, LJ16 and LJ17, LJ24 and LJ25, LJ26 and LJ27, and LJ30 and LJ31 were from mother-child pairs.
IE, infective endocarditis.
Sucrose-dependent adhesion to glass surfaces.
Sucrose-dependent cellular adhesion to glass surfaces was analyzed using a procedure described previously (12). Briefly, the test strains were grown at 37°C for 18 h at an angle of 30° in brain heart infusion broth (Difco) containing 1% sucrose. After incubation, the culture tubes were vigorously vortexed for 3 s, and the nonadhesive cells were transferred to fresh tubes. The cells remaining on the glass surfaces (adhesive cells) were removed using a rubber scraper and suspended in 3 ml of water. Both the adhesive and the nonadhesive cells were dispersed by ultrasonication, and their masses were determined from their optical densities at 550 nm (OD550). “Total cells” was defined as the OD550 of the adhesive cells plus the nonadhesive cells, and “percentage adherence” as 100 × (OD550 of the adhesive cells)/(OD550 of the total cells). All assays were performed three times, and the means and standard deviations were calculated. Statistical analysis was performed using Prism4 software (version 4.0c; GraphPad Software, Inc., San Diego, CA).
Housekeeping loci used for MLST.
Eight housekeeping gene loci were used in this study. The following sequences were obtained from the genome sequence of S. mutans UA159 (NC_004350): tkt, which encodes transketolase (30); glnA, which encodes glutamine synthetase subunit 1a (30); gltA, which encodes glutamate synthase (23); glk, which encodes glucose kinase (23); aroE, which encodes shikimate 5-dehydrogenase (15); gyrA, which encodes DNA gyrase subunit A (7); murI, which encodes glutamate racemase (31); and lepC, which encodes signal peptidase I (39) (Table 1).
DNA isolation and sequencing.
Chromosomal DNA was prepared from all isolates as described previously (5). Internal fragments of the MLST loci were PCR amplified (AmpliTaq Gold, 10 × PCR buffer, and 2 mM GeneAmp deoxynucleotide triphosphate [dNTP] mix; Applied Biosystems) with the following cycling parameters: an initial denaturation at 94°C for 5 min and then 30 cycles consisting of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 7 min. The amplified fragments were visualized after agarose gel electrophoresis on 1.5% agarose in the presence of 1 μg/ml ethidium bromide. They were then extracted from the agarose gel and cloned into the pGEM-T Easy vector (Promega). These plasmids were sequenced on both strands using vector-specific primers and BigDye Terminator version 3.1 (Applied Biosystems) by use of an ABI Prism 3110 genetic analyzer. In-frame internal fragments of the genes were selected for use in the MLST scheme.
Confirmation of serotypes.
Serotyping was performed with the immunodiffusion method using rabbit antisera specific for serotypes c, e, f, and k, and with PCR methods using serotype-specific sets of primers, as described previously (22, 33). To determine serotypes c, e, and f, PCR amplification (Takara ExTaq, 10 × buffer, and 2.5 mM dNTPs; Takara) was performed with the following cycling parameters: initial denaturation at 96°C for 2 min and then 25 PCR cycles of 15 s of denaturation at 96°C, 30 s of annealing at 61°C, and 1 min of extension at 72°C. To identify serotype k, PCR amplification (AmpliTaq Gold, 10 × PCR buffer, and 2 mM GeneAmp dNTP mix; Applied Biosystems) was performed with the following cycling parameters: an initial denaturation at 94°C for 5 min and then 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 7 min. The amplified fragments were separated by agarose gel electrophoresis on 1.5% agarose.
Alleles and ST assignments.
For each locus, distinct allele sequences were assigned arbitrary allele numbers, with no weighting given to the degree of sequence divergence among the alleles. For each isolate, the alleles at each of the eight loci, defining the allelic profile or sequence type (ST), were analyzed with the nonredundant databases (http://linux.mlst.net/nrdb/). The STs were assigned arbitrary numbers in the order of their description. STs were grouped into lineages or clonal complexes with the program eBURST version 3, written and developed by E. Feil (http://eburst.mlst.net/), and the START version 2 package of programs, developed by K. Jolley (http://pubmlst.org/software/analysis/start2/) (11). Two or more independent isolates that shared identical alleles at six or more loci were defined as the members of a lineage. Each lineage was called a “group,” designated according to the ST identified as the putative ancestral type by use of eBURST.
Computational analyses.
The degree of clonality within the data set was estimated by calculating the index of association (IA) (34) between all of the STs in the data set by use of LIAN, version 3.5, written by B. Haubold and R. R. Hudson (http://adenine.biz.fh-weihenstephan.de/cgi-bin/lian/lian.cgi.pl) (10). We determined the number of polymorphic nucleotide sites and calculated the dN/dS ratio (where dN values represent nonsynonymous base substitutions and dS values represent synonymous base substitutions). We also performed phylogenic analyses of both individual genes and concatenated 3351-bp sequences by use of the DNAML, SeqBoot, and drawtree programs in the PHYLIP software package, written by J. Felsenstein (version 3.66; http://evolution.genetics.washington.edu/phylip.html), with the unweighted pair group method using arithmetic means.
Analysis of cnm genes of S. mutans strains.
The prevalence of the cnm gene was evaluated by Southern blot analysis. Total genomic DNA from each strain was digested with HindIII (New England Biolabs, Beverly, MA). The DNA fragments were separated by 0.7% agarose gel electrophoresis and transferred to a nylon membrane (Hybond-N; Amersham Pharmacia Biotech). A 1,617-bp cnm gene fragment containing the whole open reading frame was amplified by PCR from the genomic DNA of S. mutans NN2072 with the primers cnm-F (5′-ATGAAAAGAAAAGGTTTACGAAGAC-3′) and cnm-R (5′-TCAGCTATGATATTTACGGTAAAC-3′), designed based on the sequence of strain Z1 (GenBank accession no. AB102689) (29). The gene fragment was then labeled using a digoxigenin High-Prime DNA labeling and detection starter kit (Roche) according to the manufacturer's instructions. The blotted membrane was prehybridized and then hybridized according to the protocol described by the manufacturer. After hybridization, the membrane was washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% sodium dodecyl sulfate at room temperature and then washed twice in 0.5× SSC containing 0.1% sodium dodecyl sulfate for 15 min at 65°C. The washed membrane was visualized with the immunological method described by the manufacturer (Roche).
RESULTS
Development of an MLST scheme for S. mutans.
Chromosomal DNA was obtained from 102 isolates. The eight housekeeping gene loci were amplified from 102 strains. The sequences of the eight loci were determined and allelic profiles were assigned. The alleles defined for the MLST scheme were based on sequences of between 389 (gltA) and 462 (glnA) nucleotides. Between 14 (tkt) and 23 (gltA and lepC) alleles per locus were identified. The proportion of variable nucleotide sites present in the selected housekeeping genes ranged from 3.24% (glnA) to 5.43% (gltA) (Table 2) . The proportion of nucleotide changes that altered the amino acid sequence (dN) and the proportion of silent changes (dS) were calculated for each gene, and the dN/dS ratios for all eight loci were calculated. All ratios were substantially less than 1 (Table 2). For the 102 S. mutans isolates, the mean number of alleles per locus was about 18, providing the theoretical potential to distinguish more than 1.2 × 1010 different allelic types. We also compared a phylogenetic tree based on individual sequences with a phylogenetic tree based on the concatenated sequences of all eight alleles. However, no phylogenies constructed with these two data sets were incompatible (data not shown). Therefore, our MLST scheme for S. mutans showed a high discriminatory capacity.
TABLE 2.
Primer sequences and characteristics of housekeeping gene loci included in the S. mutans MLST scheme
| Locus | Gene locus tag and putative function of gene products | Primer names | Primer sequences (5′ to 3′) | Size of sequence fragment (bp) | No. of alleles identified | No. (%) of polymorphic nucleotides | dN/dS | GenBank accession no. |
|---|---|---|---|---|---|---|---|---|
| tkt | Smu.291, transketolase | tktF | ACC CGG GTG TTG TCA TGG GCG CTG C | |||||
| tktR | CAT AGG ATG ACG CTT CGC CAG AAA C | 432 | 14 | 16 (3.70) | 0.1032 | AB281702-AB281802 | ||
| glnA | Smu.364, glutamine | glnA-F | CCT TGG GGA GAT GAA AAC GGA GCC G | |||||
| synthetase | glnA-R | TGG CCA TAA AGG TTG CAT ACA AAC C | 462 | 16 | 15 (3.24) | 0.0628 | AB281803-AB281903 | |
| gltA | Smu.365, glutamate | gltA-F | TGC CTT AAC GAT GTT AGA GAG AAT G | |||||
| synthase | gltA-R | AAA GAC TAT CTT CAA AAG CAC ACC C | 387 | 23 | 21 (5.43) | 0.2082 | AB281904-AB282004 | |
| glk | Smu.542, glucose | glk-F | GAC AAG TTC AGG AGA AAT GGG CTA T | |||||
| kinase | glk-R | CAG CAA CTC CAT GAA TAA GAT TGC C | 402 | 19 | 16 (3.98) | 0.6333 | AB282005-AB282105 | |
| aroE | Smu.778, Shikimate | aroE-F | GAT GAA GTA ACG AAA GCA GCA GAT T | |||||
| 5-dehydrogenase | aroE-R | TGC CAT CCA TAC CAA CAT TGG TCG C | 396 | 21 | 19 (4.80) | 0.4467 | AB282207-AB282307 | |
| gyrA | Smu.1114, DNA gyrase | gyrA-F | TAC AGG TGA TGT CAT GGG TAA ATA C | |||||
| subunit A | gyrA-R | CCG GGT AGT ACT TCC ATT AGG TCA C | 432 | 17 | 17 (3.94) | 0.1520 | AB282106-AB282206 | |
| murI | Smu.1718, glutamate | murI-F | TCC GGA GTG GGC GGT TTA ACG GTC G | |||||
| racemase | murI-R | TCA ACA ATA GGA ACA AAT TTG GGG C | 423 | 15 | 17 (4.02) | 0.0544 | AB282308-AB282408 | |
| lepC | Smu.1874, signal | lepC-F | CCG CGT CTC TTT ATC TGG TTT CTT G | |||||
| peptidase I | lepC-R | GAC AAT GCG ATC ATC ACC TAA AAG C | 417 | 23 | 19 (4.56) | 0.1305 | AB282409-AB282509 |
Relatedness of S. mutans isolates.
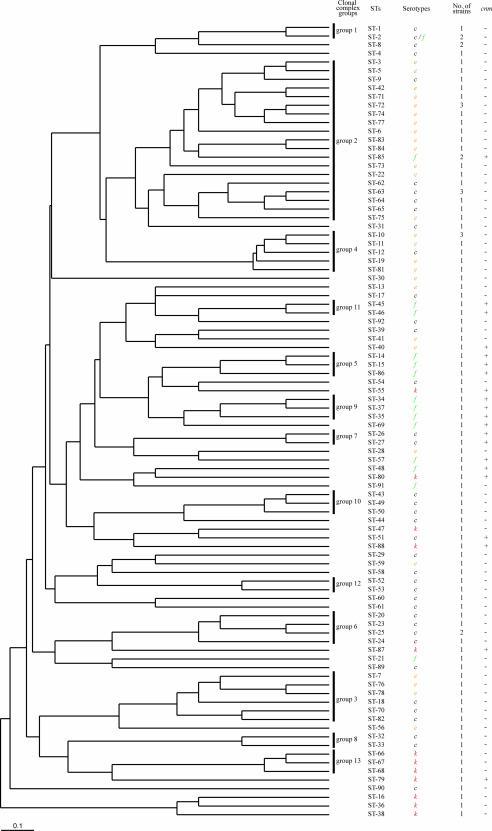
Figure 1 shows a dendrogram constructed from the matrix of pairwise allelic differences between the STs of all 102 isolates. Even though the genetic variation at the MLST loci is relatively low (average, 4.2% nucleotide sites), these isolates were resolved into 92 STs, 85 of which (92.4%) were identified only once, indicating that the S. mutans population displays diversification with little genetic variation. Other STs contained between two and three members. The assignment of STs to lineages with BURST revealed that 36 STs were both unique and unrelated to any other STs, whereas the remaining 65 were assigned to 13 lineages (Fig. 1). Group 2 was the largest lineage and contained 24 isolates representing 19 STs. The remaining 12 groups contained from two to six member STs.
FIG. 1.
Dendrogram of 102 isolates of S. mutans based on an MLST scheme with cluster analysis by the unweighted pair group method using arithmetic means. Clonal complex groups, STs, serotypes, and distribution of the collagen-binding protein gene (cnm) were determined as described in Materials and Methods.
We also tried to determine the chronological relationships of the serotypes. However, no significant differences were observed in the distributions of the serotypes. Moreover, no geographical relationships were identified among the isolates.
Evidence of recombination.
The extent of recombination within the S. mutans population was assessed by determining the standardized IA (34) with LIAN. The IA for the complete data set was 0.0931 when randomized data sets (1,000 trials) were used. This value is significantly greater than zero, which is the value expected for a population in linkage equilibrium. However, in populations in which recombination is sufficient to randomize the alleles at different loci over long time periods, the appearance of multiple isolates with similar genotypes can result from the recent expansion of clones. Therefore, the SplitsTree program was used to detect recombination among the various STs. Allelic profile data were converted into distance matrix values by use of START, and the resulting nexus file was analyzed with the split decomposition method. SplitsTree analysis of the 102 S. mutans strains yielded a very low fit value (fit = 22.2), which may have resulted from the program's inability to analyze this large amount of information (14). We also analyzed several small subsets of randomly selected strains, which considerably improved the fit values. One example of a SplitsTree analysis displaying 10 parallelograms and a high fit value of ca. 55 was based on 18 randomly selected S. mutans isolates (data not shown).
Relationships between STs and serotypes.
We then determined the serotypes of all the isolates by immunodiffusion and PCR-based typing (Table 3) . Serotype c was dominant (43 isolates), followed by e (31 isolates), f (16 isolates), and k (12 isolates). In general, the serotypes appeared to be associated with the overall genotype defined by the STs, and only ST2 contained two serotypes (c and f). As stated above, closely related STs constituted clonal complexes. However, groups 1, 2, 3, and 4 contained isolates of more than one serotype, whereas the other groups contained only one serotype. Serotype c, the major serotype of S. mutans clinical isolates, was widely distributed among the clonal complex groups, such as in groups 1, 2, 3, 4, 6, 7, 8, 10, and 12, and in many singletons, indicating that serotype c is the ancestral serotype of S. mutans. Group 2 contained 15 of the 31 serotype e isolates (ST3, 5, 6, 22, 42, 71, 72, 73, 74, 75, 77, 83, and 84), and groups 3 and 4 contained three and six serotype e isolates, respectively. Serotype f isolates were found mainly in three groups (groups 5, 9, and 11), and approximately half the serotype f isolates occurred in these groups. Another three STs (ST48, 57, and 91) were not categorized within the same clonal complex but had closely related sequence homologies (Fig. 1). Only three STs (ST2, 21, and 85) that were serotype f were located separately from the other serotype f groups. Serotype k, newly identified in the last decade, was distributed among closely related STs (ST16, 36, 38, 66, 67, 68, and 79). These results indicate that the ancestral strain of S. mutans was serotype c and that the other serotypes have branched from the serotype c groups continuously during evolution up until the present.
TABLE 3.
Allelic profiles of isolates belonging to the 92 STs identified in this study
| Strain | Serotype | ST | Allelic assignment (arbitrary allele no.)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| tkt | glnA | gltA | glk | aroE | murI | lepC | gyrA | |||
| UA159 | c | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| MT8148 | c | 63 | 4 | 1 | 1 | 3 | 2 | 3 | 1 | 1 |
| MT4065 | c | 64 | 4 | 1 | 1 | 3 | 2 | 3 | 1 | 14 |
| MT4071 | c | 92 | 14 | 2 | 4 | 1 | 2 | 3 | 1 | 1 |
| MT4076 | c | 2 | 1 | 1 | 1 | 13 | 1 | 1 | 1 | 1 |
| MT4078 | c | 20 | 1 | 5 | 9 | 1 | 4 | 3 | 11 | 9 |
| MT4083 | c | 26 | 1 | 9 | 1 | 1 | 2 | 5 | 3 | 10 |
| MT4087 | c | 29 | 1 | 15 | 15 | 3 | 4 | 2 | 19 | 1 |
| MT4093 | c | 24 | 1 | 8 | 9 | 1 | 4 | 2 | 20 | 1 |
| MT4112 | c | 61 | 3 | 14 | 1 | 17 | 20 | 15 | 21 | 1 |
| MT4118 | c | 65 | 4 | 1 | 1 | 18 | 2 | 3 | 1 | 1 |
| MT4164 | c | 23 | 1 | 8 | 9 | 1 | 4 | 2 | 11 | 9 |
| MT4117 | e | 81 | 6 | 2 | 7 | 3 | 2 | 7 | 1 | 1 |
| MT4119 | e | 59 | 3 | 10 | 15 | 3 | 21 | 2 | 1 | 1 |
| MT4217 | e | 6 | 1 | 1 | 1 | 19 | 2 | 4 | 7 | 3 |
| MT4245 | e | 72 | 6 | 1 | 1 | 3 | 2 | 4 | 1 | 3 |
| MT4274 | e | 30 | 1 | 16 | 23 | 6 | 2 | 4 | 22 | 15 |
| MT4278 | e | 84 | 6 | 5 | 15 | 3 | 2 | 4 | 1 | 3 |
| MT4279 | e | 83 | 6 | 5 | 9 | 3 | 2 | 4 | 1 | 3 |
| MT4293 | e | 73 | 6 | 1 | 1 | 3 | 14 | 4 | 1 | 16 |
| MT4368 | e | 10 | 1 | 2 | 7 | 3 | 2 | 7 | 1 | 1 |
| MT4369 | e | 11 | 1 | 2 | 7 | 3 | 2 | 7 | 1 | 17 |
| MT4251 | f | 85 | 7 | 2 | 11 | 3 | 2 | 4 | 1 | 3 |
| MT4333 | f | 46 | 3 | 2 | 5 | 1 | 2 | 3 | 23 | 1 |
| MT4348 | f | 15 | 1 | 3 | 8 | 1 | 6 | 10 | 3 | 1 |
| NN2098 | c | 31 | 2 | 1 | 1 | 2 | 2 | 2 | 1 | 2 |
| NN2099 | c | 44 | 3 | 2 | 2 | 3 | 3 | 3 | 2 | 1 |
| NN2092 | c | 9 | 1 | 2 | 1 | 3 | 2 | 4 | 1 | 3 |
| NN2093 | c | 54 | 3 | 3 | 3 | 1 | 4 | 3 | 3 | 1 |
| NN2025 | c | 70 | 5 | 4 | 1 | 4 | 5 | 5 | 4 | 4 |
| NN2004 | c | 33 | 2 | 2 | 4 | 5 | 4 | 6 | 1 | 1 |
| NN2085 | c | 39 | 2 | 5 | 5 | 6 | 2 | 3 | 3 | 1 |
| NN2087 | e | 56 | 3 | 6 | 6 | 4 | 2 | 5 | 5 | 4 |
| NN2089 | e | 42 | 3 | 1 | 1 | 1 | 2 | 4 | 1 | 3 |
| NN2037 | e | 10 | 1 | 2 | 7 | 3 | 2 | 7 | 1 | 1 |
| NN2044 | e | 22 | 1 | 7 | 1 | 3 | 2 | 3 | 1 | 4 |
| NN2054 | e | 19 | 1 | 5 | 7 | 3 | 2 | 7 | 1 | 1 |
| NN2076 | e | 75 | 6 | 1 | 1 | 3 | 2 | 8 | 1 | 1 |
| NN2042 | e | 10 | 1 | 2 | 7 | 3 | 2 | 7 | 1 | 1 |
| NN2053 | e | 76 | 6 | 1 | 6 | 4 | 5 | 5 | 1 | 4 |
| NN2072 | f | 69 | 4 | 3 | 8 | 3 | 2 | 5 | 1 | 1 |
| NN2165 | f | 57 | 3 | 8 | 1 | 1 | 6 | 3 | 3 | 1 |
| NN2007 | f | 35 | 2 | 3 | 8 | 1 | 2 | 9 | 1 | 1 |
| NN2117 | f | 48 | 3 | 2 | 9 | 1 | 7 | 4 | 6 | 1 |
| NN2138-2 | f | 45 | 3 | 2 | 5 | 1 | 2 | 3 | 7 | 1 |
| NN2168M-5 | c | 49 | 3 | 2 | 10 | 3 | 8 | 3 | 5 | 1 |
| NN2121 | f | 85 | 7 | 2 | 11 | 3 | 2 | 4 | 1 | 3 |
| NN2431M-2 | f | 86 | 8 | 3 | 8 | 1 | 6 | 10 | 1 | 1 |
| NN2011 | k | 47 | 3 | 2 | 8 | 7 | 10 | 5 | 3 | 1 |
| NN2111 | k | 67 | 4 | 2 | 12 | 1 | 7 | 5 | 8 | 6 |
| NN2323M-1 | k | 66 | 4 | 2 | 12 | 1 | 4 | 5 | 8 | 6 |
| NN2193-1 | k | 16 | 1 | 3 | 13 | 8 | 11 | 2 | 9 | 7 |
| NN2105 | k | 68 | 4 | 2 | 12 | 1 | 4 | 5 | 10 | 6 |
| LJ1 | c | 17 | 1 | 3 | 15 | 1 | 2 | 3 | 12 | 1 |
| LJ2 | e | 40 | 2 | 9 | 5 | 1 | 4 | 3 | 7 | 1 |
| LJ3 | e | 5 | 1 | 1 | 1 | 3 | 2 | 4 | 1 | 3 |
| LJ4 | e | 7 | 1 | 1 | 6 | 4 | 5 | 5 | 1 | 8 |
| LJ7 | f | 14 | 1 | 3 | 8 | 1 | 4 | 10 | 3 | 1 |
| LJ11 | c | 25 | 1 | 8 | 14 | 1 | 4 | 2 | 11 | 9 |
| LJ12 | c | 25 | 1 | 8 | 14 | 1 | 4 | 2 | 11 | 9 |
| LJ13 | c | 58 | 3 | 10 | 15 | 4 | 13 | 11 | 13 | 1 |
| LJ14 | c | 52 | 3 | 2 | 15 | 3 | 1 | 11 | 1 | 3 |
| LJ16 | c | 43 | 3 | 2 | 1 | 3 | 8 | 3 | 5 | 1 |
| LJ17 | c | 63 | 4 | 1 | 1 | 3 | 2 | 3 | 1 | 1 |
| LJ18 | e | 77 | 6 | 1 | 16 | 3 | 2 | 4 | 1 | 3 |
| LJ19 | e | 71 | 6 | 1 | 1 | 1 | 2 | 4 | 1 | 3 |
| LJ20 | c | 27 | 1 | 9 | 1 | 1 | 2 | 12 | 3 | 10 |
| LJ22 | c | 82 | 6 | 4 | 1 | 4 | 5 | 5 | 1 | 4 |
| LJ23 | k | 88 | 10 | 2 | 5 | 7 | 6 | 5 | 11 | 1 |
| LJ24 | f | 34 | 2 | 3 | 8 | 1 | 2 | 5 | 3 | 1 |
| LJ25 | c | 89 | 11 | 11 | 9 | 9 | 3 | 3 | 14 | 1 |
| LJ26 | c | 51 | 3 | 2 | 10 | 10 | 2 | 5 | 3 | 1 |
| LJ27 | c | 90 | 12 | 2 | 1 | 11 | 14 | 13 | 15 | 11 |
| LJ30 | c | 8 | 1 | 1 | 8 | 3 | 1 | 14 | 1 | 1 |
| LJ31 | c | 8 | 1 | 1 | 8 | 3 | 1 | 14 | 1 | 1 |
| LJ32 | f | 37 | 2 | 3 | 17 | 1 | 2 | 5 | 3 | 1 |
| LJ15 | c | 62 | 4 | 1 | 1 | 2 | 15 | 3 | 1 | 1 |
| LJ29 | c | 12 | 1 | 2 | 7 | 3 | 16 | 7 | 1 | 1 |
| LJ34 | e | 3 | 1 | 1 | 1 | 3 | 2 | 3 | 1 | 3 |
| LJ36 | e | 72 | 6 | 1 | 1 | 3 | 2 | 4 | 1 | 3 |
| LJ50 | e | 78 | 6 | 1 | 19 | 4 | 5 | 5 | 1 | 4 |
| LJ59 | e | 72 | 6 | 1 | 1 | 3 | 2 | 4 | 1 | 3 |
| LJ64 | e | 74 | 6 | 1 | 1 | 3 | 2 | 4 | 4 | 3 |
| SA22 | f | 21 | 1 | 5 | 9 | 12 | 17 | 3 | 16 | 1 |
| SA31 | k | 36 | 2 | 3 | 14 | 8 | 11 | 2 | 9 | 12 |
| SA51 | f | 2 | 1 | 1 | 1 | 13 | 1 | 1 | 1 | 1 |
| SA53 | k | 55 | 3 | 3 | 8 | 1 | 1 | 3 | 1 | 1 |
| SA72 | k | 38 | 2 | 3 | 18 | 8 | 11 | 2 | 17 | 3 |
| SA12 | c | 50 | 3 | 2 | 10 | 3 | 8 | 3 | 5 | 13 |
| SA13 | c | 18 | 1 | 3 | 20 | 4 | 5 | 5 | 1 | 4 |
| SA14 | c | 53 | 3 | 2 | 15 | 14 | 1 | 11 | 1 | 12 |
| SA15 | e | 41 | 2 | 12 | 5 | 15 | 2 | 3 | 18 | 1 |
| SA16 | e | 28 | 1 | 13 | 1 | 1 | 18 | 3 | 3 | 1 |
| SA17 | c | 60 | 3 | 14 | 1 | 16 | 2 | 2 | 19 | 1 |
| SA18 | c | 32 | 2 | 2 | 4 | 5 | 4 | 5 | 1 | 4 |
| TW295 | k | 80 | 6 | 2 | 5 | 1 | 9 | 4 | 1 | 1 |
| TW871 | k | 79 | 6 | 2 | 1 | 1 | 9 | 5 | 7 | 5 |
| TW964 | f | 91 | 13 | 5 | 21 | 1 | 6 | 4 | 1 | 1 |
| TW1378 | e | 13 | 1 | 2 | 22 | 1 | 19 | 3 | 13 | 1 |
| V1 | c | 63 | 4 | 1 | 1 | 3 | 2 | 3 | 1 | 1 |
| P1 | c | 4 | 1 | 1 | 1 | 13 | 7 | 3 | 11 | 1 |
| OR22P1 | k | 87 | 9 | 8 | 14 | 1 | 12 | 4 | 11 | 1 |
Association between lineages and diseases.
Sucrose-dependent adhesion to glass surfaces is an important feature in the cariogenicity of S. mutans. However, the serotype-specific adhesion mechanism has still not been investigated. Therefore, we determined the in vitro adhesion capacities of S. mutans isolates. Serotype c isolates showed a slightly increased rate (84.01% ± 11.81%), and serotype k isolates showed a low rate (78.01% ± 6.94%) compared with those of the other serotype strains. Serotypes e and f showed moderate rates of 81.49% ± 14.75% and 82.34% ± 6.39%, respectively. However, there was no statistically significant difference among the isolates. This observation indicates that the phylogenetic differences determined with the eight housekeeping gene loci do not reflect the adhesion capacities of the S. mutans isolates. Three clinical isolates (MT4078, serotype c; MT4293, serotype e; and V1, serotype c) showed significantly lower adhesion than did the other isolates. However, we identified no relationship between serotype, phylogenetic position, regional specificity, and years of isolation.
We next determined the relationship between serotype and systemic infection, because S. mutans frequently causes bacteremia and infective endocarditis. However, the blood-derived isolates were widely distributed between the various STs and clonal complex groups (Fig. 1), and no significant relationship between STs and the blood-derived isolates was observed. Therefore, we conclude that the S. mutans blood isolates are not a specific strain and that each isolate has the potential to induce systemic infections such as bacteremia or infective endocarditis.
Strain-specific collagen-binding adhesin (encoded by cnm) is a recently identified wall-anchored protein. However, its prevalence among S. mutans isolates has not been determined. To clarify the prevalence of the cnm gene, we analyzed the distribution of cnm-positive isolates by PCR and Southern blot analysis. Of the 102 strains examined, 22 were cnm positive (21.6%; see also data in Fig. 1). The cnm-positive isolates were distributed in ST85; in ST45 and ST46 (group 11); in ST40; in ST14, 15, and 86 (group 5); in ST55; in ST34, 37, and 35 (group 9); in ST69; in ST26 and 27 (group 7); in ST57; in ST48; in ST80; in ST51; in ST88; in ST87; and in ST79 (Fig. 1). The predominant cnm-positive isolates were of serotype f (13 cnm positive of 18 total [81.3%]), followed by serotype k (5 cnm-positive of 12 total [41.7%]). Three isolates of serotype c (3 [ST26, 27, and 51] of 43 total [7.0%]) and 1 isolate of serotype e (1 [ST40] of 31 total [3.2%]) had the cnm gene. The prevalence of cnm gene in each serotype is statistically significant (between serotype c and serotype f, a P value of <0.0001; between serotypes c and k, a P value of 0.087; between serotypes e and f, a P value of <0.0001; between serotypes e and k, a P value of 0.0087; and between serotypes e and k, a P value of 0.0042 [all by Fisher's exact probability test]). These isolates were closely related to the cnm-positive serotype f and k strains on phylogenetic analysis. Interestingly, the distribution of cnm-positive strains was limited to closely related groups. These observations indicate that some clinical isolates have acquired the cnm gene, probably by horizontal gene transfer, during evolution.
Streptococcus mutans transmission from mother to child.
In the six mother-child pairs of S. mutans isolates (Table 4), the STs of two pairs (LJ11 and 12 and LJ30 and 31) were completely identical, indicating that these S. mutans isolates were vertically transmitted from mother to child. Of the other four pairs, the LJ3-LJ4 and LJ16-LJ17 pairs had three and four matches, respectively, in their ST profiles. However, on the MLST-based dendrogram and sequence-based phylogenetic tree, these two pairs were grouped onto different branches. The LJ24-LJ25 and LJ26-LJ27 pairs had only one allele match, and the LJ24-LJ25 members showed different serotypes. Although only a limited number of mother-child pairs were analyzed in this study, the MLST method allowed the strict discrimination of the strains from both individuals in each mother-child pair.
TABLE 4.
Mother-child transmission of S. mutans by MLST analysisa
| Strain | Serotype | ST | Allelic assignment
|
Patient age | Concatenated sequence homology (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| tkt | glnA | gltA | glk | aroE | murI | lepC | gyrA | |||||
| LJ3 | e | 5 | 1 | 1 | 1 | 3 | 2 | 4 | 1 | 3 | 37 yr | 99.4 |
| LJ4 | e | 7 | 1 | 1 | 6 | 4 | 5 | 5 | 1 | 8 | 4 yr 9 mo | |
| LJ11 | c | 25 | 1 | 8 | 14 | 1 | 4 | 2 | 11 | 9 | 38 yr | 100 |
| LJ12 | c | 25 | 1 | 8 | 14 | 1 | 4 | 2 | 11 | 9 | 7 yr 3 mo | |
| LJ16 | c | 43 | 3 | 2 | 1 | 3 | 8 | 3 | 5 | 1 | 33 yr | 99.6 |
| LJ17 | c | 63 | 4 | 1 | 1 | 3 | 2 | 3 | 1 | 1 | 3 yr 7 mo | |
| LJ24 | f | 34 | 2 | 3 | 8 | 1 | 2 | 5 | 3 | 1 | 31 yr | 99.1 |
| LJ25 | c | 89 | 11 | 11 | 9 | 9 | 3 | 3 | 14 | 1 | 5 yr 4 mo | |
| LJ26 | c | 51 | 3 | 2 | 10 | 10 | 2 | 5 | 3 | 1 | 37 yr | 99.2 |
| LJ27 | c | 90 | 12 | 2 | 1 | 11 | 14 | 13 | 15 | 11 | 9 yr 11 mo | |
| LJ30 | c | 8 | 1 | 1 | 8 | 3 | 1 | 14 | 1 | 1 | 35 yr | 100 |
| LJ31 | c | 8 | 1 | 1 | 8 | 3 | 1 | 14 | 1 | 1 | 2 yr 2 mo | |
For each pair of rows, the top row gives mother data and the bottom row gives child data.
DISCUSSION
In developing the MLST in this study, target genes were selected based on the complete genome sequence of S. mutans UA159 (1) and the ongoing complete genome sequencing of S. mutans NN2025 in our laboratories, on the genomic information from other bacteria, and on the published MLST schemes for other streptococci, such as S. pyogenes (4), S. uberis (44), S. suis (13), and S. pneumoniae (3), together with their locations in the genome. Finally, eight housekeeping genes were selected for this study. The eight loci were not subject to positive selection, as demonstrated by the dN/dS ratio for each locus, which was substantially less than 1 (Table 2). The mean number of alleles identified per locus was 18.5, suggesting that S. mutans represents a genetically diverse species. The housekeeping genes tkt (transketolase), glnA (glutamine synthetase 1a subunit), gyrA (DNA gyrase subunit A), murI (glutamate racemase), and lepC (signal peptidase I) were chosen because these genes are conserved across the genus Streptococcus and other genera. We also included three other genes, aroE (shikimate 5-dehydrogenase), glk (glucose kinase), and gltA (glutamate synthase), whose products are housekeeping proteins with sequence homologies that are specific to S. mutans strains.
Seven genes have usually been used in recent MLST schemes, because it is important that the selection of the genes to be sequenced in an MLST scheme developed for a particular bacterial species should be optimized to save time and cost (40). However, the discrimination of each serotype, especially serotypes f and k, was not definitive in our MLST when seven genes were used in the analysis of S. mutans. In addition, the combined analysis of housekeeping genes and some virulence genes has been also used to identify virulent or pandemic clones (35, 40). In this study, we used eight housekeeping loci because the virulence genes of S. mutans are not as genetically diverse as those of other organisms. For example, glucosyltransferases synthesize water-soluble and -insoluble glucans from a sucrose substrate, and these activities are important in the colonization and biofilm formation of S. mutans (27). Chia et al. (2) reported the existence of DNA polymorphisms in the 5′ regions of the gtfB and gtfC genes. However, only four or five variants have been observed in this region (2). This observation has been confirmed by use of restriction fragment length polymorphisms of the gtfB gene (36). Therefore, these putative virulence genes of S. mutans are not suitable for the analysis of the evolution or population biology of the species because their heterogeneity is low. The mean number of alleles identified per locus was 18.5, providing the theoretical potential to distinguish more than 1.2 × 1010 different genotypes. In fact, as shown in Fig. 1 and Table 3, our MLST method divided the 102 strains into 92 STs, indicating that this MLST scheme has high discriminatory power.
The cell wall antigens of S. mutans are rhamnose-glucose polysaccharides, which are composed of α1,2- and α1,3-linked rhamnan backbones and glucose side chains. Bacterial cell wall polysaccharides decorate the cell surface and often play an important role in the colonization of the bacterial ecological niche. Each serotype-specific polysaccharide has a unique linkage to its glucose side chains. Serotype c has an α1,2 linkage, serotype e has a β1,2 linkage, and serotype f has an α1,3 linkage (17). Serotype k strains have a unique “untypeable” phenotype in terms of known serotype antibodies because they lack glucose side chains linked to their rhamnose backbones (21). The genes related to serotype specificity have been determined, and each serotype has a specific and heterogenic region downstream from the conserved rgpA-rgpF operon (33). On the basis of these specific regions, Shibata et al. suggested that none of the three serotypes could be defined as the ancestral strain (33). However, in our MLST analysis, most of the serotype e and serotype f strains occur on their own branches on the dendrogram. Most serotype e strains are included in clonal complex group 2, and the root of this branch derives from serotype c (Fig. 1). The ST85 isolate in this group is serotype f, but the restriction fragment length pattern of the rgpA-rgpF region in the ST85 isolate seems to be that of serotype c (data not shown). Why these strains appear phenotypically serotype f rather than serotype c is now under investigation by complete nucleotide sequence analysis. The distribution of serotype f isolates is also restricted to limited branches derived from serotype c. These observations suggest that a serotype c strain, the dominant serotype among S. mutans isolates, is the ancestral phenotype of this organism and that serotype e and f strains acquired their strain-specific genes during evolution. It is still unclear whether these phenotypic changes are due to genetic transfer or genetic exchange. Therefore, these issues require further analysis.
Serotype k, recently identified as a new serotype of S. mutans, also has an interesting distribution in our MLST analysis. All genes for the biosynthesis of serotype-specific polysaccharides are highly conserved relative to those of serotype c, and only the mRNA expression of rgpE is diminished in serotype k strains (25). Therefore, we hypothesized that serotype k might be derived from serotype c. As we expected, many serotype k isolates were derived from the serotype c branch (Fig. 1). However, some serotype k strains seem to have been derived from serotype f (ST55 and ST80). Polysaccharide synthesis is a sequential reaction involving several gene products. Therefore, a defect in a gene or a loss of function of a gene product related to the biosynthesis of polysaccharides results in the formation of a serotype k strain. In fact, a gluA-destructive mutant of a serotype c strain has the “serotype k” phenotype (21). Therefore, it is reasonable to infer that S. mutans strains that are phenotypically “serotype k” have arisen from the genetic dysfunction of serotype c or f strains in this era. It is possible that all S. mutans strains may have the potential to become serotype k in the future.
In this study, we compared the distribution and lineage differences of Japanese isolates with those of Finnish isolates because Japan is geographically and ethnically distinct from Finland. However, we found no regional bias in the distribution of the S. mutans isolates (Fig. 1). Several lines of epidemiological evidence indicate that serotype c is predominant worldwide (70% to 80%), with serotype e the next commonest (10% to 15%) and serotype f occurring rarely (1% to 5%) (9). On the basis of these findings, we speculate that the serological differentiation of S. mutans occurred earlier than we had previously inferred. To address this question further, we are continuing to analyze worldwide serotype distributions using our MLST scheme.
Oral bacteria are considered to cause transient bacteremia, countered by professional dental treatment and daily oral care practices such as tooth brushing and flossing (32). However, the incidence of infective endocarditis estimated by a review of reports published between 1993 and 2003 was only 3.6 per 100,000 head of population per year (20). These observations suggest that some infective-endocarditis-specific or virulent strains exist and that these strains necessarily induce systemic infections. However, the specific distribution of S. mutans strains related to systemic infections was not determined in our MLST analysis.
In contrast, the distribution of the collagen-binding protein gene (cnm) was clearly predominant in the clonal complex groups of serotype f strains (Fig. 1). The gene product of cnm was first identified in the cold-agglutination phenotype of S. mutans, and thereafter the binding of collagen-binding adhesin to collagen and laminin was reported (29). The binding of collagen-binding adhesin to specific extracellular matrix molecules may be important for the initial bacterial attachment to blood vessels. However, the distribution of cnm-positive isolates does not exactly match that of isolates derived from bacteremia or infective endocarditis patients, so the contribution of collagen-binding adhesin to the progression of systemic infections is not clearly explained.
The vertical transmission of S. mutans from mother to child is thought to be the major route for early acquisition (16). In contrast, the detection of genotypes in children that are not found in their mothers or other family members indicates that S. mutans may also be acquired from other sources. In our MLST analysis, two of six mother-child pairs showed complete identity of ST profiles for the mother and the child, whereas the other pairs showed differing ST profiles (Table 4). This result indicates that our MLST scheme is useful for epidemiological studies and is suitable for the long-term monitoring of S. mutans transmission. Mattos-Graner et al. reported at least two to five different genotypes in 30% of the children tested (18). In this study, we used only one isolate each from the mother and the child. Further prospective studies involving greater numbers of S. mutans isolates are required to explore the frequencies of vertical and horizontal transmission.
In conclusion, we have devised the first clear typing system for S. mutans. In this study, we have provided new insight into the lineages of the four serotypes, their regional specificities, and the distribution of a newly identified virulence gene and have demonstrated the applicability of the scheme to epidemiological studies. The superior discriminatory capacity of MLST, compared with that of classical serotyping or DNA-based genotyping methods, may have important practical implications. We hope that this MLST scheme will now be expanded to include isolates from other countries.
Acknowledgments
This study was supported by the 21st Century COE program “Origination of Frontier BioDentistry” at Osaka University Graduate School of Dentistry, supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan; Grant-in-Aid for Scientific Research (B) 16390605 from the Japan Society for the Promotion of Science; Grant-in-Aid for Exploratory Research 17659647; Grant-in-Aid for Young Scientists (A) 18689050 Grant-in-Aid for Scientific Research on Priority Areas “Applied Genomics” from the Ministry of Education, Culture, Sports, Science and Technology; and PRESTO, the Japan Science and Technologies Agency.
Footnotes
Published ahead of print on 13 June 2007.
REFERENCES
- 1.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chia, J. S., T. Y. Hsu, L. J. Teng, J. Y. Chen, L. J. Hahn, and C. S. Yang. 1991. Glucosyltransferase gene polymorphism among Streptococcus mutans strains. Infect. Immun. 59:1656-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 4.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69:2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujiwara, T., T. Hoshino, T. Ooshima, S. Sobue, and S. Hamada. 2000. Purification, characterization, and molecular analysis of the gene encoding glucosyltransferase from Streptococcus oralis. Infect. Immun. 68:2475-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujiwara, T., K. Nakano, M. Kawaguchi, T. Ooshima, S. Sobue, S. Kawabata, I. Nakagawa, and S. Hamada. 2001. Biochemical and genetic characterization of serologically untypable Streptococcus mutans strains isolated from bacteremia. Eur. J. Oral Sci. 109:330-334. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez, I., M. Georgiou, F. Alcaide, D. Balas, J. Linares, and A. G. de la Campa. 1998. Fluoroquinolone resistance mutations in the parC, parE, and gyrA genes of clinical isolates of viridans group streptococci. Antimicrob. Agents Chemother. 42:2792-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grönroos, L., M. Saarela, J. Matto, U. Tanner-Salo, A. Vuorela, and S. Alaluusua. 1998. Mutacin production by Streptococcus mutans may promote transmission of bacteria from mother to child. Infect. Immun. 66:2595-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamada, S., and H. D. Slade. 1980. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol. Rev. 44:331-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haubold, B., and R. R. Hudson. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Linkage analysis. Bioinformatics 16:847-848. [DOI] [PubMed] [Google Scholar]
- 11.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 12.Kawabata, S., and S. Hamada. 1999. Studying biofilm formation of mutans streptococci. Methods Enzymol. 310:513-523. [DOI] [PubMed] [Google Scholar]
- 13.King, S. J., J. A. Leigh, P. J. Heath, I. Luque, C. Tarradas, C. G. Dowson, and A. M. Whatmore. 2002. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J. Clin. Microbiol. 40:3671-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotetishvili, M., O. C. Stine, A. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2002. Multilocus sequence typing for characterization of clinical and environmental Salmonella strains. J. Clin. Microbiol. 40:1626-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, H. Y., and J. C. Cote. 2006. Phylogenetic analysis of γ-proteobacteria inferred from nucleotide sequence comparisons of the house-keeping genes adk, aroE and gdh: comparisons with phylogeny inferred from 16S rRNA gene sequences. J. Gen. Appl. Microbiol. 52:147-158. [DOI] [PubMed] [Google Scholar]
- 16.Li, Y., and P. W. Caufield. 1998. Arbitrarily primed polymerase chain reaction fingerprinting for the genotypic identification of mutans streptococci from humans. Oral Microbiol. Immunol. 13:17-22. [DOI] [PubMed] [Google Scholar]
- 17.Linzer, R., M. S. Reddy, and M. J. Levine. 1986. Immunochemical aspects of serotype carbohydrate antigens of Streptococcus mutans, p. 29-38. In S. Hamada, S. M. Michalek, H. Kiyono, L. Manaker, and J. R. McGhee (ed.), Molecular microbiology and immunology of Streptococcus mutans. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 18.Mattos-Graner, R. O., Y. Li, P. W. Caufield, M. Duncan, and D. J. Smith. 2001. Genotypic diversity of mutans streptococci in Brazilian nursery children suggests horizontal transmission. J. Clin. Microbiol. 39:2313-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mineyama, R., S. Yoshino, and N. Maeda. 24 July 2006, posting date. DNA fingerprinting of isolates of Streptococcus mutans by pulsed-field gel electrophoresis. Microbiol. Res. doi: 10.1016/j.micres.2006.06.014. [DOI] [PubMed]
- 20.Moreillon, P., and Y. A. Que. 2004. Infective endocarditis. Lancet 363:139-149. [DOI] [PubMed] [Google Scholar]
- 21.Nakano, K., R. Nomura, I. Nakagawa, S. Hamada, and T. Ooshima. 2004. Demonstration of Streptococcus mutans with a cell wall polysaccharide specific to a new serotype, k, in the human oral cavity. J. Clin. Microbiol. 42:198-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakano, K., R. Nomura, N. Shimizu, I. Nakagawa, S. Hamada, and T. Ooshima. 2004. Development of a PCR method for rapid identification of new Streptococcus mutans serotype k strains. J. Clin. Microbiol. 42:4925-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nesbo, C. L., S. L'Haridon, K. O. Stetter, and W. F. Doolittle. 2001. Phylogenetic analyses of two “archaeal” genes in Thermotoga maritima reveal multiple transfers between Archaea and Bacteria. Mol. Biol. Evol. 18:362-375. [DOI] [PubMed] [Google Scholar]
- 24.Nomura, R., K. Nakano, H. Nemoto, K. Fujita, S. Inagaki, T. Takahashi, K. Taniguchi, M. Takeda, H. Yoshioka, A. Amano, and T. Ooshima. 2006. Isolation and characterization of Streptococcus mutans in heart valve and dental plaque specimens from a patient with infective endocarditis. J. Med. Microbiol. 55:1135-1140. [DOI] [PubMed] [Google Scholar]
- 25.Nomura, R., K. Nakano, and T. Ooshima. 2005. Molecular analysis of the genes involved in the biosynthesis of serotype specific polysaccharide in the novel serotype k strains of Streptococcus mutans. Oral Microbiol. Immunol. 20:303-309. [DOI] [PubMed] [Google Scholar]
- 26.Ooshima, T., A. Izumitani, S. Sobue, and S. Hamada. 1983. Cariostatic effects of palatinose on experimental dental caries in rats. Jpn. J. Med. Sci. Biol. 36:219-223. [DOI] [PubMed] [Google Scholar]
- 27.Ooshima, T., M. Matsumura, T. Hoshino, S. Kawabata, S. Sobue, and T. Fujiwara. 2001. Contribution of three glucosyltransferases to sucrose-dependent adherence of Streptococcus mutans. J. Dent. Res. 80:1672-1677. [DOI] [PubMed] [Google Scholar]
- 28.Saarela, M., S. Alaluusua, T. Takei, and S. Asikainen. 1993. Genetic diversity within isolates of mutans streptococci recognized by an rRNA gene probe. J. Clin. Microbiol. 31:584-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato, Y., K. Okamoto, A. Kagami, Y. Yamamoto, T. Igarashi, and H. Kizaki. 2004. Streptococcus mutans strains harboring collagen-binding adhesin. J. Dent. Res. 83:534-539. [DOI] [PubMed] [Google Scholar]
- 30.Schenk, G., R. Layfield, J. M. Candy, R. G. Duggleby, and P. F. Nixon. 1997. Molecular evolutionary analysis of the thiamine-diphosphate-dependent enzyme, transketolase. J. Mol. Evol. 44:552-572. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt, D. M., B. K. Hubbard, and J. A. Gerlt. 2001. Evolution of enzymatic activities in the enolase superfamily: functional assignment of unknown proteins in Bacillus subtilis and Escherichia coli as l-Ala-d/l-Glu epimerases. Biochemistry 40:15707-15715. [DOI] [PubMed] [Google Scholar]
- 32.Seymour, R. A., R. Lowry, J. M. Whitworth, and M. V. Martin. 2000. Infective endocarditis, dentistry and antibiotic prophylaxis; time for a rethink? Br. Dent. J. 189:610-616. [DOI] [PubMed] [Google Scholar]
- 33.Shibata, Y., K. Ozaki, M. Seki, T. Kawato, H. Tanaka, Y. Nakano, and Y. Yamashita. 2003. Analysis of loci required for determination of serotype antigenicity in Streptococcus mutans and its clinical utilization. J. Clin. Microbiol. 41:4107-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, J. M., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan, C. B., M. A. Diggle, and S. C. Clarke. 2005. Multilocus sequence typing: data analysis in clinical microbiology and public health. Mol. Biotechnol. 29:245-254. [DOI] [PubMed] [Google Scholar]
- 36.Toi, C. S., P. Cleaton-Jones, and P. Fatti. 2005. Characterization of Streptococcus mutans diversity by determining restriction fragment-length polymorphisms of the gtfB gene of isolates from 5-year-old children and their mothers. Antonie Leeuwenhoek 88:75-85. [DOI] [PubMed] [Google Scholar]
- 37.Tsukioka, Y., Y. Yamashita, Y. Nakano, T. Oho, and T. Koga. 1997. Identification of a fourth gene involved in dTDP-rhamnose synthesis in Streptococcus mutans. J. Bacteriol. 179:4411-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsukioka, Y., Y. Yamashita, T. Oho, Y. Nakano, and T. Koga. 1997. Biological function of the dTDP-rhamnose synthesis pathway in Streptococcus mutans. J. Bacteriol. 179:1126-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Roosmalen, M. L., N. Geukens, J. D. Jongbloed, H. Tjalsma, J. Y. Dubois, S. Bron, J. M. van Dijl, and J. Anne. 2004. Type I signal peptidases of gram-positive bacteria. Biochim. Biophys. Acta 1694:279-297. [DOI] [PubMed] [Google Scholar]
- 40.Urwin, R., and M. C. Maiden. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11:479-487. [DOI] [PubMed] [Google Scholar]
- 41.Yamashita, Y., Y. Shibata, Y. Nakano, Y. Tsuda, N. Kido, M. Ohta, and T. Koga. 1999. A novel gene required for rhamnose-glucose polysaccharide synthesis in Streptococcus mutans. J. Bacteriol. 181:6556-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashita, Y., Y. Tsukioka, Y. Nakano, K. Tomihisa, T. Oho, and T. Koga. 1998. Biological functions of UDP-glucose synthesis in Streptococcus mutans. Microbiology 144:1235-1245. [DOI] [PubMed] [Google Scholar]
- 43.Yamashita, Y., Y. Tsukioka, K. Tomihisa, Y. Nakano, and T. Koga. 1998. Genes involved in cell wall localization and side chain formation of rhamnose-glucose polysaccharide in Streptococcus mutans. J. Bacteriol. 180: 5803-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zadoks, R. N., Y. H. Schukken, and M. Wiedmann. 2005. Multilocus sequence typing of Streptococcus uberis provides sensitive and epidemiologically relevant subtype information and reveals positive selection in the virulence gene pauA. J. Clin. Microbiol. 43:2407-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]