Abstract
Mycobacterium avium subsp. paratuberculosis, the etiological agent of paratuberculosis, affects a wide range of domestic ruminants and has been suggested to be involved in Crohn's disease in humans. Most available methods for identifying and differentiating strains of this difficult species are technically demanding and have limited discriminatory power. Here, we report the identification of novel PCR-based typing markers consisting of variable-number tandem repeats (VNTRs) of genetic elements called mycobacterial interspersed repetitive units (MIRUs). Eight markers were applied to 183 M. avium subsp. paratuberculosis isolates from bovine, caprine, ovine, cervine, leporine, and human origins from 10 different countries and to 82 human isolates of the closely related species M. avium from France. Among the M. avium subsp. paratuberculosis isolates, 21 patterns were found by MIRU-VNTR typing, with a discriminatory index of 0.751. The predominant R01 IS900 restriction fragment length polymorphism type, comprising 131 isolates, was divided into 15 MIRU-VNTR types. Among the 82 M. avium isolates, the eight MIRU-VNTR loci distinguished 30 types, none of which was shared by M. avium subsp. paratuberculosis isolates, resulting in a discriminatory index of 0.889. Our results suggest that MIRU-VNTR typing is a fast typing method that, in combination with other methods, might prove to be optimal for PCR-based molecular epidemiological studies of M. avium/M. avium subsp. paratuberculosis pathogens. In addition, presumably identical M. avium subsp. paratuberculosis 316F vaccine strains originating from the Weybridge laboratory and from different commercial batches from Mérial actually differed by one or both typing methods. These results indicate a substantial degree of genetic drift among different vaccine preparations, which has important implications for prophylactic approaches.
Mycobacterium avium subsp. paratuberculosis is the etiological agent of a severe gastroenteritis in ruminants known as Johne's disease, or paratuberculosis, since 1895 (11). Paratuberculosis is prevalent in domestic animals worldwide and has a significant impact on the economy. Recent studies have also described M. avium subsp. paratuberculosis isolation from wildlife (16), including rabbits (8). In addition, it has been suggested that M. avium subsp. paratuberculosis may be involved in Crohn's disease, a chronic enteritis in humans, but evidence for a causal link remains controversial (23). Therefore, paratuberculosis is considered a public health concern. Control of this disease requires a better knowledge of the causative agent and of its epidemiology, interspecies transmission, and biodiversity within M. avium subsp. paratuberculosis strains.
Study of M. avium subsp. paratuberculosis is hampered by the difficulty of growing and manipulating the organism in a laboratory setting. M. avium subsp. paratuberculosis is an extremely slow-growing organism and requires the addition of the iron chelator mycobactin for in vitro growth (2), and most bovine strains require 4 to 6 months of incubation. M. avium subsp. paratuberculosis strains are very difficult to isolate from sheep and humans and may require years to produce colonies. Therefore, small numbers of M. avium subsp. paratuberculosis isolates have been maintained in available collections, which has limited biodiversity studies.
Another limiting factor has been the lack of convenient discriminatory typing methods. The most widely used method to type M. avium subsp. paratuberculosis isolates is restriction fragment length polymorphism (RFLP), with detection of polymorphisms by hybridization to IS900 (IS900 RFLP typing) (24). As it is applicable only to cultivable strains, this method is slow and technically demanding. Moreover, it requires analysis of complex banding patterns and has limited discriminatory power. Therefore, rapid and discriminatory molecular typing methods need to be assessed as alternatives for studying the diversity of M. avium subsp. paratuberculosis strains. Motiwala et al. have recently reviewed the current genotyping methods used for determining genetic diversity within a population of M. avium subsp. paratuberculosis isolates (17).
Tandem-repeat (TR) sequences represent one of the rare categories of polymorphic structures in the genomes of highly monomorphic species, such as Bacillus anthracis and Yersinia pestis (12). Variable-number TRs (VNTRs), in particular those of genetic elements called mycobacterial interspersed repetitive units (MIRUs), have been discovered and used for typing of various mycobacterial species, including the Mycobacterium tuberculosis complex, Mycobacterium marinum, and Mycobacterium ulcerans (21, 25-27). Recently, partial genome screenings have identified a limited number of MIRU-VNTR loci in the M. avium-Mycobacterium intracellulare complex, providing very limited discrimination among M. avium subsp. paratuberculosis isolates (4, 18).
The aim of this study was to identify novel MIRU-VNTR loci based on an exhaustive screening of TR loci in the M. avium subsp. paratuberculosis genome and to study their variability in a large collection of M. avium subsp. paratuberculosis and M. avium isolates obtained from different hosts and from different geographic origins. The discrimination provided by the novel MIRU-VNTR loci was compared to that achieved by IS900 RFLP and IS1245 RFLP typing.
MATERIALS AND METHODS
Strains.
M. avium subsp. paratuberculosis isolates were isolated on Herrold's egg yolk medium containing mycobactin J, amphotericin B, and nalidixic acid (Becton Dickinson, Le Pont de Claix, France) according to the method of Whipple et al. (29). Mycobacterial isolates were subcultured in Middlebrook 7H9 broth supplemented with Middlebrook albumin-dextrose-catalase enrichment medium (Becton Dickinson, Le Pont de Claix, France) and 2 μg/ml of mycobactin J (Institut Pourquier, Montpellier, France) when required. A panel of 183 M. avium subsp. paratuberculosis isolates was assembled (see Table S1 in the supplemental material) from 10 countries and different host species. The M. avium isolates included in the present study were obtained from the Institute Pasteur of Paris, Laboratoire de Référence des Mycobactéries, Paris, France (see Table S6 in the supplemental material). These isolates were all isolated from blood samples recovered from 93 AIDS patients over several months (14) and were typed by serotyping, by IS1245 RFLP analysis, in some cases by RFLP analysis with plasmids pVT2 and pLR7 as probes, and by pulsed-field gel electrophoresis (20).
The M. avium subsp. paratuberculosis vaccine strains analyzed in the present study were obtained from the Veterinary Laboratories Agency Weybridge laboratory and from various batches of the vaccine Néoparasec (Mérial, Bourgelat, France).
Preparation of mycobacterial DNA.
Mycobacterial DNA was obtained according to the method of Baulard et al. (3). M. avium DNA for PCR amplifications was obtained from strains preserved at −20°C in Youmans medium as follows: 0.2 ml of the medium was centrifuged for 10 min at 6,000 × g, and the pellet was washed twice with 0.2 ml of Tris-EDTA buffer and then resuspended in 0.2 ml of Tris-EDTA buffer. The bacteria were heat killed for 30 min at 95°C, and the DNA from the supernatant was directly used as a template.
Molecular identification of M. avium and M. avium subsp. paratuberculosis.
All M. avium subsp. paratuberculosis isolates were screened for the presence or absence of IS900 and IS901 insertion sequences. Synthetic oligonucleotides (Sigma), described by Sanderson et al. (22) for IS900 primers and by Inglis et al. (10) for IS901 primers, were used.
IS900 RFLP typing.
IS900 RFLP typing of M. avium subsp. paratuberculosis DNA was performed as previously described by van Soolingen et al. (28), with some modifications. The IS900 DNA probe was prepared by PCR amplification of a 707-bp fragment of the IS900 insertion sequence specific for M. avium subsp. paratuberculosis using the primers described by Overduin et al. (18). PCRs were performed starting from 10 ng of chromosomal DNA of M. avium subsp. paratuberculosis strain ATCC 19698 by using a Bio-Rad iCycler thermal cycler. The PCR product was purified on Qiaquick spin columns (QIAGEN) according to the manufacturer's instructions. The probe was biotin labeled with the NEBlot Phototope kit (New England Biolabs) by following the instructions of the manufacturer.
Digestion was performed with 3 μg of DNA prepared as described above and 7 U of BstEII (Promega) at 37°C for at least 4 h. Fragments were resolved by agarose gel electrophoresis and transferred onto Immobilon-S nylon membranes (Millipore) by vacuum transfer with the Vacu-Gene system (Pharmacia LKB Biotechnology). Detection of DNA fragments hybridizing with the biotinylated probe was performed with the Phototope-Star detection kit for nucleic acids (New England Biolabs), according to the manufacturer's instructions. A photobiotinylated mixture of HindIII-digested lambda DNA and HaeIII-digested φX174 DNA at a concentration of 100 ng/μl (New England Biolabs) was used as a molecular size marker.
Analysis of RFLP patterns was performed according to the methods of Overduin et al. (18) and other studies (5, 19). Conserved bands of 8.8, 5.2, 3.0, 2.4, 2.1, and 1.6 kb in the IS900 RFLP pattern were used as internal standards for the normalization of RFLP patterns.
Identification of TR and MIRU loci.
The published genomic sequence of M. avium subsp. paratuberculosis strain K10 (13) (GenBank accession number NC_002944 [http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=genome&gi=380]) was used to identify MIRU and potential VNTR sequences. TRs were identified by using the Tandem Repeats Finder software of the Laboratory for Biocomputing and Informatics, Boston University (http://tandem.bu.edu/trf/trf.submit.options.html), under the default settings of the program. MIRU loci were identified by searching sequences homologous to those of previously described MIRU loci in the M. tuberculosis H37Rv chromosome (27) using the BLAST 2.2.11 software at the NCBI website (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi).
MIRU-VNTR typing.
Primers designed to target flanking regions of the MIRU-VNTRs and the conditions of the PCR amplification are listed in Table 1 and Table S2 in the supplemental material. The PCR mixture was composed as follows using the Go Taq Flexi DNA polymerase (Promega). Five microliters from fivefold-diluted DNA solution was added to a final volume of 25 μl containing 0.1 μl of Go Taq Flexi DNA polymerase (5 U/μl), 5 μl of betaine (Sigma), or 1 μl of dimethyl sulfoxide (Sigma); 0.2 mM (each) dATP, dCTP, dGTP, and dTTP (Promega); 5 μl of 5× PCR buffer supplied by the manufacturer; 1 μM of primers; and 1.5 mM of MgCl2. The primers were designed using Oligo 5.0 software (National Biosciences). The reactions were carried out using an iCycler thermal cycler (Bio-Rad). PCR conditions were as follows: 1 cycle of 5 min at 94°C; 40 cycles of 30 s at 94°C, 30 s at 58°C, and 30 s at 72°C; and 1 cycle of 7 min at 72°C. To detect differences in repeat numbers, the PCR products were analyzed by electrophoresis using 1.5% agarose gels (agarose electrophoresis grade; Invitrogen).
TABLE 1.
Polymorphic TRs, positions, and primer sequences
| TR no. | Position of primer on M. avium subsp. paratuberculosis genome
|
Predicted size of PCR product (bp) | Primer
|
Tma (°C) | Buffer usedb
|
|||
|---|---|---|---|---|---|---|---|---|
| Start | Stop | Forward | Reverse | 1 μl DMSO | 5 μl betaine | |||
| 32 | 1125707 | 1126004 | 298 | CCACAGGGTTTTTGGTGAAG | GGAAATCCAACAGCAAGGAC | 55 | + | + |
| 292 | 3253590 | 3253889 | 300 | CTTGAGCAGCTCGTAAAGCGT | GCTGTATGAGGAAGTCTATTCATGG | 58 | + | − |
| X3 | 4441875 | 4442070 | 196 | AACGAGAGGAAGAACTAAGCCG | TTACGGAGCAGGAAGGCCAGCGGG | 58 | − | − |
| 25 | 3665598 | 3665947 | 350 | GTCAAGGGATCGGCGAGG | TGGACTTGAGCACGGTCAT | 58 | + | − |
| 3 | 131320 | 131527 | 208 | CATATCTGGCATGGCTCCAG | ATCGTGTTGACCCCAAAGAAAT | 60 | + | − |
| 7 | 3711417 | 3711619 | 203 | GACAACGAAACCTACCTCGTC | GTGAGCTGGCGGCCTAAC | 60 | + | − |
| 10 | 4279553 | 4279855 | 303 | GACGAGCAGCTGTCCGAG | GAGAGCGTGGCCATCGAG | 60 | − | + |
| 47 | 4128604 | 4128821 | 217 | CGTTGCGATTTCTGCGTAGC | GGTGATGGTCGTGGTCATCC | 64 | + | − |
Tm, melting temperature.
DMSO, dimethyl sulfoxide. +, used; −, not used.
Calculation of discriminatory power.
The discriminatory index (DI) described by Hunter and Gaston (9) was used as a numerical index for the discriminatory power of each typing method. The DI was calculated using the following formula:
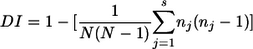 |
where N is the total number of strains in the typing scheme, s is the total number of distinct patterns discriminated by each typing method and strategy, and nj is the number of strains belonging to the jth pattern.
RESULTS
IS900 RFLP typing.
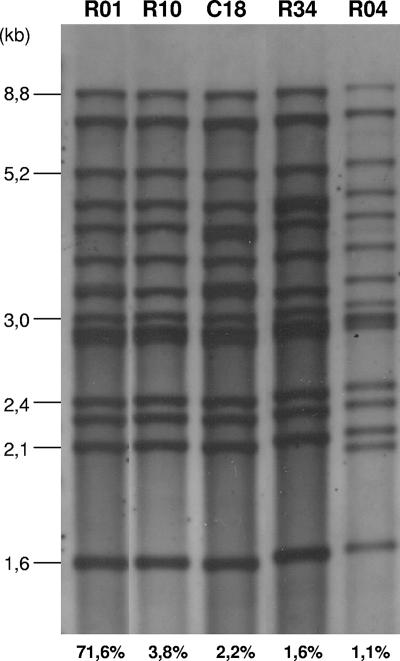
For relevant study of M. avium subsp. paratuberculosis TR variability, a representative panel of isolates from diverse geographic regions and host origins was assembled based on a preliminary IS900 RFLP typing analysis. All of the M. avium subsp. paratuberculosis isolates analyzed in this study were found to be positive for IS900 and negative for IS901 by PCR, confirming them as M. avium subsp. paratuberculosis strains. Over 183 M. avium subsp. paratuberculosis isolates were subjected to IS900 RFLP typing. Twenty-six different RFLP types were found (see Tables S1 and S3 in the supplemental material). The vast majority (131 isolates; 72%) were of type R01 (Fig. 1). Interestingly, all caprine strains were grouped in this profile. The R09 type was represented among 5.5% of the isolates, followed by R10 (3.8%); C18 (2.2%); R13, R24, and R34 (1.6%); and R04, R20, and R27 (1.1%) (Fig. 1). The other 16 profiles were found in single isolates. Thus, 167 isolates belonged to 10 cluster patterns, whereas 16 patterns were unique (see Table S3 in the supplemental material).
FIG. 1.
Selected IS900 RFLP profiles represented in our collection of M. avium subsp. paratuberculosis strains. The percentages indicate the proportion of each IS900 RFLP profile in our collection. R types are designated according to the nomenclature of the National Institute of Public Health and the Environment, Bilthoven, The Netherlands.
In silico identification and characterization of TRs and MIRU loci.
The genome sequence of M. avium subsp. paratuberculosis strain K10 was analyzed for the presence of TRs. Over 363 TR sequences were identified in the genome. We focused on TRs of the minisatellite category, defined by a repeat unit size in the range of 10 to 100 bp, as their corresponding allelic differences can be easily resolved by agarose gel electrophoresis. Thirty-three TRs present in more than two copies and with 85% or more nucleotide identity among individual repeat units in the reference strain were selected for experimental analysis. The use of these two criteria was based on the observation that the presence of at least two identical or nearly identical repeats is necessary and sufficient to generate TR variability in the case of M. tuberculosis MIRU minisatellites (27).
In addition, two MIRU loci were identified in M. avium subsp. paratuberculosis strain K10 by BLAST searches using as templates the sequences of the flanking genes of two polymorphic MIRU-VNTR loci in M. tuberculosis. These two MIRU loci were called M. avium subsp. paratuberculosis SenX3-RegX3 and M. avium subsp. paratuberculosis 2920c-2921c. The repeat units of these MIRUs in the M. avium subsp. paratuberculosis K10 genome have a length of 53 bp and are present with copy numbers of two and three in the SenX3-RegX3 and 2920c-2921c loci, respectively. These loci containing TRs of MIRUs were added to the above selection of TR loci for further experimental analysis.
Polymorphism in repeat numbers among M. avium subsp. paratuberculosis isolates.
The polymorphism of the 35 TR loci selected by in silico analysis was initially investigated using a subset of M. avium subsp. paratuberculosis isolates selected for diversity based on IS900 RFLP types and geographic and host origins (see Table S1 in the supplemental material). Only the eight TR and MIRU loci that showed size polymorphism after PCR among the isolates in this subset were used for typing the total collection of 183 isolates.
Twenty-one different MIRU-VNTR types were found in the total collection (see Tables S1 and S4 in the supplemental material). Patterns INMV1 and INMV2 represented the majority of the isolates (36% and 34%, respectively), followed by 11 patterns representing from 1 to 5.5% of the isolates. In total, MIRU-VNTR grouped 175 isolates into 13 clusters, whereas 8 MIRU-VNTR patterns were unique (see Table S4 in the supplemental material). All ovine strains in our collection have the same type: INMV2.
Comparison of IS900 RFLP and MIRU-VNTR typing and a combination of the two methods.
Interestingly, the major RFLP type R01, representing 131 isolates, could be subdivided into 15 different MIRU-VNTR types. Likewise, the seven isolates with identical RFLP types (R10) were divided into five different VNTR types. RFLP types R09 and C18 could be divided into three VNTR types, while RFLP types R13, R24, and R27 could each be divided into two different VNTR types.
Conversely, several major and minor MIRU-VNTR types were also subdivided by IS900 RFLP typing. For instance, MIRU-VNTR type INMV2, comprising 66 isolates of M. avium subsp. paratuberculosis, and MIRU-VNTR type INMV1, comprising 62 isolates, were subdivided into 11 and 10 IS900 RFLP types, respectively. At the other extreme, the minor MIRU-VNTR types INMV5 to -8, INMV11, and INMV13 could each be divided into two RFLP types.
In total, the combination of the two methods distinguished 51 distinct patterns, including 18 cluster patterns comprising 150 isolates and 33 unique patterns (Table 2; see Tables S5 and S6 in the supplemental material). Therefore, a maximal DI (9) of 0.855 was achieved for the 183 isolates by using IS900 RFLP and MIRU-VNTR typing in combination compared to 0.483 for IS900 RFLP typing alone and 0.751 for VNTR typing alone.
TABLE 2.
Discriminatory powers of IS900 RFLP and MIRU-VNTR typing used alone and in combination
| Typing method | No. of different patterns | No. of clusters | No. of clustered isolates | No. of unique isolates | No. of isolates in each cluster | DI |
|---|---|---|---|---|---|---|
| RFLP | 26 | 10 | 167 | 16 | 2-131 | 0.483 |
| MIRU-VNTR | 21 | 13 | 175 | 8 | 2-66 | 0.751 |
| RFLP + MIRU-VNTR | 51 | 18 | 150 | 33 | 2-53 | 0.855 |
Polymorphism among 316F vaccine strains.
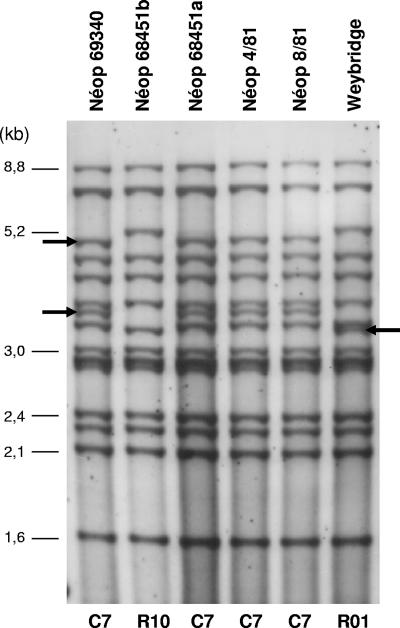
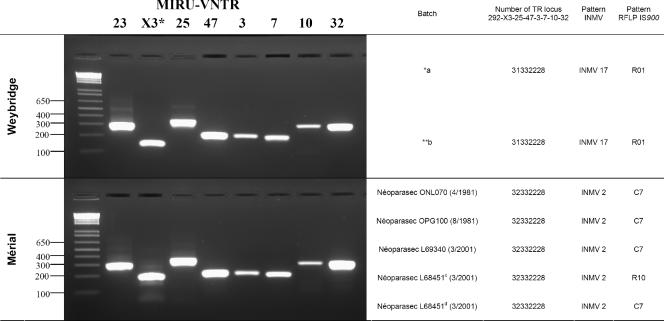
The M. avium subsp. paratuberculosis strain 316F is one of the strains used for vaccination against paratuberculosis. In this study, we analyzed the clonal identity between 316F vaccine batches from various origins or from the same origin by using both IS900 RFLP and MIRU-VNTR typing. The results shown in Fig. 2 and Fig. 3 demonstrate that Weybridge-316F differs from Mérial-316F by both genotyping methods. The two Weybridge-316F batches analyzed by MIRU-VNTR typing displayed the same INMV17 pattern, but it differed from that of the all Mérial-316F batches by a variation in the single SenX3-RegX3 locus (Fig. 3). By IS900 RFLP analysis, the two Weybridge-316F batches displayed identical R01 patterns, which clearly differed by three bands from the C7 pattern detected for four Mérial-316F batches in accordance with that described in the past (6). In addition, a fifth analyzed Mérial-316F vial displayed an R10 pattern differing from C7 by two bands (Fig. 2). Furthermore, two isolates from two different Mérial vials identified by identical batch numbers (L68451) produced in the same year, 2001, showed the same MIRU-VNTR type but two RFLP profiles differing by two IS900 bands, suggesting a degree of clonal heterogeneity among commercial preparations of the vaccine strain.
FIG. 2.
IS900 RFLP profiles of M. avium subsp. paratuberculosis 316F strains. Profiles from five different cultures of Mérial-316F (Néoparasec) corresponding to four batches, Néop 69340, Néop 68451, Néop 4/81, and Néop 8/81, as well as from one culture from a 316F strain from Weybridge, are represented. Néop 68451a and Néop 68451b correspond to cultures from two different Mérial-316F vials identified by the same batch number, 68451. R and C types are designated according to the nomenclature of the National Institute of Public Health and the Environment, Bilthoven, The Netherlands, and Collins et al. (5) and Pavlik et al. (19), respectively. The arrows indicate polymorphic bands among the different profiles.
FIG. 3.
MIRU-VNTR profiles of M. avium subsp. paratuberculosis 316F strains from Weybridge laboratory and Mérial. The PCR products were analyzed by electrophoresis using agarose gels, as described in Materials and Methods. The positions of size standard bands and designations of MIRU-VNTR loci are indicated on the left and at the top, respectively. a, analysis of Weybridge 316F strain; b, analysis of Mérial 316F strain. A large asterisk indicates the locus (X3) that varies between the two strains. *a, provided by P. Willemsen (Central Institute for Animal Disease Control, Department of Bacteriology and TSEs, 8203 AA Lelystad, The Netherlands); **b, provided by K. Stevenson (Moredun Research Institute, Pentlands Science Park, Bush Loan, Penicuik EH26 0PZ, Scotland, United Kingdom). The batches marked c and d were cultured from two different Néoparasec vials identified by the same batch number on the same date.
INVM polymorphism in M. avium.
In order to study the levels of polymorphism of the eight MIRU-VNTR loci in other members of the M. avium-M. intracellulare complex, 82 M. avium isolates previously serotyped and typed by IS1245 RFLP were typed using the same primers as those targeting the eight MIRU-VNTR loci of M. avium subsp. paratuberculosis. This analysis (see Tables S6 and S7 in the supplemental material) showed that MIRU-VNTR typing may be applied to M. avium isolates using the same conditions as those defined for M. avium subsp. paratuberculosis. Thirty MIRU-VNTR types were found for the 82 M. avium isolates (see Table S7 in the supplemental material), yielding a DI of 0.889. None of the M. avium MIRU-VNTR profiles matched those of M. avium subsp. paratuberculosis isolates.
DISCUSSION
VNTRs of the minisatellite class are valuable markers used for genotyping several mycobacterial species (4, 18, 21, 25, 26). Two previous studies (4, 18) identified a few VNTR loci in M. avium subsp. paratuberculosis isolates based on partial screenings of the M. avium subsp. paratuberculosis K10 genome. In this study, we performed an exhaustive screening of potential VNTR loci in this genome based on in silico identification of TRs in the whole M. avium subsp. paratuberculosis K10 genome and experimental testing of the polymorphism of the most interesting TR candidates using a reference set of M. avium subsp. paratuberculosis isolates with diverse IS900 RFLP types and geographic and host origins. By this means, we have identified eight VNTR loci, seven of which are novel and one of which (senX3-regX3) has been previously identified by Bull et al. (4).
When used alone, this eight-locus-based typing system distinguished slightly fewer types of M. avium subsp. paratuberculosis isolates in this collection than IS900 RFLP (21 versus 26), but the MIRU-VNTR types were more equally distributed in this M. avium subsp. paratuberculosis collection than the IS900 RFLP types (i.e., with the single R01 type comprising 71.5% of the isolates). Interestingly, this R01 RFLP type, representing the vast majority of the M. avium subsp. paratuberculosis isolates found in this study and in the other studies, could be successfully divided into 15 subgroups by MIRU-VNTR typing. On the other hand, 10 VNTR types could also be subdivided by IS900 RFLP, six of which were subdivided into only two IS900 RFLP subgroups. Therefore, the highest resolution was achieved when the two typing methods were combined.
We analyzed the eight MIRU-VNTR loci in different batches of 316F M. avium subsp. paratuberculosis vaccine in order to investigate both the degree of clonality of the presumably identical corresponding strains and the clonal stability of MIRU-VNTR markers. The stability of M. tuberculosis complex MIRU-VNTR loci was analyzed in a similar manner using the genealogically distant Mycobacterium bovis BCG strain (27). Rather surprisingly, both typing methods distinguished Weybridge-316F batches from Mérial-316F batches. In the case of MIRU-VNTR typing, the batches from Weybridge and Mérial differed by a single locus, namely, senX3-regX3. Interestingly, the same locus was shown to display some degree of VNTR polymorphism among BCG sister strains cultivated separately for more than 30 years (27). This single MIRU-VNTR locus difference was corroborated by differences of three IS900 RFLP bands between batches from Weybridge and four batches from Mérial. These results confirm the IS900 RFLP profiles of the vaccine strains described by Collins et al. (6). Furthermore, a polymorphism involving two other IS900 RFLP single-band differences was detected, not only between different Mérial batches, but also between two cultures from two different Mérial vials identified by the same batch number. In the latter case at least, the vials can be assumed to originate from the same seed stock. In contrast, the eight MIRU-VNTR loci remained unchanged among these different commercial preparations. These observations suggest that there has been a substantial degree of genetic drift between the Weybridge- and Mérial-316F strains, which most likely results from separate culturing after the exchange of the presumed original strain decades ago. The outcome of this process has been two closely related but now clearly distinct clones in the Weybridge and commercial preparations, as judged by two fully independent genotyping methods. Remarkably, this genetic drift appears to be ongoing for the commercial vaccine, as indicated by IS900 RFLP observed between preparations from different batches or even from the same batch. Because MIRU-VNTR types remained the same among tested commercial preparations, this degree of clonal heterogeneity appears to be less than that observed between these commercial preparations and those from Weybridge. The conservation of the eight MIRU-VNTR loci among these apparent clonal variants with slightly different IS900 RFLP types thus suggests a slightly lower evolutionary rate for these eight-locus-based genotypes than those of IS900 RFLP fingerprints.
Traditional techniques or comparative genomics used for studying the genetic structures of M. avium subsp. paratuberculosis and M. avium populations has shown the very close relatedness, as well as the distinctiveness, of these mycobacterial species (13). Consistent with the first feature, our results show that the flanking sequences and the polymorphisms of the eight MIRU-VNTR loci are sufficiently conserved between the two species to use the same PCR primers and loci for MIRU-VNTR typing of M. avium isolates. In accordance with the second feature, the MIRU-VNTR types identified for the M. avium isolates were all distinct from any of those identified for M. avium subsp. paratuberculosis. Interestingly, 30 MIRU-VNTR types were obtained for the 82 M. avium isolates, although they all came from a single country (France) and host (human), while only 21 types were obtained for the 183 M. avium subsp. paratuberculosis isolates from different hosts and settings. Although these results, based on limited samples, must be considered preliminary, this higher degree of MIRU-VNTR diversity among M. avium isolates is consistent with the higher genetic diversity in M. avium strains seen by using other markers (20).
In addition, the use of these markers could shed new light on molecular studies of M. avium subsp. paratuberculosis epidemiology. For instance, it is interesting to note that two M. avium subsp. paratuberculosis strains in our series that shared a rare MIRU-VNTR pattern (INMV9) and an IS900 RFLP pattern (R01) were isolated from humans and cattle from the same geographical origin (France), raising the question of a common source.
In conclusion, we have described here the identification of novel MIRU-VNTR markers for more specific differentiation of M. avium subsp. paratuberculosis isolates. Our preliminary analyses suggest that MIRU-VNTR typing provides us with a discriminatory power close to that obtained with the IS900 RFLP method. Parts of the respective discriminatory powers provided by these two independent methods are nonredundant, resulting in higher resolution when the two typing approaches are combined. This result should be verified with a larger panel of isolates with different IS900 RFLP patterns and geographic origins and from hosts other than cattle, such as sheep. However, it is already clear that these markers constitute very useful additional tools for typing M. avium subsp. paratuberculosis (as well as M. avium), especially because MIRU-VNTR typing is PCR based. As suggested by previous results (4, 18), the MIRU-VNTR loci could be further subjected to DNA sequence analysis to detect possible sequence polymorphisms among repeat units, in addition to the variation in the number of repeats among M. avium subsp. paratuberculosis isolates. This potential supplementary polymorphism and the addition of the few nonredundant VNTR loci described by the authors of the previous studies may further improve the DI of this typing method. According to the results for allelic diversity (Table 3), some markers are more polymorphic than others. These markers must be applied in priority for genotyping. Recently, a multilocus short sequence repeat sequencing approach was described for discriminatory genotyping of M. avium subsp. paratuberculosis strains (1, 7). Eventually, a combination of this method with VNTR-MIRU-based typing might prove to be optimal for PCR-based molecular epidemiological studies of this pathogen. Last but not least, the phenomena of genetic drift and clonal heterogeneity discovered among vaccine preparations from different origins, or even a single origin, have implications that must be taken into account for evaluating and ensuring the stability of protective effects of paratuberculosis vaccine preparations over time and across settings.
TABLE 3.
MIRU-VNTR allelic distribution among M. avium subsp. paratuberculosis and M. avium isolates
| Isolate and locus | No. of isolates with the specified MIRU allele
|
Allelic diversity (h)a | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| M. avium subsp. paratuberculosis | ||||||||||||
| 32 | 177 | 5 | 1 | 0.59 | ||||||||
| 292 | 6 | 108 | 69 | 0.51 | ||||||||
| 7 | 12 | 164 | 3 | 2 | 2 | 0.19 | ||||||
| 10 | 19 | 164 | 0.18 | |||||||||
| 25 | 176 | 1 | 6 | 0.07 | ||||||||
| 47 | 5 | 178 | 0.05 | |||||||||
| X3 | 3 | 179 | 1 | 0.04 | ||||||||
| 3 | 1 | 182 | 0.005 | |||||||||
| M. avium | ||||||||||||
| X3 | 1 | 31 | 10 | 26 | 12 | 2 | 0.72 | |||||
| 47 | 63 | 19 | 0.35 | |||||||||
| 25 | 2 | 66 | 4 | 7 | 3 | 0.33 | ||||||
| 32 | 1 | 6 | 67 | 8 | 0.3 | |||||||
| 292 | 8 | 1 | 70 | 3 | 0.27 | |||||||
| 10 | 7 | 75 | 0.15 | |||||||||
| 7 | 2 | 80 | 0.04 | |||||||||
| 3 | 82 | 0 | ||||||||||
MIRU-VNTR allelic distribution was calculated as described by Mazars et al. (15).
Supplementary Material
Acknowledgments
We thank Raúl Barletta (Department of Veterinary and Biomedical Sciences, University of Nebraska, Lincoln) for providing the M. avium subsp. paratuberculosis K10 strain and Véronique Vincent (Institut Pasteur, Paris, France) and Claude Couquet (Laboratoire Départemental de Limoges, Limoges, France) for providing us M. avium subsp. paratuberculosis isolates.
This work was supported by the Institut National de la Recherche Agronomique and Agence Française de Sécurité Sanitaire des Aliments (contract AIP P00297).
Footnotes
Published ahead of print on 30 May 2007.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Amonsin, A., L. L. Li, Q. Zhang, J. P. Bannantine, A. S. Motiwala, S. Sreevatsan, and V. Kapur. 2004. Multilocus short sequence repeat sequencing approach for differentiating among Mycobacterium avium subsp. paratuberculosis strains. J. Clin. Microbiol. 42:1694-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barclay, R., and C. Ratledge. 1983. Iron-binding compounds of Mycobacterium avium, Mycobacterium intracellulare, Mycobacterium scrofulaceum, and mycobactin-dependent Mycobacterium paratuberculosis and M. avium. J. Bacteriol. 153:1138-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baulard, A., L. Kremer, and C. Locht. 1996. Efficient homologous recombination in fast-growing and slow-growing mycobacteria. J. Bacteriol. 178:3091-3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bull, T. J., K. Sidi-Boumedine, E. J. McMinn, K. Stevenson, R. Pickup, and J. Hermon-Taylor. 2003. Mycobacterial interspersed repetitive units (MIRU) differentiate Mycobacterium avium subspecies paratuberculosis from other species of the Mycobacterium avium complex. Mol. Cell. Probes 17:157-164. [DOI] [PubMed] [Google Scholar]
- 5.Collins, D. M., S. Cavaignac, and G. W. de Lisle. 1997. Use of four DNA insertion sequences to characterize strains of the Mycobacterium avium complex isolated from animals. Mol. Cell. Probes 11:373-380. [DOI] [PubMed] [Google Scholar]
- 6.Collins, D. M., D. M. Gabric, and G. W. de Lisle. 1990. Identification of two groups of Mycobacterium paratuberculosis strains by restriction endonuclease analysis and DNA hybridization. J. Clin. Microbiol. 28:1591-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghadiali, A. H., M. Strother, S. A. Naser, E. J. Manning, and S. Sreevatsan. 2004. Mycobacterium avium subsp. paratuberculosis strains isolated from Crohn's disease patients and animal species exhibit similar polymorphic locus patterns. J. Clin. Microbiol. 42:5345-5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greig, A., K. Stevenson, D. Henderson, V. Perez, V. Hughes, I. Pavlik, M. E. Hines II, I. McKendrick, and J. M. Sharp. 1999. Epidemiological study of paratuberculosis in wild rabbits in Scotland. J. Clin. Microbiol. 37:1746-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inglis, N. F., K. Stevenson, R. C. Davies, D. G. Heaslip, and J. M. Sharp. 2001. Unique expression of a highly conserved mycobacterial gene in IS901+ Mycobacterium avium. Microbiology 147:1557-1564. [DOI] [PubMed] [Google Scholar]
- 11.Johne, H. A., and L. Frothinghan. 1895. Ein eigenthuemlicher tall van tuberckulose beim rind. Dtsch. Z. Tiermed. Pathol. 21:438-454. [Google Scholar]
- 12.Le Fleche, P., Y. Hauck, L. Onteniente, A. Prieur, F. Denoeud, V. Ramisse, P. Sylvestre, G. Benson, F. Ramisse, and G. Vergnaud. 2001. A tandem repeats database for bacterial genomes: application to the genotyping of Yersinia pestis and Bacillus anthracis. BMC Microbiol. 1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, L., J. P. Bannantine, Q. Zhang, A. Amonsin, B. J. May, D. Alt, N. Banerji, S. Kanjilal, and V. Kapur. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. USA 102:12344-12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.May, T., F. Brel, C. Beuscart, V. Vincent, C. Perronne, T. Doco-Lecompte, T. Saint-Marc, B. Dautzenberg, J. Grosset, et al. 1997. Comparison of combination therapy regimens for treatment of human immunodeficiency virus-infected patients with disseminated bacteremia due to Mycobacterium avium. Clin. Infect. Dis. 25:621-629. [DOI] [PubMed] [Google Scholar]
- 15.Mazars, E., S. Lesjean, A. L. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Motiwala, A. S., A. Amonsin, M. Strother, E. J. Manning, V. Kapur, and S. Sreevatsan. 2004. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis isolates recovered from wild animal species. J. Clin. Microbiol. 42:1703-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Motiwala, A. S., L. Li, V. Kapur, and S. Sreevatsan. 2006. Current understanding of the genetic diversity of Mycobacterium avium subsp. paratuberculosis. Microbes Infect. 8:1406-1418. [DOI] [PubMed] [Google Scholar]
- 18.Overduin, P., L. Schouls, P. Roholl, A. van der Zanden, N. Mahmmod, A. Herrewegh, and D. van Soolingen. 2004. Use of multilocus variable-number tandem-repeat analysis for typing Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 42:5022-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pavlik, I., A. Horvathova, L. Dvorska, J. Bartl, P. Svastova, R. du Maine, and I. Rychlik. 1999. Standardisation of restriction fragment length polymorphism analysis for Mycobacterium avium subspecies paratuberculosis. J. Microbiol. Methods 38:155-167. [DOI] [PubMed] [Google Scholar]
- 20.Picardeau, M., A. Varnerot, T. Lecompte, F. Brel, T. May, and V. Vincent. 1997. Use of different molecular typing techniques for bacteriological follow-up in a clinical trial with AIDS patients with Mycobacterium avium bacteremia. J. Clin. Microbiol. 35:2503-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roring, S., A. Scott, D. Brittain, I. Walker, G. Hewinson, S. Neill, and R. Skuce. 2002. Development of variable-number tandem repeat typing of Mycobacterium bovis: comparison of results with those obtained by using existing exact tandem repeats and spoligotyping. J. Clin. Microbiol. 40:2126-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanderson, J. D., M. T. Moss, M. L. Tizard, and J. Hermon-Taylor. 1992. Mycobacterium paratuberculosis DNA in Crohn's disease tissue. Gut 33:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sartor, R. B. 2005. Does Mycobacterium avium subspecies paratuberculosis cause Crohn's disease? Gut 54:896-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevenson, K., V. M. Hughes, L. de Juan, N. F. Inglis, F. Wright, and J. M. Sharp. 2002. Molecular characterization of pigmented and nonpigmented isolates of Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 40:1798-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stragier, P., A. Ablordey, W. M. Meyers, and F. Portaels. 2005. Genotyping Mycobacterium ulcerans and Mycobacterium marinum by using mycobacterial interspersed repetitive units. J. Bacteriol. 187:1639-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Supply, P., J. Magdalena, S. Himpens, and C. Locht. 1997. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol. Microbiol. 26:991-1003. [DOI] [PubMed] [Google Scholar]
- 27.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762-771. [DOI] [PubMed] [Google Scholar]
- 28.van Soolingen, D., P. E. de Haas, P. W. Hermans, and J. D. van Embden. 1994. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol. 235:196-205. [DOI] [PubMed] [Google Scholar]
- 29.Whipple, D. L., D. R. Callihan, and J. L. Jarnagin. 1991. Cultivation of Mycobacterium paratuberculosis from bovine fecal specimens and a suggested standardized procedure. J. Vet. Diagn. Investig. 3:368-373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.