Abstract
Adeno-associated virus type 5 (AAV5) is unique among human AAV serotypes in that it uses a polyadenylation site [(pA)p] within the single small intron in the center of the genome. We previously reported that inhibition of polyadenylation at (pA)p, necessary for read-through of P41-generated capsid gene pre-mRNAs which are subsequently spliced, requires binding of U1 snRNP to the upstream donor. Inhibition was reduced as the distance between the cap site and the donor was increased (increasing the size of the 5′ exon). Here, we have demonstrated that U1-70K is a key component of U1 snRNP that mediates inhibition of polyadenylation at (pA)p. Furthermore, introduction of a U-rich stretch, predicted to target TIA-1 and thus increase the affinity of U1 snRNP binding to the intervening donor site, significantly augmented inhibition of (pA)p, while depletion of TIA-1 by siRNA increased (pA)p read-through. Finally, artificially tethering the cap binding complex (CBC) components CBP80 and CBP20 upstream of the intron donor increased inhibition of polyadenylation at (pA)p. Our results suggest that interaction with the CBC strengthens U1 snRNP binding to the downstream intron donor in a manner inversely proportional to the size of the 5′ exon, thus governing the competition between intron splicing and polyadenylation at (pA)p. This competition must be optimized to program both the levels of polyadenylation of P7- and P19-generated RNA at (pA)p required to produce proper levels of the essential Rep proteins and the splicing of P41-generated RNAs to produce the proper ratio of capsid proteins during AAV5 infection.
Alternative polyadenylation of pre-mRNAs is an important mechanism that contributes to the diversity of expression of the human genome. Recent reports have demonstrated that more than half of the genes in the human genome have multiple polyadenylation sites [poly(A) sites] (56, 60). A functional poly(A) signal is composed of the highly conserved CPSF-binding hexanucleotide AAUAAA, found 10 to 30 nucleotides (nt) upstream of the cleavage site; a less highly conserved U/GU-rich element located downstream of the cleavage site which interacts with CstF; the pre-mRNA cleavage site itself engaged by CFI and CFII, which becomes the point of poly(A) addition; and in some cases an enhancing upstream region, whose nature is less well defined (see reference 61 for a review). The nucleotide composition of the poly(A) site, functioning as a cis-acting signal and binding site, governs the efficiency of polyadenylation and thus is one factor that governs alternative polyadenylation.
The U1 small nuclear ribonucleoprotein particle (snRNP), which facilitates splicing of pre-mRNAs via interaction at the 5′ splice site, has also been reported to be involved in regulation of polyadenylation of some viral and cellular genes, in some cases enhancing and in others inhibiting efficient poly(A) site usage (4, 5, 7, 16-19, 58). U1 snRNP contains a single small RNA and 10 protein subunits (36). The 164-nt-long RNA subunit of U1 snRNA has a trimethyl-guanosine cap at its 5′ end and folds into a secondary structure consisting of four stem-loop motifs. The single-stranded 5′ end of U1 snRNA is complementary to the conserved sequence at the 5′ splice site and plays an important role in recognition of the 5′ splice site (34, 52, 53, 64). Seven Sm proteins (B or B′, D1, D2, D3, E, F, and G) assemble to form a heptameric ring known as the core domain around the U1 snRNA Sm site (24, 49). U1 snRNP also contains three additional U1-specific proteins: U1A, U1-70K, and U1C. U1-70K and U1A can bind to stem-loop I and stem-loop II in U1 snRNA, respectively (39, 48, 50). Unlike U1A and U1-70K, however, U1C alone does not bind U1 snRNA. U1C binding requires both the U1 snRNP core domain and an N-terminal fragment of U1-70K (residues 1 to 97) (39), and it has been suggested that U1-70K mediates the binding of U1C to the U1 snRNP complex.
It has been postulated that 5′-terminal exons are recognized via interactions between factors recognizing the 5′-terminal cap structure and downstream 5′ splice site (6). In the nucleus, the cap structure interacts with the nuclear cap binding complex (CBC), a heterodimeric complex that consists of small (CBP20) and large (CBP80) protein subunits. The CBC has been shown to promote early steps in splicing, i.e., U1 snRNP recruitment and subsequent commitment complex formation (1, 8, 14, 15, 27, 28). CBP80 requires CBP20 to interact directly with the cap structure (m7GpppG) (32), and the N-terminal portion of CBP80 constitutes part of the binding groove for the conformationally sensitive loop of CBP20. The CBC has been shown to facilitate U1 snRNP recruitment to the cap-proximal donor site (28). This evidence, and the demonstration that CBC binds directly to the U1 snRNP in yeast (14), led to a model in which the CBC stimulates splicing of 5′-terminal exons by promoting U1 snRNP binding to the 5′ splice donor site (26). However, in the mammalian system, there is yet no evidence of a direct interaction between CBC and U1 snRNP (28).
U1 snRNP, specifically the U1A protein, has been shown to play an important role in governing the coupling of splicing and polyadenylation in some systems (7, 18, 20, 42, 43). The U1A protein has been shown to interact with the 160-kDa subunit of CPSF and activate polyadenylation (31). Polyadenylation of simian virus 40 late RNA is activated via interaction between U1A and CPSF following binding of U1 snRNP to a sequence element upstream of the target poly(A) site (9, 21, 30, 31, 59). Conversely, U1 snRNP has also been shown to inhibit polyadenylation within the upstream human immunodeficiency virus long terminal repeat via binding to the 5′ splice site downstream of the poly(A) site (5). U1 snRNP binding to a 5′ splice site also results in inhibition of polyadenylation of bovine papillomavirus (BPV) late pre-mRNA via a direct interaction with poly(A) polymerase (17, 19), and this represents a key regulation step for BPV late gene expression which is limited to terminally differentiated keratinocytes in infected epithelium.
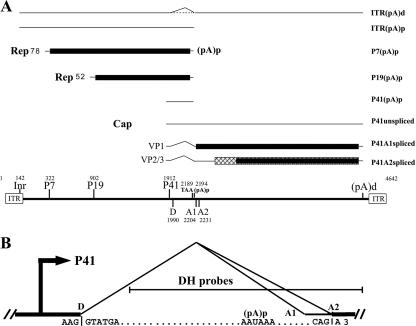
Adeno-associated virus type 5 (AAV5), a member of the Dependovirus genus of the subfamily Parvovirinae (11, 37), is unique among AAV serotypes in that it uses a polyadenylation site in the center of the genome (45), within the sole functional intron (Fig. 1A) that has a nonconsensus donor site (AAG/GTATGA, compared to the consensus sequence MAG/GURAGU, in which M is A/C and R is A/G) (Fig. 1B). The great majority of transcripts generated from the upstream P7 and P19 promoters are polyadenylated at a site in the central intron [(pA)p]; however, most of the viral transcripts generated by the proximal P41 promoter read through (pA)p and are polyadenylated at the distal polyadenylation site at the 3′ end of the genome [(pA)d] and subsequently spliced (44, 45). Polyadenylation at (pA)p increases as the distance between the RNA initiation site and the intron and (pA)p site is increased (47). The steady-state level of RNAs polyadenylated at (pA)p is independent of the promoter used and of the nature of the intervening 5′ exon sequence but is dependent upon competition with splicing, inhibition by U1 snRNP binding to the intron donor, and the intrinsic efficiency of the cleavage/polyadenylation reaction (47). Each of these determinants shows a marked dependence on the distance between the RNA initiation site and the intron and (pA)p (47). Unlike for other reported systems, inhibition of (pA)p by U1 snRNP binding to the intron donor is decreased as the distance between the donor and (pA)p is increased (47). Because splicing and polyadenylation of P41-generated RNA compete for the same pool of precursor pre-mRNA molecules (47), the result of this competition must be optimized to program both the proper levels of polyadenylation of P7- and P19-generated RNAs at (pA)p and the splicing of P41-generated RNAs which are required during AAV5 infection.
FIG. 1.
Transcription map of AAV5. (A) The AAV5 genome is depicted at the bottom of the figure and includes the locations of the viral promoters (Inr, P7, P19, and P41), the small intron donor (D), acceptors (A1 and A2), the termination site for the Rep proteins, the internal polyadenylation site (pA)p, and the inverted terminal repeats (ITRs). The major transcript classes and the proteins that they encode are shown above. The different ORFs that are used are shown in different shading patterns. It is not clear whether a portion of Inr-initiated RNA is spliced or not; this is indicated by a dashed line. (B) The AAV5 intron is shown in more detail, with sequences of the donor, poly(A) signal, and acceptor. The DH probes used for subsequent RPAs are also shown.
In this report, we have continued our analysis of how U1 snRNP governs the choice between splicing and polyadenylation of AAV5 RNA. We have demonstrated that U1-70K is the main component of U1 snRNP that mediates inhibition of polyadenylation at (pA)p. Additionally, we have shown that interaction with the CBC governs the competition between intron splicing and polyadenylation at (pA)p, likely by strengthening U1 snRNP binding to the downstream intron donor in a manner inversely proportional to the size of the 5′ exon.
MATERIALS AND METHODS
Cells, viruses, and siRNA transfection.
293 human kidney cells (ATCC CRL-1573) and HeLa (human cervix) cells (ATCC CCL-2) were maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum at 37°C in 5% CO2. Human adenovirus type 5 (Ad5) was propagated and assayed in 293 cells as previously reported (46). Small interfering RNAs (siRNAs) used to target the TIA-1 gene were either AACACAACAAATTGGCCAGTA (validated TIA-1 siRNA; QIAGEN) or a pool of three siRNAs (CCAAGCAACUGUACUGUAUTT, CGGUUCAAUUCCCAUGAAATT, and CUCAGAUCCUUGUUUCAAATT) from Santa Cruz Biotechnology (Santa Cruz, CA). A random siRNA (CGUGAUUGCGAGACUCUGATT) (sc-44231; Santa Cruz Biotechnology) showing no significant homology to any known protein sequence (as analyzed using BLAST) was used as a control (35). siRNAs were transfected into 293 and HeLa cells at a final concentration of 50 nM with HiPerfect transfection reagent (QIAGEN), following the reverse transfection method as described by the supplier.
Plasmid constructs. (i) U1 snRNA mutants.
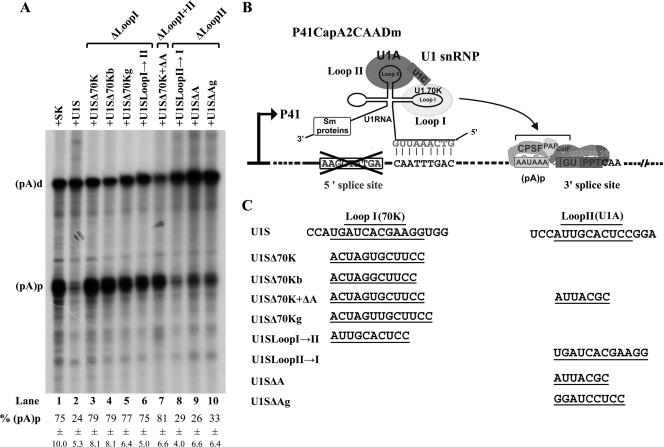
Mutations in stem-loop I and/or stem-loop II, diagramed in Fig. 3C, were introduced into the U1S clone (47) to generate stem-loop I mutants U1SΔ70K, U1SΔ70Kb, U1SΔ70Kg, and U1SLoopI→II; stem-loop II mutants U1SΔA, U1SΔAg, and U1SLoopII→I; and stem-loop I and II mutant U1SΔ70K+ΔA.
FIG. 3.
Mutations in stem-loop I but not stem-loop II of U1 RNA abolished inhibition of polyadenylation of P41-generated RNAs at (pA)p. (A) RPA, using the DHA2CAA probe, of RNA isolated from 293 cells transfected with reporter plasmid P41CapA2CAADm, together with either the control plasmid SK or various U1S and U1S mutants as described in Materials and Methods and as indicated on the top of each lane. Bands representing RNAs that either read through or are polyadenylated at (pA)p are designated (pA)d or (pA)p, respectively. Quantifications of the ratios of P7-generated RNA at (pA)p to total P7-generated RNA are shown as percent (pA)p and are averages of the results for at least three individual experiments, with standard deviations underneath. (B) Diagram of the putative interaction of the U1S snRNA with the P41CapA2CAADm reporter plasmid. The curved arrow indicates that the U1 snRNP has an inhibitory effect on polyadenylation at (pA)p. (C) The individual mutations for stem-loop I mutants U1SΔ70K, U1SΔ70Kb, U1SΔ70Kg, and U1SLoopI→II; stem-loop II mutants U1SΔA, U1SΔAg, and U1SLoopII→I; and stem-loop I and II mutant U1SΔ70K+ΔA are indicated.
(ii) U-stretch-induced constructs.
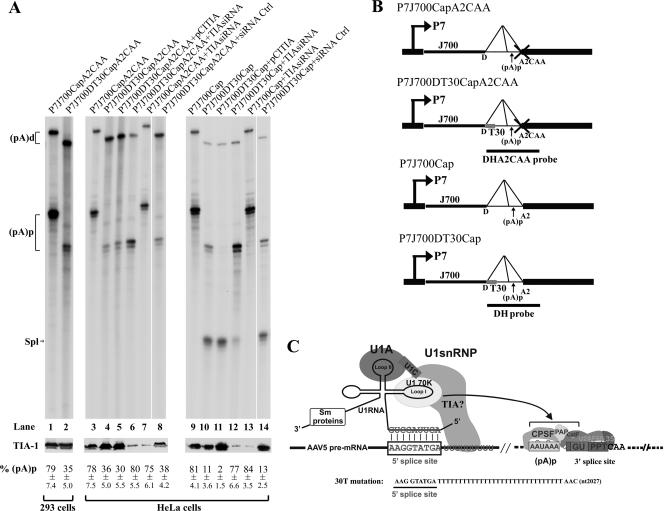
A stretch of 30 thymidines was introduced immediately downstream between the AAV5 donor site (AAG/GTA TGA) and nt 2057, which is diagramed in Fig. 4C, in plasmids P7J700CapA2CAA and P7J700Cap (in which there are approximately 700 nt between the promoter and the donor site [47]) to generate P7J700DT30CapA2CAA and P7J700DT30Cap, respectively. These mutants are diagramed in detail in Fig. 4.
FIG. 4.
Introduction of a putative TIA-1 binding site immediately downstream of the donor site inhibited polyadenylation of the P7-generated RNAs at (pA)p, and modulation of TIA-1 levels in HeLa cells affects the relative ratios of polyadenylation and splicing of P7-generated RNAs. Results are shown for RPAs, using either probe DHA2CAA or probe DH as indicated, of RNA isolated from 293 cells (lanes 1 and 2) or HeLa cells (lanes 3 to 14) transfected with either the splicing-defective plasmid P7J700CapA2CAA (lanes 1, 3, and 7; diagramed in panel B) or P7J700DT30CapA2CAA (lanes 2, 4 to 6, and 8; diagramed in panel B) or the splicing-competent plasmid P7J700Cap (lanes 9 and 13; diagramed in panel B) or P7J700DT30Cap (lanes 10 to 12 and 14; diagramed in panel B), either together with the TIA-1 expression plasmid pCITIA-1 (lanes 5 and 11) or pretransfected with TIA-1 siRNA (lanes 6, 7, 12, and 13) or a control siRNA (lanes 8 and 14). HeLa cells were coinfected with human Ad5 at a multiplicity of infection sufficient to achieve full activity of the AAV5 P7 promoter. Bands representing RNAs that either read through or are polyadenylated at (pA)p are designated (pA)d or (pA)p, respectively. Bands representing RNAs that are spliced are designated Spl. Quantifications of the ratios of P7-generated RNA at (pA)p to total P7-generated RNA are shown as percent (pA)p and are averages of the results for at least three individual experiments, with standard deviations underneath. Shown are expression levels of the TIA-1 protein, as monitored by immunoblotting using a polyclonal anti-human TIA-1 antibody as shown below each RNase protection gel. The predicted interactions between the U1 snRNP and the putative TIA-1 binding site and the mutation introduced into P7J700CapDT30CAA are illustrated in panel C. The curved arrow indicates that the U1 snRNP has an inhibitory effect on polyadenylation at (pA)p. The (pA)p and (pA)d bands protected by RNAs generated from all the plasmids that have the T30 mutation downstream of the donor site were smaller than those protected by RNA generated from the wild-type plasmids. Because the DHA2CAA probe starts a few nucleotides after the donor site, the 30 T mutations present after the donor site are predicted to make both the (pA)p and the (pA)d bands ∼20 nt smaller.
(iii) MS2BS constructs.
A prokaryotic sequence of 160 nt from PBR322 was inserted into the Mfe I site (nt 1976) of P7J700CapA2CAA (47) to generate the intermediate plasmid P7J700MfeJ160CapCAA. Three repeated MS2 binding sites (MS2BS) were amplified from plasmid dsxΔE-Δ5′SS+3xMS2 (a gift from Ben Blencowe, University of Toronto) (33) and subsequently inserted into the MfeI site of the aforementioned plasmid P7J700MfeJ160CapA2CAA, in either a sense or a reverse orientation, to create P7J700MS2BSJ160CapA2pCAA or P7J700MS2BS(−)J160CapA2CAA, respectively.
(iv) cDNA clones for protein expression in vivo.
pcDNA-HAMS2-SRm160 was first created by introducing a hemagglutinin (HA) tag at the amino terminus encoded by the MS2-coding sequence in the MS2 expression vector pcDNA3-fMS2-SRm160, also a gift from Ben Blencowe (54). The MS2-coding sequence in these constructs was humanized to improve expression. Replacement of the SRm160 open reading frame (ORF) with the CBP80 ORF or the CBP20 ORF was next performed to generate pMS2CBP20 or pMS2CBP80, respectively. All other cDNA expression clones were constructed by insertion of the HA tag into the amino terminus of the ORF encoding CBP20, CBP80, U1A, U1C, and U1-70K, in pCI (Promega). The U1-70K ORF was a gift from Robert W. Hoffman, University of Miami; the TIA-1 ORF was a gift from Juan Valcárcel at the Centre de Regulació Genòmica, Barcelona, Spain; and all other human ORF clones were purchased from OpenSystems (Huntsville, AL).
Plasmid transfection.
All plasmids were transfected using Lipofectamine as previously described (46). For experiments with siRNA, plasmids were transfected 1 day after siRNA transfection. To achieve normal AAV5 expression levels in HeLa cells, Ad5 was coinfected at a multiplicity of infection of 5 immediately after plasmid transfection. Transfected cells were maintained at 37°C in 5% CO2 for 36 to 48 h.
RNA isolation and RPA.
Total RNA was isolated using either guanidine isothiocyanate followed by CsCl ultracentrifugation as previously described (51) or a total RNA isolation kit (Invitrogen), following protocols supplied by the company. RNase protection assays (RPAs), using homologous probes, were performed on 10 μg of total RNA as previously described (38, 51). The DH and DHA2CAA probes were generated from linearized templates by in vitro transcription (44). RNA hybridizations were done in substantial probe excess, and signals were quantified with Fuji FLA 3000 and Fuji Multi Gauge v2.3 software (FUJIFILM Medical Systems U.S.A., Inc.). Relative molar ratios of individual species of RNAs were determined after adjustment for the number of 32P-labeled uridines in each protected fragment as previously described (51).
Immunoblot analysis.
Ten percent of transfected cells were typically harvested for immunoblot analysis as previously described (29), using monoclonal antibody HA-7 (Sigma, St. Louis, MO) to detect all HA-tagged proteins and a polyclonal antibody [TIA-1(C-20); Santa Cruz Biotechnology, Inc.] to detect TIA-1 accumulation.
RESULTS
The U1 stem-loop I-binding protein U1-70K, but not the stem-loop II-binding protein U1A, plays the primary role in inhibiting polyadenylation of AAV5 pre-mRNAs at (pA)p.
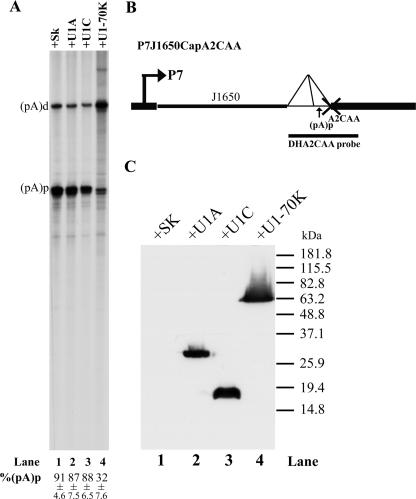
We have previously demonstrated that U1 snRNP binding to the central intron donor site plays an important role in inhibition of polyadenylation of AAV5 pre-mRNAs at (pA)p (47). We first sought to determine if one of the U1 unique protein(s) was necessary for this inhibition. To this end, U1A, U1-70K, and U1C were individually overexpressed in 293 cells together with a previously described AAV5 reporter plasmid, P7J1650CapA2CAA (diagramed in Fig. 2B). In this reporter plasmid, the distance between the promoter and the intron [with the internal (pA) site] is sufficiently large that polyadenylation of P7-generated transcripts at (pA)p is highly efficient. In addition, the 3′ splice site CAG\ has been destroyed by mutation to CAA\, which allowed us to evaluate polyadenylation at (pA)p in the absence of the competing splicing of the small intron (44). (This applies to all constructs containing the CAA\ mutation used in this study and described below.)
FIG. 2.
Overexpression of U1-70K decreased polyadenylation of AAV5 RNA at (pA)p. (A) RPA, using the DHA2CAA probe, of RNA isolated from 293 cells transfected with P7J1650CapA2CAA, together with either the control plasmid SK (pBluescript, Stratagene) (lane 1) or plasmids expressing the U1A, U1C, or U1-70K proteins (pCIHAU1A [lane 2], pCIHAU1C [lane 3], or pCIHAU1-70K [lane 4], respectively). Bands representing RNAs that either read through or are polyadenylated at (pA)p are designated (pA)d or (pA)p, respectively. Quantifications of the ratios of P7-generated RNA at (pA)p to total P7-generated RNA [(pA)p+(pA)d] are shown as percent (pA)p and are averages of the results for at least three individual experiments with standard deviations. (B) Map of plasmid P7J1650CapA2CAA and the location of the DHA2CAA probe. (C) Immunoblot analysis, using an anti-HA antibody, showing similar levels of expression of the various U1 proteins.
In the absence of ectopic expression of any of the U1snRNA-specific binding proteins, approximately 90% of the RNAs generated from P7J1650CapA2CAA were polyadenylated at (pA)p [expressed as percent (pA)p] (Fig. 2A, lane 1) as previously described (47). Overexpression of the U1-70K protein, however, reduced polyadenylation at (pA)p threefold, to approximately 32% (Fig. 2A, lane 4), while overexpression of either U1A or U1C had little effect on polyadenylation at (pA)p in these assays (Fig. 2A, lanes 2 and 3). These results, in which expression of the U1-70K protein untargeted and by itself could inhibit polyadenylation at (pA)p, suggested that perhaps this was the constituent of U1 snRNP likely to be involved in inhibition of (pA)p.
To further investigate whether the U1-70K protein was the component of U1 snRNP that inhibits (pA)p, we examined the effect on polyadenylation of a targeted U1 snRNP into which mutations in the stem-loop I and stem-loop II region of the U1 snRNA, previously shown to abolish binding of the U1-70K and U1A proteins, respectively, were introduced. Plasmid P41CapA2CAADm, which was previously used in retargeting experiments to demonstrate the requirement of U1 snRNP binding to AAV5 pre-mRNAs for inhibition of polyadenylation at (pA)p (47), was employed for these experiments. The native 5′ splice site of this reporter construct has been destroyed, allowing targeting of an ectopically expressed U1 snRNP engineered to base pair to a site 22 nt downstream of the authentic 5′ donor (U1S). The retargeted U1S snRNP has been shown to inhibit polyadenylation of the P41-generated mRNAs at (pA)p at levels comparable to that of wild-type U1 snRNP binding to the wild-type donor (47) (Fig. 3A, compare lane 2 with lane 1). U1S was then further mutagenized to destroy stem-loop I and stem-loop II, either individually or simultaneously, using mutations that were previously used by others to probe binding of U1 snRNA-specific proteins and were reported to leave the overall structure and binding of U1 snRNA intact (3, 19). All stem-loop I mutants tested (U1Δ70K, U1SΔ70Kb, U1SΔ70Kg, and U1SloopI→II) inhibited (pA)p significantly less well than wild-type U1S RNA (Fig. 3A, lanes 3 to 6, compare to lane 2), as did the combined stem-loop I and stem-loop II mutant U1SΔ70K+ΔA (Fig. 3A, lane 7). Conversely, all loop II mutants tested (U1SLoop II→I, U1SΔA, and U1SΔAg) retained their abilities to inhibit (pA)p to levels similar to that for the wild-type U1S RNA (Fig. 3A, lanes 8 to 10, compare to lane 2). The stem-loop 1 mutations (U1SΔ70K [3], U1SΔ70Kb [3], U1SΔ70Kg [19], and U1SLoopI→II [19]) have been shown to abolish binding of U1-70K but not U1A or U1C to U1 snRNA, while the stem-loop II mutants (U1SLoopII→I [19], U1SΔA [3], and U1SΔAg [19]) have been shown to abolish binding of U1A but not U1-70K or U1C to U1 RNA. These results, together with results derived from the U1-70K overexpression described above, suggested that the U1-70K protein was the key component of U1 snRNP responsible for inhibition of polyadenylation of AAV5 pre-mRNAs at (pA)p.
U1 snRNP binding to the intervening donor site governs inhibition of (pA)p.
As noted above, the AAV5 intron donor has a less-than-fully consensus sequence (AAG/GTATGA, compared to the consensus sequence AAG/GTAAGT). We have previously shown that making this sequence even less consensus (in the parent construct P41CapA2CAA, described above) reduced inhibition of polyadenylation of (pA)p following transfection, presumably by reducing the ability of U1 snRNP to bind the donor site (47). Surprisingly, making this 5′ splice site sequence consensus in a parent construct in which the promoter was at a distance from which RNAs were efficiently polyadenylated did not result in the reestablishment of inhibition (data not shown). Because these mutations also failed to increase levels of splicing (data not shown), we concluded that this simple change could not functionally enhance U1 snRNP interaction at this site. Thus, we searched for additional ways to increase the interaction of U1 snRNP with the 5′ donor.
TIA-1 is an apoptosis-induced protein (57) which plays a role in RNA processing. One of the functions of TIA-1 has been shown to be the enhancement, following interaction with an intronic splicing enhancer (characterized as a U-stretch sequence), of U1 snRNP binding to an adjacent 5′ splice donor site (23) via protein-protein interactions involving the glutamine-rich domain of TIA-1 and the U1-specific protein U1C (10, 14, 27). Thus, we evaluated polyadenylation at (pA)p following attempts to modulate U1 snRNP binding to the AAV5 donor by enriching the local concentration of the endogenous TIA-1 protein by introducing a stretch of 30 uridines immediately downstream of the donor as diagramed in Fig. 4C. In addition, TIA-1 levels were also varied by either overexpressing TIA-1 or reducing its steady-state level via siRNA.
For these experiments, we started with plasmid P7J700CapA2CAA, a splicing-deficient construct which was used previously to preliminarily characterize the role that the size of the 5′ exon played in polyadenylation at (pA)p (Fig. 4A) (47). In this construct, the promoter is located at an intermediate distance from the intron and (pA)p site (approximately 700 nt), such that nearly 80% of the P7-generated pre-mRNAs were polyadenylated at (pA)p (47) (Fig. 4A, lane 1). Introducing 30 uridines immediately downstream of the donor site (P7J700DT30CapA2CAA) significantly reduced polyadenylation of P7-generated RNAs at (pA)p, from approximately 79% to 35% (Fig. 4A, compare lanes 1 and 2) in 293 cells, suggesting that enriching U-rich binding proteins (likely TIA-1) adjacent to the donor could promote inhibition of polyadenylation at (pA)p of these P7-generated RNAs.
We found that HeLa cells were more amenable to siRNA manipulation of TIA-1 levels than were 293 cells. Transfection of both parent P7J700CapA2CAA and the uridine-containing P7J700DT30CapA2CAA into these cells generated RNAs which were polyadenylated at (pA)p at levels similar to that seen in 293 cells (78% and 36%, respectively) (Fig. 4A, lanes 3 and 4). When the levels of TIA-1 were reduced by specific siRNA treatment, inhibition of polyadenylation at (pA)p of RNAs generated by P7J700DT30A2CapCAA was reduced such that polyadenylation at (pA)p increased from 36% to 80% (Fig. 4A, compare lane 4 to lane 6; the steady-state levels of TIA-1 as assayed by immunoblot analysis are shown underneath). Inhibition of (pA)p polyadenylation of RNAs generated by the non-uridine-stretch-containing P7J700CapA2CAA in cells treated with the specific TIA siRNA remained high (Fig. 4A, lane 7), and a random siRNA was unable to relieve inhibition of (pA)p polyadenylation of RNA generated by P7J700DTCapA2CAA (Fig. 4A, lane 8). These results suggested that for RNAs generated by the P7J700DT30CapA2CAA plasmid, the targeted TIA-1 protein enhanced the inhibition of polyadenylation at (pA)p, presumably via enhancement of U1 binding. These results also precluded any effect in cis due merely to the extended U-rich region which had been introduced. Surprisingly, overexpression of TIA-1 did not significantly increase inhibition of (pA)p polyadenylation of P7J700DT30CapA2CAA RNA (Fig. 4A, lane 5), perhaps because the endogenous levels were already saturating for the inhibitory effect.
Targeting of TIA-1 could also bring about an increase in splicing of RNAs generated by otherwise identical plasmids containing a splicing-competent intron in which the 3′ splice site remained wild type (Fig. 4B). RNAs generated by P7J700DT30Cap, which included the potential TIA-1 binding site (U stretch), were spliced to a significantly greater level, presumably mediated by TIA-1 recruitment of U1, than those generated by the parent, P7J700Cap (Fig. 4, compare lanes 9 and 10). Reduction of TIA-1 levels by siRNA reduced the accumulated levels of spliced P7 RNAs generated from P7J700DT30Cap (Fig. 4, lane 12), while overexpression of TIA-1, in this case, enhanced the levels of spliced RNAs generated by this plasmid close to their predicted maximum (Fig. 4, lane 11). The specific TIA siRNA had little effect on processing of RNAs generated by the non-uridine stretch-containing P7J700Cap plasmid (Fig. 4, lane 13), and a random siRNA had little effect on the processing fates of RNA generated by P7J700DTCap (Fig. 4, lane 14). Taken together, these results suggested that when TIA-1 was involved in promoting U1 snRNP binding to their donor site, this interaction mediated the choice between competing polyadenylation at (pA)p (inhibited by U1 snRNP binding) and read-through and subsequent splicing (enhanced by U1 snRNP binding).
The CBC is involved in determining the strength of U1 snRNP binding.
As mentioned above (cf. reference 47), the distance between the AAV5 RNA initiation site, which binds to the CBC, and the donor site, which binds to the U1 snRNP, governs the relative efficiencies of polyadenylation at (pA)p versus read-through and subsequent splicing (47). The CBC has been shown to facilitate U1 snRNP recruitment to the cap-proximal donor site (28), and the CBC has been shown to bind directly to U1 snRNP in yeast (14), leading to a model in which the CBC stimulates splicing of 5′-terminal exons by promoting U1 snRNP binding to the 5′ splice donor site (26). Thus, it seemed feasible that perhaps the CBC might influence the choice between polyadenylation and read-through with subsequent splicing of AAV5 pre-mRNAs by stabilizing binding of U1 snRNP to the donor of this 5′ exon in a distance-dependent manner.
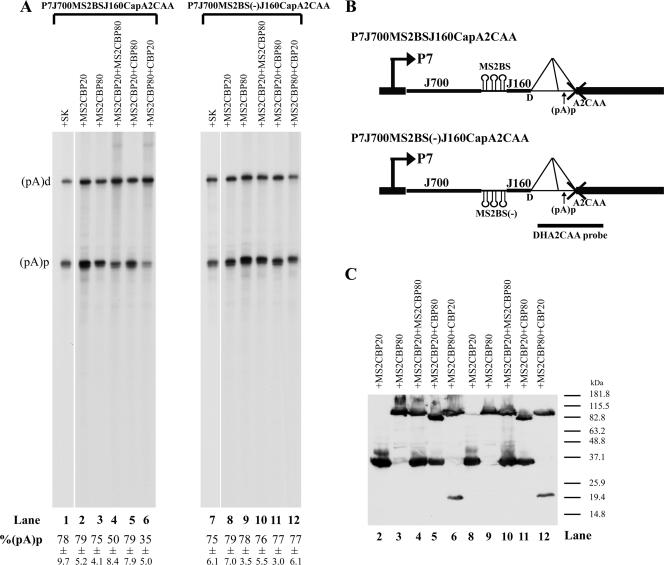
To evaluate the role of the CBC in the inhibition of polyadenylation at (pA)p, we used derivatives of the P7J700CapA2CAA plasmid, described above, in which the promoter is approximately 700 nt upstream of the donor. As shown above (Fig. 4, lane 1), approximately 80% of the RNA generated by the promoter at this distance was polyadenylated at (pA)p (Fig. 5, lane 1). If this was because the CBC was too far away to stabilize (pA)p-inhibiting U1 snRNP binding, we would predict that we could bypass this distance-dependent lack of stabilization and regain inhibition of (pA)p by targeting CBC components closer to the donor, using the MS2-phage coat protein system. Three MS2BS were introduced into P7J700CapA2CCA in either a sense (binding, 3XMS2BS) or an antisense [nonbinding, 3XMS2BS(−)] orientation, separated from the donor by a 160-nt spacer (P7J700MS2BSJ160CapA2CAA or P7J700MS2BS(−)J160CapA2CAA, respectively) (Fig. 5B). RNAs generated by these constructs transfected alone were polyadenylated at (pA)p to levels similar to that for the parent construct (Fig. 5, lanes 1 and 7). However, when the reporter construct containing the MS2BS in the sense direction (Fig. 5A, left) but not the antisense direction (Fig. 5A, right) was cotransfected together with both CBP20 fused to the MS2 coat protein binding peptide (MS2CBP20) and CBP80 fused to the MS2 coat protein binding peptide (MS2CBP80) (Fig. 5A, lane 4) or MS2CBP80 and native CBP20 (Fig. 5A, lane 6) but not MS2CBP20 or MS2CBC80 alone (Fig. 5A, lane 2 or 3, respectively), polyadenylation of the P7-generated RNAs at (pA)p was significantly reduced (nearly 40%). This suggested that the CBC formed by CBP20 and CBP80 could inhibit polyadenylation of (pA)p when targeted near the intron donor. Interestingly, the CBC complex formed by MS2CBP20 and CBP80 was unable to confer inhibition of polyadenylation at (pA)p (Fig. 5A, lane 5). It is unclear why this is the case, but perhaps while targeting of untagged CBP20 can be achieved by tagged CBP80, the reverse is not true. Alternatively, it may be that fusion of the MS2 domain on CBP20 interfered with its ability to bind the cap structure. Figure 5C demonstrates that all recombinant proteins were expressed to similar levels in these experiments.
FIG. 5.
Retargeting of the CBC to a location closer to the donor reestablished inhibition of polyadenylation of P7-generated RNAs at (pA)p. (A) RPA, using the DHA2CAA probe, of RNA isolated from 293 cells transfected with reporter plasmid P7JMS2BSJ160CapCAA (left) or control plasmid P7JMSBS2(−)J160CapCAA (right) (which are both diagramed in panel B), together with either the empty vector control SK (lanes 1 and 7) or cDNA expression plasmid MS2CBP20 (lanes 2 and 8), MS2CBP80 (lanes 3 and 9), MS2CBP20 plus MS2CBP80 (lanes 4 and 10), MS2CBP20 plus CBP80 (lanes 5 and 11), or MS2CBP80 plus CBP20 (lanes 6 and 12). Bands representing RNAs that either read through or are polyadenylated at (pA)p are designated (pA)d or (pA)p, respectively. Quantifications of the ratios of P7-generated RNA at (pA)p to total P7-generated RNA are shown as percent (pA)p and are averages of the results for at least three individual experiments, with standard deviations underneath. (C) Immunoblot analysis, using an anti-HA antibody, showing similar levels of expression of the various transfected cDNA plasmids.
DISCUSSION
In previous reports, we have shown that the processing of AAV5 RNA is governed by competition between splicing and alternative polyadenylation for the available pools of AAV5 pre-mRNA (47). Inhibition of internal polyadenylation of AAV5 RNA at (pA)p, which lies within the central viral intron, is required for read-though and eventual splicing of the P41-generated, capsid-encoding RNAs and is accomplished by the U1 snRNP in a manner inversely related to the distance between the RNA initiation site and the intron and (pA)p. Here, we show, using specific mutations of U1 snRNA as well as knock-down and overexpression assays, that this inhibition is mediated by the U1 snRNP 70K protein. Additionally, targeting of TIA-1 to the intron donor enhanced inhibition of (pA)p, suggesting that this inhibition was governed by the affinity of U1 binding to the intron donor. In a transcription unit in which the 5′ exon is large, targeting of the CBC closer to the donor site led to increased inhibition of polyadenylation at (pA)p.
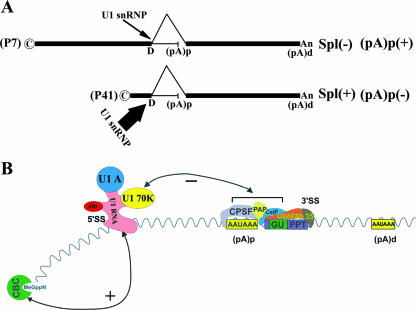
Based on previous results and the experimental results presented here, the simplest model for explaining how the competition between splicing and alternative polyadenylation in AAV5 is governed is that strong binding of U1 snRNP to the intron donor facilitates both splicing of the intron and inhibition of polyadenylation at (pA)p (portrayed in Fig. 6). That polyadenylation at (pA)p is inhibited when the promoter is closer to the intron and (pA)p (favoring read-through and subsequent splicing) and that polyadenylation is favored when the promoter is a greater distance away are consistent with a model in which U1 snRNP binding is stronger when the promoter is close and is weaker when the distance is large. The CBC bound to pre-mRNA (22, 62) has been suggested to affect both splicing and polyadenylation. We have shown here that targeted CBC can reestablish inhibition of polyadenylation at (pA)p of RNAs generated by a promoter at a distance (Fig. 5). This suggests that during expression of the wild-type virus, CBC interaction can facilitate association of U1 snRNP with the less-than-fully consensus AAV5 pre-mRNA donor site when the cap site is very close (78 nt for the capsid gene promoter P41 [Fig. 1]) but not so at a distance (1,668 and 1,088 nt for the large and small Rep gene promoters, respectively). Therefore, it seems likely that that interaction with the CBC strengthens U1 snRNP binding to the downstream intron donor in a manner inversely proportional to the size of the 5′ exon and thus governs the competition between intron splicing and polyadenylation at (pA)p (Fig. 6).
FIG. 6.
Model for the distance-dependent processing of AAV5 RNA. (A) This diagram shows the processing fate of the AAV5 P7- and P41-generated RNAs. P7-generated RNAs, which have a large 5′ exon, are polyadenylated at high efficiency at the internal (pA)p site; few molecules read through and are ultimately spliced. The opposite is true for P41-generated RNAs. Potential strength of U1 snRNP interaction at the upstream donor, postulated to govern the outcome of the competition between splicing and polyadenylation, is indicated by the width of the arrow. (B) This diagram depicts how the U1 snRNP binding at the intron donor may exert its negative effect on polyadenylation at (pA)p. The U1-70K protein is required for this effect, although the specific target for this inhibition is not yet known. Also shown is the binding of splicing factor U2AF(65+35) at the overlapping 3′ splice site. Stabilization of U1 snRNP to the intron 5′ splice site, which facilitates splicing and simultaneously inhibition of (pA)p, may occur through interaction with the CBC associated with the 5′ cap site as shown and may be more effective in this regard when the RNA is initiated closer to the donor site [which promotes splicing and (pA)p inhibition, i.e., with RNA generated from the P41 promoter] than when the RNA is initiated at a distance [such that inhibition of (pA)p is lost, favoring polyadenylation at the internal site, i.e., with RNA generated from the P7 promoter].
While the introduction of a TIA-1 binding site is only an indirect way of modulating U1 snRNP, binding of TIA-1 to adjacent uridine-rich regions has been shown to promote recruitment of U1 to a class of weak 5′ splice sites and is important for governing correct processing of RNAs generated by the Fas gene (12) and the CGPR gene (63) and for activating the 5′ splice sites of TIAR alternative exons (25). Binding of TIA-1 in the vicinity of a 5′ splice site has been shown to stabilize U1 snRNP recruitment via direct interaction with U1C (13). Thus, increased inhibition of polyadenylation at (pA)p resulting from the introduction of a uridine stretch downstream of the AAV5 intron donor (Fig. 4) was most likely due to TIA-1 stabilization of U1 snRNP to that site. This presumption was consistent with the observation that splicing of AAV5 RNAs, generated from plasmids containing the uridine stretch in a functional intron, was also significantly enhanced (Fig. 4).
U1 snRNP has been shown to inhibit polyadenylation of the human immunodeficiency virus poly(A) site in the 5′ long terminal repeat (4, 5, 58) and the distal poly(A) used for BPV late gene RNAs (16, 17, 19). In both cases, inhibition has been attributed to action of the U1-70K protein. U1 snRNP inhibition of AAV5 RNA polyadenylation, however, seems to be quite different from these examples. First, unlike for other reported systems, inhibition of (pA)p by U1 snRNP binding to the intron donor is decreased as the distance between the donor and (pA)p is increased. Second, in contrast to the two other examples described above, the AAV5 (pA)p signal lies within a functional intron, and its use is governed by competition between splicing and polyadenylation. The downstream element required for polyadenylation at (pA)p overlaps with the intron 3′ splice site. The cis-acting signals within the intron 3′ splice site that govern polyadenylation and splicing of AAV5 RNAs must be optimized to program both (i) the levels of polyadenylation of P7- and P19-generated RNA at (pA)p required to generate the proper levels of the essential Rep proteins and (ii) the splicing of P41-generated RNAs to generate the proper ratio of capsid proteins during AAV5 infection. It is interesting to note that although usage of (pA)p must be regulated for optimal AAV5 expression, both core elements of this signal, the CPSF- and CstF-binding sites, are highly consensus and have optimal placements relative to each other.
A similar architecture governs polyadenylation of the immunoglobulin μ gene (2): the μs poly(A) signal, utilized during μ pre-mRNA alternative processing, is in direct competition with splicing that generates the Cμ4-M1 RNA (40, 41, 55), and this competition is modulated in a tissue-dependent manner. The mechanism that controls the outcome of competition in this system has not yet been fully elucidated.
There are clearly many factors that control 5′ exon definition (6), but a result of this process needs to be stabilization of U1 snRNP to the 5′ exon donor. In general, most mammalian 5′ exons are smaller than 500 nt. However, AAV5 generates pre-mRNAs with overlapping 5′ exons of quite different sizes. Competition between splicing and polyadenylation of these various RNAs is controlled by the size of the 5′ exon, most likely by stabilization of U1snRNP by the CBC in a distance-dependent manner. A similar mechanism likely plays an important role in the definition of 5′ exons in general.
Acknowledgments
We thank Ben Blencowe, Juan Valcárcel, and Rob Hoffman for reagents and Lisa Burger for excellent technical assistance.
This work was supported by PHS grants AI56310 and AI46458 from NIAID to D.J.P.
Footnotes
Published ahead of print on 16 May 2007.
REFERENCES
- 1.Abovich, N., X. C. Liao, and M. Rosbash. 1994. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 8:843-854. [DOI] [PubMed] [Google Scholar]
- 2.Alt, F. W., A. L. Bothwell, M. Knapp, E. Siden, E. Mather, M. Koshland, and D. Baltimore. 1980. Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3′ ends. Cell 20:293-301. [DOI] [PubMed] [Google Scholar]
- 3.Ashe, M. P., A. Furger, and N. J. Proudfoot. 2000. Stem-loop 1 of the U1 snRNP plays a critical role in the suppression of HIV-1 polyadenylation. RNA 6:170-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashe, M. P., P. Griffin, W. James, and N. J. Proudfoot. 1995. Poly(A) site selection in the HIV-1 provirus: inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev. 9:3008-3025. [DOI] [PubMed] [Google Scholar]
- 5.Ashe, M. P., L. H. Pearson, and N. J. Proudfoot. 1997. The HIV-1 5′ LTR poly(A) site is inactivated by U1 snRNP interaction with the downstream major splice donor site. EMBO J. 16:5752-5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berget, S. M. 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 270:2411-2414. [DOI] [PubMed] [Google Scholar]
- 7.Boelens, W. C., E. J. Jansen, W. J. van Venrooij, R. Stripecke, I. W. Mattaj, and S. I. Gunderson. 1993. The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell 72:881-892. [DOI] [PubMed] [Google Scholar]
- 8.Colot, H. V., F. Stutz, and M. Rosbash. 1996. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev. 10:1699-1708. [DOI] [PubMed] [Google Scholar]
- 9.Cooke, C., H. Hans, and J. C. Alwine. 1999. Utilization of splicing elements and polyadenylation signal elements in the coupling of polyadenylation and last-intron removal. Mol. Cell. Biol. 19:4971-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Gatto-Konczak, F., C. F. Bourgeois, G. C. Le, L. Kister, M. C. Gesnel, J. Stevenin, and R. Breathnach. 2000. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 20:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fauquet, C. M., M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball. 2005. Virus taxonomy, VIIIth report of the ICTV. Elsevier/Academic Press, London, England.
- 12.Forch, P., O. Puig, N. Kedersha, C. Martinez, S. Granneman, B. Seraphin, P. Anderson, and J. Valcarcel. 2000. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell 6:1089-1098. [DOI] [PubMed] [Google Scholar]
- 13.Forch, P., O. Puig, C. Martinez, B. Seraphin, and J. Valcarcel. 2002. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 21:6882-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fortes, P., D. Bilbao-Cortes, M. Fornerod, G. Rigaut, W. Raymond, B. Seraphin, and I. W. Mattaj. 1999. Luc7p, a novel yeast U1 snRNP protein with a role in 5′ splice site recognition. Genes Dev. 13:2425-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fortes, P., J. Kufel, M. Fornerod, M. Polycarpou-Schwarz, D. Lafontaine, D. Tollervey, and I. W. Mattaj. 1999. Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol. Cell. Biol. 19:6543-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furth, P. A., and C. C. Baker. 1991. An element in the bovine papillomavirus late 3′ untranslated region reduces polyadenylated cytoplasmic RNA levels. J. Virol. 65:5806-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furth, P. A., W. T. Choe, J. H. Rex, J. C. Byrne, and C. C. Baker. 1994. Sequences homologous to 5′ splice sites are required for the inhibitory activity of papillomavirus late 3′ untranslated regions. Mol. Cell. Biol. 14:5278-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunderson, S. I., K. Beyer, G. Martin, W. Keller, W. C. Boelens, and L. W. Mattaj. 1994. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell 76:531-541. [DOI] [PubMed] [Google Scholar]
- 19.Gunderson, S. I., M. Polycarpou-Schwarz, and I. W. Mattaj. 1998. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol. Cell 1:255-264. [DOI] [PubMed] [Google Scholar]
- 20.Gunderson, S. I., S. Vagner, M. Polycarpou-Schwarz, and I. W. Mattaj. 1997. Involvement of the carboxyl terminus of vertebrate poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes Dev. 11:761-773. [DOI] [PubMed] [Google Scholar]
- 21.Hans, H., and J. C. Alwine. 2000. Functionally significant secondary structure of the simian virus 40 late polyadenylation signal. Mol. Cell. Biol. 20:2926-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Izaurralde, E., J. Lewis, C. McGuigan, M. Jankowska, E. Darzynkiewicz, and I. W. Mattaj. 1994. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78:657-668. [DOI] [PubMed] [Google Scholar]
- 23.Izquierdo, J. M., N. Majos, S. Bonnal, C. Martinez, R. Castelo, R. Guigo, D. Bilbao, and J. Valcarcel. 2005. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol. Cell 19:475-484. [DOI] [PubMed] [Google Scholar]
- 24.Kambach, C., S. Walke, R. Young, J. M. Avis, E. de la Fortelle, V. A. Raker, R. Luhrmann, J. Li, and K. Nagai. 1999. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96:375-387. [DOI] [PubMed] [Google Scholar]
- 25.Le, G. C., F. Lejeune, D. Galiana, L. Kister, R. Breathnach, J. Stevenin, and F. Del Gatto-Konczak. 2001. TIA-1 and TIAR activate splicing of alternative exons with weak 5′ splice sites followed by a U-rich stretch on their own pre-mRNAs. J. Biol. Chem. 276:40638-40646. [DOI] [PubMed] [Google Scholar]
- 26.Le, H. H., A. Nott, and M. J. Moore. 2003. How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 28:215-220. [DOI] [PubMed] [Google Scholar]
- 27.Lewis, J. D., D. Gorlich, and I. W. Mattaj. 1996. A yeast cap binding protein complex (yCBC) acts at an early step in pre-mRNA splicing. Nucleic Acids Res. 24:3332-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis, J. D., E. Izaurralde, A. Jarmolowski, C. McGuigan, and I. W. Mattaj. 1996. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 10:1683-1698. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Z., J. Qiu, F. Cheng, Y. Chu, Y. Yoto, M. G. O'Sullivan, K. E. Brown, and D. J. Pintel. 2004. Comparison of the transcription profile of simian parvovirus with that of the human erythrovirus B19 reveals a number of unique features. J. Virol. 78:12929-12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lutz, C. S., and J. C. Alwine. 1994. Direct interaction of the U1 snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes Dev. 8:576-586. [DOI] [PubMed] [Google Scholar]
- 31.Lutz, C. S., K. G. Murthy, N. Schek, J. P. O'Connor, J. L. Manley, and J. C. Alwine. 1996. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 10:325-337. [DOI] [PubMed] [Google Scholar]
- 32.Mazza, C., A. Segref, I. W. Mattaj, and S. Cusack. 2002. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 21:5548-5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCracken, S., M. Lambermon, and B. J. Blencowe. 2002. SRm160 splicing coactivator promotes transcript 3′-end cleavage. Mol. Cell. Biol. 22:148-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michaud, S., and R. Reed. 1991. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 5:2534-2546. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell, K. J., T. Tsuboi, and G. A. Rutter. 2004. Role for plasma membrane-related Ca2+-ATPase-1 (ATP2C1) in pancreatic beta-cell Ca2+ homeostasis revealed by RNA silencing. Diabetes 53:393-400. [DOI] [PubMed] [Google Scholar]
- 36.Muto, Y., K. D. Pomeranz, C. Oubridge, H. Hernandez, C. V. Robinson, D. Neuhaus, and K. Nagai. 2004. The structure and biochemical properties of the human spliceosomal protein U1C. J. Mol. Biol. 341:185-198. [DOI] [PubMed] [Google Scholar]
- 37.Muzyczka, N., and K. I. Berns. 2001. Parvoviridae: the viruses and their replication, p. 2327-2359. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott-Raven Publishers, Philadephia, PA.
- 38.Naeger, L. K., R. V. Schoborg, Q. Zhao, G. E. Tullis, and D. J. Pintel. 1992. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 6:1107-1119. [DOI] [PubMed] [Google Scholar]
- 39.Nelissen, R. L., C. L. Will, W. J. van Venrooij, and R. Luhrmann. 1994. The association of the U1-specific 70K and C proteins with U1 snRNPs is mediated in part by common U snRNP proteins. EMBO J. 13:4113-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson, M. L., E. R. Gimmi, and R. P. Perry. 1991. The developmentally regulated shift from membrane to secreted μ mRNA production is accompanied by an increase in cleavage-polyadenylation efficiency but no measurable change in splicing efficiency. Mol. Cell. Biol. 11:2324-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peterson, M. L., and R. P. Perry. 1989. The regulated production of μm and μs mRNA is dependent on the relative efficiencies of μs poly(A) site usage and the Cμ4-to-M1 splice. Mol. Cell. Biol. 9:726-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phillips, C., S. Jung, and S. I. Gunderson. 2001. Regulation of nuclear poly(A) addition controls the expression of immunoglobulin M secretory mRNA. EMBO J. 20:6443-6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phillips, C., N. Pachikara, and S. I. Gunderson. 2004. U1A inhibits cleavage at the immunoglobulin M heavy-chain secretory poly(A) site by binding between the two downstream GU-rich regions. Mol. Cell. Biol. 24:6162-6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu, J., R. Nayak, and D. J. Pintel. 2004. Alternative polyadenylation of adeno-associated virus type 5 RNA within an internal intron is governed by both a downstream element within the intron 3′ splice acceptor and an element upstream of the P41 initiation site. J. Virol. 78:83-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiu, J., R. Nayak, G. E. Tullis, and D. J. Pintel. 2002. Characterization of the transcription profile of adeno-associated virus type 5 reveals a number of unique features compared to previously characterized adeno-associated viruses. J. Virol. 76:12435-12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiu, J., and D. J. Pintel. 2002. The adeno-associated virus type 2 Rep protein regulates RNA processing via interaction with the transcription template. Mol. Cell. Biol. 22:3639-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiu, J., and D. J. Pintel. 2004. Alternative polyadenylation of adeno-associated virus type 5 RNA within an internal intron is governed by the distance between the promoter and the intron and is inhibited by U1 small nuclear RNP binding to the intervening donor. J. Biol. Chem. 279:14889-14898. [DOI] [PubMed] [Google Scholar]
- 48.Query, C. C., R. C. Bentley, and J. D. Keene. 1989. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell 57:89-101. [DOI] [PubMed] [Google Scholar]
- 49.Raker, V. A., G. Plessel, and R. Luhrmann. 1996. The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J. 15:2256-2269. [PMC free article] [PubMed] [Google Scholar]
- 50.Scherly, D., W. Boelens, N. A. Dathan, W. J. van Venrooij, and I. W. Mattaj. 1990. Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B′′ and their cognate RNAs. Nature 345:502-506. [DOI] [PubMed] [Google Scholar]
- 51.Schoborg, R. V., and D. J. Pintel. 1991. Accumulation of MVM gene products is differentially regulated by transcription initiation, RNA processing and protein stability. Virology 181:22-34. [DOI] [PubMed] [Google Scholar]
- 52.Seraphin, B., and M. Rosbash. 1989. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell 59:349-358. [DOI] [PubMed] [Google Scholar]
- 53.Siliciano, P. G., and C. Guthrie. 1988. 5′ splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 2:1258-1267. [DOI] [PubMed] [Google Scholar]
- 54.Szymczyna, B. R., J. Bowman, S. McCracken, A. Pineda-Lucena, Y. Lu, B. Cox, M. Lambermon, B. R. Graveley, C. H. Arrowsmith, and B. J. Blencowe. 2003. Structure and function of the PWI motif: a novel nucleic acid-binding domain that facilitates pre-mRNA processing. Genes Dev. 17:461-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takagaki, Y., R. L. Seipelt, M. L. Peterson, and J. L. Manley. 1996. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87:941-952. [DOI] [PubMed] [Google Scholar]
- 56.Tian, B., J. Hu, H. Zhang, and C. S. Lutz. 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 33:201-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tian, Q., M. Streuli, H. Saito, S. F. Schlossman, and P. Anderson. 1991. A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell 67:629-639. [DOI] [PubMed] [Google Scholar]
- 58.Vagner, S., U. Ruegsegger, S. I. Gunderson, W. Keller, and I. W. Mattaj. 2000. Position-dependent inhibition of the cleavage step of pre-mRNA 3′-end processing by U1 snRNP. RNA 6:178-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wassarman, K. M., and J. A. Steitz. 1993. Association with terminal exons in pre-mRNAs: a new role for the U1 snRNP? Genes Dev. 7:647-659. [DOI] [PubMed] [Google Scholar]
- 60.Yan, J., and T. G. Marr. 2005. Computational analysis of 3′-ends of ESTs shows four classes of alternative polyadenylation in human, mouse, and rat. Genome Res. 15:369-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63:405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng, Z. M., M. Tao, K. Yamanegi, S. Bodaghi, and W. Xiao. 2004. Splicing of a cap-proximal human papillomavirus 16 E6E7 intron promotes E7 expression, but can be restrained by distance of the intron from its RNA 5′ cap. J. Mol. Biol. 337:1091-1108. [DOI] [PubMed] [Google Scholar]
- 63.Zhu, H., R. A. Hasman, K. M. Young, N. L. Kedersha, and H. Lou. 2003. U1 snRNP-dependent function of TIAR in the regulation of alternative RNA processing of the human calcitonin/CGRP pre-mRNA. Mol. Cell. Biol. 23:5959-5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhuang, Y., and A. M. Weiner. 1986. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell 46:827-835. [DOI] [PubMed] [Google Scholar]