Abstract
Pegylated alpha interferon and ribavirin therapy for hepatitis C virus (HCV) genotype 1 infection fails for half of Caucasian American patients (CA) and more often for African Americans (AA). The reasons for these low response rates are unknown. HCV is highly genetically variable, but it is unknown how this variability affects response to therapy. To assess effects of viral diversity on response to therapy, the complete pretreatment genotype 1 HCV open reading frame was sequenced using samples from 94 participants in the Virahep-C study. Sequences from patients with >3.5 log declines in viral RNA levels by day 28 (marked responders) were more variable than those from patients with declines of <1.4 log (poor responders) in NS3 and NS5A for genotype 1a and in core and NS3 for genotype 1b. These correlations remained when all T-cell epitopes were excluded, indicating that these differences were not due to differential immune selection. When the sequences were compared by race of the patients, higher diversity in CA patients was found in E2 and NS2 but only for genotype 1b. Core, NS3, and NS5A can block the action of alpha interferon in vitro; hence, these genetic patterns are consistent with multiple amino acid variations independently impairing the function of HCV proteins that counteract interferon responses in humans, resulting in HCV strains with variable sensitivity to therapy. No evidence was found for novel HCV strains in the AA population, implying that AA patients may be infected with a higher proportion of the same resistant strains that are found in CA patients.
Chronic infection with hepatitis C virus (HCV) is a major cause of cirrhosis, liver disease, and hepatocellular carcinoma (reviewed in reference 37). About 3.1 million Americans are chronically infected with HCV, causing 8,000 to 10,000 deaths annually (3). Due to the slow progression of hepatitis C virus infections and the increasing prevalence of HCV in the American population, HCV-associated deaths are expected to more than triple over the next two decades, eventually exceeding those from AIDS (45).
HCV is a hepatotropic Flavivirus (reviewed in reference 40). The virion contains a lipid envelope with two envelope proteins surrounding a capsid. Within the capsid is a positive-polarity RNA genome about 9,600 nucleotides long that contains an open reading frame (ORF) that encodes a polyprotein of ∼3,000 amino acids (Fig. 1). The structural proteins include the core protein that forms the capsid and the E1 and E2 surface glycoproteins. The nonstructural proteins include P7 (ion channel), NS2 (protease), NS3 (protease and helicase), NS4A (cofactor for NS3), NS4B (putative organizer of the viral replicase complex), NS5A (implicated in viral replication and pathogenesis), and NS5B (RNA polymerase). An 11th viral protein, the alternate reading frame (ARF) protein, is encoded in the +1 frame within the core region and is of unknown function (7).
FIG. 1.
The HCV genome. The HCV genome contains a single major ORF flanked by untranslated regions (UTR). The 10 proteins encoded within the main ORF are indicated by alternate shading. The ARF protein is encoded in the +1 reading frame within the core region.
Six HCV genotypes that are less than 72% identical at the nucleotide level have been identified, and within these genotypes, subtypes with 75% to 86% nucleotide identities may occur (8, 55, 60-62). HCV replicates as a quasispecies rather than as a clonal population; hence, multiple HCV variants are observed within individual patients that differ from each other by a few percent at the nucleotide level. These variants are in competition with each other, and at any given time one or a few sequences are dominant because they are most fit for the prevailing conditions (35, 74). The quasispecies distribution can vary with time, either through adaptive or neutral evolution (60). Adaptive changes are due to emergence of fitter variants as conditions facing the virus change, and neutral changes result from replacement of sequences with others of equivalent fitness.
The currently recommended therapy for chronic HCV infection is a combination of pegylated alpha interferon (peginterferon) and ribavirin for 24 to 48 weeks. Interferon (or peginterferon) provides the primary antiviral effect and can eradicate HCV even when used alone (25, 38, 42, 50, 75). Ribavirin is ineffective by itself in eliminating viremia (6, 11, 13), but in combination with peginterferon it increases the clearance rate and decreases the risk of relapse (42, 50). Sustained viral response (SVR; undetectable HCV RNA for at least 24 weeks posttherapy) is the primary goal of therapy. SVR rates with peginterferon and ribavirin therapy differ between racial groups; most notably, African Americans (AA) respond to therapy only about half as well as Caucasian Americans (CA) and Asians (9, 27, 29, 44, 53).
The reasons that peginterferon and ribavirin therapy is effective in only about half of cases are unknown, as are the reasons for the exceptionally poor SVR rate for AA patients. Host factors such as differences in immune responses to the virus and environmental factors such as alcohol use and drug tolerance can contribute to failure of therapy (9). HCV genetic variation can have a major effect on efficacy of therapy, because patients infected with HCV genotype 1 have a much lower response rate (40% to 50%) than patients with genotype 2 or 3 (75% to 80%) (18, 21, 41, 63). However, the role of genetic variation within a genotype in response to therapy is less clear, because there is little evidence that response to therapy differs between subtypes of a given genotype (e.g., patients infected with genotype 1a and 1b both respond to therapy in 40% to 50% of cases, despite the subtypes differing by 10% to 12% at the amino acid level) (5, 9). This observation is equally consistent with the possibility that there is no contribution of the variation between 1a and 1b to response to therapy or that “sensitive” and “resistant” HCV variants are found with equal frequencies in genotypes 1a and 1b.
Therefore, to determine the role of HCV genotype 1 genetic variation between infected individuals and between CA and AA patients in responses to therapy, we conducted a comprehensive analysis of the association of HCV genetic variability in the viral ORF with outcome of therapy. This analysis was part of the Study of Viral Resistance to Antiviral Therapy of Chronic Hepatitis C (Virahep-C), a multicenter clinical study designed to assess rates of response of CA and AA patients to peginterferon and ribavirin therapy and to assess reasons for nonresponse (9).
MATERIALS AND METHODS
Virahep-C.
Virahep-C was a multicenter trial of peginterferon and ribavirin therapy administered to treatment-naïve participants chronically infected with HCV genotype 1 (9). Virahep-C enrolled 205 CA and 196 AA participants; the Institutional Review Boards of all centers approved the protocols, and all patients gave informed, written consent for therapy and for investigations of viral, immunological, and cell-signaling responses. Per protocol, all participants were treated with peginterferon alpha-2a (Pegasys; Roche Pharmaceuticals) (180 μg weekly self-administered by subcutaneous injection) and ribavirin (Copegus; Roche Pharmaceuticals) (1,000 mg/day for those who weighed <75 kg or 1,200 mg/day for those who weighed ≥75 kg; administered orally); treatment duration was 24 to 48 weeks for nonresponders and 48 weeks for responders. Serum HCV RNA levels were quantified using a COBAS AMPLICOR hepatitis C virus monitor test, version 2.0 (Roche Molecular Diagnostics). The primary outcome was the SVR. The SVR rate was higher among CA (52%) than AA (28%) patients, and this racial difference could not be explained by clinical factors associated with response such as age, gender, body weight, obesity, severity of the underlying hepatitis, pretreatment viral levels, or amount of drug taken. A full description of Virahep-C is provided in reference 9.
Sequencing.
Consensus sequences for the full HCV ORF were obtained by directly sequencing overlapping nested reverse transcriptase PCR amplicons as described previously (72). Briefly, HCV RNA was isolated from plasma and nested reverse transcriptase PCR was performed. Both strands of the amplified DNAs were sequenced using ABI dye-terminator technology; sequence depth averaged over four. Sequences were assembled using Vector NTI software, and base-calling errors were corrected following inspection of the chromatograms. Mixed-base positions due to the HCV quasispecies were resolved by identifying the predominant base at each position. The extreme 3′ end of the ORF could not be amplified in 13 of the 94 samples; hence, the C-terminal 56 amino acids of NS5B were excluded from the analyses to ensure that all sequences were represented equally.
Sequence analyses.
The genotype 1a and 1b samples were analyzed separately, because the sequence variation between the genotypes was anticipated to be larger than differences associated with the response class or race of the patient. Amino acid sequences were deduced from the nucleotide sequences, and all analyses were performed at the amino acid level.
Sequence alignments were done with ClustalW software. Positions that differed relative to the genotype 1a or 1b population consensus sequence were identified with Mutation Master software (70). The genotype 1a consensus sequence (data not shown) was derived from all 12 full-length ORFs in the Los Alamos National Laboratory and the European HCV databases (33) in April 2005 that were from different patients and from 5 genotype 1a ORFs that we sequenced using samples from members of non-Virahep-C cohorts. The genotype 1b population consensus sequence (data not shown) was generated from all 126 full-length ORFs from different patients in the Los Alamos and European HCV databases in January 2006. The known and predicted CD4+ and CD8+ T-cell epitope sequences were obtained from the HCV Immunology Database at the Los Alamos National Laboratory in March 2006 (73). The Shannon's entropy value (64) was calculated with Bioedit software (23). Phylogenetic trees were generated using the neighbor-joining algorithm in PHYLIP software (16). The mean genetic distance value for each group was calculated using the p-distance algorithm in the MEGA DNA analysis package (34).
Statistical analyses.
Race and sex distributions were compared across response groups by use of Pearson's chi-square test for association. Normally distributed characteristics were compared across response groups by use of analysis of variance, whereas the Kruskal-Wallis nonparametric test was used when the distributions were not normally distributed. Fisher's exact test was used to compare the proportions of unique variations between marked- and poor-response groups and between CA and AA patients. Unique variations across all three response groups were compared using the Freeman-Halton exact test. To determine whether there were racial or response differences in the number of variations (unique or total) or whether any response relationship to the number of variations differed by race, we used Poisson regression models with the number of variations as outcome and race, response groups, and their interactions as independent variables. The likelihood ratio test was used to assess statistical significance of the race and response interaction. Shannon's entropy values were compared between races and response groups separately using the Mann-Whitney rank sum test, and the average genetic distances between the groups were compared using an independent-sample t test. The level of significance (α) for statistical significance was 0.01. Statistical analyses were carried out using SAS software (SAS Institute, Inc.) or SPSS version 13.0 (SPSS, Inc.).
Nucleotide sequence accession numbers.
The sequences determined in this work have been deposited in GenBank (EF407411 to EF407504).
RESULTS
Experimental design and patient selection.
We hypothesized that HCV genetic variation results in viruses with variable sensitivity to peginterferon and ribavirin. We further hypothesized that AA patients may be infected with HCV strains that are somewhat more resistant to therapy than are those infecting CA patients. Therefore, we analyzed the pretreatment HCV sequences from 96 Virahep-C patients evenly distributed by race (CA or AA), genotype (1a or 1b), and day 28 response to therapy (marked, intermediate, or poor). The samples were stratified by day 28 response to minimize effects on suppression of viremia from factors other than biologic response to the drugs (such as amount of drug taken), to limit the effects of viral genetic adaptation to selective pressures induced by therapy, and most importantly, because all non-SVR patients are not the same. There are patients who respond well and then relapse, those who have a partial response to the drugs but never eliminate HCV, those who stop therapy because of side effects, and “null” patients who fail to respond to therapy in the first place (our “poor” category). Defining response at day 28 distinguishes the null patients from the other types of non-SVR patients, although it may not distinguish SVR patients from relapse patients. This is important because relapse patients respond to the drugs and almost achieve SVR, but it is the biological reason for the failure of therapy in the null patients which is the central focus of these analyses. In this study, “marked” responders had a decline in HCV titers of >3.5 log10 or to undetectable levels between baseline and day 28 of therapy, “intermediate” responders had declines of 1.4 to 3.5 log, and “poor” responders had declines of <1.4 log. To eliminate the effect of drug reductions on the day 28 outcome, all 96 patients received full doses of both peginterferon and ribavirin for the first 28 days. One genotype 1b CA marked responder and one genotype 1a AA intermediate responder proved to be coinfected with both genotype 1a and 1b viruses and were dropped from the analyses, yielding a final group of 94 patients.
The baseline characteristics of these 94 patients are shown in Table 1. There were no significant (P ≤ 0.05) differences among the three response groups except in baseline HCV RNA levels, platelet counts, and alpha-fetoprotein (AFP) levels. The lower platelet counts and higher AFP levels reflect slightly more severe liver disease, which is known to affect response to therapy, as do lower serum HCV RNA levels at baseline (17, 63). Lower pretreatment HCV titers in responders were also seen in the whole Virahep-C cohort (9), and although viral titer differences were associated with response to therapy, they did not account for the lower response rate in the AA patients in the full Virahep-C cohort.
TABLE 1.
Baseline characteristics of patients in the Virahep-C viral genetics study
| Characteristic | Value for group
|
P | ||
|---|---|---|---|---|
| Marked | Intermediate | Poor | ||
| No. of patients | 31 | 31 | 32 | |
| No. (%) of AA patients | 16 (51.6) | 15 (48.4) | 16 (50.0) | 0.97a |
| No. (%) of male patients | 22 (71.0) | 25 (80.7) | 21 (65.6) | 0.40a |
| Mean (SD) age (yr) of patients | 46.5 (6.2) | 48.1 (6.7) | 49.4 (8.8) | 0.30b |
| Median (25th, 75th percentile) age (yr) of patients | 46.0 (43.0, 49.0) | 48.0 (45.0, 53.0) | 50.0 (44.0, 53.5) | |
| Mean (SD) body wt (kg) of patients | 84.9 (16.8) | 88.6 (14.6) | 91.0 (13.5) | 0.28b |
| Median (25th, 75th percentile) body wt (kg) of patients | 84.8 (71.7, 98.4) | 87.5 (77.6, 97.5) | 93.0 (81.7, 103.4) | 0.003c |
| HCV RNA (log10 IU/ml) (mean SD) | 5.9 (0.9) | 6.5 (0.6) | 6.4 (0.5) | |
| Median (25th, 75th) HCV RNA (log10 IU/ml) | 5.6 (5.1, 6.6) | 6.7 (6.1, 6.9) | 6.5 (6.2, 6.7) | |
| Mean (SD) ALT (U/liter) | 79.0 (52.3) | 70.6 (36.3) | 86.3 (43.6) | 0.17c |
| Median (25th, 75th percentile) ALT (U/liter) | 56 (40, 110.5) | 62.0 (47, 84) | 71.0 (59.5, 107) | |
| Mean (SD) albumin (g/dl) | 4.1 (0.4) | 4.2 (0.3) | 4.2 (0.3) | 0.29c |
| Median (25th, 75th percentile) albumin (g/dl) | 4.1 (3.7, 4.4) | 4.3 (4.0, 4.4) | 4.2 (3.9, 4.4) | |
| Mean (SD) no. of platelets (×1,000/mm3) | 256.2 (61.2) | 222.5 (56.5) | 202.8 (59.3) | 0.002b |
| Median (25th, 75th percentile) no. of platelets (×1,000/mm3) | 268 (204, 299) | 212.0 (187, 257) | 200 (161, 251) | |
| Mean (SD) AFP (ng/ml) | 3.9 (2.1) | 11.7 (42.3) | 10.4 (6.7) | <0.0001c |
| Median (25th, 75th percentile) AFP (ng/ml) | 3.1 (2.4, 5.0) | 4.0 (2.7, 5.7) | 8.8 (5.5, 12.8) | |
| Mean (SD) Ishak necroinflammation score (0-18) | 7.2 (2.5) | 7.4 (3.0) | 7.9 (2.7) | 0.52c |
| Median (25th, 75th percentile) Ishak necroinflammation score (0-18) | 7 (6, 8) | 7.0 (5, 9) | 7 (6.5, 10) | |
| Mean (SD) Ishak fibrosis score (0-6) | 1.8 (1.3) | 1.8 (1.3) | 2.3 (1.4) | 0.15c |
| Median (25th, 75th percentile) Ishak fibrosis score (0-6) | 2 (1, 3) | 2 (1, 3) | 2 (2, 3) | |
Determined by chi-square test.
Determined by analysis of variance.
Determined by Kruskal-Wallis nonparametric test; data missing for one patient.
Use of the consensus sequence.
HCV replicates as a quasispecies rather than as a clonal population. Therefore, the virus can be represented genetically either by characterizing the quasispecies distribution or by using the consensus sequence of the quasispecies to reflect the center of the genetic distribution in each individual. We used the consensus sequence as determined by directly sequencing HCV DNA amplified from plasma (72) for three reasons. First, detailed analysis of quasispecies variation within an individual does not assess genetic variation between different individuals undergoing antiviral therapy, which is the goal of this project. Second, interpatient genetic variation is much larger than intrapatient quasispecies variation and would overwhelm intrapatient variation. Finally, characterizing the quasispecies spectrum of the entire ORF for each isolate was not feasible on the scale needed for this study.
Directly comparing the consensus sequence determined by the method employed here with a near full-length quasispecies analysis of variants found in the same patient sample explicitly demonstrated that the consensus sequence was representative of the common quasispecies variants (76). The average amino acid difference between the consensus sequence and the individual quasispecies variants was 1.8%, which was smaller than the average distance between the quasispecies variants themselves (2.3%). Phylogenetic analyses revealed that the consensus sequence was either in the center or near the base of the phylogenetic tree for each HCV gene.
Lack of clustering by response or race class.
We first tested the hypothesis that distinct viral strains were found in marked compared to poor responders or in AA compared to CA patients. We generated phylogeny trees from alignments of all 32 1a or 31 1b marked- and poor-responder amino acid sequences (intermediate responders were excluded to focus on the biological extremes of response), and similar trees were generated for race categories (AA versus CA) for all samples. No clustering of sequences by day 28 response or by race was found for any viral gene for either genotype 1a or 1b (data not shown). Therefore, the response and racial groups were not infected with distinctly different HCV strains.
Few amino acid positions differ consistently by response or race group.
To determine whether any amino acid positions differed consistently between the response and race groups, we aligned the sequences by response or race class and identified positions that were conserved in at least 60% of the sequences in a given group. Eight positions at which the respective biological classes had different conserved residues were found (Table 2). In all cases, the prevalence of the dominant amino acid in the alignment was ∼60% to ∼70%, and in all but one case, the predominant amino acid in the alignment of the other biological group was the second most prevalent amino acid. These variations were largely conservative, with the biggest differences occurring at S or P at position 401 for genotype 1a NS5A by race. The conservative nature of these variations and frequent presence of the most prevalent residue from the other biological class indicates that the variations were unlikely to play a dominant role in causing the limited reduction in viral titers in the poor responders or the low rate of response to therapy in the AA patients.
TABLE 2.
Positions at which the race or response groups are conserved at ≥60% but at which the marked-responder and poor-responder or CA and AA conserved residues are different
| Comparison category | Gene | Position (amino acid no.) | Amino acids (responder or race group) |
|---|---|---|---|
| Genotype 1a response | NS4A | 29 | V (marked), I (poor) |
| Genotype 1b response | Core | 70 | R (marked), Q (poor) |
| E1 | 139 | T (marked), A (poor) | |
| Genotype 1a race | ARF | 67 | R (CA), H (AA) |
| NS5A | 401 | S (CA), P (AA) | |
| NS5B | 98 | R (CA), K (AA) | |
| Genotype 1b race | E2 | 31 | V (CA), I (AA) |
| 66 | S (CA), A (AA) |
Diversity differences between response groups.
Because discrete differences at a few key residues were unlikely to explain the differences in response to therapy, we next assessed diversity differences between the response or race groups. These diversity analyses were designed to detect genes or regions of genes at which sequences of one group were more variable than sequences in another group (e.g., genotype 1a marked versus poor responders). The simplest way to analyze genetic diversity differences between groups was to identify the amino acids in each sequence that differed from an external reference sequence and then compare the variations in each group. As reference sequences, we employed genotype 1a and 1b population-wide consensus sequences derived from all full-ORF sequences available in the public databases that were from different individuals because they represent “typical” HCV sequences without host-specific adaptations (52). The ARF protein was omitted from this analysis due to variability in its start/stop sites, leading to difficulty in establishing a reference sequence consistent with those employed for the other genes. The intermediate responders were excluded to focus on the biological extremes of response to therapy.
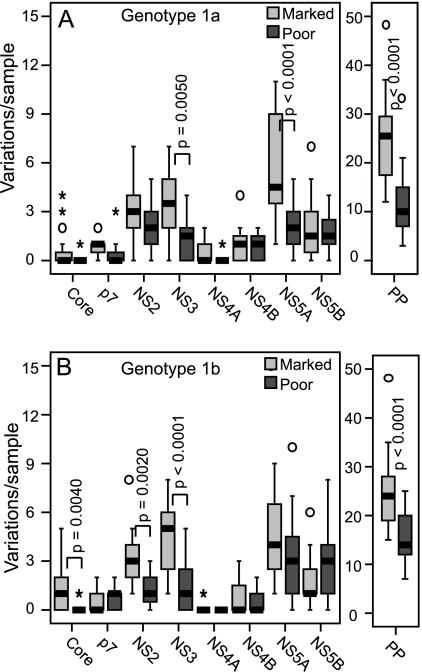
Each Virahep-C sequence was compared to the 1a or 1b reference sequence, positions of nonidentity were identified, and the numbers of variations were compared by response group. The total number of variations was generally higher in the marked than in the poor responders for both genotype 1a and genotype 1b, but only sequence variations in genotype 1a NS3 and genotype 1b NS2 and NS3 were significant at the P ≤ 0.01 level by a Poisson analysis. Next, to focus on differences that may be more strongly associated with response to therapy, we eliminated variations common to both the marked- and poor-responder groups because they were likely to be neutral. The number of unique variations in the sequences from the marked responder patients was generally higher than in those from the poor responders; these differences were statistically significant for genotype 1a E2, NS3, and NS5A (Fig. 2A) and genotype 1b core, E2, NS2, and NS3 (Fig. 2B).
FIG. 2.
Number of unique variations per sample by response. The numbers of variations relative to a population consensus that were unique to either the marked or poor responders are shown for the polyprotein (PP) and for each gene within the polyprotein. Statistical significance is shown for genes with P ≤ 0.01. (A) Genotype 1a. (B) Genotype 1b.
We extended these analyses of viral diversity between HCV-infected patients by employing other measures of genetic diversity, including the proportion of unique relative to nonunique variations assessed by the Fisher's exact test (data not shown), Shannon's entropy (a quantitative measure of diversity in an alignment [59, 64; Fig. 3]), the number and location of focal Shannon's entropy differences (Table 3), and the mean pair-wise protein distance for all proteins in an alignment (Fig. 4). For genotype 1a, samples from marked responder patients were consistently more diverse than those from poor responders in NS3 and NS5A, with E2 often approaching significance. For genotype 1b, samples from marked responders were consistently more diverse than those from poor responders in core and NS3. Together, these analyses indicate that HCV sequences from marked responders were significantly more diverse than sequences from poor responders in a subset of viral genes.
FIG. 3.
Sum of Shannon's entropy values by response. The marked- and poor-responder sequences were aligned separately, Shannon's entropy was determined for each position in the two alignments, and the entropy values were summed for each gene. Statistical significance between values for the marked- and poor-responder samples is indicated for genes where P ≤ 0.01. PP, polyprotein. (A) Genotype 1a. (B) Genotype 1b.
TABLE 3.
Number of focal entropy differences between marked and poor responders compared by response and in all samples compared by race
| Genotype or gene | No. of focal entropy differences for indicated category
|
|||||
|---|---|---|---|---|---|---|
| Responsea | Raceb | |||||
| Genotype 1a | M > P | P > M | 1a > 1b | CA > AA | AA > CA | 1a > 1b |
| Core | 1 | 1 | ||||
| ARF | ||||||
| E1 | 2 | |||||
| E2 | 2 | |||||
| P7 | 1 | 1 | ||||
| NS2 | 2 | 1 | 1 | 1 | ||
| NS3 | 3 | |||||
| NS4A | 1 | |||||
| NS4B | 1 | |||||
| NS5A | 4 | 2 | ||||
| NS5B | 1 | 1 | ||||
| Genotype 1b | M > P | P > M | 1b > 1a | CA > AA | AA > CA | 1b > 1a |
| Core | 2 | 1 | ||||
| ARF | 1 | 1 | ||||
| E1 | 2 | |||||
| E2 | 2 | 1 | 1 | |||
| P7 | 1 | |||||
| NS2 | 2 | 3 | ||||
| NS3 | 3 | 1 | ||||
| NS4A | 1 | |||||
| NS4B | 1 | 1 | ||||
| NS5A | 2 | 1 | 1 | 1 | ||
| NS5B | 1 | 1 | 2 | 1 | ||
Marked (M)- and poor (P)-responder samples only.
All samples.
FIG. 4.
Average genetic distances by response. The marked- and poor-responder sequences were aligned separately, the genetic distance between each pair of sequences in the alignments was calculated using the p-distance algorithm, and the average genetic distance for each protein in the alignment was plotted. The statistical significance of the difference between the groups is indicated for those genes where P ≤ 0.01. (A) Genotype 1. (B) Genotype 1b.
Variation in sequences from intermediate responders.
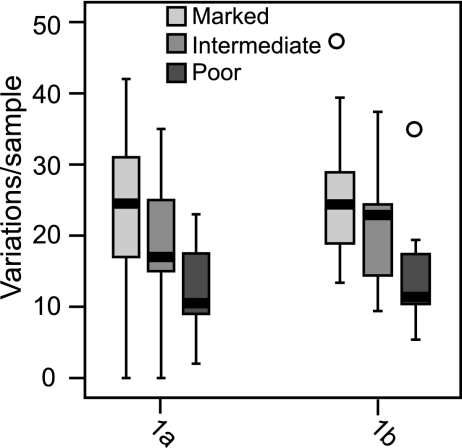
We next asked whether the intermediate samples had a distinct genetic diversity pattern or were intermediate in diversity between the marked- and poor-responder groups. We identified variations relative to the external reference sequences in the marked-, intermediate-, and poor-responder groups and then compared the numbers of these variations among all three response classes. In general, the marked responders had the most unique variations, the poor responders had the fewest, and the results for the intermediate responders were in between (Fig. 5). This pattern was supported by assessing the proportion of unique compared to nonunique variations by use of the Freeman-Halton exact test (data not shown). Therefore, there was a continuum in the degree of variation in the three response classes, with variation being highest in the marked-responder samples, lower in the intermediate-responder samples, and lowest in the poor-responder samples.
FIG. 5.
Numbers of unique variations per sample for all three response classes. The numbers of variations relative to a population consensus that were unique to the marked, intermediate, or poor-responder sequences are shown for the full genotype 1a and 1b polyproteins.
Diversity differences between isolates from AA and CA patients.
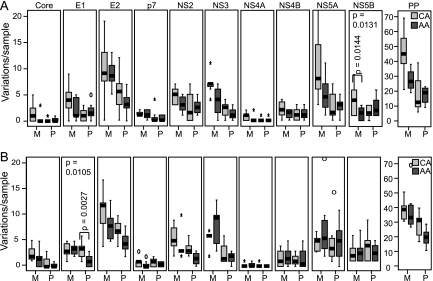
We next repeated the diversity analyses by comparing the sequences by race of the patient rather than by response to therapy to ask whether there were race-specific viral diversity differences. These analyses included all 94 sequences. When all variations were considered, there was a trend of greater diversity for the CA patients, but this was significant only for genotype 1b E1 at P ≤ 0.01. When variations unique to either the AA or CA sequences were considered to minimize neutral genetic variation, no significant differences were observed for genotype 1a (Fig. 6A). For genotype 1b the number of variations was significantly higher for CA patients in E1, E2, NS2, and NS5B (Fig. 6B).
FIG. 6.
Numbers of unique variations per sample by race. The numbers of variations relative to a population consensus that were unique to either the AA or CA sequences are shown for the polyprotein (PP) and for each gene within the polyprotein. Statistical significance is shown for genes with P ≤ 0.01. (A) Genotype 1a. (B) Genotype 1b.
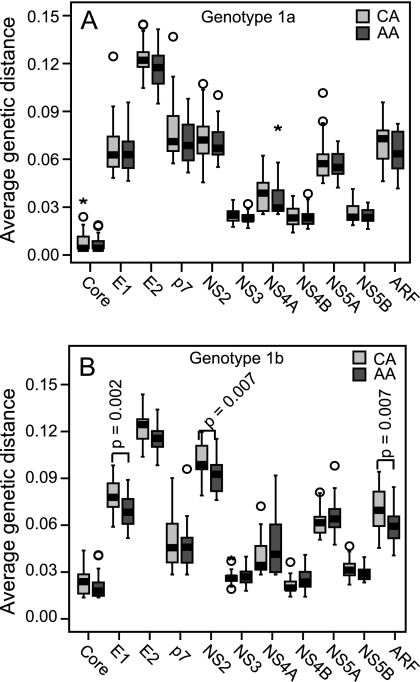
We extended these analyses of viral diversity by analyzing the proportion of unique relative to nonunique variations (data not shown), Shannon's entropy values (Fig. 7), the numbers and locations of focal Shannon's entropy differences (Table 3), and the mean pair-wise protein distance for all proteins in an alignment (Fig. 8). No significant differences were found for genotype 1a, but differences were found most consistently for genotype 1b in E1 and NS2, with the CA sequences being more diverse than the AA sequences. Therefore, in contrast to the large number of significant diversity differences observed in isolates from patients with divergent responses to therapy, significant differences were found when the sequences were compared by race of the patients exclusively in genotype 1b.
FIG. 7.
Sum of Shannon's entropy values by race. The AA and CA sequences were aligned separately, Shannon's entropy was determined for each position in the two alignments, and the entropy values were summed for each gene. Statistical significance between values for the AA and CA samples is indicated for genes where P ≤ 0.01. (A) Genotype 1a. (B) Genotype 1b.
FIG. 8.
Average genetic distances by race. The AA and CA sequences were aligned separately, the genetic distance between each pair of sequences in each of the two alignments was calculated using the p-distance algorithm, and the average genetic distance for each protein in the alignment was plotted. The significance of the difference between the groups was determined using an independent sample t test and is indicated for those genes where P ≤ 0.01. (A) Genotype 1. (B) Genotype 1b.
Influence of race on variation associated with response.
We next asked whether the race of the patient influenced variation associated with response to therapy. We first examined all variations relative to the reference consensus sequences in samples from the marked and poor responders and asked whether the AA and CA samples had the same pattern of variation within each response class. For genotype 1a, no gene results were significant at P ≤ 0.01, but those for NS5B approached significance (P = 0.014), with the marked-responder CA samples being more diverse than the marked-responder AA samples. For genotype 1b, only NS4A results achieved significance, with the marked-responder CA samples having fewer variations than the marked-responder AA samples. When this analysis was limited to variations unique to either the marked- or poor-responder samples to minimize neutral genetic variation, genotype 1a NS5B results approached significance (CA marked responders > AA marked responders), and for genotype 1b E1 the CA poor-responder samples were more diverse than the AA poor-responder samples (Fig. 9). Finally, we stratified the marked-responder and poor-responder samples by both response and race and identified variations that were unique to each of the four race/response groups. Using a log-likelihood ratio test to determine whether the variations were distributed differently among the groups, we found no significant differences for either genotype 1a or 1b in any of the genes (data not shown). Therefore, the effect of race on variations associated with response to therapy was relatively small. However, we may have underestimated the effect of race on variation associated with response because the power of this analysis was lower than when race and response were assessed separately.
FIG. 9.
Effect of race on genetic variation associated with response to therapy. Variations relative to a population consensus that were unique to the marked- and poor-responder samples were divided by race, and a log-likelihood test was performed to determine whether race affected the diversity associated with response to therapy. Statistical significance is shown for domains with P ≤ 0.01; the upper P value is for the log-likelihood interaction of race and response, and the lower P value is for the Poisson analysis of the indicated response groups. M, marked; P, poor; PP, polyprotein. (A) Genotype 1a. (B) Genotype 1b.
Variation outside regions of immunological pressure.
Interpatient HCV diversity differences associated with response could be due to preexisting variation leading to resistant and sensitive isolates, and/or they could be due to stronger immune pressures in the marked responders inducing more escape mutations (10, 20, 52, 67, 68). The latter possibility is plausible because anti-HCV CD4+ T-cell responses were stronger in Virahep-C patients who had sustained clearance of the virus than in those who in whom HCV infection was not eradicated (Rosen et al., manuscript in press). We therefore asked whether immune pressures were sufficient to produce the diversity differences associated with response to therapy. We removed the E1 and E2 glycoproteins from the analysis because they are subject to humoral selective pressures and then used a Poisson analysis to ask whether there were diversity differences in the remaining genes outside of all known or predicted T-cell epitopes. This strategy overestimates the number of T-cell epitopes because HLA restriction is not considered, but it eliminates almost all targets of cell-mediated selective immune pressures in the nonepitope sequences. When all variations outside T-cell epitopes were considered, NS3 and the full polyprotein from marked responders were significantly more diverse than those from poor responders for both genotypes 1a and 1b (data not shown). When variations unique to the marked or poor responders outside of the T-cell epitopes were considered, the diversity differences were significant in the nonepitope regions of genotype 1a NS3 and NS5A (Fig. 10A). For genotype 1b, the diversity differences remained significant in the core, NS2, and NS3 (Fig. 10B). Very similar results were obtained when the proportion of unique versus nonunique variations in the nonepitope sequences from the marked- and poor-responder patients was assessed using Fisher's exact test (data not shown). Therefore, significant diversity differences were present in sequences that were very unlikely to be under immune selection; hence, these differences must be due to allelic variation. The correlation of these allelic variations with response to therapy directly implies that genetic diversity differences modulated sensitivity of the isolates to therapy with peginterferon and ribavirin.
FIG. 10.
Numbers of unique variations in the nonepitope regions by response. Variations relative to the population consensus sequence that were not in any known or predicted T-cell epitope were compared in the marked- and poor-responder samples. Statistical significance is indicated where P ≤ 0.01. PP, polyprotein. (A) Genotype 1a. (B) Genotype 1b.
Variation in NS3.
We next examined the pattern of genetic variation in NS3 more closely because of its strong association with response to therapy for both genotypes 1a and 1b. The N-terminal domain of NS3 contains a protease that cleaves NS3-NS5B from the polyprotein and also degrades cellular signal transduction molecules to block the type 1 interferon response to double-stranded RNA (reviewed in reference 30). The C-terminal domain contains an RNA helicase activity that presumably functions during RNA replication. We predicted that variations correlating with response to therapy would be concentrated in the protease domain because of its role in blocking induction of alpha interferon. Unexpectedly, significantly higher diversity in the marked responders was found exclusively in the helicase domain for both genotype 1a (Fig. 11A and B) and genotype 1b (Fig. 11C and D). Similar results were obtained when the proportion of unique versus nonunique variations was assessed using Fisher's exact test (data not shown).
FIG. 11.
Numbers of variations in NS3 protease and helicase domains by response. Variations relative to a population consensus sequence in the NS3 protease and helicase domains were compared in the marked- and poor-responder samples. Statistical significance is shown for domains with P ≤ 0.01. (A) All variations in genotype 1a NS3 domains. (B) Variations unique to the marked- or poor-responder sequences for genotype 1a NS3 domains. (C) All variations in genotype 1b NS3 domains. (D) Variations unique to the marked- or poor-responder sequences for genotype 1b NS3 domains.
Variation in NS5A.
Variation in NS5A correlated highly with response to therapy for genotype 1a. NS5A has three proposed structural domains (I, II, and III) separated by two low-complexity sequences (LCS I and II) (66), and within these structural domains are many putative functional motifs (39). Domain I contains the membrane anchor and a Zn++ binding site, domain II contains the interferon-sensitivity-determining region (ISDR) and an overlapping protein kinase R (PKR) binding site, and domain III contains a polyproline cluster and a variable region (V3) for which higher diversity has been correlated with response to therapy (14, 36, 46). Genetic diversity associated with response to therapy was widely spread throughout genotype 1a NS5A, and consequently the differences in the numbers of total or unique variations between the marked and poor responders did not achieve significance for any of the individual domains at P ≤ 0.01 (data not shown). However, genotype 1a marked-responder sequences had a significantly higher proportion of unique variations relative to total variations than the poor-responder sequences for domain I, domain III, and V3 by Fisher's exact test (data not shown). Finally, the ISDR has been widely studied as a potential genetic modulator of interferon sensitivity (47, 71). In this study, higher variation in the ISDR/PKR binding site correlated with response to therapy only for genotype 1a CA patients (Fig. 12).
FIG. 12.
Variations in the genotype 1a NS5A ISDR/PKR binding domain. Sequences encompassing the ISDR/PKR binding site in NS5A from CA and AA marked (M) and poor (P) responders were aligned relative to the genotype 1a population consensus sequence (Ref). Positions where the Virahep-C sequences differ from the reference sequence are indicated. Numbers at the top indicate amino acid positions in NS5A.
DISCUSSION
The effects of genetic diversity of HCV on the success of antiviral therapy are complex for many reasons. First, two drugs are used, both drugs are likely to have more than one effect, and peginterferon at least does not act directly on any of the HCV proteins. Second, viral genetic variability is only one factor in response; host genetics and immune function are also important. Third, even closely related HCV subtypes such as 1a and 1b differ by ∼10% to ∼12% at the amino acid level; hence, isolates from different genotypes or subtypes may not be affected the same way at the molecular level by the drugs even when overall response rates to therapy are similar. Finally, all HCV isolates from treated individuals are wild-type strains that established chronic infections; hence, stark differences between isolates are not expected in parameters essential for viral replication or immune evasion. Rather, differences in susceptibility to therapy between isolates from different individuals are likely to be due to quantitative differences in viral functions, with some isolates being somewhat more sensitive than others. Therefore, variations at simple genetic motifs are unlikely to correlate highly with response. Consistent with this prediction, we found only three positions at which variation showed ≥60% correlation with response to therapy but found many diversity differences that were highly correlated with day 28 virological response.
The diversity data summarized in Table 4 led to three conclusions. First, there were strong associations between response to therapy and genetic diversity of the consensus sequences from infected individuals for a few viral genes but little to no association for the other genes. Higher variation was found by multiple analytical methods in sequences from the marked compared to poor responders in genotype 1a NS3 and NS5A and in genotype 1b core and NS3. Associations with response to therapy were also seen with E1, E2, and NS2, depending on the analytical method used. Second, the patterns of association between genetic diversity and response to therapy were not the same for genotypes 1a and 1b. This implies that the two genotypes may be affected by therapy through somewhat different molecular mechanisms. Finally, diversity differences correlating with the race of the patient were seen only for genotype 1b.
TABLE 4.
Summary of HCV genetic diversity differences by response and race
| Gene | HCV genetic diversity difference by category
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Responsea
|
Racea
|
|||||||||
| No. of unique variationsb | Proportion of unique variationsc | Total entropyd | Focal entropye | Genetic distancef | No. of unique variationsb | Proportion of unique variationsc | Total entropye | Focal entropyd | Genetic distancef | |
| Core | 1b | 1b | 1b | 1b | ||||||
| ARF | 1b | 1b | ||||||||
| E1 | 1a | 1b | 1b | 1b | 1b | |||||
| E2 | 1a, 1b | 1a | 1b | 1b | ||||||
| P7 | ||||||||||
| NS2 | 1b | 1b | 1b | 1b | 1b | 1b | ||||
| NS3 | 1a, 1b | 1a, 1b | 1a, 1b | 1a, 1b | 1a, 1b | |||||
| NS41a | ||||||||||
| NS4B | ||||||||||
| NS5A | 1a | 1a | 1a | 1a, 1b | 1b | |||||
| NS5B | 1b | 1b | 1b | |||||||
1a, P ≤ 0.01 for genotype 1a; 1b, P ≤ 0.01 for genotype 1b.
Number of unique variations per gene (marked versus poor responders or AA versus CA patients) relative to a population consensus sequence (Poisson analysis).
Observed number of variations relative to a population consensus per gene compared to the expected number (Fisher's exact test).
Sum of Shannon's entropy (Kolmogorov-Smirnov test).
Number of focal Shannon's entropy differences (three or more per gene).
Average genetic distance (t test).
The diversity differences between infected individuals correlating with response to therapy may be due to either allelic variation leading to differential sensitivity to therapy and/or to differential accumulation of escape mutations due to differing immune pressures in the marked and poor responders. Immune escape mutations can reduce viral fitness (10, 52, 68), and our data do not preclude such effects in this cohort. However, genetic variability was also significantly higher for the marked compared to poor responders in sequences that are very unlikely to be targets of cellular or humoral immune selective pressures (Fig. 10). These include nonepitope regions in core, NS3, and NS5A where the correlation between genetic diversity and response to therapy was strongest. Therefore, selection of immune escape variants cannot explain all of the correlation of pretreatment viral genetic variations with response to therapy. This implies that the higher diversity in the marked samples was due to preexisting allelic variation directly associated with response to therapy. It is well accepted that HCV isolates can differ in sensitivity to alpha interferon-based therapy at the level of the major genotypes (e.g., genotypes 2 and 3 are more sensitive than genotype 1 [18, 21, 41, 63]). However, the idea that interferon-sensitive and -resistant isolates exist within the major genotypes is controversial. The strong association of viral diversity with early response to therapy within both genotypes 1a and 1b reported here indicates that even within the major genotypes, viral genetic variability leads to differential sensitivity to peginterferon and ribavirin therapy.
A possible confounding issue concerning the relevance of pretherapy HCV sequences to the outcome of therapy is that HCV replicates as a quasispecies and hence can evolve rapidly in response to the selective pressures induced by antiviral therapy. Viral evolution during the first month of therapy could skew our results by misclassifying sensitive HCV sequences into the poor-responder group or resistant sequences into the marked-responder group. Misclassifying resistant sequences into the marked-responder group was not a major problem in this study because 28 of the 31 marked responders achieved SVR; hence, the large majority of HCV quasispecies variants in the marked responders were sensitive to therapy. It is possible that some of the pretreatment sequences in the poor-responder group were sensitive to the drugs and were rapidly supplanted by resistant strains during therapy. To evaluate the degree of viral evolution during therapy, we are sequencing the full ORFs at 24 weeks posttherapy from the 1a patients who did not achieve SVR. Posttherapy sequence data have been obtained for the full NS3 gene and for NS5A amino acids 1 to 328 (of 448) from all 24 1a non-SVR patients for whom a follow-up week 24 blood sample is available. In NS3 the median numbers of variations relative to the reference sequence were 18 prior to therapy and 19 after therapy. When the pre- and posttherapy NS3 sequences from the same patient were compared, the median number of amino acid changes per patient was only 1. In the first 328 amino acids of NS5A, the median number of total variations relative to the reference sequence in both the pre- and posttherapy sequences was 14, and the median number of amino acid changes between the pre- and posttherapy sequences from the same patient was again just 1. Therefore, the pre- and posttherapy sequences were very similar in NS3 and NS5A, the two genotype 1a genes in which high pretherapy diversity correlated with suppression of viremia. This indicates that viral evolution during therapy does not alter the observation that higher sequence diversity correlates with successful suppression of HCV during therapy.
Alpha interferon causes the large majority of the decline in viral titers by day 28 during combination therapy (12); hence, our marked- and poor-response categories were defined primarily by the response to peginterferon. Alpha interferon is a key player in the innate immune system, and HCV has evolved strategies to inhibit its effects (reviewed in references 22, 26, 48, 51, 54, and 60). Therefore, the variable sensitivity to peginterferon of HCV isolates from different individuals is likely due to variations in the sequences and functions of viral proteins that dampen host antiviral responses. The strongest correlations between response to therapy and viral variation were in NS3 and NS5A for genotype 1a and in core and NS3 for 1b. Importantly, all of these genes can actively block the action of alpha interferon in vitro (reviewed in reference 19). Therefore, the genetic data reported here provide strong support for a role for core, NS3, and NS5A (and, to a lesser extent, E2) in antagonizing the type 1 interferon response induced pharmacologically during treatment of HCV infections in humans.
Core residues R70 and M91 have been associated with response to therapy (2), but few other studies have examined the role of diversity in the core in the outcome of therapy. We observed a similar association of R70 with marked response for genotype 1b but not 1a; however, M91 was highly dominant in both the marked- and poor-responder sequences. E2 has been more thoroughly studied, because it has a PKR-phosphorylation homology domain (PePHD) which inhibits PKR in vitro (65). Increased diversity in the PePHD has been correlated with response to therapy in studies of patients infected with genotype 3 (58) but not in other studies that included genotypes 1 to 3 (1, 4, 49, 56, 57). We found that by some measures, E2 from marked responders was more diverse than E2 from poor responders for genotype 1a, and this difference approached significance for genotype 1b. However, these differences clustered in the N-terminal third of E2, and there were almost no variations among the isolates in the PePHD for either genotype 1a or 1b.
We observed a strong correlation between HCV diversity in NS3 between infected individuals and response to therapy, and to our knowledge variation in NS3 has not previously been associated with response to alpha interferon-based therapy in humans. The NS3 protease can block induction of type 1 interferon in response in vitro by cleaving signal transduction molecules downstream of the double-stranded RNA sensors Rig-I and TLR3 (reviewed in reference 30). In our sequences, diversity differences in NS3 correlating with response to therapy mapped exclusively to the helicase domain rather than the protease domain (Fig. 11). This implies that variation in NS3 correlating with response to therapy may affect the helicase activity rather than the protease activity or that the helicase domain may modulate the activity of the protease domain allosterically; interdomain communication within NS3 has been reported previously (32). These results also raise the possibility that variability in the viral replication rate modulated by variation in helicase activity may correlate with sensitivity to antiviral therapy and/or that the helicase may limit accumulation of double-stranded RNA that can trigger innate immune responses.
We found significant differences between viral sequences from marked and poor responders in NS5A, but only in genotype 1a. The functions of NS5A during HCV infection are unknown, but NS5A can attenuate the type 1 interferon response in vitro (19, 24, 39). Variation within a short region of NS5A termed the ISDR (15) has been associated with interferon sensitivity during antiviral therapy in some populations (47, 71). We observed higher variability in the ISDR of marked responders relative to the poor responders, but only in genotype 1a CA patients (Fig. 12). The role of genetic variation in NS5A outside the ISDR or the overlapping PKR binding site in response to therapy has been investigated in relatively few studies (14, 36, 46, 69). These studies found greater diversity in responders to alpha interferon-based therapy, especially in the carboxy-terminal half of the protein which includes the V3 hypervariable region (28). The Virahep-C samples were more diverse in the marked responders primarily in the carboxy-terminal half of the protein, including the V3 region (data not shown), so our results agree with previous studies of the full NS5A gene. Because most of the variability associated with response in NS5A in our samples was outside the ISDR/PKR binding site, variation in the ISDR may have modulated sensitivity to therapy in some patients, but this is not the only contribution that diversity in NS5A could have made to response of HCV to therapy.
Two recent studies examined diversity differences in E2 and NS5A between CA and AA patients (31, 36). In E2, sequences from CA patients were more diverse than those from AA patients for both genotype 1a and genotype 1b. This diversity was very significant in genotype 1b, with the majority of the difference clustering in the middle third of the protein. In NS5A, higher viral V3 variability was found for CA than for AA patients, and this increased variability correlated with response to therapy (36). We found a trend to higher diversity in the CA compared to the AA sequences throughout the viral ORF. For genotype 1a, these differences did not achieve statistical significance for any gene, but for 1b significant differences in E1, NS2, and NS5B were found by at least three analytical methods. The association between diversity and race for NS2 is novel, but the implications of this association are not clear because the role(s) of NS2 in HCV biology is not well known.
This study is the most comprehensive analysis to date of HCV genetic variation in the AA population. Our data do not support the idea that unusual genotype 1a HCV strains infecting AA patients exist, but there may be minor strain differences for genotype 1b between the AA and CA populations that could contribute to the poor response of AA patients to therapy. However, this equivocal conclusion does not preclude a role for viral diversity contributing to the unusually poor response of AA patients to therapy. We selected equal numbers of marked, intermediate, and poor responders in the AA and CA populations for sequence analysis, despite significantly lower response rates to therapy for the Virahep-C AA patients (28% versus 52% SVR rates) (9). The genetic similarity of HCV strains in the AA and CA patients across the three response classes and the intermingling of the AA and CA sequences in phylogenetic analyses (data not shown) indicate that very similar HCV strains circulate in the two racial groups; hence, the higher proportion of poor responders in the AA population implies that a higher proportion of relatively resistant HCV variants may circulate in the AA population, even for genotype 1a. Therefore, viral sequence variation may contribute to the unusually low response of AA patients to therapy, but the mechanism would be circulation in the AA population of a higher proportion of the same resistant strains found in the CA population rather than circulation of novel resistant strains that are absent from the CA population. Genotype 1b is more common in the AA population than in the CA population (53), so there is a precedent for differential circulation of HCV strains in the two racial groups.
The goal of antiviral therapy for HCV is SVR, the clearance of detectable viral RNA from serum for at least 6 months posttherapy. Large declines in viral titers by day 28 (such as occurred in the marked responders) correlate well with SVR (17, 43), with a major confounding factor being the failure or inability of the patients to take sufficient drug amounts during the demanding 48-week treatment regimen. A complete analysis of how viral genetic diversity is related to SVR and other clinical variables will be presented elsewhere, but we have performed an initial characterization of how pretreatment HCV genetic diversity correlates with SVR. The pretherapy sequences were stratified as SVR or non-SVR, and then the total and unique numbers of variations relative to the population consensus reference sequence, the total Shannon's entropy values, and the mean genetic distances in the two groups were compared. For both genotype 1a and genotype 1b, the polyproteins of the SVR sequences were significantly more diverse than those of non-SVR samples at P ≤ 0.01. For genotype 1a, NS3 and NS5A were significantly more diverse in the SVR samples by at least three of the four analytical methods. For genotype 1b, core was significantly more diverse in SVR patients by all four measures, and E2, NS2, and NS3 were more diverse by three of the four measures. Therefore, our basic observation that pretherapy sequence diversity was higher in the day 28 marked responders than in the poor responders for a few select viral genes is also true when the sequences are analyzed with respect to the ultimate goal of therapy, SVR.
Taking the data together, the focal distribution of viral genetic diversity at the patient level correlating with response to therapy and the inability of immune selection to account for all of this diversity lend strong support to the hypothesis that HCV genetic variation modulates efficacy of antiviral therapy in human patients. Because most of the reduction in viral levels at day 28 was due to peginterferon therapy, these data imply that the genes for which diversity differences correlate with early response to therapy function at least in part to blunt the type 1 interferon response in humans. This is consistent with the in vitro immune evasion activities reported for core, NS3, and NS5A (19). The consistently higher diversity in isolates from the marked responders (for whom the virus failed to counteract the drugs) compared to the poor responders (for whom the drugs were ineffective) implies that the functions of the proteins which counteract the effects of therapy can be impaired by amino acid differences at multiple locations. In essence, diversity would correlate with clearance because there are only a few ways to optimize activity of the viral proteins but many ways to interfere with their function.
Acknowledgments
We thank the participants of Virahep-C for their invaluable commitment of time and effort. The members of the Virahep-C study group are listed in reference 9.
Virahep-C was funded by the National Institute of Diabetes and Digestive and Kidney Diseases and the National Cancer Institute with further support under a Cooperative Research and Development Agreement with Roche Laboratories, Inc. Funding for this viral genetics project was through U01 DK60345; other grants supporting Virahep-C were U01 DK60329, U01 DK60340, U01 DK60324, U01 DK60344, U01 DK60327, U01 DK60335, U01 DK60352, U01 DK60342, U01 DK60309, U01 DK60346, U01 DK60349, and U01 DK60341. Support through the National Center for Research Resources General Clinical Research Centers Program was through grants M01 RR00645, M02 RR000079, M01 RR16500, M01 RR000042, and M01 RR00046.
Footnotes
Published ahead of print on 23 May 2007.
REFERENCES
- 1.Abid, K., R. Quadri, and F. Negro. 2000. Hepatitis C virus, the E2 envelope protein, and alpha-interferon resistance. Science 287:1555. [DOI] [PubMed] [Google Scholar]
- 2.Akuta, N., F. Suzuki, H. Sezaki, Y. Suzuki, T. Hosaka, T. Someya, M. Kobayashi, S. Saitoh, S. Watahiki, J. Sato, M. Matsuda, M. Kobayashi, Y. Arase, K. Ikeda, and H. Kumada. 2005. Association of amino acid substitution pattern in core protein of hepatitis C virus genotype 1b high viral load and non-virological response to interferon-ribavirin combination therapy. Intervirology 48:372-380. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong, G. L., A. Wasley, E. P. Simard, G. M. McQuillan, W. L. Kuhnert, and M. J. Alter. 2006. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann. Intern. Med. 144:705-714. [DOI] [PubMed] [Google Scholar]
- 4.Berg, T., M. A. Mas, M. Hohne, B. Wiedenmann, U. Hopf, and E. Schreier. 2000. Mutations in the E2-PePHD and NS5A region of hepatitis C virus type 1 and the dynamics of hepatitis C viremia decline during interferon alfa treatment. Hepatology 32:1386-1395. [DOI] [PubMed] [Google Scholar]
- 5.Berg, T., M. von Wagner, S. Nasser, C. Sarrazin, T. Heintges, T. Gerlach, P. Buggisch, T. Goeser, J. Rasenack, G. R. Pape, W. E. Schmidt, B. Kallinowski, H. Klinker, U. Spengler, P. Martus, U. Alshuth, and S. Zeuzem. 2006. Extended treatment duration for hepatitis C virus type 1: comparing 48 versus 72 weeks of peginterferon-alfa-2a plus ribavirin. Gastroenterology 130:1086-1097. [DOI] [PubMed] [Google Scholar]
- 6.Bodenheimer, H. C., Jr., K. L. Lindsay, G. L. Davis, J. H. Lewis, S. N. Thung, and L. B. Seeff. 1997. Tolerance and efficacy of oral ribavirin treatment of chronic hepatitis C: a multicenter trial. Hepatology 26:473-477. [DOI] [PubMed] [Google Scholar]
- 7.Branch, A. D., D. D. Stump, J. A. Gutierrez, F. Eng, and J. L. Walewski. 2005. The hepatitis C virus alternate reading frame (ARF) and its family of novel products: the alternate reading frame protein/F-protein, the double-frameshift protein, and others. Semin. Liver Dis. 25:105-117. [DOI] [PubMed] [Google Scholar]
- 8.Bukh, J., R. Miller, and R. Purcell. 1995. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin. Liver Dis. 15:41-63. [DOI] [PubMed] [Google Scholar]
- 9.Conjeevaram, H. S., M. W. Fried, L. J. Jeffers, N. A. Terrault, T. E. Wiley-Lucas, N. Afdhal, R. S. Brown, S. H. Belle, J. H. Hoofnagle, D. E. Kleiner, and C. D. Howell for the Virahep-C Study Group. 2006. Peginterferon and ribavirin treatment in African American and Caucasian American patients with hepatitis C genotype 1. Gastroenterology 131:470-477. [DOI] [PubMed] [Google Scholar]
- 10.Cox, A. L., T. Mosbruger, Q. Mao, Z. Liu, X. H. Wang, H. C. Yang, J. Sidney, A. Sette, D. Pardoll, D. L. Thomas, and S. C. Ray. 2005. Cellular immune selection with hepatitis C virus persistence in humans. J. Exp. Med. 201:1741-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Bisceglie, A. M., H. S. Conjeevaram, M. W. Fried, R. Sallie, Y. Park, C. Yurdaydin, M. Swain, D. E. Kleiner, K. Mahaney, and J. H. Hoofnagle. 1995. Ribavirin as therapy for chronic hepatitis C. a randomized, double-blind, placebo-controlled trial. Ann. Intern. Med. 123:897-903. [DOI] [PubMed] [Google Scholar]
- 12.Dixit, N. M., J. E. Layden-Almer, T. J. Layden, and A. S. Perelson. 2004. Modelling how ribavirin improves interferon response rates in hepatitis C virus infection. Nature 432:922-924. [DOI] [PubMed] [Google Scholar]
- 13.Dusheiko, G., J. Main, H. Thomas, O. Reichard, C. Lee, A. Dhillon, S. Rassam, A. Fryden, H. Reesink, M. Bassendine, G. Norkrans, T. Cuypers, N. Lelie, P. Telfer, J. Watson, C. Weegink, P. Sillikens, and O. Weiland. 1996. Ribavirin treatment for patients with chronic hepatitis C: results of a placebo-controlled study. J. Hepatol. 25:591-598. [DOI] [PubMed] [Google Scholar]
- 14.Duverlie, G., H. Khorsi, S. Castelain, O. Jaillon, J. Izopet, F. Lunel, F. Eb, F. Penin, and C. Wychowski. 1998. Sequence analysis of the NS5A protein of European hepatitis C virus 1b isolates and relation to interferon sensitivity. J. Gen. Virol. 79(Pt. 6):1373-1381. [DOI] [PubMed] [Google Scholar]
- 15.Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, N. Izumi, F. Marumo, and C. Sato. 1995. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J. Clin. Investig. 96:224-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 17.Ferenci, P. 2004. Predictors of response to therapy for chronic hepatitis C. Semin. Liver Dis. 24(Suppl. 2):25-31. [DOI] [PubMed] [Google Scholar]
- 18.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 19.Gale, M., Jr., and E. M. Foy. 2005. Evasion of intracellular host defence by hepatitis C virus. Nature 436:939-945. [DOI] [PubMed] [Google Scholar]
- 20.Gaudieri, S., A. Rauch, L. P. Park, E. Freitas, S. Herrmann, G. Jeffrey, W. Cheng, K. Pfafferott, K. Naidoo, R. Chapman, M. Battegay, R. Weber, A. Telenti, H. Furrer, I. James, M. Lucas, and S. A. Mallal. 2006. Evidence of viral adaptation to HLA class I-restricted immune pressure in chronic hepatitis C virus infection. J. Virol. 80:11094-11104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hadziyannis, S. J., H. Sette, Jr., T. R. Morgan, V. Balan, M. Diago, P. Marcellin, G. Ramadori, H. Bodenheimer, Jr., D. Bernstein, M. Rizzetto, S. Zeuzem, P. J. Pockros, A. Lin, and A. M. Ackrill. 2004. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann. Intern. Med. 140:346-355. [DOI] [PubMed] [Google Scholar]
- 22.Hahn, Y. S. 2003. Subversion of immune responses by hepatitis C virus: immunomodulatory strategies beyond evasion? Curr. Opin. Immunol. 15:443-449. [DOI] [PubMed] [Google Scholar]
- 23.Hall, T. A. 2006. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 24.He, Y., and M. G. Katze. 2002. To interfere and to anti-interfere: the interplay between hepatitis C virus and interferon. Viral Immunol. 15:95-119. [DOI] [PubMed] [Google Scholar]
- 25.Heathcote, E. J., M. L. Shiffman, W. G. Cooksley, G. M. Dusheiko, S. S. Lee, L. Balart, R. Reindollar, R. K. Reddy, T. L. Wright, A. Lin, J. Hoffman, and J. De Pamphilis. 2000. Peginterferon alfa-2a in patients with chronic hepatitis C and cirrhosis. N. Engl. J. Med. 343:1673-1680. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann, W. P., S. Zeuzem, and C. Sarrazin. 2005. Hepatitis C virus-related resistance mechanisms to interferon alpha-based antiviral therapy. J. Clin. Virol. 32:86-91. [DOI] [PubMed] [Google Scholar]
- 27.Howell, C., L. Jeffers, and J. H. Hoofnagle. 2000. Hepatitis C in African Americans: summary of a workshop. Gastroenterology 119:1385-1396. [DOI] [PubMed] [Google Scholar]
- 28.Inchauspe, G., S. Zebedee, D. H. Lee, M. Sugitani, M. Nasoff, and A. M. Prince. 1991. Genomic structure of the human prototype strain H of hepatitis C virus: comparison with American and Japanese isolates. Proc. Natl. Acad. Sci. USA 88:10292-10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeffers, L. J., W. Cassidy, C. D. Howell, S. Hu, and K. R. Reddy. 2004. Peginterferon alfa-2a (40 kd) and ribavirin for black American patients with chronic HCV genotype 1. Hepatology 39:1702-1708. [DOI] [PubMed] [Google Scholar]
- 30.Johnson, C. L. and M. Gale, Jr. 2006. CARD games between virus and host get a new player. Trends Immunol. 27:1-4. [DOI] [PubMed] [Google Scholar]
- 31.Keenan, E. D., S. D. Rouster, N. J. Shire, P. S. Horn, and K. E. Sherman. 2004. Complexity and diversity of hepatitis C virus RNA in African Americans and whites: analysis of the envelope-coding domain. J. Infect. Dis. 190:511-514. [DOI] [PubMed] [Google Scholar]
- 32.Kuang, W. F., Y. C. Lin, F. Jean, Y. W. Huang, C. L. Tai, D. S. Chen, P. J. Chen, and L. H. Hwang. 2004. Hepatitis C virus NS3 RNA helicase activity is modulated by the two domains of NS3 and NS4A. Biochem. Biophys. Res. Commun. 317:211-217. [DOI] [PubMed] [Google Scholar]
- 33.Kuiken, C., K. Yusim, L. Boykin, and R. Richardson. 2005. The Los Alamos hepatitis C sequence database. Bioinformatics 21:379-384. [DOI] [PubMed] [Google Scholar]
- 34.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 35.Kurosaki, M., N. Enomoto, F. Marumo, and C. Sato. 1994. Evolution and selection of hepatitis C virus in patients with chronic hepatitis C. Virology 205:161-169. [DOI] [PubMed] [Google Scholar]
- 36.Layden-Almer, J. E., C. Kuiken, R. M. Ribeiro, K. J. Kunstman, A. S. Perelson, T. J. Layden, and S. M. Wolinsky. 2005. Hepatitis C virus genotype 1a NS5A pretreatment sequence variation and viral kinetics in African American and white patients. J. Infect. Dis. 192:1078-1087. [DOI] [PubMed] [Google Scholar]
- 37.Lemon, S. M., C. Walker, M. J. Alter, and M. Yi. 2007. Hepatitis C virus, p. 1253-1304. In D. M. Knipe, P. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 38.Lindsay, K. L., C. Trepo, T. Heintges, M. L. Shiffman, S. C. Gordon, J. C. Hoefs, E. R. Schiff, Z. D. Goodman, M. Laughlin, R. Yao, and J. K. Albrecht. 2001. A randomized, double-blind trial comparing pegylated interferon alfa-2b to interferon alfa-2b as initial treatment for chronic hepatitis C. Hepatology 34:395-403. [DOI] [PubMed] [Google Scholar]
- 39.Macdonald, A., and M. Harris. 2004. Hepatitis C virus NS5A: tales of a promiscuous protein. J. Gen. Virol. 85:2485-2502. [DOI] [PubMed] [Google Scholar]
- 40.Major, M. E., B. Rehermann, and S. M. Feinstone. 2001. Hepatitis C viruses, p. 1127-1161. In D. M. Knipe, P. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 41.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 42.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, and J. K. Albrecht. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 43.Mihm, U., E. Herrmann, C. Sarrazin, and S. Zeuzem. 2006. Review article: predicting response in hepatitis C virus therapy. Aliment. Pharmacol. Ther. 23:1043-1054. [DOI] [PubMed] [Google Scholar]
- 44.Muir, A. J., J. D. Bornstein, P. G. Killenberg, and the Atlantic Coast Hepatitis Treatment Group. 2004. Peginterferon alfa-2b and ribavirin for the treatment of chronic hepatitis C in blacks and non-Hispanic whites. N. Engl. J. Med. 350:2265-2271. [DOI] [PubMed] [Google Scholar]
- 45.National Institutes of Health Consensus Development Conference Panel. 1997. Statement: management of hepatitis C. Hepatology 26:2S-10S. [DOI] [PubMed] [Google Scholar]
- 46.Nousbaum, J., S. J. Polyak, S. C. Ray, D. G. Sullivan, A. M. Larson, R. L. Carithers, Jr., and D. R. Gretch. 2000. Prospective characterization of full-length hepatitis C virus NS5A quasispecies during induction and combination antiviral therapy. J. Virol. 74:9028-9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pascu, M., P. Martus, M. Hohne, B. Wiedenmann, U. Hopf, E. Schreier, and T. Berg. 2004. Sustained virological response in hepatitis C virus type 1b infected patients is predicted by the number of mutations within the NS5A-ISDR: a meta-analysis focused on geographical differences. Gut 53:1345-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pavio, N., and M. M. Lai. 2003. The hepatitis C virus persistence: how to evade the immune system. J. Biosci. 28:287-304. [DOI] [PubMed] [Google Scholar]
- 49.Polyak, S. J., J. B. Nousbaum, A. M. Larson, S. Cotler, R. L. Carithers, Jr., and D. R. Gretch. 2000. The protein kinase-interacting domain in the hepatitis C virus envelope glycoprotein-2 gene is highly conserved in genotype 1-infected patients treated with interferon. J. Infect. Dis. 182:397-404. [DOI] [PubMed] [Google Scholar]
- 50.Poynard, T., P. Marcellin, S. S. Lee, C. Niederau, G. S. Minuk, G. Ideo, V. Bain, J. Heathcote, S. Zeuzem, C. Trepo, J. Albrecht, and the International Hepatitis Interventional Therapy Group (IHIT). 1998. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. Lancet 352:1426-1432. [DOI] [PubMed] [Google Scholar]
- 51.Racanelli, V., and B. Rehermann. 2003. Hepatitis C virus infection: when silence is deception. Trends Immunol. 24:456-464. [DOI] [PubMed] [Google Scholar]
- 52.Ray, S. C., L. Fanning, X. H. Wang, D. M. Netski, E. Kenny-Walsh, and D. L. Thomas. 2005. Divergent and convergent evolution after a common-source outbreak of hepatitis C virus. J. Exp. Med. 201:1753-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reddy, K. R., J. H. Hoofnagle, M. J. Tong, W. M. Lee, P. Pockros, E. J. Heathcote, D. Albert, T. Joh, and the Consensus Interferon Study Group. 1999. Racial differences in response to therapy with interferon in chronic hepatitis C. Hepatology 30:787-793. [DOI] [PubMed] [Google Scholar]
- 54.Rehermann, B., and M. Nascimbeni. 2005. Immunology of hepatitis B virus and hepatitis C virus infection. Nat. Rev. Immunol. 5:215-229. [DOI] [PubMed] [Google Scholar]
- 55.Robertson, B., G. Myers, C. Howard, T. Brettin, J. Bukh, B. Gaschen, T. Gojobori, G. Maertens, M. Mizokami, O. Nainan, S. Netesov, K. Nishioka, T. Shin-i, P. Simmonds, D. Smith, L. Stuyver, and A. Weiner. 1998. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. Arch. Virol. 143:2493-2503. [DOI] [PubMed] [Google Scholar]
- 56.Saito, T., T. Ito, H. Ishiko, M. Yonaha, K. Morikawa, A. Miyokawa, and K. Mitamura. 2003. Sequence analysis of PePHD within HCV E2 region and correlation with resistance of interferon therapy in Japanese patients infected with HCV genotypes 2a and 2b. Am. J. Gastroenterol. 98:1377-1383. [DOI] [PubMed] [Google Scholar]
- 57.Sarrazin, C., M. Bruckner, E. Herrmann, B. Ruster, K. Bruch, W. K. Roth, and S. Zeuzem. 2001. Quasispecies heterogeneity of the carboxy-terminal part of the E2 gene including the PePHD and sensitivity of hepatitis C virus 1b isolates to antiviral therapy. Virology 289:150-163. [DOI] [PubMed] [Google Scholar]
- 58.Sarrazin, C., I. Kornetzky, B. Ruster, J. H. Lee, B. Kronenberger, K. Bruch, W. K. Roth, and S. Zeuzem. 2000. Mutations within the E2 and NS5A protein in patients infected with hepatitis C virus type 3a and correlation with treatment response. Hepatology 31:1360-1370. [DOI] [PubMed] [Google Scholar]
- 59.Shannon, C. E., and W. Weaver. 1963. The mathematical theory of communication. University of Illinois Press, Champaign, IL.
- 60.Simmonds, P. 2004. Genetic diversity and evolution of hepatitis C virus—15 years on. J. Gen. Virol. 85:3173-3188. [DOI] [PubMed] [Google Scholar]
- 61.Simmonds, P., J. Bukh, C. Combet, G. Deleage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspe, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J. M. Pawlotsky, F. Penin, E. Sablon, I. Shin, L. J. Stuyver, H. J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962-973. [DOI] [PubMed] [Google Scholar]
- 62.Simmonds, P., E. C. Holmes, T. A. Cha, S. W. Chan, F. McOmish, B. Irvine, E. Beall, P. L. Yap, J. Kolberg, and M. S. Urdea. 1993. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J. Gen. Virol. 74:2391-2399. [DOI] [PubMed] [Google Scholar]
- 63.Strader, D. B., T. Wright, D. L. Thomas, and L. B. Seeff. 2004. Diagnosis, management, and treatment of hepatitis C. Hepatology 39:1147-1171. [DOI] [PubMed] [Google Scholar]
- 64.Strait, B. J., and T. G. Dewey. 1996. The Shannon information entropy of protein sequences. Biophys. J. 71:148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taylor, D. R., S. T. Shi, P. R. Romano, G. N. Barber, and M. M. Lai. 1999. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285:107-110. [DOI] [PubMed] [Google Scholar]
- 66.Tellinghuisen, T. L., J. Marcotrigiano, A. E. Gorbalenya, and C. M. Rice. 2004. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J. Biol. Chem. 279:48576-48587. [DOI] [PubMed] [Google Scholar]
- 67.Tester, I., S. Smyk-Pearson, P. Wang, A. Wertheimer, E. Yao, D. M. Lewinsohn, J. E. Tavis, and H. R. Rosen. 2005. Immune evasion versus recovery after acute hepatitis C virus infection from a shared source. J. Exp. Med. 201:1725-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Timm, J., G. M. Lauer, D. G. Kavanagh, I. Sheridan, A. Y. Kim, M. Lucas, T. Pillay, K. Ouchi, L. L. Reyor, J. S. Zur Wiesch, R. T. Gandhi, R. T. Chung, N. Bhardwaj, P. Klenerman, B. D. Walker, and T. M. Allen. 2004. CD8 epitope escape and reversion in acute HCV infection. J. Exp. Med. 200:1593-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vuillermoz, I., E. Khattab, E. Sablon, I. Ottevaere, D. Durantel, C. Vieux, C. Trepo, and F. Zoulim. 2004. Genetic variability of hepatitis C virus in chronically infected patients with viral breakthrough during interferon-ribavirin therapy. J. Med. Virol. 74:41-53. [DOI] [PubMed] [Google Scholar]
- 70.Walewski, J. L., J. A. Gutierrez, W. Branch-Elliman, D. D. Stump, T. R. Keller, A. Rodriguez, G. Benson, and A. D. Branch. 2002. Mutation Master: profiles of substitutions in hepatitis C virus RNA of the core, alternate reading frame, and NS2 coding regions. RNA 8:557-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Witherell, G. W., and P. Beineke. 2001. Statistical analysis of combined substitutions in nonstructural 5A region of hepatitis C virus and interferon response. J. Med. Virol. 63:8-16. [PubMed] [Google Scholar]
- 72.Yao, E., and J. E. Tavis for the Virahep-C Study Group. 2005. A general method for nested RT-PCR amplification and sequencing the complete HCV genotype 1 open reading frame. Virol. J. 2:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yusim, K., R. Richardson, N. Tao, A. Dalwani, A. Agrawal, J. Szinger, R. Funkhouser, B. Korber, and C. Kuiken. 2005. Los Alamos hepatitis C immunology database. Appl. Bioinformatics 4:217-225. [DOI] [PubMed] [Google Scholar]
- 74.Zeuzem, S. 2000. Hepatitis C virus: kinetics and quasispecies evolution during antiviral therapy. Forum 10:32-42. [PubMed] [Google Scholar]
- 75.Zeuzem, S., S. V. Feinman, J. Rasenack, E. J. Heathcote, M. Y. Lai, E. Gane, J. O'Grady, J. Reichen, M. Diago, A. Lin, J. Hoffman, and M. J. Brunda. 2000. Peginterferon alfa-2a in patients with chronic hepatitis C. N. Engl. J. Med. 343:1666-1672. [DOI] [PubMed] [Google Scholar]
- 76.Zhou, D., X. Fan, D. Tan, Y. Xu, J. E. Tavis, and A. M. Di Bisceglie. 2007. Separation of near full-length hepatitis C virus quasispecies variants from a complex population. J. Virol. Methods 141:220-224. [DOI] [PubMed] [Google Scholar]