Abstract
Human immunodeficiency virus type 1 (HIV-1) envelope (gp120) binding to DC-SIGN, a C-type lectin that can facilitate HIV infection in cis and in trans, is largely dependent on high-mannose-content moieties. Here, we delineate the N-linked glycosylation (N-glycan) sites in gp120 that contribute to optimal DC-SIGN binding. Soluble DC-SIGN was able to block 2G12 binding to gp120, but not vice versa, suggesting that DC-SIGN binds to a more flexible combination of N-glycans than 2G12. Consistent with this observation, HIV strain JRCSF gp120 prebound to 2G12 was 10-fold more sensitive to mannan competition than gp120 that was not prebound in a DC-SIGN cell surface binding assay. The analysis of multiple mutant forms of the 2G12 epitope revealed one triple glycosylation mutant form, termed 134mut (carrying N293Q, N382Q, and N388Q mutations), that exhibited a significant increase in sensitivity to both mannan competition and endoglycosidase H digestion compared to that of the 124mut form (carrying N293Q, N328Q, and N388Q mutations) and wild-type gp120 in a DC-SIGN binding assay. Importantly, no such differences were observed when binding to Galanthus nivalis was assessed. The 134mut form of gp120 also exhibited decreased binding to DC-SIGN in the context of native envelope spikes on a virion, and virus bearing 134mut exhibited less efficient DC-SIGN-mediated infection in trans. Significantly, 124mut and 134mut differed by only one glycosylation site mutation in each construct, and both 124mut and 134mut viruses exhibited wild-type levels of infectivity when used in a direct infection assay. In summary, while DC-SIGN can bind to a flexible combination of N-glycans on gp120, its optimal binding site overlaps with specific N-glycans within the 2G12 epitope. Conformationally intact envelopes that are DC-SIGN binding deficient can be used to probe the in vivo biological functions of DC-SIGN.
DC-SIGN (dendritic cell-specific ICAM-3-grabbing nonintegrin) is a calcium-dependent lectin that has been detected on subsets of dendritic cells (DCs) and macrophages in various tissues (references 19 and 47 and reviewed in references 14 and 59). DCs come in many specialized subtypes, some of which are thought to play an important role during the early steps of human immunodeficiency virus (HIV) dissemination after sexual transmission (44, 56). DC-SIGN has been implicated as a player in this process largely due to a body of in vitro (59) and ex vivo (23, 26, 31) data, but other C-type lectins are likely to be involved as well (18, 51, 59). DC-SIGN binds to the highly glycosylated HIV envelope (Env) gp120 in a CD4-independent fashion and can efficiently transfer the virus to CD4+ permissive T cells, thereby facilitating viral infection in trans (19, 20, 29, 38, 48, 52). However, DC-facilitated infection in trans likely occurs in two phases: a short-term phase (<18 to 24 h) that relies on DC-SIGN and other lectin receptors that can bind and transfer the primary source of HIV that is originally deposited (11, 54) and a long-term phase (>24 h) that is dependent on the de novo infection of DCs and results in the transfer of progeny virus (36, 54). DC-SIGN can also markedly enhance the efficiency of HIV infection in cis when DC-SIGN is present on cells that express limiting levels of CD4 and a coreceptor (11, 28, 36). Indeed, it was shown recently that the cis enhancement effect of DC-SIGN contributes to the efficient de novo infection of DCs that results in the aforementioned long-term transfer of virus to T cells (11).
In addition to DC-SIGN, other C-type lectins have been shown to bind and transfer HIV (18, 52, 53), and there is controversy as to whether even DC-SIGN itself is essential for DC-mediated viral transfer (reviewed in references 14 and 59). In particular, the in vivo significance of DC-SIGN in the mucosal transmission of HIV remains to be determined. However, several lines of evidence suggest that DC-SIGN may play contributory roles in this process. The addition of a single N-linked glycosylation (N-glycan) site in the V2 loop of SF162 leads to a gain of DC-SIGN binding function and correlates with increased efficiency of mucosal transmission of simian-human immunodeficiency virus (SHIV) 162P3 (31), a mucosally transmitted pathogenic SHIV variant whose parental virus lacking that N-glycan site is nonpathogenic and poorly transmissible (31). Furthermore, while most studies on the binding and transfer of HIV have been performed with monocyte-derived DCs (MDDCs), DC-SIGN+ cells isolated directly from the vaginal mucosa (26) or the rectal mucosa (23) have been shown to bind HIV and efficiently transfer the virus to CD4+ T cells in a manner that is dependent to some degree on DC-SIGN. Identifying the N-glycan sites on gp120 that result in optimal DC-SIGN binding may shed further light on the viral attachment process, suggest avenues for therapeutic development, and provide further insight into strategies for vaccine development, especially with regard to selectively deglycosylated Envs that may elicit antibodies to block gp120-DC-SIGN interactions (41). More importantly, the identification of a DC-SIGN binding-deficient envelope that is conformationally intact and borne by virus that is fully infectious may represent a formal tool that can be used to discern the biological relevance of DC-SIGN in HIV transmission.
DC-SIGN is composed of a cytoplasmic domain, a transmembrane domain, an extracellular neck domain of eight tandem 23-amino-acid-residue repeats, and a carbohydrate recognition domain (CRD). The neck region mediates the tetramerization of DC-SIGN, and indeed, DC-SIGN can be found as tetramers in vitro (16, 46) and on the surfaces of DCs (5, 49). The CRD region of DC-SIGN binds to high-mannose-content N-glycans (22, 32), and it is the tetramerization of DC-SIGN that results in the high-avidity binding to cognate ligands (32). The tetrameric nature of DC-SIGN binding likely puts some constraints on the spacing of glycans that results in optimal DC-SIGN binding (16, 39). Initial structural data indicated that DC-SIGN binds to an internal trimannose structure found only in high-mannose oligosaccharides but not in complex glycans (17). However, further carbohydrate profiling studies have found that DC-SIGN can bind to a wider range of glycan ligands, including fucosylated glycans such as Lewis X found in other pathogens and in human milk (1, 22, 35, 55). Interestingly, although DC-SIGN can bind to a wider array of glycan ligands than its closely related homolog, L-SIGN/DC-SIGNR (22), our previous biochemical data indicated that gp120-DC-SIGN interaction on cell lines and DCs is dependent solely on high-mannose glycans, as endoglycosidase H (Endo H) treatment of gp120 completely abolishes binding to DCs and DC-SIGN+ cell lines (25).
On gp120, DC-SIGN preferentially binds to high-mannose structures found on N-glycan sites (19, 25, 49). We previously reported our initial efforts to map the DC-SIGN binding determinants in gp120. The high-mannose N-glycans were found to cluster on the immunologically silent face of gp120 (62). We reported that no single glycosylation site is critical for DC-SIGN binding and that two reagents that bind to high-mannose glycans on gp120, 2G12 and cyanovirin (CVN), do not block gp120 binding to DC-SIGN (25). 2G12 is a monoclonal antibody whose carbohydrate-dependent epitope involves distinct N-glycan sites on the silent face of gp120 (13, 42, 43, 50). CVN is a well-characterized lectin from blue-green algae that binds to terminal mannose residues (10). Our mutational and biochemical data initially suggested that the DC-SIGN binding determinants on gp120 did not involve the 2G12 epitope. Here, we report our continued efforts to identify the N-glycan sites that are involved in the optimal gp120-DC-SIGN interaction. We have developed two complementary but fundamentally different assays to discern if DC-SIGN binding to gp120 is flexible and to identify the N-glycan sites that may give rise to high-mannose N-glycans that are most optimally spaced to interact efficiently with DC-SIGN.
MATERIALS AND METHODS
Viruses and cells.
DC-SIGN-positive, green fluorescent protein-positive cell lines were established by retroviral transduction and single-cell cloning for high-level-expression clones as previously described (28, 62). Green fluorescent protein-positive cell lines were confirmed to be DC-SIGN positive by costaining with a monoclonal antibody against DC-SIGN (DC028) (4, 28). Pseudotyped HIV-luciferase reporter constructs were made as previously described (45). The BC7 cell line (a kind gift from Jim Hoxie, University of Pennsylvania) is a CD4-negative clone of SupT1 cells and was previously used to obtain CD4-independent viruses (15). The RajiB cell line is a CD4-negative B-cell line (60). The TZM-bl (catalog no. 8129) indicator cell line is a CD4+ CCR5+ HeLa cell line acquired from the AIDS Research and Reference Reagent Program (www.aidsreagent.org). The 293 CD4+ CCR5+ cell line is a dually inducible cell line in which the expression of CD4 and CCR5 can be simultaneously or independently regulated with tetracycline and ponasterone, respectively. Detailed properties of this cell line will be reported elsewhere (S. Nguyen et al., unpublished data). Human monocytes for in vitro differentiation of MDDCs were isolated from peripheral blood obtained from healthy donors through the Virology Core of the University of California—Los Angeles (UCLA) AIDS Institute. Purified monocytes were obtained using the RosetteSep monocyte enrichment cocktail according to the guidelines of the manufacturer (StemCell Technologies, Canada). Monocytes were plated in 12-well plates (3 × 105 cells/well) and differentiated for 5 days in RPMI 1640 containing 10% fetal bovine serum plus 50 ng/ml (>500 U/ml) granulocyte-macrophage colony-stimulating factor and 100 ng/ml (>500 U/ml) interleukin-4 (PeproTech, Rocky Hill, NJ).
Plasmids and proteins.
A NotI-XhoI fragment of the gp120-Fc gene (the gp120 gene from the JR-CSF clone) was subcloned from pRSC-GS-Rev-gp120-Fc/RRE into pCR3 (Invitrogen, Carlsbad, CA) to maintain the natural signal leader sequence present in the gp120 gene. This process also resulted in the placement of the gp120-Fc gene downstream of a T7 promoter. All glycosylation mutations (Asn-X-Ser/Thr to Ala/Glu-X-Ser/Thr) were made using the pCR3-gp120-Fc plasmid by QuikChange (Stratagene, La Jolla, CA) site-directed mutagenesis and later confirmed with sequencing. A vaccinia virus-driven T7 polymerase system was used for the production of recombinant gp120-Fc. Briefly, 293T cells were infected with vTF1.1 (a recombinant vaccinia virus strain expressing T7 polymerase) at a multiplicity of infection of 5 for 1 h. Subsequently, vaccinia virus-infected cells were transfected with 10 μg of pCR3-gp120-Fc by calcium phosphate precipitation. Four hours posttransfection, cells were washed twice with 1× phosphate-buffered saline (PBS) and re-placed into serum-free medium (Dulbecco's modified Eagle's medium). Forty-eight hours posttransfection, the supernatant was harvested, filtered through a 0.22-μm-pore-size filter, and stored at 4°C in the presence of a complete EDTA-free protease inhibitor cocktail (Roche, Indianapolis, IN) until further use. The gp120-Fc thus produced was dimeric as judged by its migration (∼300 kDa) upon native sodium dodecyl sulfate-polyacrylamide gel electrophoresis (25). The recombinant soluble extracellular domain (ECD) of DC-SIGN was produced exactly as described previously and purified by metal chelation affinity chromatography (49). The purified ECD was checked for conformational integrity by binding to a panel of conformational anti-DC-SIGN antibodies (monoclonal antibodies 506 and 612) (4) and wild-type gp120-Fc. The 2G12 (catalog no. 1476), F105 (catalog no. 857), and 1b12 (catalog no. 2640) monoclonal antibodies were obtained from the AIDS Research and Reference Reagent Program (www.aidsreagent.org).
Fc ELISA.
Equal amounts of glycosylation mutant forms of gp120-Fc, as determined by an Fc enzyme-linked immunosorbent assay (ELISA), were tested for their ability to bind to DC-SIGN. One hundred microliters (1:500 dilution) of a monoclonal anti-human immunoglobulin G1 (IgG1) Fc-specific biotin-conjugated antibody (Caltag, Burlingame, CA) was added to Reacti-Bind streptavidin-coated clear wells with Superblock blocking buffer (Pierce, Rockford, IL). The plates were incubated for 2 h at room temperature and washed three times with wash buffer (Tris-buffered saline [TBS], 2% bovine serum albumin [BSA], 0.05% Tween 20). One hundred microliters of each gp120-Fc mutant construct was then added for 1 h at room temperature, and the plates were washed three times with wash buffer, followed by the addition of 100 μl (1:5,000) of a polyclonal anti-human IgG Fc horseradish peroxidase (HRP)-conjugated antibody (Pierce, Rockford, IL) for 30 min at room temperature. After the final three washes, samples were detected with a 1-Step Ultra 3,3′,5,5′-tetramethylbenzidine (TMB) ELISA (Pierce, Rockford, IL). Purified human IgG was used to generate a standard curve.
Cell surface DC-SIGN-gp120 binding and competition assays.
BC7 DC-SIGN-expressing cells or parental BC7 cells (2.5 × 105) were resuspended in 0.1 mg of gp120-Fc/ml for 30 min on ice and washed twice with DC-SIGN binding buffer (1× TBS, 2 mM CaCl2, 2.5% calf serum, 0.05% sodium azide) before the subsequent detection of gp120-Fc binding by using a goat phycoerythrin-conjugated anti-human Fc antibody (Caltag, Burlingame, CA). Mannan competition was evaluated by preincubating the DC-SIGN-expressing cells with the amounts of mannan (Sigma, St. Louis, MO) indicated in the figure legends for 15 min before the addition of gp120-Fc. 2G12 competition was evaluated by preincubating gp120-Fc with 2G12 for 15 min before adding the supernatant to the cells. Endo H digestion of gp120-Fc was performed by incubating gp120-Fc with the amounts of Endo H (New England Biolabs, Cambridge, MA) indicated in the figure legends for 2 h at 37°C. The extent of deglycosylation was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of untreated and Endo H-treated gp120-Fc (data not shown).
Galanthus nivalis and soluble DC-SIGN (ECD)-gp120 binding and competition assays.
For G. nivalis binding and competition studies, Reacti-Bind streptavidin-coated clear strip plates with blocker BSA (Pierce, Rockford, IL) were washed three times with wash buffer (TBS, 0.1% BSA, 0.05% Tween 20), and 0.2 μg of biotinylated G. nivalis (Vector Laboratories, Burlingame, CA) was added to each well (25). For ECD studies, a MaxiSorp high-level-binding flat-bottom 96-well plate (Nunc, Rochester, NY) was coated with 300 nM soluble DC-SIGN (ECD) and incubated overnight at 4°C. Each well was then washed three times with the wash buffer described above and blocked with TBS with 5% BSA and 1 mM CaCl2. Each mannan competition and Endo H treatment assay was performed essentially as described for the cell surface assay. After the preincubation of the plate with mannan or the pretreatment of the gp120-Fc with Endo H, gp120-Fc binding to G. nivalis or ECD was detected by using 100 μl (1:5,000) of a polyclonal anti-human IgG Fc HRP-conjugated antibody (Pierce, Rockford, IL) for 30 min at room temperature. After the final three washes, samples were detected with a 1-Step Ultra TMB ELISA (Pierce, Rockford, IL).
Virion binding assay.
For the virion binding assay, a plate was coated with 500 nM ECD and blocked as described above. Virus equivalent to 5 ng of p24, as determined by a commercially available p24 ELISA kit (PerkinElmer, Boston, MA), was spin inoculated (2,000 rpm at 4°C for 2 h) onto the plate. The wells were washed five times with a wash buffer (ice-cold PBS, 2.5% fetal bovine serum, and 1 mM CaCl2). The viruses were then lysed with 0.5% Triton X and assayed for p24 content by using the p24 ELISA kit. RajiB DC-SIGN+ cells were seeded into a V-bottom 96-well tissue culture plate (Corning Inc., Corning, NY) by spinning at 1,250 rpm for 10 min. Cells were resuspended in 100 μl containing viruses equivalent to 20 ng of p24 and incubated at 37°C for 2 h. The cells were then washed five times with a wash buffer, lysed, and assayed for p24 content. MDDCs were prepared as described previously and incubated with 250 μl containing viruses equivalent to 5 ng of p24 for viral transfer, and the binding assay was performed as described above.
Virus transfer.
The 293 CD4+ CCR5+ (dually inducible) cells were seeded into a 24-well tissue culture plate (Becton Dickinson and Company, Franklin Lakes, NJ) at a concentration of approximately 1.25 × 105 cells per well, and the plate was incubated for 24 h at 37°C. Cells were then induced with 0.1 μg of tetracycline/ml and 4 μM ponasterone (Invitrogen, Carlsbad, CA) for 18 h at 37°C to allow for maximal expression of CD4 and CCR5 before virus-bound RajiB DC-SIGN+ cells or MDDCs were added. For virus-bound RajiB DC-SIGN+ cells, the infection was allowed to proceed for 6 to 8 h, at which point the cells were lysed and treated with proteinase K before DNA extraction using the DNeasy tissue kit (QIAGEN, Valencia, CA). The extent of infection, as indicated by the copy number of viral cDNA, was quantified by the quantitative PCR (Q-PCR; see below). To determine the luciferase activity as an indication of viral transfer by MDDCs, the infection was allowed to proceed for 48 h and the cells were lysed. Fifty microliters of cell lysate was added with an equal volume of luciferase substrate (Promega, Madison, WI) in a flat-bottom, nonbinding-surface, black polystyrene plate (Corning Inc., Corning, NY).
Virus infections.
Approximately 1.25 × 105 TZM cells per well were seeded into a 24-well tissue culture plate. HIV type 1 (HIV-1) equivalent to 5 ng of p24 was added in the presence or absence of 2G12 or ECD as indicated (see Fig. 1), and the plate was spun at 2,000 rpm and 37°C for 2 h. The infection was allowed to proceed for 48 h, and the level of infection was quantified by the amount of luciferase activity by using the luciferase assay system (Promega, Madison, WI).
FIG. 1.
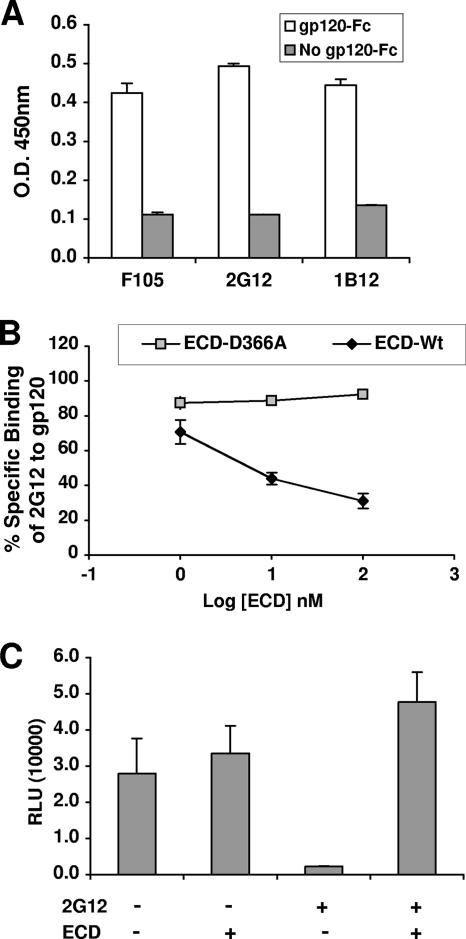
DC-SIGN binding to gp120 masks the 2G12 epitope. (A) gp120-Fc used in the binding assay was conformationally intact. Various conformational antibodies (1b12, 2G12, and F105) were tested for gp120-Fc binding in an ELISA. Antibody binding to gp120-Fc was detected by HRP-conjugated antibody against the kappa light chains (on the conformational antibodies). Error bars represent standard deviations of results from a representative experiment done in triplicate. O.D. 450nm, optical density at 450 nm. (B) Soluble DC-SIGN blocks 2G12 binding to gp120. 2G12 was tested for gp120-Fc binding in the absence or presence of increasing concentrations (1 to 100 nM) of the ECD of DC-SIGN in an ELISA, as described in Materials and Methods. 2G12 binding to gp120-Fc was detected by HRP-conjugated antibody against the kappa light chain (on 2G12). ECD carrying the D366A mutation (ECD-D366A) is incapable of binding gp120 (49) and served as a negative specificity control. The optical density obtained by 2G12 binding to gp120-Fc was normalized to the optical density obtained in the absence of ECD (set at 100%). ECD-wt, wild-type ECD. (C) Soluble DC-SIGN reverses the inhibitory effect of 2G12 on HIV infection. JRCSF (R5) virus equivalent to 5 ng of p24 was used to infect TZM cells (CD4+ CCR5+ HeLa indicator cells) in the presence of ECD (500 nM) or 2G12 (400 nM) alone or in combination. TZM cells have the luciferase gene under the control of the HIV long terminal repeat promoter. Data shown are averages ± standard errors of results from at least three independent experiments. RLU, relative light units; +, present; −, absent.
Q-PCR.
A Q-PCR was set up to detect early reverse transcriptase products in the R5/U5 region of the long terminal repeat. Endogenous beta-globin levels were also measured in order to normalize R5/U5 values. For each reaction, 2 μl of DNA from the DNeasy tissue kit was added to a PCR master mix (QIAGEN, Valencia, CA), which consisted of 3 μM forward and reverse primers, 5 μM probe, 50 mM MgCl2, and DNase-free water in a total volume of 25 μl. A DNA Engine Opticon system (QIAGEN, Valencia, CA) was used to carry out the following reaction steps: (i) an initial 10 min at 95°C and then (ii) 95°C for 15 s and (iii) 65°C for 1 min, with steps ii and iii repeated 40 times. The primers (MWG Biotechnology, High Point, NC) and probes (PE Applied Biosystems, Foster City, CA) used were as follows: for HIV, forward primer AA51 (5′-CAAGTAGTGTGTGCCCGTCTGT-3′), reverse primer SR1 (5′-CTGCTAGAGATTTTCCACACTGAC-3′), and a probe (5′-TGTGACTCTGGTAACTAGAGATCCCTCAGACCC-3′) modified with 6-carboxyfluorescein reporter dye at the 5′ end and 6-carboxytetramethylrhodamine quencher dye at the 3′ end, and for beta-globin, forward primer BGF1 (5′-CAACCTCAAACAGACACCATGG-3′), reverse primer BGR1 (5′-TCCACGTTCACCTTGCCC-3′), and a probe (5′-CTCCTGAGGAGAAGTCTGCCGTTACTGCC-3′) modified with 6-carboxyfluorescein reporter dye at the 5′ end and 6-carboxytetramethylrhodamine quencher dye at the 3′ end.
F105-, 1b12-, and 2G12-gp120 binding ELISA.
One hundred microliters of neat gp120-Fc supernatant was added to Dynex Immulon 2 plates (Fisher Scientific, Tustin, CA), and the plates were incubated for 2 h at 37°C. Each plate was then blocked with 300 μl of blocking buffer (PBS, 0.1% Tween, 1% BSA) at room temperature for 1 h and washed three times with 1× PBS-0.1% BSA. F105, 1b12, and 2G12 (conformational monoclonal antibodies against gp120) at 10 μg/ml were added, followed by incubation at room temperature for 1 h. F105, 1b12, and 2G12 are all IgG1 antibodies with kappa light chains; therefore, monoclonal antibody binding to gp120-Fc was detected with 100 μl of a goat anti-human kappa antibody conjugated with HRP (Caltag, Burlingame, CA) at a dilution of 1:1,000 after incubation for 30 min at room temperature. The plates were again washed three times and developed with 1-Step Ultra TMB substrate (Pierce, Rockford, IL). The reaction was stopped by the addition of 2 M H2SO4, and the plates were read at 450 nm in a microtiter plate reader (Dynex Technologies, Chantilly, VA).
gp120 competition ELISA.
One hundred microliters of neat gp120-Fc supernatant was added to Dynex Immulon 2 plates (Fisher Scientific, Tustin, CA), and the plates were incubated for 2 h at 37°C. Each plate was then blocked with 300 μl of blocking buffer (PBS, 0.1% Tween, 1% BSA) at room temperature for 1 h and washed three times with 1× PBS-0.1% BSA. A wide range of ECD concentrations (1 to 100 nM) was preincubated with gp120-Fc before 100 μl of 2G12 (400 nM) was added and the mixture was incubated at room temperature for 1 h. The plates were washed three times, and 100 μl of a goat anti-human kappa antibody conjugated with HRP (Caltag, Burlingame, CA) was added as described for the monoclonal antibody-gp120 binding ELISA.
Structural modeling.
The crystal structures of gp120 (27) and the CRD of DC-SIGN (16) were downloaded from the Protein Data Bank (www.rcsb.org/pdb). Sixteen amino acids within the CRD region responsible for the carbohydrate binding were extracted using a WordPad program. Each structure was then superimposed using the Win ONO program, and coordinates were saved as a Pdb file. The final model was created after uploading both structures onto the PyMOL viewer.
RESULTS
DC-SIGN binding to gp120 masks the 2G12 epitope.
2G12 is a broadly neutralizing monoclonal antibody whose carbohydrate-dependent epitope can be disrupted by mutations of specific N-linked glycosylation (N-glycan) sites on the silent face of gp120 that are known to give rise to high-mannose glycans (43, 50). We previously showed that the epitopes for DC-SIGN and 2G12 on gp120 are distinct and nonoverlapping (25). This conclusion was based primarily on the observations that 2G12 does not block gp120 binding to DC-SIGN and that gp120 mutant forms lacking one or more of the N-glycan sites involved in the 2G12 epitope still bind to DC-SIGN. However, in the case of CVN, another high-mannose binding lectin that can bind gp120 and block viral fusion, it was found that 2G12 does not inhibit CVN binding to gp120. In contrast, CVN can clearly inhibit 2G12 binding to gp120, suggesting that CVN binding to gp120 is flexible and involves a larger set of optimally spaced glycans. Thus, we investigated if DC-SIGN binding is similarly flexible and whether DC-SIGN can similarly inhibit 2G12 binding to gp120.
To do this, we first developed a gp120-2G12 binding assay in which recombinant gp120-Fc (from strain JRCSF) was captured onto an ELISA plate and 2G12 binding to gp120-Fc was detected with an HRP-conjugated antibody against the kappa light chain of 2G12. Figure 1A shows that the gp120-Fc used in our binding assay was conformationally intact, as it was recognized by two other conformational antibodies (F105 and 1b12) in addition to 2G12 (both F105 and 1b12 also had kappa light chains). Figure 1B shows that 2G12 binding to gp120 was inhibited by the soluble recombinant ECD of DC-SIGN in a dose-dependent fashion. A gp120 binding mutant form of DC-SIGN (the ECD with the D366A mutation) in amounts equal to those of the wild-type form had no effect on 2G12 binding to gp120, underscoring the specificity of the observed inhibition. The D366A mutation abolishes the calcium-chelating site in DC-SIGN and thereby eliminates its ability to bind gp120 (49).
To confirm that DC-SIGN binding to gp120 can similarly inhibit 2G12 binding in the context of trimeric envelope spikes on virions, we determined if soluble DC-SIGN could reverse 2G12's inhibition of viral infection. HIV-1 (JR-CSF) was used to infect CD4+ CCR5+ TZM cell lines in the presence of ECD or 2G12 alone or in combination. Figure 1C shows that while 2G12 inhibited HIV-1 infection, ECD alone had no effect. However, in the presence of ECD, the inhibition of viral infection by 2G12 was reversed, suggesting that DC-SIGN binding to envelope on virions prevented 2G12 binding and its subsequent inhibition of viral entry. These results suggest that the binding site of DC-SIGN on gp120 and on virions can involve the 2G12 epitope, although specific N-glycan sites in the 2G12 epitope may not be necessary or sufficient for DC-SIGN binding. Indeed, many single, double, and triple N-glycan mutations in the 2G12 epitope did not seem to affect gp120's ability to bind DC-SIGN (Table 1). The few mutations that did affect DC-SIGN binding also affected CD4 binding (Table 1), and thus, the defect in binding was likely due to the conformational perturbation of gp120.
TABLE 1.
Binding of wild-type and mutant forms of gp120-Fca
| Mutation(s) in gp120-Fc construct from strain:
|
Bindingb of gp120-Fc construct to:
|
||
|---|---|---|---|
| JR-CSF | HXB2 | DC-SIGN | CD4 |
| None (wild type) | None (wild type) | + | ++ |
| N195Q | N197Q | + | ++ |
| N228Q | N230Q | + | ++ |
| N239Q | N241Q | + | + |
| N260Q | N262Q | + | + |
| N287Q | N289Q | + | ++ |
| N293Q | N295Q | + | ++ |
| N299Q | N301Q | + | + |
| N328Q | N332Q | + | ++ |
| N336Q | N339Q | + | ++ |
| N382Q | N386Q | + | ++ |
| N388Q | N392Q | + | ++ |
| N393Q | N397Q | + | ++ |
| N239Q, N260Q | N241Q, N262Q | ± | − |
| N239Q, N299Q | N241Q, N301Q | ± | − |
| N260Q, N299Q | N262Q, N301Q | ± | − |
| N228Q, N239Q | N230Q, N241Q | + | + |
| N228Q, N260Q | N230Q, N262Q | + | + |
| N228Q, N299Q | N230Q, N301Q | + | + |
| N228Q, N287Q | N230Q, N289Q | + | ++ |
| N228Q, N293Q | N230Q, N295Q | + | ++ |
| N228Q, N328Q | N230Q, N332Q | + | ++ |
| N228Q, N382Q | N230Q, N386Q | + | ++ |
| N228Q, N336Q | N230Q, N339Q | + | ++ |
| N328Q, N382Q | N332Q, N386Q | + | ++ |
| N293Q, N388Q | N295Q, N392Q | + | ++ |
| N239Q, N260Q, N299Q | N241Q, N262Q, N301Q | ± | − |
| N293Q, N328Q, N388Q (124mut) | N295Q, N332Q, N392Q | + | ++ |
| N293Q, N382Q, N388Q (134mut) | N295Q, N386Q, N392Q | + | ++ |
Twelve N-glycan sites in JR-CSF known to give rise to high-mannose-content structures (62) were mutated singly or in combination as indicated. Equal amounts of wild-type gp120-Fc and the indicated mutant forms were added to DC-SIGN-expressing 293T cells or CD4+ SupT1 cells, and binding was detected in a FACS-based assay as described in Materials and Methods. Binding to DC-SIGN or CD4 (indicated by mean fluorescence intensities) was normalized to wild-type gp120-Fc binding, which was set at 100%. The N239Q, N260Q, and N299Q mutations, in the context of double and triple mutant forms, affected both DC-SIGN and CD4 binding and thus were likely to perturb the conformational integrity of gp120. Mutations in the 2G12 epitope, which can be disrupted by mutations in five N-glycan sites, are indicated in bold. Residue numbering in the context of the JR-CSF strain and the homologous numbering in the reference HXB2 strain are shown.
Levels of binding are indicated as follows. DC-SIGN: +, 50 to 100% of wild-type binding; ±, <50% of wild-type binding. CD4: ++, 80 to 100% of wild-type binding; +, 20 to 80% of wild-type binding; −, <20% of wild-type binding.
DC-SIGN binding to gp120 is flexible, but optimal binding involves the 2G12 epitope.
The data in Table 1 were obtained via an equilibrium binding assay. Essentially, a saturating amount of gp120-Fc was bound onto a DC-SIGN-expressing cell line, and binding was detected by flow cytometric analysis using anti-Fc secondary antibodies. Indeed, this assay showed that various gp120 N-glycan mutant forms were not affected in their ability to bind DC-SIGN in the presence or absence of 2G12 (which alone can potentially mask at least five N-glycans on gp120). Figure 2A shows a representative series of gp120 constructs carrying double and triple glycosylation mutations, in addition to wild-type JR-CSF gp120, whose binding to DC-SIGN was not affected by the presence of prebound 2G12 (fluorescence-activated cell sorter [FACS] histogram data). Taken at face value, the analysis presented in Fig. 2A suggests that 9 out of the 12 N-glycan sites on the silent face of gp120 (where the high-mannose glycans are clustered) may not be necessary for DC-SIGN binding. Alternatively, DC-SIGN binding may simply be flexible and not dependent on a particular N-glycan.
FIG. 2.
DC-SIGN binding to gp120 is flexible, but optimal binding involves the 2G12 epitope. (A) Wild-type gp120-Fc or the indicated mutant forms were prebound with 2G12 (20 μg/ml) for 15 min. Then the 2G12-gp120-Fc complex or unbound gp120-Fc was tested for DC-SIGN binding on 293T DC-SIGN+ cells in a FACS-based assay using anti-Fc secondary antibodies as a detection agent. Equal amounts (∼500 nM, as quantified by the Fc-specific ELISA described in Materials and Methods) of wild-type and mutant gp120-Fc were used. Representative histograms are shown. All the N-glycan sites known to give rise to high-mannose glycans are shown in the context of the structural model of gp120 (beige ribbon) complexed with CD4 (blue ribbon; Protein Data Bank identification no., 2B4C). N-glycan sites that contribute to the 2G12 epitope are shown as yellow space-filling residues, and the remaining N-glycan sites are depicted as red space-filling residues. The structure was produced with the PyMOL program. Mutant forms of gp120 are identified by their mutations. HuIgG, human IgG; +, prebound; −, not prebound. (B) gp120 binding to DC-SIGN is more sensitive to mannan competition when gp120 is prebound to 2G12. gp120-Fc prebound with 2G12 (20 μg/ml) and unbound gp120-Fc were tested for DC-SIGN binding on BC7 DC-SIGN+ cells in the presence of increasing concentrations of mannan (0.1 to 1,000 μg/ml). The mean fluorescence intensities of gp120-Fc binding in the presence of increasing amounts of mannan were normalized to the mean fluorescence intensity of gp120-Fc binding in the absence of mannan (set at 100%). The 50% inhibitory concentrations (IC50) for the mannan competition were obtained by nonlinear regression using the GraphPad Prism 4 program.
We therefore developed a competitive assay that might be better suited to revealing if any N-glycan sites were more optimally positioned to mediate DC-SIGN binding. We reasoned that if we blocked or mutated a particular N-glycan site on gp120 that was more critical than others to DC-SIGN binding, the presence of other N-glycan sites would be less able to compensate for the loss of this particular site. Thus, in a competitive DC-SIGN binding assay using another ligand for DC-SIGN, e.g., mannan, one may expect this particular mutant or blocked construct to be more sensitive to mannan competition than its wild-type counterpart. So although we had previously shown that 2G12 does not block gp120 binding to DC-SIGN in an equilibrium binding assay (25), we now showed that gp120-Fc (JR-CSF) prebound to 2G12 was at least 10-fold more sensitive to mannan competition than untreated gp120-Fc (Fig. 2B). We believe that this result was not due simply to a reduction in the number of N-glycans on gp120, as will be shown in the next section.
Optimal DC-SIGN binding to gp120 involves a unique glycosylation site within the 2G12 epitope.
Although DC-SIGN binding to gp120 may be flexible, optimal binding likely requires a set of N-glycans optimally spaced to interact with the tetrameric configuration of native DC-SIGN (5, 16, 49). Thus, we tested double and triple mutations in the 2G12 epitope in the mannan competition assay. We found that the 134mut triple mutant form of gp120 (carrying N293Q, N382Q, and N388Q) was more sensitive to mannan competition than the 124mut triple mutant form (carrying N293Q, N328Q, and N388Q) (Fig. 3A and B and Table 2), even though 134mut and 124mut differed in only one glycosylation site mutation in each construct (N382Q and N328Q, respectively). Indeed, 124mut, which lacks three N-glycan sites in the 2G12 epitope, was not significantly different from wild-type gp120-Fc in its sensitivity to mannan competition during cell surface DC-SIGN and soluble DC-SIGN (ECD) binding (Fig. 3A and B and Table 2). Thus, the increased sensitivity to mannan competition of 134mut was not due simply to a reduction in the number of N-glycan sites available for DC-SIGN binding; one or more N-glycan sites mutated in the 134mut form were likely positioned to be in the optimally spaced set of N-glycans required for DC-SIGN binding. Indeed, neither 124mut nor 134mut differed from wild-type gp120-Fc in sensitivity to mannan competition when binding to G. nivalis was assayed (Fig. 3C and Table 2). G. nivalis is a plant lectin that binds to terminal sugars in high-mannose structures. Thus, the differences among results presented in Fig. 3A, B, and C suggest that while other N-glycans in gp120 were able to compensate for the lost N-glycan sites in both 124mut and 134mut when binding to G. nivalis was assessed, they were less able to compensate for the loss of N-glycan sites in 134mut when DC-SIGN binding was concerned.
FIG. 3.
Optimal DC-SIGN binding to gp120 involves a unique glycosylation site within the 2G12 epitope. (A, B, and C) Equimolar amounts (quantified by an Fc-specific ELISA) of wild-type gp120-Fc and two triple glycosylation site mutant forms (124mut and 134mut) (Table 1) with mutations in the 2G12 epitope were tested for cell surface DC-SIGN (A), soluble DC-SIGN (ECD) (B), and G. nivalis (GNV) (C) binding in the presence of increasing concentrations of mannan (0.1 to 1,000 μg/ml). (D, E, and F) The same gp120-Fc constructs described above were treated with increasing concentrations of Endo H (10 to 10,000 mU) for 2 h at 37°C. Then each Endo H-treated gp120-Fc and untreated gp120-Fc construct was tested for cell surface DC-SIGN (D), ECD (E), and G. nivalis (F) binding. Cell surface DC-SIGN binding (A and D) was tested with BC7 DC-SIGN+ cells in a FACS-based assay. The mean fluorescence intensities of gp120-Fc binding to DC-SIGN (A and D) in the presence of increasing amounts of mannan or Endo H were normalized to the mean fluorescence intensity of gp120-Fc binding to DC-SIGN in the absence of mannan or Endo H (set at 100%). ECD binding (B and E) was performed using ECD captured onto an ELISA plate. The A450s of gp120-Fc binding to ECD (B and E) in the presence of increasing amounts of mannan or Endo H were normalized to the A450 of gp120-Fc binding to ECD in the absence of mannan or Endo H (set at 100%). G. nivalis binding (C and F) was performed in an ELISA using biotinylated G. nivalis captured onto streptavidin plates. The A450 of gp120-Fc binding to G. nivalis (C and F) was normalized as described above. The 50% inhibitory concentration for inhibition from mannan or Endo H treatment was obtained via nonlinear regression using the GraphPad Prism 4 program. Data shown are averages ± standard errors of results from at least three independent experiments.
TABLE 2.
50% inhibitory concentrations (IC50) corresponding to curves presented in Fig. 3
To provide further supporting evidence for this ongoing hypothesis, we developed an Endo H digestion assay. Endo H removes only accessible high-mannose glycans on glycoproteins. Increasing concentrations of Endo H were added to wild-type gp120-Fc, 124mut, or 134mut for a fixed time period, and the resultant Endo H-treated Env was then used in a cell surface DC-SIGN binding assay as described above. Again, if a particular N-glycan on gp120 was more critical for DC-SIGN binding, removal of that N-glycan at the genetic level would make the mutant gp120 more sensitive to Endo H digestion in terms of its resultant ability to bind DC-SIGN. Remarkably, Fig. 3D shows that while 124mut exhibited a 30-fold increase in sensitivity to Endo H digestion (in terms of cell surface DC-SIGN binding ability) compared to that of wild-type gp120-Fc, 134mut showed an even greater, 700-fold increase in sensitivity to Endo H digestion (Table 2). These differences were still significant but not as marked in a soluble DC-SIGN (ECD) binding assay (Fig. 3E and Table 2); 124mut and 134mut showed ∼2-fold and ∼15-fold increases in Endo H sensitivity, respectively. Importantly, when this Endo H digestion assay was used to probe gp120-Fc binding to G. nivalis, 134mut showed no difference from wild-type gp120-Fc in its sensitivity to Endo H digestion and 124mut showed even a slight decrease in sensitivity (increased resistance) to Endo H digestion (Fig. 3F and Table 2).
In summary, these results show that N-glycan sites within the 2G12 epitope, and likely one or more of those missing in 134mut, are optimally positioned to contribute significantly to DC-SIGN binding. Our data also indicate that the N-glycans on gp120 that are preferentially bound by DC-SIGN are different from those bound by at least one other high-mannose binding lectin, G. nivalis.
Specific glycosylation sites within the 2G12 epitope also contribute significantly to optimal virus binding to DC-SIGN.
We next sought to determine if the N-glycan sites in gp120 thus far implicated in optimal DC-SIGN binding were also important for DC-SIGN binding in the context of trimeric envelope spikes on virions. Pseudotyped viruses bearing wild-type JR-CSF envelope, 124mut, 134mut, or no envelope (this “bald” virus was used as a specificity control and as an indicator of the background binding mediated by nonenvelope virion-associated factors) were assayed for binding to recombinant soluble DC-SIGN in a solid-phase assay (Fig. 4A) or cell surface DC-SIGN in a solution-phase assay (Fig. 4B). In the solid-phase assay, equal amounts of the virus preparations indicated in Fig. 4A were allowed to bind to ECD that had been precaptured onto ELISA plates. The plates were then washed, and the amount of virus bound was detected by a p24 ELISA assay. We note that the 134mut pseudotyped virus bound to ECD at only 20% of the wild-type level (P, <0.0001 for the difference between levels of wild-type and 134mut binding), while 124mut viruses bound to ECD at the wild-type level. In the cell surface DC-SIGN binding assay (Fig. 4B), 134mut pseudotyped viruses also bound to DC-SIGN at significantly lower levels than wild-type-envelope pseudotyped viruses (60% of wild-type binding; P = 0.0357), although the reduction in binding to cell surface DC-SIGN was not as dramatic as the reduction in binding to ECD. Again, the 124mut viruses bound to cell surface DC-SIGN at the wild-type level (Fig. 4B). The data in Fig. 4B also suggest that virion-associated nonenvelope factors contribute significantly to cell surface binding.
FIG. 4.
Specific glycosylation sites within the 2G12 epitope also contribute significantly to optimal virus binding to DC-SIGN. (A) HIV-1 virion binding to soluble DC-SIGN (ECD). Five nanograms of wild-type JR-CSF viruses or viruses bearing the indicated mutant Envs (as pseudotyped virions) was spin inoculated onto ELISA plates precoated with 500 nM ECD. After extensive washing, the amount of virus bound was determined by a p24 ELISA. The p24 values obtained with wild-type JR-CSF were normalized to 100%. (B) RajiB DC-SIGN+ cells were pulsed with 20 ng of pseudotyped virions bearing the wild-type JR-CSF Env or the indicated mutant Envs. The amount of virus bound was determined by a p24 ELISA and normalized as described for panel A. Data shown are averages ± standard errors of results from at least three independent experiments. Equal amounts of wild-type and mutant gp120-Fc constructs were added to the assay mixture, and the Fc concentration was determined by an ELISA (as described in Materials and Methods).
Suboptimal DC-SIGN binding also leads to a decrease in DC-SIGN-mediated infection in trans.
Finally, we asked whether a decrease in the level of virus binding to DC-SIGN would translate to a decrease in the level of DC-SIGN-mediated infection in trans. We tested the ability of RajiB DC-SIGN+ cells to bind and transfer the wild-type- and mutant-envelope viruses to 293 CD4+ CCR5+ cells (Fig. 5A). The binding of virus (equivalent to 20 ng of p24) to RajiB DC-SIGN+ cells was performed as described in the legend to Fig. 4B. The virus-bound RajiB DC-SIGN+ cells were subsequently added to 293 CD4+ CCR5+ cells, and the extent of virus infection in trans was assessed 6 to 8 h later by Q-PCR measurement of early reverse transcriptase products. RajiB cells do not express CD4 (60) and cannot be infected with HIV in the presence or absence of DC-SIGN (61). Thus, the early reverse transcriptase products must come from the DC-SIGN-mediated viral transfer to the permissive target cells. Figure 5A shows that the extent of DC-SIGN-mediated infection with the 134mut virus in trans was reduced significantly (P = 0.001) compared to the extent of infection with the wild-type and 124mut viruses. To determine that this reduction in trans infection was due primarily to the suboptimal DC-SIGN binding of the 134mut envelope, and not to any secondary effects on the efficiency of viral entry due to the various N-glycan mutations, the same three virus stocks used in the trans infection assay were assessed in a direct infection assay. Indeed, Fig. 5C shows that the entry efficiency of 134mut virus equaled that of 124mut and wild-type viruses.
FIG. 5.
Suboptimal DC-SIGN binding also leads to a decrease in DC-SIGN-mediated infection in trans. (A and B) The DC-SIGN-mediated trans infection assay was set up after the virus binding assay. (A) Briefly, 20 ng of wild-type, 124mut, or 134mut pseudotype viruses was incubated with RajiB DC-SIGN+ cells, and the cells were then washed vigorously. Virus-bound cells were then added to 293 CD4+ CCR5+ cells and incubated at 37°C for 6 to 8 h. The detection of virus transfer was done by Q-PCR analysis of early reverse transcriptase products (normalized to input beta-globin gene [BetaGlbn] copy numbers) as described in Materials and Methods. (B) Five nanograms of wild-type, 124mut, or 134mut luciferase reporter viruses was incubated with MDDCs, and the cells were washed vigorously. Virus-bound MDDCs were then added to 293 CD4+ CCR5+ cells and incubated at 37°C for 48 h. Virus transfer was indicated by luciferase activity as described in Materials and Methods. The luciferase value obtained with wild-type JR-CSF was normalized at 100%. Data shown are averages ± standard errors of results from four independent experiments for two different donors. (C and D) Wild-type and mutant pseudotype viruses maintained comparable levels of entry efficiency and infectivity. (C) Five nanograms of wild-type JR-CSF virus or the viruses bearing the indicated mutant Envs (as pseudotyped virions) was added onto 293 CD4+ CCR5+ cells, and the cells were incubated at 37°C for 6 to 8 h. Early reverse transcriptase products were detected as described for panel A. (D) Five nanograms of the luciferase reporter viruses mentioned in the legend to panel B was added onto 293 CD4+ CCR5+ cells, and the cells were incubated at 37°C for 48 h. Luciferase activity was detected as described for panel B. Data shown are averages of results from two independent experiments done in triplicate. Wt, wild type; NE, no Env.
Next, we sought to determine if the 134mut virus would show the same reduction in trans infection of MDDCs, which express DC-SIGN in addition to a host of other C-type lectins. Figure 5B shows that the 134mut virus exhibited the same defect in trans infection efficiency compared to the 124mut and wild-type viruses. Again, the same three viral stocks used for the MDDC-based trans infection assay showed equal entry efficiencies when tested in a direct infection assay with 293 CD4+ CCR5+ cells (Fig. 5D). Importantly, trans infection with JR-CSF and 124mut viruses could be blocked to the background levels observed with the 134mut virus and the no-Env control by ECD (data not shown), strongly suggesting that DC-SIGN on MDDCs can mediate the trans infection of CD4+ CCR5+ cells with HIV.
DISCUSSION
We had previously suggested that gp120 binding to DC-SIGN does not involve the 2G12 epitope because 2G12 does not block gp120 binding to cell surface DC-SIGN and mutations of individual N-glycan sites within the 2G12 epitope do not affect DC-SIGN binding (25). However, our present data indicate that DC-SIGN bound to a flexible combination of N-glycans on gp120 and that this interaction, while flexible, was most efficiently mediated by specific N-glycan sites located both within and without the 2G12 epitope. While our former study used a saturation equilibrium binding assay which examined simply whether a particular mutated N-glycan site was critical for DC-SIGN binding, our present study used a combination of two different competitive assays to assess whether the absence of a particular N-glycan in gp120 could be compensated for by other N-glycans on gp120. We reasoned that gp120 forms with mutations in one or more N-glycans, optimally spaced to contribute to the largest subset of N-glycans that can be bound by DC-SIGN, would be more sensitive to mannan competition or Endo H digestion when binding to DC-SIGN was measured.
We confirm and extend our previous observations that multiple N-glycan mutations in gp120 did not affect DC-SIGN binding in the presence or absence of 2G12 (Table 1 and Fig. 2A), suggesting that DC-SIGN was able to bind to other accessible N-glycans even when the 2G12 epitope was blocked. However, we found that the soluble ECD of DC-SIGN blocked 2G12 binding to gp120, indicating that DC-SIGN bound to a larger set or a different combination of N-glycan sites than 2G12. This interaction was highly specific, as a form of ECD with a point mutation (D366A) was not able to block the 2G12-gp120 interaction. 2G12, constrained by the dimeric heavy-chain swap that gives rise to its unorthodox mode of antigen binding (13), likely has a more restricted specificity for the spacing of high-mannose N-glycan sites that give rise to its epitope. Our data are consistent with those of Binley and colleagues, who recently reported that 2G12 inhibits gp120 interaction with DC-SIGN (7), albeit at higher concentrations and in a slightly different way than in our assays. We also note that while ECD can compete with 2G12 binding to gp120, it does not have neutralizing activity in an infection assay. Competition for overlapping binding sites on gp120 does not necessarily translate to other activities (such as neutralization), as exemplified by the many CD4 binding site antibodies that can compete with one another for binding to gp120 but among which only 1b12 has unique, broadly neutralizing activities (8, 12).
Lectins, in general, are flexible in binding to their carbohydrate ligands on cognate glycoproteins (3, 25, 37). Thus, other high-mannose binding lectins, such as concanavalin A (ConA) and CVN, can also inhibit 2G12 binding to gp120, but 2G12 does not inhibit ConA and CVN binding to gp120 (25, 37). Although ConA, CVN, and DC-SIGN can inhibit 2G12 binding to gp120 under the specific conditions examined, there is a clear distinction between DC-SIGN and other high-mannose binding lectins. While both ConA and CVN can bind to Man-α-(1-2)-Man-α moieties, they show specificities for the kind of branching high-mannose structures on which these glycan moieties are presented (30, 33, 34). For example, CVN shows a high level of specificity for terminal Man-α(1-2)-Man-α moieties on Man8 or Man9 glycans but does not bind to Man7 or lower glycans (6, 9). In contrast, DC-SIGN has broader specificity and also binds to internal trisaccharides of high-mannose oligosaccharides but does not bind terminal Man-α-(1-2)-Man-α residues (17, 22). Significantly, despite the considerable flexibility of the binding epitope of DC-SIGN on gp120, we have shown that some N-glycan sites within the 2G12 epitope on gp120 are clearly more critical to DC-SIGN binding than to binding to the high-mannose binding plant lectin G. nivalis (Fig. 3). Indeed, while 134mut showed considerably greater sensitivity to mannan competition and Endo H digestion than wild-type gp120 when DC-SIGN binding was assayed, no such differences were observed when G. nivalis binding was assessed. These results suggest that the lost N-glycan sites within 134mut contribute critically to DC-SIGN binding and that N-glycan sites outside of 134mut can compensate for G. nivalis binding but not DC-SIGN binding. Nevertheless, we concede that the effects of carbohydrate removal can be indirect (such as differential accessibility or processing of the remaining glycans), but regardless of whether the effect is direct (an interpretation that we prefer and that is the most parsimonious explanation for our data) or indirect, the conclusion that the removal of specific glycans can affect DC-SIGN binding remains.
Our results are also consistent with the hypothesis that at least one other site outside of the 2G12 epitope contributes critically to optimal DC-SIGN binding. For example, Fig. 3D shows that 10 mU Endo H can decrease 134mut binding to cell surface DC-SIGN by 80% while having almost no effect on wild-type gp120 binding. Thus, some other N-glycan site(s) not mutated in 134mut is critical for compensating for the loss of N-glycan sites in 134mut during DC-SIGN binding. The removal of this compensatory N-glycan(s) by Endo H likely accounts for the marked difference in DC-SIGN binding. It remains to be seen whether the DC-SIGN binding properties of these N-glycan sites are maintained when the cognate envelopes are produced in primary cells like macrophages versus T cells, where the glycosylation machinery may differ.
The broadly neutralizing antibody 2G12 has a carbohydrate-dependent epitope that can be disrupted by mutations in at least five N-glycan sites (N293, N328, N382, N388, and N393) that give rise to high-mannose structures (13, 43, 50). Although mutagenic analysis suggests that N-linked glycans at positions 293, 328, and 388 are critical for 2G12 interactions (43), structural and modeling studies suggest that the Fab 2G12 dimer most likely binds gp120 at the N-glycan sites positioned at N328 and N388 (13, 43) (all residue numbers are given as homologous N-glycan sites in the JRCSF context as indicated in Table 1 and Fig. 2A). These are the two N-glycan sites that are represented in the 124mut and 134mut envelope proteins. Interestingly, a CVN- and ConA-resistant strain of HIV contains an elimination of the N-glycan site at position N328 (57). In our hands, site-directed mutagenesis of this single site did not affect DC-SIGN binding (Table 1 and reference 25) or binding to CVN and ConA (57). In fact, a triple mutant form of gp120 (124mut) with the N328Q mutation bound DC-SIGN at wild-type levels, whether examined in the context of soluble gp120 or virion-associated trimeric envelope spikes. In contrast, another triple mutant form (134mut), which differed from 124mut by only one N-glycan site mutation in each construct, had significant differences in DC-SIGN binding and transfer (Fig. 3 to 5). The mutations in both these triple glycosylation mutant proteins are within the putative 2G12 epitope. The locations of the respective mutated N-glycan sites can be mapped onto the structure of gp120 bound to CD4, and the remaining high-mannose N-glycan sites can also be visualized (Fig. 6B). Figure 6B shows that 124mut and 134mut necessarily give rise to differently spaced sets of N-glycan sites. We speculate that the N382 site, present in 124mut but absent in 134mut, may be more centrally positioned than other sites so as to satisfy a larger set of intramolecularly spaced N-glycans that can fit into the CRD of the DC-SIGN monomer and oligomer (see below).
FIG. 6.
Pairs of N-glycan sites in gp120 can fit the spatial requirements of the CRDs in the DC-SIGN dimer. (A) The crystal structure of the DC-SIGN CRD as a dimer is shown as a space-filling model. The 16-amino-acid loop that forms the carbohydrate binding region of each CRD of DC-SIGN is highlighted in red (Protein Data Bank identification no., 1XAR). (B) A scaled model of the dimeric CRDs of DC-SIGN is positioned next to a space-filling model of the crystal structure of the CD4-gp120 complex (the V3 loop is included; Protein Data Bank identification no., 2B4C). For clarity, only the 16-amino-acid loop that forms the carbohydrate binding region is shown (red). CD4 is in green, and gp120 is in blue. High-mannose N-glycan sites are shown in yellow, and the specific sites mutated in the 134mut and 124mut envelopes are indicated by asterisks in the respective models. The five N-glycan sites that make up the potential 2G12 epitope are those within the dotted lines. The scaled modeling of this composite structure is described in Materials and Methods.
Recent cryoelectron microscopy analysis of soluble DC-SIGN bound to dengue virus suggests that the CRD monomer can bind to two N-glycan sites spaced 18 Å apart on adjacent envelope dimers on the virion surface (39). This highly unusual mode in which two carbohydrate moieties bind to a single CRD monomer may be due to the elongated “oligosaccharide binding valley” present in the DC-SIGN CRD (17, 39, 49). All five N-glycan sites within the 2G12 epitope are approximately 19 to 23 Å apart and thus fulfill the minimal spatial requirement for at least two N-glycan sites for binding to a single DC-SIGN CRD. It is possible that 134mut disrupts or removes a larger set of N-glycan pairs that can fit into the CRD monomer than 124mut. In addition, the distance between the centers of two adjacently fitted CRDs on the dengue virus surface is 51 Å (39), which is close to the dimer distance in the modeled structure of the DC-SIGN tetramer (16). Since the nanomolar affinity of DC-SIGN-gp120 interactions likely comes from interaction with the tetrameric form of DC-SIGN, we speculate that at least two N-glycan pairs on gp120 are bound by the DC-SIGN dimer or tetramer during native interactions. Figure 6 shows a scaled model of the CRDs of a DC-SIGN dimer placed in the context of gp120. For clarity, only 16 amino acids constituting the oligosaccharide binding valley (17, 49) of the CRDs are shown; clearly, it is possible to fit the binding N-glycan pairs within a single gp120 molecule onto a DC-SIGN dimer. In toto, our data show that DC-SIGN binding to gp120, while flexible, is not completely promiscuous and requires an optimally spaced set of N-glycans that can bind to the native DC-SIGN dimer or tetramer.
While the cis enhancement effect of DC-SIGN appears to be real (11, 28), the trans enhancement effect of DC-SIGN remains controversial, especially with regard to its role in DC-mediated viral transfer to T cells (2, 11, 14, 21, 36, 58, 59). It is likely that while DC-SIGN can play a role in this process, other mannose-dependent C-type lectins (such as macrophage mannose receptor, DC and epithelial cell receptor DEC-205, and DC immunoreceptor) on DCs are also important for DC-mediated infection in trans (14, 18, 51, 59). Nevertheless, the recent discovery of DC-SIGN expression on activated B cells and of its role in relevant pathogenic processes in HIV infection, such as polyclonal Ig class switching (24) and B-cell mediated viral transfer in lymph nodes (40), suggests that the DC-SIGN binding-deficient envelope identified in the present study may be a useful tool for future functional studies of the biology of DC-SIGN. Finally, the combination of N-glycan mutations that result in deficient DC-SIGN binding (e.g., those of 134mut), when engineered in the context of a mucosally transmissible SHIV, may be used to formally test if DC-SIGN binding is indeed important for sexual mucosal transmission.
Acknowledgments
We thank members of the Lee Lab for technical assistance and review of the manuscript and Linda Baum for continued education in all issues related to glycobiology.
P.W.-P.H. is supported by the UCLA AIDS Institute, the UCLA Center for AIDS Research (AI28697), and the State of California Universitywide AIDS Research Program (CC02-LA-001). This work was supported by NIH grants RO1 AI52021 and R21 AI55305 (to B.L.), a Burroughs Wellcome Fund career development award, and a Culpepper biomedical scholar award (to B.L.). We also acknowledge the support of the UCLA AIDS Institute, the UCLA flow cytometry core (funded by NIH Center for AIDS Research grant AI28697), and the Pendelton Charitable Trust.
Footnotes
Published ahead of print on 23 May 2007.
REFERENCES
- 1.Appelmelk, B. J., I. van Die, S. J. van Vliet, C. M. Vandenbroucke-Grauls, T. B. Geijtenbeek, and Y. van Kooyk. 2003. Cutting edge: carbohydrate profiling identifies new pathogens that interact with dendritic cell-specific ICAM-3-grabbing nonintegrin on dendritic cells. J. Immunol. 170:1635-1639. [DOI] [PubMed] [Google Scholar]
- 2.Arrighi, J. F., M. Pion, M. Wiznerowicz, T. B. Geijtenbeek, E. Garcia, S. Abraham, F. Leuba, V. Dutoit, O. Ducrey-Rundquist, Y. van Kooyk, D. Trono, and V. Piguet. 2004. Lentivirus-mediated RNA interference of DC-SIGN expression inhibits human immunodeficiency virus transmission from dendritic cells to T cells. J. Virol. 78:10848-10855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balzarini, J. 2006. Inhibition of HIV entry by carbohydrate-binding proteins. Antiviral Res. 71:237-247. [DOI] [PubMed] [Google Scholar]
- 4.Baribaud, F., S. Pohlmann, T. Sparwasser, M. T. Kimata, Y. K. Choi, B. S. Haggarty, N. Ahmad, T. Macfarlan, T. G. Edwards, G. J. Leslie, J. Arnason, T. A. Reinhart, J. T. Kimata, D. R. Littman, J. A. Hoxie, and R. W. Doms. 2001. Functional and antigenic characterization of human, rhesus macaque, pigtailed macaque, and murine DC-SIGN. J. Virol. 75:10281-10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernhard, O. K., J. Lai, J. Wilkinson, M. M. Sheil, and A. L. Cunningham. 2004. Proteomic analysis of DC-SIGN on dendritic cells detects tetramers required for ligand binding but no association with CD4. J. Biol. Chem. 279:51828-51835. [DOI] [PubMed] [Google Scholar]
- 6.Bewley, C. A., and S. Otero-Quintero. 2001. The potent anti-HIV protein cyanovirin-N contains two novel carbohydrate binding sites that selectively bind to Man(8) D1D3 and Man(9) with nanomolar affinity: implications for binding to the HIV envelope protein gp120. J. Am. Chem. Soc. 123:3892-3902. [DOI] [PubMed] [Google Scholar]
- 7.Binley, J. M., S. Ngo-Abdalla, P. Moore, M. Bobardt, U. Chatterji, P. Gallay, D. R. Burton, I. A. Wilson, J. H. Elder, and A. de Parseval. 2006. Inhibition of HIV Env binding to cellular receptors by monoclonal antibody 2G12 as probed by Fc-tagged gp120. Retrovirology 3:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Botos, I., B. R. O'Keefe, S. R. Shenoy, L. K. Cartner, D. M. Ratner, P. H. Seeberger, M. R. Boyd, and A. Wlodawer. 2002. Structures of the complexes of a potent anti-HIV protein cyanovirin-N and high mannose oligosaccharides. J. Biol. Chem. 277:34336-34342. [DOI] [PubMed] [Google Scholar]
- 10.Boyd, M. R., K. R. Gustafson, J. B. McMahon, R. H. Shoemaker, B. R. O'Keefe, T. Mori, R. J. Gulakowski, L. Wu, M. I. Rivera, C. M. Laurencot, M. J. Currens, J. H. Cardellina II, R. W. Buckheit, Jr., P. L. Nara, L. K. Pannell, R. C. Sowder II, and L. E. Henderson. 1997. Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: potential applications to microbicide development. Antimicrob. Agents Chemother. 41:1521-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burleigh, L., P. Y. Lozach, C. Schiffer, I. Staropoli, V. Pezo, F. Porrot, B. Canque, J. L. Virelizier, F. Arenzana-Seisdedos, and A. Amara. 2006. Infection of dendritic cells (DCs), not DC-SIGN-mediated internalization of human immunodeficiency virus, is required for long-term transfer of virus to T cells. J. Virol. 80:2949-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burton, D. R., R. L. Stanfield, and I. A. Wilson. 2005. Antibody vs. HIV in a clash of evolutionary titans. Proc. Natl. Acad. Sci. USA 102:14943-14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, P. M. Rudd, R. A. Dwek, H. Katinger, D. R. Burton, and I. A. Wilson. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065-2071. [DOI] [PubMed] [Google Scholar]
- 14.Donaghy, H., J. Wilkinson, and A. L. Cunningham. 2006. HIV interactions with dendritic cells: has our focus been too narrow? J. Leukoc. Biol. 80:1001-1012. [DOI] [PubMed] [Google Scholar]
- 15.Endres, M. J., P. R. Clapham, M. Marsh, M. Ahuja, J. D. Turner, A. McKnight, J. F. Thomas, B. Stoebenau-Haggarty, S. Choe, P. J. Vance, T. N. Wells, C. A. Power, S. S. Sutterwala, R. W. Doms, N. R. Landau, and J. A. Hoxie. 1996. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell 87:745-756. [DOI] [PubMed] [Google Scholar]
- 16.Feinberg, H., Y. Guo, D. A. Mitchell, K. Drickamer, and W. I. Weis. 2005. Extended neck regions stabilize tetramers of the receptors DC-SIGN and DC-SIGNR. J. Biol. Chem. 280:1327-1335. [DOI] [PubMed] [Google Scholar]
- 17.Feinberg, H., D. A. Mitchell, K. Drickamer, and W. I. Weis. 2001. Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science 294:2163-2166. [DOI] [PubMed] [Google Scholar]
- 18.Figdor, C. G., Y. van Kooyk, and G. J. Adema. 2002. C-type lectin receptors on dendritic cells and Langerhans cells. Nat. Rev. Immunol. 2:77-84. [DOI] [PubMed] [Google Scholar]
- 19.Geijtenbeek, T. B., D. S. Kwon, R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, J. Middel, I. L. Cornelissen, H. S. Nottet, V. N. KewalRamani, D. R. Littman, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100:587-597. [DOI] [PubMed] [Google Scholar]
- 20.Geijtenbeek, T. B., and Y. van Kooyk. 2003. DC-SIGN: a novel HIV receptor on DCs that mediates HIV-1 transmission. Curr. Top. Microbiol. Immunol. 276:31-54. [DOI] [PubMed] [Google Scholar]
- 21.Granelli-Piperno, A., A. Pritsker, M. Pack, I. Shimeliovich, J. F. Arrighi, C. G. Park, C. Trumpfheller, V. Piguet, T. M. Moran, and R. M. Steinman. 2005. Dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin/CD209 is abundant on macrophages in the normal human lymph node and is not required for dendritic cell stimulation of the mixed leukocyte reaction. J. Immunol. 175:4265-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo, Y., H. Feinberg, E. Conroy, D. A. Mitchell, R. Alvarez, O. Blixt, M. E. Taylor, W. I. Weis, and K. Drickamer. 2004. Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat. Struct. Mol. Biol. 11:591-598. [DOI] [PubMed] [Google Scholar]
- 23.Gurney, K. B., J. Elliott, H. Nassanian, C. Song, E. Soilleux, I. McGowan, P. A. Anton, and B. Lee. 2005. Binding and transfer of human immunodeficiency virus by DC-SIGN+ cells in human rectal mucosa. J. Virol. 79:5762-5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He, B., X. Qiao, P. J. Klasse, A. Chiu, A. Chadburn, D. M. Knowles, J. P. Moore, and A. Cerutti. 2006. HIV-1 envelope triggers polyclonal Ig class switch recombination through a CD40-independent mechanism involving BAFF and C-type lectin receptors. J. Immunol. 176:3931-3941. [DOI] [PubMed] [Google Scholar]
- 25.Hong, P. W., K. B. Flummerfelt, A. de Parseval, K. Gurney, J. H. Elder, and B. Lee. 2002. Human immunodeficiency virus envelope (gp120) binding to DC-SIGN and primary dendritic cells is carbohydrate dependent but does not involve 2G12 or cyanovirin binding sites: implications for structural analyses of gp120-DC-SIGN binding. J. Virol. 76:12855-12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu, Q., I. Frank, V. Williams, J. J. Santos, P. Watts, G. E. Griffin, J. P. Moore, M. Pope, and R. J. Shattock. 2004. Blockade of attachment and fusion receptors inhibits HIV-1 infection of human cervical tissue. J. Exp. Med. 199:1065-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, C. C., M. Tang, M. Y. Zhang, S. Majeed, E. Montabana, R. L. Stanfield, D. S. Dimitrov, B. Korber, J. Sodroski, I. A. Wilson, R. Wyatt, and P. D. Kwong. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, B., G. Leslie, E. Soilleux, U. O'Doherty, S. Baik, E. Levroney, K. Flummerfelt, W. Swiggard, N. Coleman, M. Malim, and R. W. Doms. 2001. cis expression of DC-SIGN allows for more efficient entry of human and simian immunodeficiency viruses via CD4 and a coreceptor. J. Virol. 75:12028-12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lekkerkerker, A. N., Y. van Kooyk, and T. B. Geijtenbeek. 2006. Viral piracy: HIV-1 targets dendritic cells for transmission. Curr. HIV Res. 4:169-176. [DOI] [PubMed] [Google Scholar]
- 30.Loris, R., D. Maes, F. Poortmans, L. Wyns, and J. Bouckaert. 1996. A structure of the complex between concanavalin A and methyl-3,6-di-O-(alpha-d-mannopyranosyl)-alpha-d-mannopyranoside reveals two binding modes. J. Biol. Chem. 271:30614-30618. [DOI] [PubMed] [Google Scholar]
- 31.Lue, J., M. Hsu, D. Yang, P. Marx, Z. Chen, and C. Cheng-Mayer. 2002. Addition of a single gp120 glycan confers increased binding to dendritic cell-specific ICAM-3-grabbing nonintegrin and neutralization escape to human immunodeficiency virus type 1. J. Virol. 76:10299-10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell, D. A., A. J. Fadden, and K. Drickamer. 2001. A novel mechanism of carbohydrate recognition by the C-type lectins DC-SIGN and DC-SIGNR. Subunit organization and binding to multivalent ligands. J. Biol. Chem. 276:28939-28945. [DOI] [PubMed] [Google Scholar]
- 33.Moothoo, D. N., B. Canan, R. A. Field, and J. H. Naismith. 1999. Man alpha1-2 Man alpha-OMe-concanavalin A complex reveals a balance of forces involved in carbohydrate recognition. Glycobiology 9:539-545. [DOI] [PubMed] [Google Scholar]
- 34.Moothoo, D. N., and J. H. Naismith. 1998. Concanavalin A distorts the beta-GlcNAc-(1→2)-Man linkage of beta-GlcNAc-(1→2)-alpha-Man-(1→3)-[beta-GlcNAc-(1→2)-alpha-Man-(1→6)]-Man upon binding. Glycobiology 8:173-181. [DOI] [PubMed] [Google Scholar]
- 35.Naarding, M. A., I. S. Ludwig, F. Groot, B. Berkhout, T. B. Geijtenbeek, G. Pollakis, and W. A. Paxton. 2005. Lewis X component in human milk binds DC-SIGN and inhibits HIV-1 transfer to CD4+ T lymphocytes. J. Clin. Investig. 115:3256-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nobile, C., C. Petit, A. Moris, K. Skrabal, J. P. Abastado, F. Mammano, and O. Schwartz. 2005. Covert human immunodeficiency virus replication in dendritic cells and in DC-SIGN-expressing cells promotes long-term transmission to lymphocytes. J. Virol. 79:5386-5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pashov, A., S. MacLeod, R. Saha, M. Perry, T. C. VanCott, and T. Kieber-Emmons. 2005. Concanavalin A binding to HIV envelope protein is less sensitive to mutations in glycosylation sites than monoclonal antibody 2G12. Glycobiology 15:994-1001. [DOI] [PubMed] [Google Scholar]
- 38.Pohlmann, S., F. Baribaud, and R. W. Doms. 2001. DC-SIGN and DC-SIGNR: helping hands for HIV. Trends Immunol. 22:643-646. [DOI] [PubMed] [Google Scholar]
- 39.Pokidysheva, E., Y. Zhang, A. J. Battisti, C. M. Bator-Kelly, P. R. Chipman, C. Xiao, G. G. Gregorio, W. A. Hendrickson, R. J. Kuhn, and M. G. Rossmann. 2006. Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN. Cell 124:485-493. [DOI] [PubMed] [Google Scholar]
- 40.Rappocciolo, G., P. Piazza, C. L. Fuller, T. A. Reinhart, S. C. Watkins, D. T. Rowe, M. Jais, P. Gupta, and C. R. Rinaldo. 2006. DC-SIGN on B lymphocytes is required for transmission of HIV-1 to T lymphocytes. PLoS Pathog. 2:e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 42.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. Ollmann Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sewell, A. K., and D. A. Price. 2001. Dendritic cells and transmission of HIV-1. Trends Immunol. 22:173-175. [DOI] [PubMed] [Google Scholar]
- 45.Sharron, M., S. Pohlmann, K. Price, E. Lolis, M. Tsang, F. Kirchhoff, R. W. Doms, and B. Lee. 2000. Expression and coreceptor activity of STRL33/Bonzo on primary peripheral blood lymphocytes. Blood 96:41-49. [PubMed] [Google Scholar]
- 46.Snyder, G. A., J. Ford, P. Torabi-Parizi, J. A. Arthos, P. Schuck, M. Colonna, and P. D. Sun. 2005. Characterization of DC-SIGN/R interaction with human immunodeficiency virus type 1 gp120 and ICAM molecules favors the receptor's role as an antigen-capturing rather than an adhesion receptor. J. Virol. 79:4589-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soilleux, E. J., L. S. Morris, G. Leslie, J. Chehimi, Q. Luo, E. Levroney, J. Trowsdale, L. J. Montaner, R. W. Doms, D. Weissman, N. Coleman, and B. Lee. 2002. Constitutive and induced expression of DC-SIGN on dendritic cell and macrophage subpopulations in situ and in vitro. J. Leukoc. Biol. 71:445-457. [PubMed] [Google Scholar]
- 48.Su, S. V., K. B. Gurney, and B. Lee. 2003. Sugar and spice: viral envelope-DC-SIGN interactions in HIV pathogenesis. Curr. HIV Res. 1:87-99. [DOI] [PubMed] [Google Scholar]
- 49.Su, S. V., P. Hong, S. Baik, O. A. Negrete, K. B. Gurney, and B. Lee. 2004. DC-SIGN binds to HIV-1 glycoprotein 120 in a distinct but overlapping fashion compared with ICAM-2 and ICAM-3. J. Biol. Chem. 279:19122-19132. [DOI] [PubMed] [Google Scholar]
- 50.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turville, S., J. Wilkinson, P. Cameron, J. Dable, and A. L. Cunningham. 2003. The role of dendritic cell C-type lectin receptors in HIV pathogenesis. J. Leukoc. Biol. 74:710-718. [DOI] [PubMed] [Google Scholar]
- 52.Turville, S. G., J. Arthos, K. M. Donald, G. Lynch, H. Naif, G. Clark, D. Hart, and A. L. Cunningham. 2001. HIV gp120 receptors on human dendritic cells. Blood 98:2482-2488. [DOI] [PubMed] [Google Scholar]
- 53.Turville, S. G., P. U. Cameron, A. Handley, G. Lin, S. Pohlmann, R. W. Doms, and A. L. Cunningham. 2002. Diversity of receptors binding HIV on dendritic cell subsets. Nat. Immunol. 3:975-983. [DOI] [PubMed] [Google Scholar]
- 54.Turville, S. G., J. J. Santos, I. Frank, P. U. Cameron, J. Wilkinson, M. Miranda-Saksena, J. Dable, H. Stossel, N. Romani, M. Piatak, Jr., J. D. Lifson, M. Pope, and A. L. Cunningham. 2004. Immunodeficiency virus uptake, turnover, and 2-phase transfer in human dendritic cells. Blood 103:2170-2179. [DOI] [PubMed] [Google Scholar]
- 55.van Liempt, E., C. M. Bank, P. Mehta, J. J. Garcia-Vallejo, Z. S. Kawar, R. Geyer, R. A. Alvarez, R. D. Cummings, Y. Kooyk, and I. van Die. 2006. Specificity of DC-SIGN for mannose- and fucose-containing glycans. FEBS Lett. 580:6123-6131. [DOI] [PubMed] [Google Scholar]
- 56.Wilkinson, J., and A. L. Cunningham. 2006. Mucosal transmission of HIV-1: first stop dendritic cells. Curr. Drug Targets 7:1563-1569. [DOI] [PubMed] [Google Scholar]
- 57.Witvrouw, M., V. Fikkert, A. Hantson, C. Pannecouque, B. R. O'Keefe, J. McMahon, L. Stamatatos, E. de Clercq, and A. Bolmstedt. 2005. Resistance of human immunodeficiency virus type 1 to the high-mannose binding agents cyanovirin N and concanavalin A. J. Virol. 79:7777-7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, L., A. A. Bashirova, T. D. Martin, L. Villamide, E. Mehlhop, A. O. Chertov, D. Unutmaz, M. Pope, M. Carrington, and V. N. KewalRamani. 2002. Rhesus macaque dendritic cells efficiently transmit primate lentiviruses independently of DC-SIGN. Proc. Natl. Acad. Sci. USA 99:1568-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu, L., and V. N. KewalRamani. 2006. Dendritic-cell interactions with HIV: infection and viral dissemination. Nat. Rev. Immunol. 6:859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu, L., T. D. Martin, M. Carrington, and V. N. KewalRamani. 2004. Raji B cells, misidentified as THP-1 cells, stimulate DC-SIGN-mediated HIV transmission. Virology 318:17-23. [DOI] [PubMed] [Google Scholar]
- 61.Wu, L., T. D. Martin, Y. C. Han, S. K. Breun, and V. N. KewalRamani. 2004. Trans-dominant cellular inhibition of DC-SIGN-mediated HIV-1 transmission. Retrovirology 1:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu, X., C. Borchers, R. J. Bienstock, and K. B. Tomer. 2000. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry 39:11194-11204. [DOI] [PubMed] [Google Scholar]