Abstract
CD4+ T-cell depletion during acute human immunodeficiency virus infection occurs predominantly in the gastrointestinal mucosa. Using experimental data on SIVmac251 viral load in blood and CD4+ T cells in the jejunum, we modeled the kinetics of CD4+ T-cell infection and death and estimated the viral infectivity. The infectivity of SIVmac251 is higher than previously estimated for SHIV89.6P infection, but this higher infectivity is offset by a lower average peak viral load in SIVmac251. Thus, the dynamics of target cell infection and death are remarkably similar between a CXCR4- and a CCR5-tropic infection in vivo.
Depletion of CD4+ T cells during acute human immunodeficiency virus (HIV) infection occurs throughout the body but is greater in the gastrointestinal mucosa than in peripheral blood, lymph nodes, or other organs (6, 13, 21). Similarly, in simian immunodeficiency virus (SIV) infection of macaques, extensive CD4+ T-cell depletion in mucosal tissues is observed (relative to peripheral blood) within the first few weeks (10, 15, 19, 20, 25-27). Targeting mucosal CD4+ T cells is thought to be due to the relatively high expression of CCR5 and T-cell activation at this site and is independent of the route of challenge. Recently it has been shown that CD4+ T-cell depletion in the gut during acute SIV infection can be reduced by vaccination (18) and that preservation of CD4+ T cells is associated with improved disease outcome (14). Thus, understanding the determinants of CD4+ T-cell infection and killing in the gut may have important implications for both vaccination and therapy for HIV.
We recently demonstrated a correlation between peak viral load and CD4+ T-cell depletion in peripheral blood following SHIV89.6P infection (10). This relationship was predicted by mathematical models of CD4+ T-cell infection and death and allowed estimation of the infectivity of virus. The relationship between viral infectivity and peak viral load allows prediction of the impact of a given reduction in viral load (for example, through vaccination) on the extent of CD4+ T-cell depletion during acute infection (10). SHIV89.6P is a CXCR4-tropic virus, and thus differs from CCR5-tropic SIVs and HIV in the range of CD4+ T cells it targets. In SHIV89.6P infection, extensive CD4+ T-cell depletion is observed in peripheral blood, whereas significantly less depletion is observed in peripheral blood with CCR5-tropic SIV and HIV infections. There are two likely factors contributing to the observed reduction in CD4+ T-cell depletion in SIV: (i) reduced infectivity of virus for its target CD4+ T cells and (ii) a reduced proportion of the total CD4+ T-cell pool available for infection (i.e., only CCR5+ cells are targets for infection). By focusing on the dynamics of CD4+ T-cell infection and death in the gut, we are able to directly estimate the infectivity of SIV for its target cells. This study demonstrates that SIV is equally infectious for its target cells and that the reduced depletion of CD4+ T cells can be explained simply on the basis of a reduced target cell range.
MATERIALS AND METHODS
Animals.
In this study, we analyze published data (18, 19) on CD4+ T-cell kinetics in the jejunum following SIV infection. Briefly, 20 rhesus macaques (Macaca mulatta) were challenged intravenously with 100 monkey infectious doses of uncloned pathogenic SIVmac251; 6 of these macaques received prior vaccination with plasmid DNA encoding SIV envelope, Gag, and Pol, and were boosted with recombinant adenovirus encoding the same antigens. Plasma and jejunum tissue samples were collected at various time points by biopsy or necropsy, and CD4+ T-cell percentages and plasma viral loads were determined (18, 19).
Mathematical model.
Here, we use the same mathematical model of CD4+ T-cell infection and death that we used previously to model the depletion of CD4+ T cells over time and estimate the infectivity of SHIV89.6P in blood (10). Briefly, assuming that production and natural death of CD4+ T cells are low compared with the loss due to viral infection during acute SIV, we ignore the relatively small magnitudes of uninfected cell turnover. Thus, the standard model of HIV infection dynamics reduces to:
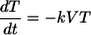 |
(1) |
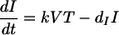 |
(2) |
That is, uninfected CD4+ T cells (T) decay as a function of viral levels (V) and become infected cells (I), determined by the infectivity of virus (k). Once infected, CD4+ T cells die at a rate dI (1.49 day−1, as estimated independently) (5). The total number of CD4 cells present at any time is:
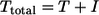 |
(3) |
This model assumes mass-action kinetics for cell infection and that the system is completely closed (that is, we neglect any lymphocyte redistribution as a possible cause of CD4 decline). We also use data for viral load in peripheral blood as a substitute for viral load in the gut.
Bootstrapping and confidence intervals.
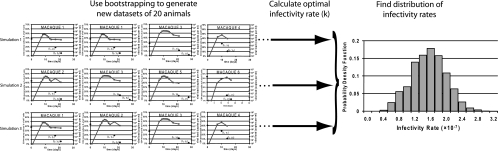
The analysis of data from the jejunum is more complex than that of peripheral blood, since each animal has just two samples taken during infection (in addition to a baseline sample), whereas for peripheral blood data it is more practical to obtain considerably more samples postchallenge. To obtain an accurate estimate of the distribution for viral infectivity given the data, we applied a bootstrapping approach (7, 11). Bootstrapping provides a reliable way to construct a confidence interval for a parameter by resampling from the original data to create replicate data sets. We performed 10,000 simulations, executed with Matlab (version 7; Mathworks, MA). For each simulation, we (i) sampled 20 animals (with replacement) from the pool of 20 macaques to generate a new data set (there are ∼7 × 1010 unique sampling combinations), (ii) used the plasma viral load measured in each animal to predict the infection and death of jejunum CD4+ memory T cells in each sampled animal over the duration of infection [a piecewise linear expression of the log-transformed viral load data was used for viral time courses, V(t), in the mathematical model], and (iii) obtained an optimal infectivity rate (k) for the population of all animals that produced the best fit of the predicted total CD4+ T cells to the observed CD4+ T cells over the entire generated data set for the simulation. This procedure is illustrated schematically in Fig. 1. To obtain the optimal infectivity rate, we set up an array containing all data points from the sampled monkeys (the array contained replicated data points associated with animals selected multiple times in the generated data set); this data array contained time points and numbers of CD4+ memory T cells for the data during infection, linked to the corresponding macaque. We also established an algorithm to compute an additional array of the same length, containing the predicted total CD4+ T cells from the mathematical model, Ttotal, corresponding to the respective macaque and time point. The predicted CD4+ T-cell number, Ttotal (as defined in equations 1 to 3), strictly depends on an (initially unknown) infectivity rate, the viral load time course (from the measured viral load data) that corresponds specifically to the macaque in question, and the baseline percentage of CD4+ T cells in each animal (which was used as the initial conditions in the mathematical model). We then used Matlab's built-in optimization function “lsqcurvefit” to determine the optimal infectivity rate that minimizes the residual difference (sum of squares) between the model-predicted CD4+ T cells and the generated data array. To ensure the robustness of our procedure, we also performed the optimization with different algorithms and optimization tools, and we concluded that the small discrepancies with these approaches were not significantly different. The 10,000 simulations each generated an optimal infectivity, and these infectivity rates were sorted and a 95% confidence interval was calculated using the bias- corrected and accelerated method (described in detail in references 7 and 12).
FIG. 1.
Schematic diagram of our bootstrapping methodology to obtain an empirical probability distribution for the viral rate of infectivity. We performed 10,000 simulations, and for each simulation, we sampled 20 animals (with replacement) from the pool of 20 macaques to generate a new data set. From each generated data set, we calculated an optimal infectivity rate for the population of animals in the given data set, using the plasma viral load measured in each animal and our mathematical model to predict the infection and death of jejunum CD4+ memory T cells in each sampled animal. We then determined the empirical distribution of the viral infectivity rate from the 10,000 simulations.
RESULTS
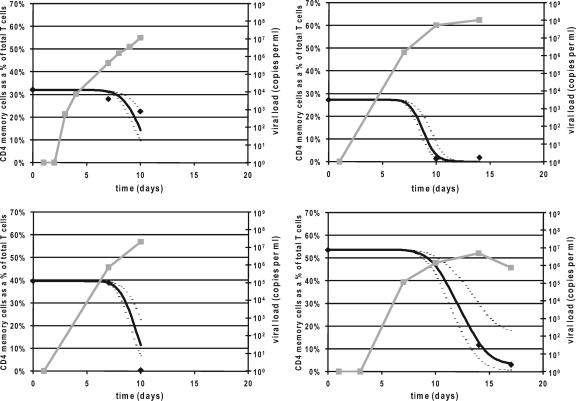
We calculated a mean infectivity of 1.45 × 10−7 ml copy−1 day−1 (95% confidence interval, 0.58 to 2.33 × 10−7 ml copy−1 day−1). Figure 2 shows simulations for four animals using this infectivity rate (along with the experimental data). This infectivity parameter provides insights into the relationship between peak viral load and CD4+ T-cell depletion in jejunum (10) and allows us to predict the impact of reductions in viral load (Fig. 3A). The average peak viral load for unvaccinated macaques infected with SIVmac251 is approximately 4.57 × 107 copies ml−1 (22). In the absence of interventions, this results in almost complete depletion of CD4+ memory T cells in the jejunum as observed experimentally and predicted by our fits (Fig. 3A). Indeed, our modeling suggests that peak viral loads in control animals are considerably greater than the “threshold” level required to deplete the majority of CD4+ memory T cells (a threefold reduction in peak viral load would result in ∼94% depletion of CD4 cells in the gut, and even a 10-fold reduction in peak viral load would still result in ∼60% depletion) (Fig. 3A).
FIG. 2.
CD4+ T-cell depletion in the jejunum. The data of percentage of T cells in the jejunum that are CD4+ memory cells for four typical monkeys are shown (dark diamonds), along with our model prediction of the CD4+ T cells based on our mean estimate of the infectivity across all animals (black curve). Upper and lower 95% confidence interval values for the rate of infectivity across all animals are also shown (dashed curves). The viral load for each animal is plotted in gray.
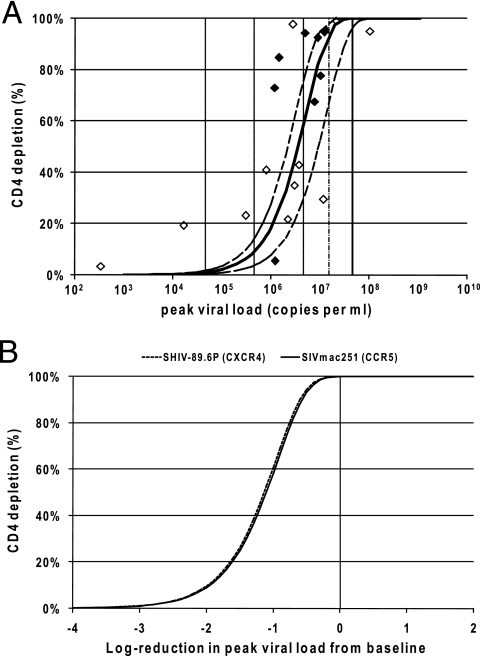
FIG. 3.
(A) The predicted relationship between peak viral load and CD4+ T-cell depletion in acute SIVmac251 infection (mean, solid curve; 95% confidence interval, dashed curves) is shown. The diamond points represent the experimental data from the 20 macaques (filled diamonds correspond to monkeys in which the peak viral load may not have been reached and the unfilled diamonds correspond to monkeys in which the peak viral load had been reached). The solid black line segment refers to the geometric mean peak viral load in control animals challenged with SIVmac251. The vertical lines indicate the effects of different reductions in peak viral load (the thin line curve refers to a threefold reduction in viral load, and the dashed curves refer to 1-log10, 2-log10, and 3-log10 reductions in viral load). The predicted relationship is calculated by the following equation: proportion of CD4 depleted 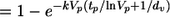 , where tp is the time of viral peak, Vp is the viral peak value, and dv is the rate of postpeak viral decay (1.49 day−1) (5), and the proportion depleted is calculated by the equation (baseline CD4 − minimum CD4)/baseline CD4. Note that for illustration, the viral peak for each monkey is used to provide a single data value (diamonds), but all viral load values for each animal are used in the simulations in calculating predicted infection dynamics and optimal infectivity rates. (B) Relationships between CD4+ T-cell depletion and reduction in peak viral load are compared between the CCR5-tropic SIVmac251 (solid curve) and CXCR4-tropic SHIV89.6P (dashed curve) viruses. The curves are aligned to show the effect of reducing the viral load from the average peak viral load in controls (zero reduction, represented by the solid line, is associated with the average viral peak in each of the respective viral infections). The rate of infectivity, viral decay rate, and time of viral peak resulting from infection with each virus contributes to the relationship between viral peak and depletion of CD4+ T cells; our presented relationships were found to be robust to changes in the viral decay rate and time of viral peak (but primarily influenced by the rate of viral infectivity).
, where tp is the time of viral peak, Vp is the viral peak value, and dv is the rate of postpeak viral decay (1.49 day−1) (5), and the proportion depleted is calculated by the equation (baseline CD4 − minimum CD4)/baseline CD4. Note that for illustration, the viral peak for each monkey is used to provide a single data value (diamonds), but all viral load values for each animal are used in the simulations in calculating predicted infection dynamics and optimal infectivity rates. (B) Relationships between CD4+ T-cell depletion and reduction in peak viral load are compared between the CCR5-tropic SIVmac251 (solid curve) and CXCR4-tropic SHIV89.6P (dashed curve) viruses. The curves are aligned to show the effect of reducing the viral load from the average peak viral load in controls (zero reduction, represented by the solid line, is associated with the average viral peak in each of the respective viral infections). The rate of infectivity, viral decay rate, and time of viral peak resulting from infection with each virus contributes to the relationship between viral peak and depletion of CD4+ T cells; our presented relationships were found to be robust to changes in the viral decay rate and time of viral peak (but primarily influenced by the rate of viral infectivity).
The rate of viral infectivity is higher for CCR5-tropic SIVmac251 infection than for CXCR4-tropic SHIV89.6P infection (1.45 × 10−7 ml copy−1 day−1 compared with 4.40 × 10−8 ml copy−1 day−1, a ∼3.3-fold greater infection rate). However, the peak viral load in blood for CCR5-tropic SIVmac251 infections (4.57 × 107 copies ml−1) is approximately 2.7 times lower than for CXCR4-tropic SHIV89.6P infection (mean of 1.22 × 108 copies ml−1) (10, 22). Although the infection rates differ considerably, surprisingly, we find that if we align the infection curves to account for the average peak viral load in control animals in the respective models, reducing the viral load is expected to have the same outcome, in terms of CD4+ T-cell depletion, in SHIV89.6P as in SIVmac251 infection (Fig. 3B). In both systems, we predict that a 1-log10 reduction in peak viral load will result in ∼60% CD4 depletion, and a 2-log10 reduction in viral load will result in depletion of only ∼10% of CD4 cells (Fig. 3B). These reductions in viral loads are achievable in many animal models with current vaccination strategies (2-4, 16-18, 24).
DISCUSSION
The current study compares depletion of total CD4+ T cells in peripheral blood for the CXCR4-tropic SHIV89.6P with depletion of memory CD4+ T cells from the jejunum in the CCR5-tropic SIVmac251. This is necessary because CCR5-tropic viruses target only a subset of activated memory (CCR5+) T cells. However, in each case we have compared viral load with “target cell depletion” and have found similar outcomes. Thus, the observed differences in CD4+ T-cell depletion in the blood between the two viruses can be attributed solely to the effects of their different target cell ranges, as each infects and depletes a similar proportion of its target cells.
Understanding the relationship between viral infectivity and peak viral load provides insights into the level of viral control needed to prevent CD4+ T-cell depletion in acute SIV infection. We have previously demonstrated an ∼10-day delay in the growth of CD8+ T-cell numbers and the control of virus in SHIV infection, suggesting that cytotoxic T lymphocytes do too little too late to control the establishment of chronic viral infection (8, 9). More recently, this has been confirmed in SIV infection (1, 23). The results of our modeling suggest that, as long as vaccination controls peak viral loads, this delay in viral control will have relatively little effect on the ability of vaccines to prevent CD4+ T-cell depletion. Surprisingly, we have found that the same reduction in peak viral load in CCR5-tropic SIVmac251 will have a similar effect in preserving target cells to that seen in CXCR4-tropic SHIV89.6P. Future work should address whether the balance between infectivity and viral load is similar in HIV infection and thus whether vaccination will have the same impact on preserving CD4+ T cells in HIV infection.
Acknowledgments
We thank Ruy Ribeiro for useful comments during preparation of the manuscript and Tony Pettitt for advice concerning the bootstrap.
This work was supported by the James S. McDonnell Foundation 21st Century Research Award/Studying Complex Systems and the Australian National Health and Medical Research Council. M.P.D. is a Sylvia and Charles Viertel Senior Medical Research Fellow. D.P.W. is a UNSW Vice-Chancellor's Research Fellow.
Footnotes
Published ahead of print on 16 May 2007.
REFERENCES
- 1.Abdel-Motal, U. M., J. Gillis, K. Manson, M. Wyand, D. Montefiori, K. Stefano-Cole, R. C. Montelaro, J. D. Altman, and R. P. Johnson. 2005. Kinetics of expansion of SIV gag-specific CD8+ T lymphocytes following challenge of vaccinated macaques. Virology 333:226-238. [DOI] [PubMed] [Google Scholar]
- 2.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2002. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Vaccine 20:1949-1955. [DOI] [PubMed] [Google Scholar]
- 3.Baba, T. W., V. Liska, R. Hofmann-Lehmann, J. Vlasak, W. Xu, S. Ayehunie, L. A. Cavacini, M. R. Posner, H. Katinger, G. Stiegler, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, Y. Lu, J. E. Wright, T. C. Chou, and R. M. Ruprecht. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200-206. [DOI] [PubMed] [Google Scholar]
- 4.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, M. G. Lewis, E. A. Emini, J. W. Shiver, and N. L. Letvin. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 5.Brandin, E., R. Thorstensson, S. Bonhoeffer, and J. Albert. 2006. Rapid viral decay in simian immunodeficiency virus-infected macaques receiving quadruple antiretroviral therapy. J. Virol. 80:9861-9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenchley, J. M., T. W. Schacker, L. E. Ruff, D. A. Price, J. H. Taylor, G. J. Beilman, P. L. Nguyen, A. Khoruts, M. Larson, A. T. Haase, and D. C. Douek. 2004. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 200:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpenter, J., and J. Bithell. 2000. Bootstrap confidence intervals: when, which, what? A practical guide for medical statisticians. Stat. Med. 19:1141-1164. [DOI] [PubMed] [Google Scholar]
- 8.Davenport, M. P., R. M. Ribeiro, and A. S. Perelson. 2004. Kinetics of virus-specific CD8+ T cells and the control of human immunodeficiency virus infection. J. Virol. 78:10096-10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davenport, M. P., L. Zhang, A. Bagchi, A. Fridman, T.-M. Fu, W. Schleif, J. W. Shiver, R. M. Ribeiro, and A. S. Perelson. 2005. High-potency human immunodeficiency virus vaccination leads to delayed and reduced CD8+ T-cell expansion but improved virus control. J. Virol. 79:10059-10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davenport, M. P., L. Zhang, J. W. Shiver, D. R. Casmiro, R. M. Ribeiro, and A. S. Perelson. 2006. Influence of peak viral load on the extent of CD4+ T-cell depletion in simian HIV infection. J. Acquir. Immune Defic. Syndr. 41:259-265. [DOI] [PubMed] [Google Scholar]
- 11.Davison, A. C., and D. V. Hinkley. 1997. Bootstrap methods and their application. Cambridge University Press, New York, NY.
- 12.Efron, B. 1987. Better bootstrap confidence-intervals. J. Am. Stat. Assoc. 82:171-185. [Google Scholar]
- 13.Guadalupe, M., E. Reay, S. Sankaran, T. Prindiville, J. Flamm, A. McNeil, and S. Dandekar. 2003. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J. Virol. 77:11708-11717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Letvin, N. L., J. R. Mascola, Y. Sun, D. A. Gorgone, A. P. Buzby, L. Xu, Z. Y. Yang, B. Chakrabarti, S. S. Rao, J. E. Schmitz, D. C. Montefiori, B. R. Barker, F. L. Bookstein, and G. J. Nabel. 2006. Preserved CD4+ central memory T cells and survival in vaccinated SIV-challenged monkeys. Science 312:1530-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, Q., L. Duan, J. D. Estes, Z. M. Ma, T. Rourke, Y. Wang, C. Reilly, J. Carlis, C. J. Miller, and A. T. Haase. 2005. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 434:1148-1152. [DOI] [PubMed] [Google Scholar]
- 16.Lifson, J. D., J. L. Rossio, R. Arnaout, L. Li, T. L. Parks, D. K. Schneider, R. F. Kiser, V. J. Coalter, G. Walsh, R. J. Imming, B. Fisher, B. M. Flynn, N. Bischofberger, M. Piatak, Jr., V. M. Hirsch, M. A. Nowak, and D. Wodarz. 2000. Containment of simian immunodeficiency virus infection: cellular immune responses and protection from rechallenge following transient postinoculation antiretroviral treatment. J. Virol. 74:2584-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mascola, J. R., G. Stiegler, T. C. VanCott, H. Katinger, C. B. Carpenter, C. E. Hanson, H. Beary, D. Hayes, S. S. Frankel, D. L. Birx, and M. G. Lewis. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207-210. [DOI] [PubMed] [Google Scholar]
- 18.Mattapallil, J. J., D. C. Douek, A. Buckler-White, D. Montefiori, N. L. Letvin, G. J. Nabel, and M. Roederer. 2006. Vaccination preserves CD4 memory T cells during acute simian immunodeficiency virus challenge. J. Exp. Med. 203:1533-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattapallil, J. J., D. C. Douek, B. Hill, Y. Nishimura, M. Martin, and M. Roederer. 2005. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature 434:1093-1097. [DOI] [PubMed] [Google Scholar]
- 20.Mattapallil, J. J., Z. Smit-McBride, M. McChesney, and S. Dandekar. 1998. Intestinal intraepithelial lymphocytes are primed for gamma interferon and MIP-1β expression and display antiviral cytotoxic activity despite severe CD4+ T-cell depletion in primary simian immunodeficiency virus infection. J. Virol. 72:6421-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mehandru, S., M. A. Poles, K. Tenner-Racz, A. Horowitz, A. Hurley, C. Hogan, D. Boden, P. Racz, and M. Markowitz. 2004. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J. Exp. Med. 200:761-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker, R. A., M. M. Regan, and K. A. Reimann. 2001. Variability of viral load in plasma of rhesus monkeys inoculated with simian immunodeficiency virus or simian-human immunodeficiency virus: implications for using nonhuman primate AIDS models to test vaccines and therapeutics. J. Virol. 75:11234-11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reynolds, M. R., E. Rakasz, P. J. Skinner, C. White, K. Abel, Z. M. Ma, L. Compton, G. Napoe, N. Wilson, C. J. Miller, A. Haase, and D. I. Watkins. 2005. CD8+ T-lymphocyte response to major immunodominant epitopes after vaginal exposure to simian immunodeficiency virus: too late and too little. J. Virol. 79:9228-9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 25.Veazey, R. S., M. DeMaria, L. V. Chalifoux, D. E. Shvetz, D. R. Pauley, H. L. Knight, M. Rosenzweig, R. P. Johnson, R. C. Desrosiers, and A. A. Lackner. 1998. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 280:427-431. [DOI] [PubMed] [Google Scholar]
- 26.Veazey, R. S., K. G. Mansfield, I. C. Tham, A. C. Carville, D. E. Shvetz, A. E. Forand, and A. A. Lackner. 2000. Dynamics of CCR5 expression by CD4+ T cells in lymphoid tissues during simian immunodeficiency virus infection. J. Virol. 74:11001-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veazey, R. S., I. C. Tham, K. G. Mansfield, M. DeMaria, A. E. Forand, D. E. Shvetz, L. V. Chalifoux, P. K. Sehgal, and A. A. Lackner. 2000. Identifying the target cell in primary simian immunodeficiency virus (SIV) infection: highly activated memory CD4+ T cells are rapidly eliminated in early SIV infection in vivo. J. Virol. 74:57-64. [DOI] [PMC free article] [PubMed] [Google Scholar]