Abstract
Respiratory syncytial virus (RSV), a member of the Paramyxoviridae family, encodes a small hydrophobic (SH) protein of unknown function. Parainfluenza virus 5 (PIV5), a prototypical paramyxovirus, also encodes an SH protein, which inhibits tumor necrosis factor alpha (TNF-α) signaling. In this study, recombinant PIV5 viruses without their own SH but containing RSV SH (from RSV strain A2 or B1) in its place (PIV5ΔSH-RSV SH) and RSV lacking its own SH (RSVΔSH) were generated and analyzed. The results indicate that the SH protein of RSV has a function similar to that of PIV5 SH and that it can inhibit TNF-α signaling.
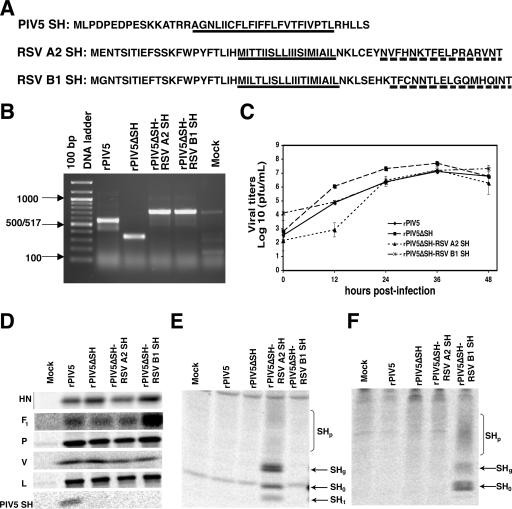
Human respiratory syncytial virus (RSV) is the leading cause of lower respiratory tract infections in infants and young children (17). RSV, along with the prototype paramyxovirus parainfluenza virus 5 (PIV5; formerly known as simian virus 5), is a member of the Paramyxoviridae family, which includes important human and animal pathogens. Both RSV and PIV5 encode small hydrophobic (SH) proteins, which are type II transmembrane proteins. The SH protein of RSV contains 64 (RSV subgroup A) or 65 (RSV subgroup B) amino acid residues (Fig. 1A) (3-5, 14). Some studies have suggested that the RSV SH protein may have a role in viral fusion (9, 19) or in changing membrane permeability (15). However, RSV lacking the SH gene (RSVΔSH) is viable, causes syncytium formation, and grows as well as the wild-type virus (1, 10, 11), indicating that the SH protein is not necessary for virus entry into host cells or syncytium formation (19). RSVΔSH is attenuated in animals, indicating that RSV plays an important role in viral pathogenesis (1). Interestingly, recombinant PIV5 lacking the SH gene (rPIV5ΔSH) has a similar phenotype: it has normal growth in vitro, but it is attenuated in vivo (7). Studies of rPIV5ΔSH have shown that the SH protein is necessary for the inhibition of tumor necrosis factor alpha (TNF-α)-induced apoptosis in L929 cells (12). Recent work suggests that the SH protein of mumps virus is a functional counterpart of the PIV5 SH protein (22), even though the PIV5 and mumps SH proteins have no sequence homology. We hypothesized that the SH protein of RSV may be functionally similar to other SH proteins from members of the Paramyxoviridae family. To test this hypothesis, recombinant viruses that contained the RSV SH gene of strain A2 or B1 in place of the PIV5 SH gene were produced and confirmed by reverse transcription (RT)-PCR (Fig. 1B). The rPIV5 and rPIV5ΔSH viruses grow to similar titers, although rPIV5ΔSH virus grows slightly faster in the first stages of infection (Fig. 1C) (6, 22). Growth of the rPIV5ΔSH-RSV SH recombinant viruses was comparable to that of rPIV5 and rPIV5ΔSH up to 2 days postinfection (dpi). Occasionally, a delay in the growth of one or both of the recombinant viruses was observed, but by 24 or 36 h the viruses had always reached titers comparable to that of the wild-type virus (Fig. 1C). The plaques formed by the rPIV5, rPIV5ΔSH, and rPIV5ΔSH-RSV SH viruses in BHK cells were of a similar size and morphology (data not shown). Radioimmunoprecipitation analyses showed that synthesis of the PIV5 V, P, and L proteins was similar in HeLa cells infected by rPIV5, rPIV5ΔSH, or rPIV5ΔSH-RSV SH (Fig. 1D). The levels of HN and F1 proteins were somewhat variable but were generally equal to or greater in rPIV5ΔSH-RSV SH-infected cells than in rPIV5-infected cells.
FIG. 1.
Generation and analysis of PIV5ΔSH-RSV SH. (A) Sequences of the SH proteins of PIV5 and RSV strains A2 and B1. The predicted transmembrane domains of the proteins are underlined with solid lines. The amino acid sequences used to generate the RSV A2 and B1 SH antibodies are underlined with dashed lines. (B) Confirmation of the generation of the PIV5ΔSH-RSV SH viruses. MDBK cells were infected at an MOI of 5, and RNA was extracted 1 dpi as previously described (8). RT-PCR, using primers BH191 and BH 194, amplified the region surrounding the SH gene in rPIV5-, rPIV5ΔSH-, rPIV5ΔSH-RSV A2 SH-, and rPIV5ΔSH-RSV B1 SH-infected cells. Lane 1 is the 100-bp DNA ladder. (C) Growth kinetics of rPIV5 and rPIV5ΔSH-RSV SH. The growth rates of recombinant viruses are shown. MDBK cells were infected at an MOI of 4. Samples from the media were taken at 0, 12, 24, 36, and 48 h postinfection, and triplicate samples were used for plaque assays. The error bars represent the standard errors of the means. (D) Expression of PIV5 proteins. HeLa cells were infected at an MOI of 10. At 1 dpi, the cells were labeled with 35S-Met and 35S-Cys, and proteins were immunoprecipitated using PIV5-specific antibodies. (E and F) Expression of the RSV SH protein by the PIV5ΔSH-RSV SH viruses. HeLa cells were infected as before, and antibodies that recognized the C terminus of the RSV SH protein were used for immunoprecipitation. (E) Anti-RSV A2 SH antibody. (F) Anti-RSV B1 antibody.
The SH protein from strain A2 is found in four different forms in infected cells: SH0, SHg, SHp, and SHt. SH0, the 7.5-kDa nonglycosylated form, is the full-length unmodified protein and is the most common form expressed (16). SHg is the 13- to 15-kDa N-linked glycosylated form of the protein and is the precursor of SHp. SHp (21 to 40 kDa) is a polylactosaminoglycan-modified form of the protein, and SHt (4.8 kDa) is a truncated form of SH0 that is generated by translation initiation at the second AUG of the SH sequence (14). Similarly, different glycosylated and nonglycosylated forms of the B1 SH protein have been detected in infected cells (4). To examine the expression of the RSV SH proteins encoded by recombinant viruses, RSV SH antibodies against the SH protein of strain A2 or B1 of RSV were generated, using the C-terminal 17 amino acids of each protein (Fig. 1A). These antisera specifically recognized glycosylated and nonglycosylated RSV SH from each strain from either rPIV5ΔSH-RSV A2 SH- or rPIV5ΔSH-RSV B1 SH-infected cells by radioimmunoprecipitation (Fig. 1E and F).
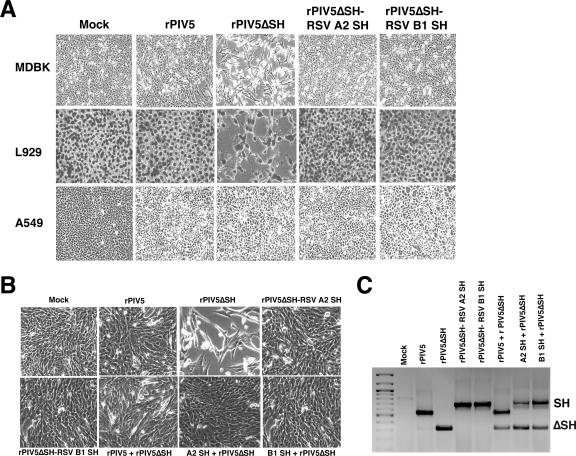
Previous studies demonstrated that rPIV5 infection does not cause a significant cytopathic effect (CPE) in MDBK, HeLa, A549, or L929 cells, whereas rPIV5ΔSH infection causes a severe CPE in MDBK and L929, but not HeLa or A549, cells (7, 12, 22). To determine if the RSV SH protein was able to replace the PIV5 SH protein in blocking cell death, MDBK, L929, and A549 cells were infected with rPIV5, rPIV5ΔSH, or rPIV5ΔSH-RSV A2 (or B1) SH at a multiplicity of infection (MOI) of 5. Consistent with previous work (7, 12, 22), rPIV5ΔSH caused a notable CPE in MDBK and L929 cells but not in A549 cells (Fig. 2A). In contrast, cells infected with the RSV SH recombinant viruses showed no visible CPE, similar to those infected with rPIV5. Since the only difference between the rPIV5ΔSH virus and the RSV SH recombinant virus is the replacement of the PIV5 SH protein with the RSV SH protein, these data suggest that the RSV SH protein was able to take the place of the PIV5 SH protein in preventing the cells from dying. To determine if the RSV SH protein could prevent apoptosis induced by rPIV5ΔSH infection, MDBK cells were infected with rPIV5ΔSH-RSV A2 (or B1) SH and, at 1 dpi, were coinfected with rPIV5ΔSH. Cells that were coinfected with rPIV5ΔSH-RSV A2 (or B1) SH had a minimal CPE and a phenotype that was more similar to the rPIV5-plus-rPIV5ΔSH-infected cells (Fig. 2B). To ensure that the cells were indeed coinfected, RT-PCRs using RNA from infected cells were performed to detect the presence of genomic RNA from rPIV5ΔSH and rPIV5 or rPIV5ΔSH-RSV SH (Fig. 2C). Thus, expression of the RSV SH protein prevented the rPIV5ΔSH-infected cells from undergoing apoptosis.
FIG. 2.
The PIV5ΔSH-RSV SH viruses inhibit apoptosis induced by PIV5ΔSH. (A) MDBK, L929, and A549 cells were mock infected or infected with rPIV5, rPIV5ΔSH, rPIV5ΔSH-RSV A2 SH, or rPIV5ΔSH-RSV B1 SH at an MOI of 5. The cells were photographed at 3 (L929 cells) or 4 (MDBK and A549 cells) dpi, using a Nikon Eclipse TE300 inverted microscope. (B) MDBK cells were mock infected or infected with rPIV5, rPIV5ΔSH-RSV A2 SH, or rPIV5ΔSH-RSV B1 SH at an MOI of 5. One day after being infected with these viruses, the cells were infected with rPIV5ΔSH at an MOI of 5. The cells were photographed, using a Nikon Eclipse TE300 inverted microscope, 3 days after being infected with the rPIV5ΔSH virus. (C) Confirmation of coinfection. RT-PCRs were carried out as for Fig. 1B. PCR products corresponding to ΔSH or SH are indicated.
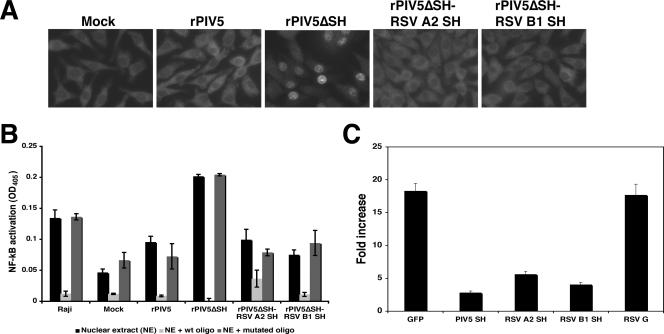
The absence of SH protein during PIV5 infection induces an increased production of TNF-α and activation of NF-κB, resulting in the translocation of the p65 subunit of NF-κB into the nucleus of rPIV5ΔSH-infected L929 cells (12). As expected, nuclear localization of p65 was observed in rPIV5ΔSH-infected cells and not in rPIV5-infected cells (Fig. 3A). Little if any p65 was found in the nuclei of rPIV5ΔSH-RSV A2 (or B1) SH-infected cells. While 30% of cells showed nuclear p65 after rPIV5ΔSH infection, only 1 to 3% of cells showed nuclear p65 after rPIV5ΔSH-RSV A2 (or B1) SH infection. These results were further confirmed, using an NF-κB binding enzyme-linked immunosorbent assay (ELISA) using immobilized DNA oligomers (Fig. 3B).
FIG. 3.
Activation of NF-κB by recombinant PIV5 and inhibition of TNF-α-mediated NF-κB activation. (A) Activation of NF-κB. L929 cells were mock infected or infected with rPIV5, rPIV5ΔSH, rPIV5ΔSH-RSV A2 SH, or rPIV5ΔSH-RSV B1 SH. At 1 dpi, the cells were fixed with 0.5% formaldehyde and permeabilized, using a 0.1% saponin-phosphate-buffered saline solution. The cells were incubated overnight, first with mouse antibody against the p65 subunit of NF-κB and then with a fluorescein isothiocyanate-labeled anti-mouse immunoglobulin G antibody. Fluorescence was observed, using an Olympus BX-60 digital microscope with Image Pro Plus software. (B) Activation of NF-κB using ELISA. L929 cells were mock infected or infected with rPIV5, rPIV5ΔSH, rPIV5ΔSH-RSV A2 SH, or rPIV5ΔSH-RSV B1 SH at an MOI of 10. At 1 dpi, nuclear extracts were obtained as described by Lin et al. (12). One microgram of protein was used for the ELISA-based NF-κB transcription assay from Active Motif (TransAM NF-κB family kit; Active Motif, Carlsbad, CA) according to the manufacturer's instructions. (C). The RSV SH protein inhibits TNF-α-induced NF-κB activation. L929F cells were transfected with pCAGGS-GFP, pCAGGS-PIV5 SH, pCAGGS-RSV A2 SH, pCAGGS-RSV B1 SH, or pCAGGS RSV G. All samples were also transfected with pκB-TATA-Luc (18) and phRL-TK (Promega), which were used to normalize transfection efficiencies among the different plasmid mixtures. At 1 day posttransfection, the media were replaced with Opti-MEM or Opti-MEM and TNF-α (10 ng/ml) and incubated for 4 h at 37°C and 5% CO2. Luciferase activity was measured, using a Veritas microplate luminometer (Turner Biosystems) for samples treated with TNF-Opti-MEM or Opti-MEM alone. Luciferase activity was measured as a ratio of firefly luciferase activity to Renilla luciferase activity. Fold increase, ratio of the amount of luciferase activity of TNF-α-treated cells to that of untreated cells; OD405, optical density at 405 nm; wt, wild type. Error bars represent the standard errors of the means.
Although biologically detectable levels of TNF-α are produced after PIV5 infection, rPIV5ΔSH infection induces a significantly larger amount of the cytokine (12). Previous work from our laboratory indicates that the SH protein of PIV5 is able to block TNF- α signaling. To study whether the RSV SH protein has a similar function, L929F cells were transfected with a luciferase gene under the control of an NF-κB-responsive promoter along with a plasmid containing the gene for RSV A2 (or B1) SH. Cells were also transfected with a plasmid containing the Renilla luciferase gene under the control of the herpes simplex virus thymidine kinase promoter as a transfection control, as previously described (22). At 1 day posttransfection, the media were replaced with Opti-MEM or Opti-MEM and TNF-α (10 ng/ml) and the cells were incubated for another 4 h. Samples were then examined for dual luciferase activities. The RSV SH protein from both strains inhibited NF-κB activation by TNF-α (Fig. 3C). As previously observed (22), cells transfected with PIV5 SH also inhibited TNF from activating NF-κB. As a control, the RSV G protein did not inhibit TNF-α-induced NF-κB activation.
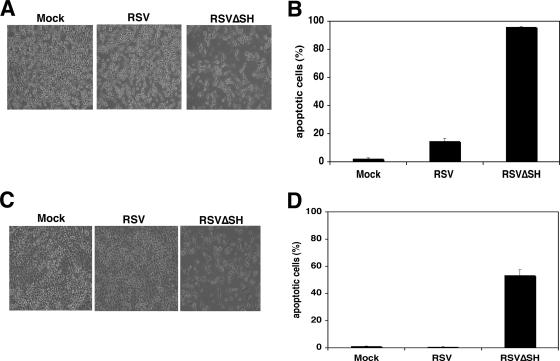
To determine whether the RSV SH protein had a role in inhibiting apoptosis during RSV infection, we generated a recombinant RSV lacking SH (RSVΔSH) by deleting the sequences in an antigenome cDNA from the M gene end (GE) signal through the SH GE, juxtaposing the 3′ untranslated region of the M gene with the SH GE. Recombinant RSV was then recovered as previously described (2). Infection of L929 cells with RSV resulted in noticeable CPE 1 dpi compared to that for mock-infected cells (Fig. 4A). However, more notable CPE was observed in RSVΔSH-infected cells at the same time point. To determine whether the cell death observed after RSVΔSH infection was due to apoptosis, a terminal deoxynucleotidyltransferase-mediated dUTP-fluoroscein isothiocyanate nick end labeling (TUNEL) assay was performed (12). As shown in Fig. 4B, only 15% of the RSV-infected cells were apoptotic by 1 dpi, compared to 95% of the RSVΔSH-infected cells at this time point. Thus, while RSV infection was capable of inducing apoptosis in L929 cells, RSVΔSH infection caused significantly more apoptosis in this cell line. To determine if the increased cell death caused by RSVΔSH was cell type specific, A549 cells, a lung epithelial cell line, were tested with RSV and RSVΔSH. The results, shown in Fig. 4C and D, indicate that while little or no CPE was observed in the mock- or RSV-infected cells, considerable CPE and apoptosis were observed in the RSVΔSH-infected cells 3 dpi, confirming the role of SH in apoptosis. These results support the hypothesis that the paramyxovirus SH proteins play a role in blocking cell death (22). However, it is not clear whether the inhibition of apoptosis by the RSV SH protein during RSV infection is due to inhibition of the TNF-α pathway. A549 cells, which can produce TNF-α but are not sensitive to TNF-α-induced death (13, 20), also displayed an increased level of apoptosis after RSVΔSH infection compared with that for wild-type infection, suggesting that, while the RSV SH protein may play a role in the TNF-α pathway, it may inhibit apoptosis by an alternative mechanism as well.
FIG. 4.
RSVΔSH virus causes accelerated apoptosis. (A) Induction of cell death by RSVΔSH. The RSVΔSH (A2 strain) we generated was slightly different from that used in other studies (1, 19, 21). L929 cells were mock infected or infected with RSV or RSVΔSH at an MOI of 3. Cells were photographed 1 dpi, using a Nikon Eclipse TE300 inverted microscope. (B) L929 cells were mock infected or infected with RSV or RSVΔSH at an MOI of 1. At 1 dpi, the cells were collected and fixed with 0.5% formaldehyde, and DNA fragmentation was measured with a TUNEL assay. Error bars represent the standard errors of the means. (C). RSVΔSH accelerated cell death in A549 cells. A549 cells were mock infected or infected with RSV or RSVΔSH at an MOI of 3. The cells were photographed 3 dpi. (D) RSVΔSH accelerated apoptosis in A549 cells. A549 cells were mock infected or infected with RSV or RSVΔSH at an MOI of 1. At 1 dpi, the cells were collected for a TUNEL assay. Error bars represent the standard errors of the means.
Acknowledgments
We thank Shao-Cong Sun for providing pNF-κB-TATA-F-Luc, Peter Collins for a reverse genetics system to make the recombinant virus, and Brian Murphy for RSV. We appreciate Ping Wang, Laurie Shuman, and Rebecca Wilson and other members of Biao He's laboratory for discussions and technical help.
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases to B.H. (R01 AI 051372).
Footnotes
Published ahead of print on 9 May 2007.
REFERENCES
- 1.Bukreyev, A., S. S. Whitehead, B. R. Murphy, and P. L. Collins. 1997. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J. Virol. 71:8973-8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins, P. L., M. G. Hill, E. Camargo, H. Grosfeld, R. M. Chanock, and R. M. Murphy. 1995. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc. Natl. Acad. Sci. USA 92:11563-11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins, P. L., and G. Mottet. 1993. Membrane orientation and oligomerization of the small hydrophobic protein of human respiratory syncytial virus. J. Gen. Virol. 74:1445-1450. [DOI] [PubMed] [Google Scholar]
- 4.Collins, P. L., R. A. Olmsted, and P. R. Johnson. 1990. The small hydrophobic protein of human respiratory syncytial virus: comparison between antigenic subgroups A and B. J. Gen. Virol. 71:1571-1576. [DOI] [PubMed] [Google Scholar]
- 5.Collins, P. L., and G. W. Wertz. 1985. The 1A protein gene of human respiratory syncytial virus: nucleotide sequence of the mRNA and a related polycistronic transcript. Virology 141:283-291. [DOI] [PubMed] [Google Scholar]
- 6.He, B., G. P. Leser, R. G. Paterson, and R. A. Lamb. 1998. The paramyxovirus SV5 small hydrophobic (SH) protein is not essential for virus growth in tissue culture cells. Virology 250:30-40. [DOI] [PubMed] [Google Scholar]
- 7.He, B., G. Y. Lin, J. E. Durbin, R. K. Durbin, and R. A. Lamb. 2001. The SH integral membrane protein of the paramyxovirus simian virus 5 is required to block apoptosis in MDBK cells. J. Virol. 75:4068-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He, B., R. G. Paterson, C. D. Ward, and R. A. Lamb. 1997. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology 237:249-260. [DOI] [PubMed] [Google Scholar]
- 9.Heminway, B. R., Y. Yu, Y. Tanaka, K. G. Perrine, E. Gustafson, J. M. Bernstein, and M. S. Galinski. 1994. Analysis of respiratory syncytial virus F, G, and SH proteins in cell fusion. Virology 200:801-805. [DOI] [PubMed] [Google Scholar]
- 10.Jin, H., H. Zhou, X. Cheng, R. Tang, M. Munoz, and N. Nguyen. 2000. Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2-2 genes are attenuated in vitro and in vivo. Virology 273:210-218. [DOI] [PubMed] [Google Scholar]
- 11.Karron, R. A., D. A. Buonagurio, A. F. Georgiu, S. S. Whitehead, J. E. Adamus, M. L. Clements-Mann, D. O. Harris, V. B. Randolph, S. A. Udem, B. R. Murphy, and M. S. Sidhu. 1997. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc. Natl. Acad. Sci. USA 94:13961-13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin, Y., A. C. Bright, T. A. Rothermel, and B. He. 2003. Induction of apoptosis by paramyxovirus simian virus 5 lacking a small hydrophobic gene. J. Virol. 77:3371-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mikamo, H., A. K. Johri, L. C. Paoletti, L. C. Madoff, and A. B. Onderdonk. 2004. Adherence to, invasion by, and cytokine production in response to serotype VIII group B streptococci. Infect. Immun. 72:4716-4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olmsted, R. A., and P. L. Collins. 1989. The 1A protein of respiratory syncytial virus is an integral membrane protein present as multiple, structurally distinct species. J. Virol. 63:2019-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez, M., B. Garcia-Barreno, J. A. Melero, L. Carrasco, and R. Guinea. 1997. Membrane permeability changes induced in Escherichia coli by the SH protein of human respiratory syncytial virus. Virology 235:342-351. [DOI] [PubMed] [Google Scholar]
- 16.Rixon, H. W., G. Brown, J. Aitken, T. McDonald, S. Graham, and R. J. Sugrue. 2004. The small hydrophobic (SH) protein accumulates within lipid-raft structures of the Golgi complex during respiratory syncytial virus infection. J. Gen. Virol. 85:1153-1165. [DOI] [PubMed] [Google Scholar]
- 17.Selwyn, B. J. 1990. The epidemiology of acute respiratory tract infection in young children: comparison of findings from several developing countries. Rev. Infect. Dis. 12(Suppl. 8):S870-S888. [DOI] [PubMed] [Google Scholar]
- 18.Sun, S. C., J. Elwood, C. Beraud, and W. C. Greene. 1994. Human T-cell leukemia virus type I Tax activation of NF-κB/Rel involves phosphorylation and degradation of IκBα and RelA (p65)-mediated induction of the c-rel gene. Mol. Cell. Biol. 14:7377-7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Techaarpornkul, S., N. Barretto, and M. E. Peeples. 2001. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J. Virol. 75:6825-6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webster, J. C., R. M. Huber, R. L. Hanson, P. M. Collier, T. F. Haws, J. K. Mills, T. C. Burn, and E. A. Allegretto. 2002. Dexamethasone and tumor necrosis factor-α act together to induce the cellular inhibitor of apoptosis-2 gene and prevent apoptosis in a variety of cell types. Endocrinology 143:3866-3874. [DOI] [PubMed] [Google Scholar]
- 21.Whitehead, S. S., A. Bukreyev, M. N. Teng, C. Y. Firestone, M. St. Claire, W. R. Elkins, P. L. Collins, and B. R. Murphy. 1999. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J. Virol. 73:3438-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson, R. L., S. M. Fuentes, P. Wang, E. C. Taddeo, A. Klatt, A. J. Henderson, and B. He. 2006. Function of small hydrophobic proteins of paramyxovirus. J. Virol. 80:1700-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]