Abstract
The heterodimer Ku70/80 Ku is the DNA-binding component of the DNA-PK complex required for the nonhomologous end-joining pathway. It participates in numerous nuclear processes, including telomere and chromatin structure maintenance, replication, and transcription. Ku interacts with retroviral preintegration complexes and is thought to interfere with the retroviral replication cycle, in particular the formation of 2-long terminal repeat (LTR) viral DNA circles, viral DNA integration, and transcription. We describe here the effect of Ku80 on both provirus integration and the resulting transgene expression in cells transduced with retroviral vectors. We found that transgene expression was systematically higher in Ku80-deficient xrs6 cells than in Ku80-expressing CHO cells. This higher expression was observed irrespective of the presence of the viral LTR and was also not related to the nature of the promoter. Real-time PCR monitoring of the early viral replicative steps demonstrated that the absence of Ku80 does not affect the efficiency of transduction. We analyzed the transgene distributions localization in nucleus by applying a three-dimensional reconstruction model to two-dimensional fluorescence in situ hybridization images. This indicated that the presence of Ku80 resulted in a bias toward the transgenes being located at the periphery of the nucleus associated with their being repressed; in the absence of this factor the transgenes tend to be randomly distributed and actively expressed. Therefore, although not strictly required for retroviral integration, Ku may be involved in targeting retroviral elements to chromatin domains prone to gene silencing.
The integration of proviral DNA into the chromosomes of infected cells is an essential step in retroviral replication and consequently in the use of retroviral vectors for gene therapy. Integration is carried out by the viral integrase, which is responsible for the two catalytic steps: 3′ processing and strand transfer leading to the integrated provirus (7). In cells either infected by retroviruses or transduced by retrovirus-derived vectors, the viral/transgene cDNA is integrated by a preintegration complex (PIC) that contains the proviral cDNA, integrase, and viral and cellular factors. In particular, two cellular proteins—the barrier-to-integration factor and the lens-epithelium-derived growth factor (LEDGF/p75)—are required for the engagement of the PIC on chromatin, leading to efficient integration (16, 27, 38). Several factors of the cellular repair pathways may be involved in the integration process. Poly(ADP-ribose)-polymerase (PARP) (18, 24), DNA-dependent protein kinase (DNA-PK) (13), Ataxia-telangiectasia mutated and Rad3-related kinase (12) facilitate integration, whereas Rad52 (33), XPB, and XPD (56) reduce the efficiency of integration. Another repair factor, Rd18 is involved in integrase stability (42). Finally, Ku interacts with both Moloney murine leukemia virus and human immunodeficiency virus type 1 (HIV-1) PICs, suggesting that at some point during integration, Ku is directly associated with the integration machinery (35). Ku is an heterodimer containing the Ku70 and Ku80 DNA-binding subunits and it recruits kinase activity to form the DNA-PK involved in the nonhomologous end-joining pathway of double-strand break repair (36). Ku binds DNA extremities and other DNA structures, including gaps, nicks, and hairpins, with a high affinity (6, 17, 40). Ku also binds specific internal DNA sequences in a variety of genes, such as c-myc, collagen III, tartrate-resistant acid phosphatase, osteocalcin, and Wilson disease genes (22, 23, 43-45, 47, 55). It participates in numerous nuclear processes, including telomere and chromatin structure maintenance, replication, and transcription. Thus, it is viewed as a DNA caretaker, protecting cells against genome instability (14, 53, 54). Ku is also required for V(D)J recombination, which is an event closely related to integration, and for Drosophila mobile DNA transposition and yeast Ty retrotransposition (5, 15). Previous reports have shown that Ku is important for forming 2-long terminal repeat (LTR) circles from unintegrated linear retroviral cDNA. By preventing accumulation of linear viral DNA in the cell nucleus, Ku may help prevent apoptosis after retroviral transduction of DNA-PK-deficient murine scid cells (13, 29, 35). Changes in the levels of Ku80 in cells are associated with changes in the expression of an integrated provirus (28). The possible interaction of Ku with the PIC while influencing transcription of integrated provirus led us to investigate whether Ku80 is involved in the targeting of integration into the cell chromatin. We monitored both the integration efficiency and the subsequent level of transgene expression after retroviral transduction according to the cellular Ku80 content. We also assessed the distribution of transgenes in the nucleus by applying a three-dimensional (3D) reconstruction model to 2D fluorescence in situ hybridization (FISH) images. We report that the presence of Ku80 does not affect transgene integration quantitatively but results in a bias toward a peripheral distribution associated with a repressed state; in absence of Ku80 the distribution tends to be random, and the transgene expression is increased.
MATERIALS AND METHODS
Plasmids and vectors.
The pHR′CMVlacZ (pHR′βgal) lentiviral shuttle vector encoding lacZ was a gift from Didier Trono. The pRRLSIN.ppT.PGK/GFP (pSINGfp) lentiviral shuttle vector containing gfp and pdeltaR8.74, containing the HIV gag-pol gene, was from the University of Turin Medical School. pRRLSIN.CMVlacZ (pSINβgal) was constructed by replacing the Acc65I-NotI fragment of pSINGfp by the Acc65I-NotI fragment of pHR′βgal. pMDG plasmid encoding the vesicular stomatitis virus envelope glycoprotein (VSV-G) was a gift from D. Trono. pNL-Luc-E−R+ lacking the HIV-1 envelope gene and in which the viral nef is replaced by the luciferase reporter gene (11) was a gift from Nathaniel Landau.
Cells and viruses.
Chinese hamster ovary (CHO) and X-ray sensitive-6 (xrs6) cells were from the American Type Culture Collection. xrs6 cells expressing human Ku80 (xrs6-Ku) were obtained after stable transfection of xrs6 cells with human Ku80 as previously described (28). CHO and derived cells were cultured in minimal essential medium supplemented with 10% fetal bovine serum, penicillin, streptomycin, and nonessential amino acids. Human 293T cells were used for retrovector production and were cultured in Dulbecco modified Eagle medium supplemented as described previously (30). Retroviral vectors were produced by triple transfection of 293T cells with the following plasmids: a vector plasmid encoding β-galactosidase (pHR′βgal or pSINβgal) or green fluorescent protein (pSINGfp), pdeltaR8.74, and pMDG. A total of 5 × 106 293T cells were transfected on day 1 with 5 μg of the vector plasmid, 1.75 μg of the plasmid encoding gag-pol, and 3.25 μg of pMDG by using Superfect transfection reagent (QIAGEN SA, Courtabeuf, France). Pseudotyped HIV-1 particles bearing the luciferase reporter gene were produced by cotransfecting 293T cells with 7.5 μg of both the proviral pNL-Luc-E−R+ and the VSV-G expression vector pCMV-G. Cells were washed 3 h after transfection. Fresh medium was added to the cells on day 2. Supernatants were harvested on days 3 and 4, centrifuged at 2,000 rpm, and filtered through 0.45-μm-pore-size filters. The retrovirus titers were evaluated by using either X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining or counting gfp-expressing cells after infection of NIH 3T3 cells. For transduction assays, CHO, xrs6 and xrs6-Ku cell lines were plated on day 1 at 2,500 cells per well in a 96-well plate and transduced. The β-galactosidase (β-Gal) assay was carried out with 100 μl of lysis buffer per well and 25 μl of lysate in 100 μl of CPRG reagent. A FACScalibur (Becton Dickinson) cell sorter was used for fluorescence-activated cell sorting analysis of cells pooled from three independent wells.
Luciferase activity quantification in cell lysates and living cells.
A total of 2 × 104 cells were plated on the center of glass-bottom dishes (MatTek Corp.) and infected with 4.2 ng of VSV-G pseudotyped HIV-1 particles bearing the luciferase reporter gene. After 3 h of incubation at 37°C, cells were washed and cultured in complete medium for 3 days before analysis. Mock infections with equivalent amounts of p24 from supernatants from 293T cells transfected only with pNL-Luc-E−R+ were performed in parallel as controls. To quantify luciferase activity in cell lysates, cells on MatTek plates were lysed with 100 μl of luciferase cell culture lysis reagent (Promega France, Charbonnières, France), and 10 μl of the lysate was used for luminescence quantification with the Promega Luciferase reporter 1000 assay system in a Veritas microplate luminometer (Turner BioSystems).
The technique for quantification of bioluminescence in infected living cells has been described in detail elsewhere (46). Briefly, the system, custom-built by ScienceWares, includes a fully automated inverted microscope (200 M; Carl Zeiss, Germany) and is housed in a light-tight dark box. Low-level light emission was collected by using an Imaging Photon Detector (IPD 3; Photek, Ltd., East Sussex, United Kingdom), and bright-field images were obtained with a charge-coupled device camera (Coolsnap HQ; Roper Scientific). Observations were made with a Plan-Neofluar 25X (oil, NA = 0.8) objective (Carl Zeiss). The data acquisition software allowed superposition of photon events with bright-field images and selective quantification of photons in delimited surfaces. Living cells on MatTek plates were analyzed in phosphate-buffered saline (PBS) plus 5% fetal calf serum at room temperature in the presence of endotoxin-free beetle d-luciferin (Promega) (final concentration, 1 mM).
X-ray resistance assay.
CHO, xrs6, or xrs6-Ku cells were plated on day 1 in a 96-well plate and were irradiated on day 2 with at a dose of 1, 2, or 4 Gy. Cell viability was monitored on day 5 by MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay as previously described (28).
Quantification of cDNAs.
CHO and derived cells (2 × 10 5) were spread on glass coverslips in six-well plates. At various times after transduction, genomic DNA was isolated by using the QIAamp blood DNA minikit (QIAGEN SA, France). Primers for total DNA amplification, AA55M/LM667, and the primers for the detection of 2-LTR circles, HIV F and HIV R, were as described previously (8). The primers GAPDH1 (CTTGGCAGCGCCAGTGGATGCAG) and GAPDH2 (CTTCACCACCATGGAGAAGGC) were used to amplify the housekeeping GAPDH gene. The genomic template was amplified in triplicate in a volume of 25 μl containing QPCR master mix PCR Sybergreen qPCR kit (Finnzymes, Espoo, Finland) and 0.5 μM concentrations of each primer. Thermal amplification was carried out on a Opticon 2 DNA engine (Bio-Rad, Marnes-la-Coquette, France) using cycling profiles described previously (8). Each sample was examined in triplicate for each set of primers. Copy numbers were determined by reference to a standard curve prepared by amplification of various quantities between 1 and 106 copies of cloned DNA with matching sequences. Integrated transgene was quantified by real-time B2A-LTR nested PCR in triplicate. We used LM667 and B2A5 Chinese hamster primer TTCACAACTCTCCGTGGATGGTGG, and nested PCR was performed on a 1/10 dilution of the reaction mixture for the first-round PCR products. The results were normalized according to GAPDH amplification and expressed as a relative number of integrated copies with respect to the amplification in transduced CHO cells.
FISH.
Cells were incubated with 75 mM KCl at 37°C for 20 min and then with methanol-acetic acid (3:1) for 10 min on ice. Nuclei were recovered by centrifugations at 400, 700, and 1,100 × g, and plated on glass slides. The slides were incubated successively with RNase A (10 μg/ml, 30 min, 37°C) and 0.01% pepsin in 10 mM HCl (20 min, 37°C) and finally fixed by incubation in paraformaldehyde (4%, 15 min). A SacI-SacI (2,796 bp) fragment of the gfp gene was labeled by nick translation using digoxigenin-coupled nucleotides (Roche), desalted using a G-50 spin column (GE Healthcare GmbH, Munziger, Germany), and resuspended at a final concentration 0.5 ng/μl in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer containing formamide and dextran sulfate (PM 8000). The probe was hybridized by incubation at 75°C for 3 min and then at 37°C for 24 h. The slides were then washed for 5 min successively in each of 2× SSC buffer at 37°C, 0.1× SSC at 60°C, and in 1× PBS at room temperature and were incubated at room temperature for 1 h with 100 μl of 1% blocking reagent in maleic acid buffer. The slides were then incubated for 1 h with 100 μl of anti-digoxigenin antibody coupled to horseradish peroxidase (150 U/ml; Roche, Meylan, France), washed in 1× PBS, incubated at room temperature for 1 h with Alexa Fluor 488 tyramide (Invitrogen, Cergy, France) in amplification buffer, and washed with 1× PBS at 37°C. Nuclear DNA was labeled with propidium iodide. Nuclear imaging was performed with confocal microscopy (LEICA TCS SP2) using a plan Apochromat 63 × 1.32 oil immersion objective and LEICA confocal software. Alexa Fluor 488 nm and propidium iodide were simultaneously excited with an argon ion laser (20 mW, 488 nm).
Image analysis.
Distances were measured by mathematical morphology transformations using a SAMBA IPS image analysis system (Unilog, Meylan, France). The FISH markers were extracted by top-hat transformation, and the nuclei were segmented by propidium iodide fluorescence imaging. Two binary images were obtained: one for markers and one for nuclei. The image of distances from the nuclear border was determined by iterative erosion of nuclei image. The markers positions were determined by the intersection of the marker image with the distance image. This method allows nuclei included in the frame analysis to be studied independent of their shape. To compensate for the differences in the sizes of nuclei, the measurements were systematically normalized by dividing each distance value by the radius of the corresponding nucleus.
Model distribution of distances.
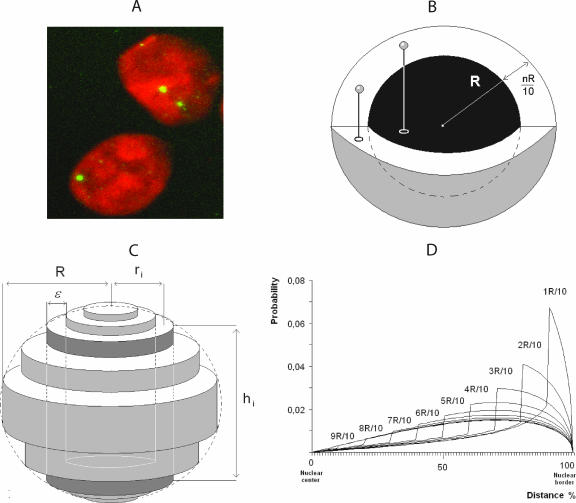
Model curves used for comparison with experimentally derived distances were obtained by stereological model. The nuclei in a 3D representation were assimilated to a spherical model formed by a set of concentric cylinders. The volume of one elementary cylinder (vi) is related to the probability (Pi) of the presence of the markers by Pi = vi/Σvi. If R is the nuclear radius, ri is the mean cylinder radius, ɛ is the thickness of cylinder, and hi is the height, then vi = π(ri +ɛ/2)2hi − π(ri − ɛ/2)2hi = 4 π ɛ ri(R − ri2)1/2. This equation allows tracing the random distribution in a sphere and the peripheral distribution in shells of different thickness. With this model, the function obtained is independent of homothetic spreading of nuclei. FISH spots at one setting of the focus adjustment (equatorial plan) were analyzed. More than 150 nuclei were evaluated per case.
Statistical analysis. (i) Bioluminescence assay.
To compare the data sets obtained for the quantitative analysis of bioluminescence in cell lysates, an independent sample t test with a probability of error threshold of 0.05 was performed. For the photon emission from individual cells, a Mann-Whitney U test with a probability of error threshold of 0.05 was performed since we compared populations of positive cells that were selected from a whole population. The statistical calculations and graphs were done with SigmaPlot software (Science Products).
(ii) FISH spot distribution analysis.
The nuclear radius was divided into 10 classes. Each distance measured between the nuclear border and the transgene FISH signals was categorized into one of these 10 classes. The χ2 minimization was used to test two hypotheses. (i) The distribution of the distances measured in xrs6 and xrs6-Ku cells were similar way to those measured for the CHO cells. (ii) The distribution of the distances measured in both cell lines were similar to that of points distributed in theoretical peripheral or random distributions. In both cases, distributions were considered different when the χ2 exceeded the value given for nine degrees of liberty with a probability of error threshold of 0.05.
RESULTS
Transgene overexpression after transduction of Ku-deficient cells by retroviral vectors.
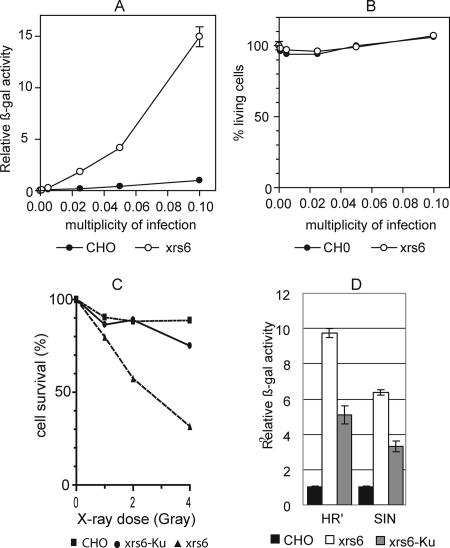
Depletion of cellular Ku80 stimulates HIV-1 expression in U1 cells bearing a silenced provirus, suggesting that Ku plays a role in the regulation of retroviral transcription (28). To evaluate the influence of Ku on the expression of a transgene after retroviral transduction, both CHO and the Ku80-lacking xrs6 cell lines were transduced with a pHR-βgal retroviral vector. This vector encodes the β-Gal transgene under the control of the cytomegalovirus (CMV) promoter. Transgene expression was evaluated 72 h posttransduction by measuring the β-Gal activity in the cell lysates according to the multiplicity of infection (MOI). Transgene expression was significantly higher in the Ku80-deficient xrs6 cells than in the Ku80-proficient CHO cells (Fig. 1A). At an MOI of 0.1, the level of expression in xrs6 cells was more than 10-fold than that in CHO cells. There was no significant cytotoxicity as assessed by an MTT assay at 72 h postinfection, indicating that the differences in transgene expression were not due to a loss of transduced cells (Fig. 1B). To determine whether this difference was due to the absence of Ku80, we constructed the xrs6-Ku cell line reexpressing human Ku80 by stable transfection. Ku80 reexpression was verified by Western blotting. Only xrs6 cells that were transduced with pCDNA3-Ku encoding human Ku80 produced wild-type levels of Ku80 (data not shown). Since Ku80 is strictly required for repairing lethal double-strand breaks in mammalian cell lines, its expression can be verified by inducing double-strand breaks by irradiation and monitoring the cell viability (49, 51, 52). The absence of Ku80 was associated with lower cell viability. In contrast, the reexpression of human Ku80 in polyclonal xrs6-Ku cells restored resistance to X-rays to a level similar to that of CHO cells (Fig. 1C). This demonstrates that heterologous Ku was functional in these cells. β-Gal expression was significantly lower in xrs6 cells expressing than not expressing the Ku80 protein (Fig. 1D), thereby confirming that the lack of Ku80 was responsible for the higher transgene expression in xrs6 cells.
FIG. 1.
Transduction by retroviral vectors leads to transgene expression enhancement in Ku80-expressing cells. (A) Expression of β-Gal 3 days after transduction. CHO and xrs6 cells were transduced by pHR′-LacZ lentiviral vectors at several MOIs. The results are expressed as the β-Gal activity. (B) Cytotoxicity evaluated by a MTT assay after CHO and xrs6 cell transduction. (C) Percentage of CHO, xrs6, and xrs6-Ku cells surviving 2 days after X-ray irradiation. Cells were plated at 5,000 cells per well in a 96-well plate and exposed to ionizing radiation 1 day later. Cell viability was evaluated by an MTT assay. (D) CHO, xrs6, and xrs6-Ku cells were transduced by LacZ encoding HR′ and SIN lentiviral vectors at an MOI of 0.1. The β-Gal activity was evaluated 3 days after transduction. The results are expressed as the relative activity of β-Gal activity in xrs6 and xrs6-Ku cells relative to the activity in transduced CHO cells (defined as 1). Bars represent the standard errors obtained from three independent experiments.
The results were similar in transduction experiments with the lentiviral HR′ shuttle plasmid, which contains the full HIV-1 5′LTR, the psi sequence and 360 bp of the gag gene upstream from the CMV transgene expression cassette; similar results were also obtained with a lacZ-encoding SIN lentiviral vector, which contains a 400-bp deletion within the HIV-1 U3 LTR region (58) (Fig. 1D). Therefore, the expression enhancement observed in absence of Ku was not related to either the retroviral vector used or the presence of the HIV-1 LTR.
Stimulation of transgene expression in xrs6 cells is independent of the promoter.
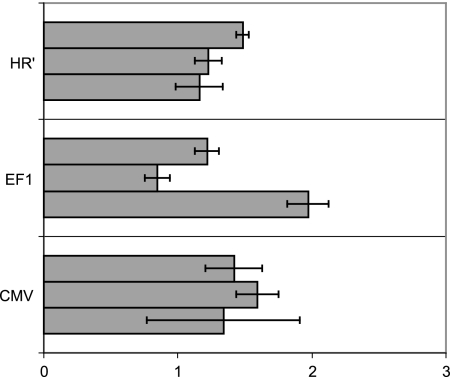
Ku80 has been implicated in the regulation of transcription of a variety of genes in either a positive or a negative way, through direct interaction with the promoter. Therefore, it may have a similar effect on the CMV promoter, which directs the expression of the β-Gal in both HR′ and SIN vectors. To test this possibility, we used transient-transfection experiments of CHO and xrs6 cells with two different plasmids allowing expression of the β-Gal under the control of either the EF1 or the CMV promoter. The HR′ shuttle plasmid encoding LacZ under the CMV promoter was also tested. None of the construct showed a significant difference in transgene expression between CHO and xrs6 cells (Fig. 2). Although there was some experimental variation, the enhancement of transgene expression was not comparable to that associated with retroviral transduction. We conclude that the increase in transgene expression was not related to direct Ku-dependent transcriptional control.
FIG. 2.
Transgene expression enhancement in xrs6 cells is not dependent on promoter regulation. Transgene expression in xrs6 and CHO cells after transfection with various plasmids encoding the LacZ transgene under EF1, CMV, or LTR(HR′) promoters was determined. Transgene expression is reported as the β-Gal activity in xrs6 cells relative to the activity in transfected CHO cells (defined as 1). Three independent experiments were performed for each construct in triplicate.
Ku80 deficiency promotes a stimulation of transgene expression in individual transduced cells.
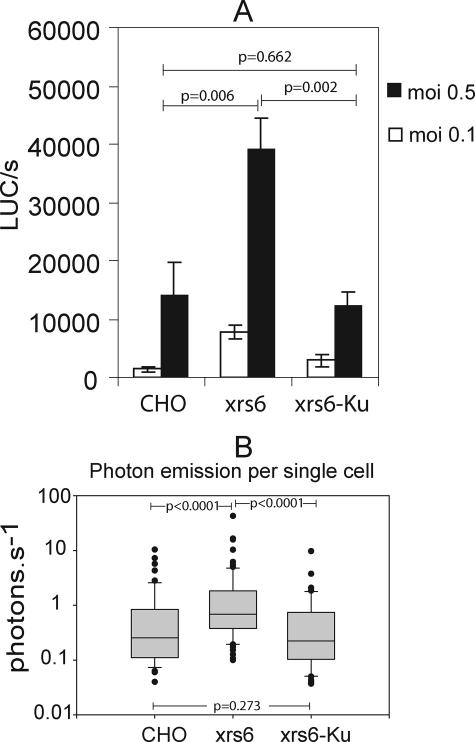
Our first experiment did not allow discrimination between specific enhancement of transgene expression in transduced cells or a better transduction efficiency of the cell population overall. To resolve this, we used a recently developed method that allows real-time quantitative imaging of a luciferase transgene expression by measuring photon emission in individual cells (46). CHO, xrs6, and xrs6-Ku cells were transduced with an NL4-3Δenv HIV-1-derived vector pseudotyped with VSV-G envelope and containing the luciferase gene inserted into the nef gene. Three days after transduction at two different MOIs, the efficiency of transduction was evaluated by measuring luciferase activity in whole-cell lysates and in intact individual cells. In agreement with the results of LacZ transduction, the luciferase activity in xrs6 cell lysates was much higher than that of CHO cell lysates (Fig. 3A). We then analyzed the photon emission from individual cells (Fig. 3B). The higher mean bioluminescence was due to a significantly greater light emission by individual transduced xrs6 cells. Therefore, the increase in transgene expression in the whole-cell population was presumably due to a stimulation of transgene expression in individual transduced cells rather than to a better transduction efficiency in xrs6 cells.
FIG. 3.
Measurement of transgene expression by bioluminescence emission in single cells. Cells were infected with NL4-3Δenv-luc VSV-G virus, and the luciferase activity was quantified both in cell lysates and in isolated cells. (A) Overall, luciferase activity in lysates following infection at MOIs of 0.1 and 0.5. The results are expressed in light units, and values reported are given as the means for three independent experiments. Errors bars represent the standard deviation. Comparisons among data sets obtained at an MOI of 0.5 were performed by an independent sample t test with a probability of error threshold of 0.05. P values obtained for an MOI of 0.1 were comparable. (B) Quantification of photon emission per second in single cells (n = 50/per cell type). Lines within the boxes indicate the means. The lowest boundaries of the boxes indicate the 25th percentile, and the upper boundaries indicate the 75th percentile. Whiskers above and below the boxes indicate the 9th and 10th percentiles. All of the outlying points are plotted. Comparisons among data sets were performed by Mann-Whitney U test, with a probability of error threshold of 0.05. All of the statistical calculations and graphs were done with SigmaPlot software (Science Products).
Ku80 does not affect integration quantitatively.
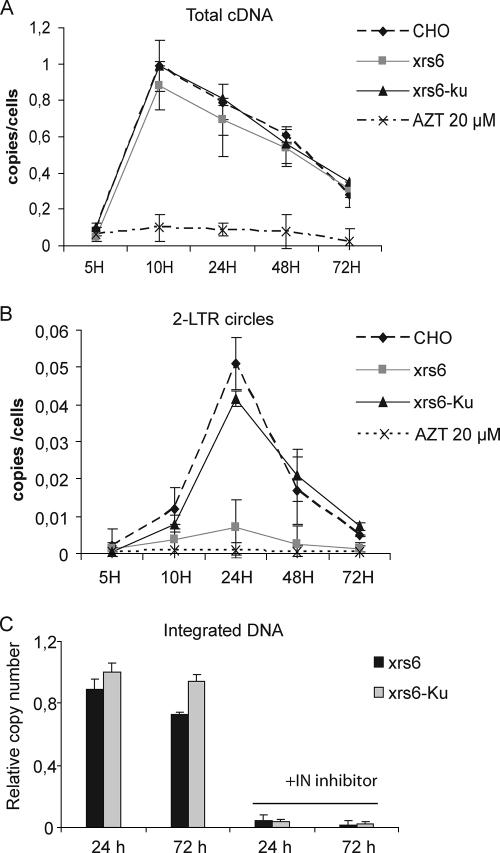
To determine whether the higher transgene expression in individual transduced cells was due to a difference during the early viral replicative steps, we measured the levels of total cDNA, 2-LTR DNA circles, and integrated forms of transgene DNA by real-time PCR in cell extracts from CHO, xrs6, and Ku-complemented cells. Cells were transduced with the pHR′-lacZ lentiviral vector at an MOI of 0.1 and harvested 5, 10, 24, 48, and 72 h later. The level of cDNA synthesis was maximal at 10 h posttransduction in the three cell lines and decreased subsequently (Fig. 4A), in agreement with a previous report (39). In control cells treated with zidovudine, the signal was weak, confirming that we were measuring exclusively de novo synthesized DNA. Thus, the absence of Ku from xrs6 cells did not affect the kinetics of total cDNA synthesis. There were sevenfold more 2-LTR circle forms 24 h posttransduction in CHO cells than in xrs6 cells (Fig. 4B). The accumulation of 2-LTR circles was restored by Ku reexpression in xrs6 cells, confirming the role of Ku in cDNA circularization (29, 35, 57).
FIG. 4.
Ku80 affect 2-LTR circle formation but not integration. CHO and derived cells were transduced by pHR′LacZ lentiviral at an MOI of 0.1. After 5, 10, 24, 48 and 72 h, DNA was extracted and subjected to real-time PCR analysis. Total cDNA, 2-LTR circles DNA, and integrated cDNA were quantified and normalized with respect to amplification of the endogenous GAPDH gene. Error bars represent the standard deviations of triplicate experiments. (A) Kinetics of cDNA synthesis; AZT at 20 μM was used in the controls. (B) 2-LTR circle DNA. (C) Kinetic analysis of integrated viral DNA. A control experiment was performed by treating cells with the integrase inhibitor L-708,906 at 25 μM. The relative copy number refers to the number of transgene copies in CHO cells normalized to 1.
We counted integration events using real-time PCR. To specifically amplify the integrated transgenes while avoiding possible interference from nonintegrated viral DNA, we adapted the quantitative Alu-PCR method to the B2A repeats, which are equivalent to Alu sequences in Chinese hamster cells (8, 26). CHO, xrs6, xrs6-Ku cells were transduced with the lentiviral pHR′-LacZ at an MOI of 0.1, and total DNA was prepared 24 and 72 h later. The sequences between the viral LTR and B2A sequences were amplified. Then, a nested amplification specific for the integrated transgene was carried out with LTR-specific primers (8). Transgene amplifications were normalized with respect to amplification of the endogenous GAPDH gene. A control experiment was performed by treating cells with the integrase inhibitor L-708,906 (25). The results are reported as the number of transgene copies in the genomic DNA of CHO, xrs6, and xrs6-Ku cells relative to the copy number in CHO (defined as 1). Integrated transgenes were readily detected in the three cell lines, whereas integrated events were barely detectable in the presence of the integrase inhibitor, indicating that only integrated transgene DNA was amplified (Fig. 4C). The amounts of integrated provirus into the three cell lines were not significantly different, thereby demonstrating that the efficiency of retroviral transgene integration was not dependent on the presence of Ku80 in the cells. These findings indicate that the stimulation of transgene expression was not due to a difference in the transduction efficiency in the absence of Ku80.
Ku80 regulates transgene spatial position within the nucleus.
The increased expression of transgenes in the absence of Ku80 is not related to differences in preintegrative and integrative steps of viral transduction or to changes in promoter activity. One possible mechanism is the modulation by Ku80 of provirus localization within the nucleus, with integration in a more favorable context in the absence of Ku80. To investigate this possibility, CHO, xrs6, and xrs6-Ku cells were transduced with a SIN vector encoding a green fluorescent protein (GFP) transgene. GFP-expressing cells were selected by fluorescence-activated cell sorting and grown for 2 months to ensure that they contained only integrated transgene cDNA (50). We then studied the distribution of the transgenes by 2D FISH. The nuclei were stained with propidium iodide, and individual integrated events were revealed by a transgene-specific Alexa Fluor 488-labeled probe (Fig. 5A). First, we quantified the number of proviruses in more than 150 randomly chosen cells by using the SAMBA IPS image analysis system (37). We found that xrs6 cells and CHO cells contained comparable numbers of integrated copies of the transgene than CHO cells, confirming that there was no difference in integration efficiency as observed on day 3 by real-time PCR.
FIG. 5.
FISH analysis of retroviral vector integration. (A) Imaging of nuclei from transduced cells. Cells were infected with the lentiviral vector pSINgfp and cultured for 2 months before FISH analysis. Nuclei were counterstained with propidium iodide. Individual integrated events are revealed by hybridization with a specific Alexa Fluor 488-labeled probe. (B) Peripheral distributions were modeled by random distribution in shells whose thickness was nR/10 (1 < n < 10). (C) Reconstruction of nuclear volume from a 2D image. The sphere (R radius) was fitted by a set of cylinders (each cylinder was defined as the mean radius = ri, height = hi, and thickness = ɛ, with ɛ = R/10). In the example shown in the figure, hi and ri refer to the third cylinder starting from the center of the sphere. (D) Model of signal distribution in shells of different thicknesses relative to the distance from nuclear border.
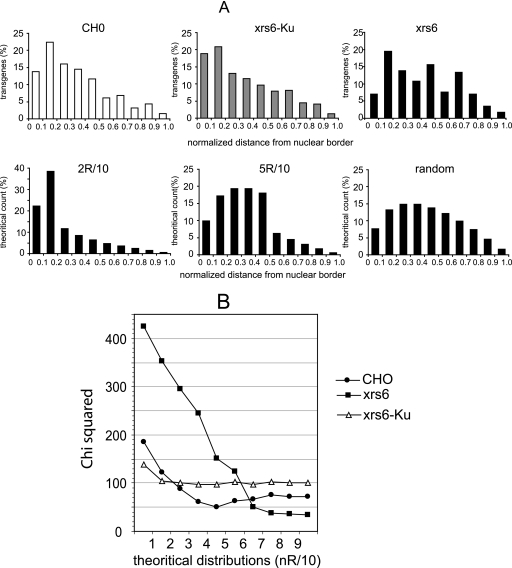
Due to the experimental process, 2D FISH degrades the nucleus 3D architecture. It is therefore not possible directly to compare 3D distributions of FISH spots obtained from confocal images. However, it was recently demonstrated that 2D projections obtained in such conditions can be used to reconstruct the 3D repartition of the FISH spots (37). A mathematical model was built that yields the theoretical distributions of distances between the FISH spots and the nucleus border in 2D images as a function of their 3D distribution (Fig. 5B, C, and D). This allows experimental distributions of spots to be compared and fitted to theoretical distributions corresponding to particular positioning. The distances between the FISH spots and the border of the nucleus were measured for 393, 456, and 382 integrated events from CHO, xrs6, and xrs6-Ku cell lines, respectively. The counting of events was performed by an automatic method (see Materials and Methods) in order to avoid a likely bias due to any subjective attribution of spots. Although such automatic counting precluded the possibility of working with strictly equal numbers of counted events, the number of counts was kept as close as possible to ensure the robustness of the statistics. Distances were ranked in 10 classes according to the distance expressed as a fraction of the nucleus radius, R (Fig. 6A). We then performed a chi-square analysis, which allows an overall comparison of the distributions by comparing their shapes and also the distance apart between these and theoretical ones. The χ2 minimization revealed a significant difference between the distribution of distances obtained in CHO and xrs6 cells (χ2 = 18.6 > 16.9, which is the χ2 value obtained for nine degrees of liberty with a probability of error threshold of 0.05). In the same way, the distributions of the FISH signals in xrs6-Ku and xrs6 cells were significantly different (χ2 = 25.0 > 16.9), suggesting that Ku80 expression in xrs6 cells affected the positioning of the transgenes. Finally, the distributions of the FISH signals in CHO and xrs6-Ku cells were not significantly different (χ2 = 5.0 < 16.9), confirming the Ku80-related positioning effect. Altogether, these results indicate that the transgene distribution was modulated by the Ku80 content in the cells.
FIG. 6.
FISH analysis of retroviral vector integration. (A) Histograms of signal distribution as measured from the border of the nuclei. The upper panels show the distribution of the FISH signals as measured for gfp transgene in CHO, xrs6, and xrs6-Ku cells. The lower panels show examples of theoretical distributions corresponding to 3D peripheral localization in a shell at 2/10 and 5/10 radius from the border of the nucleus and to a random 3D distribution. (B) Comparison of observed transgene localization with theoretical distributions. Distributions of FISH signals corresponding to the gfp transgene were compared to theoretical 3D distribution by χ2 minimization.
To determine whether there was a correlation between the transgene expression and the positioning of the transgenes in CHO and xrs6 cell lines, we compared the experimental distribution to several theoretical distributions by performing a χ2 minimization. Figure 6B shows the evolution of the χ2 test when fitting the distributions from the most peripheral model (i.e., spots are distributed mainly in a peripheral shell of 1/R thickness) to a completely random distribution. Minimization of the χ2 value indicates that although transgene distribution tended to be random in xrs6 cells, it was not in CHO cells. Indeed, there was a minimum value for CHO cells, suggesting a nonrandom distribution with a bias toward a peripheral distribution. The best fit was obtained for a theoretical distribution of transgenes located in a shell within a distance of 0.5 nuclear radius from the cell border. Finally, the χ2 analysis of the fitting of xrs6-Ku distributions to theoretical distributions shows that the distributions in xrs6-Ku cells did not tend toward randomness, as was the case in xrs6 cells, but more likely reflects an intermediary situation between CHO and xrs6 cells. This situation mirrors the partial restoration of gene silencing in xrs6 cells when Ku80 is reexpressed (see Fig. 2D).
DISCUSSION
In a previous study using U1 cells, a clone cell line in which a single the expression of the single provirus is repressed due to a mutation in Tat, we observed that proviral latency can be partly overcome by depleting Ku80 in these cells: this suggested that Ku participates in maintaining the inactive state of the provirus (28). Previous studies comparing GFP transduction of Ku-deficient and Ku-proficient cells failed to detect a difference of transduction efficiency as measured as the number of GFP-expressing cells (2, 3). These results were interpreted as indicating that there was no interaction between Ku and the integration machinery. However, the effects of Ku on provirus integration were not evaluated and the use of gfp-coding vectors allowed measurement of the number of transduced cells but not the transgene expression of individual cells. Here, we investigated the effect of Ku80 on both the integration of retroviral vectors and the resulting expression in single cells of either lacZ or luc transgenes encoded by these vectors. The transgenes were more strongly expressed after transduction of Ku80-deficient xrs6 cells than of Ku80-proficient CHO cells. This effect was directly related to the absence of Ku, since the reexpression of human Ku80 in xrs6 cells partially restrained transgene expression. Single-cell imaging demonstrated that transgene expression was higher in the Ku-deficient cell population overall. However, a novel real-time B2A-LTR nested PCR demonstrated that similar amount of transgene had integrated into transduced CHO and xrs6 cells. We previously suggested that Ku might have a sequence-specific effect on the negative element region of HIV LTR (28). This was based on similarity between a sequence found in HIV LTR and a previously described consensus Ku binding sequence. However, the study described above with the SIN lentivector, which lacks this sequence, argues against a specific role of this particular element, although interaction between Ku80 and a different sequence cannot be excluded. Nevertheless, a specific promoter regulatory effect can be ruled out because the CMV promoter activity directing transgene expression in HR′ and SIN vectors was not affected.
FISH experiments showed that the spatial distribution of transgenes in transduced xrs6 cells was significantly different from that in Ku-proficient CHO cells. Although our analysis cannot describe complex distributions, it indicated that the transgene distribution was not random in CHO cells but had a clear tendency to a more peripheral distribution. In contrast, the transgenes were more randomly distributed in xrs6 cells. This strongly suggests that Ku is involved in targeting proviruses to perinuclear regions of the genome, which would facilitate transcriptional silencing (1). This could be achieved through the interaction between Ku80 (35) and the PIC. This is consistent with the role of Ku which involves recruiting DNA-PK, upon binding to DNA, thereby inducing a shift of the local equilibrium from acetylated histones to deacetylated histones, resulting in an inactive chromatin conformation (4). Furthermore, the homologue of human Ku in yeast, yKu, organizes peripheral heterochromatin-rich domains of the nucleus (21).
Our results are also consistent with findings for PARP-1, another DNA damage sensor. As for Ku, it was first suggested that PARP-1 was required for retroviral integration, but transduction experiments in PARP-1-deficient cells suggest otherwise (2, 18). It was subsequently proposed that PARP-1 participates in the targeting of lentiviral elements to particular domains of chromosomal DNA (32). These results may reflect two aspects of a single mechanism as PARP-1 and Ku interact in vivo and are thought to cooperate in the maintenance of chromatin structure and function (19, 20).
The involvement of double-strand break repair pathway factors in the mechanism of repression of mobile elements has also been proposed in Chlamydomonas; in this organism, the suppression of two factors potentially involved in repairing double-strand breaks stimulates the expression of endogenous retromobile elements (31). Transgene expression was also studied in a cellular model of ligase IV defect to determine whether the effect seen with Ku80 can be reproduced with a catalytic factor of the nonhomologous end-joining pathway. The levels of LacZ expression were comparable in the ligase IV-defective and control cell lines (35) (and data not shown). We concluded that the enhancement of transgene expression after retroviral transduction in Ku-deficient cells was not associated with defective double-strand break repair activity but rather with the structural role of Ku.
Our observations were limited to Ku-deficient rodent cells. Ku is an essential nuclear factor for human cells, and complete Ku depletion is lethal (34). Nevertheless, partial depletion of Ku by an antisense strategy resulted in a diminution of the number of proviruses detected by nonquantitative PCR in human CEM infected cells, suggesting that Ku also interferes with the integration process in human cells (29). It remains to be determined whether this interaction affects provirus localization. Recent investigations addressing the integration of HIV-1-based vectors have shown that they integrate preferentially into transcriptional active regions of the genome (41, 48), a finding inconsistent with Ku targeting proviruses to silent chromatin. However, PIC in human cells may divert other cellular factors that counteract Ku-mediated targeting. A good candidate factor would be the integrase ligand LEDGF/p75, recently shown to be needed for site-targeting of PIC within chromatin (9, 10, 16).
Finally, the successful use of retroviral vectors in gene therapy will require sustained expression of transgene over time. Preventing interaction between PIC and Ku-rich regions may thus help overcome any positioning effect.
Acknowledgments
We thank Hervé Leh for extensive advice and Marie-Anne Nicola from the Plate-forme d'imagerie dynamique at Institut Pasteur for technical support.
C.M. and A.S.-C. were the recipients of fellowships from the Région Ile-de-France and Sidaction, respectively. L.J.-H. was funded by the Agence Nationale de Recherche sur le Sida (ANRS). E.G. received a fellowship from the EEC project TRIoH. This study was funded by the EEC TRIoH project (FP6 503480), by a grant from the ANRS, and by CNRS funding of J.-F. Mouscadet's group.
Footnotes
Published ahead of print on 16 May 2007.
REFERENCES
- 1.Andrulis, E. D., A. M. Neiman, D. C. Zappulla, and R. Sternglanz. 1998. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394:592-595. [DOI] [PubMed] [Google Scholar]
- 2.Ariumi, Y., P. Turelli, M. Masutani, and D. Trono. 2005. DNA damage sensors ATM, ATR, DNA-PKcs, and PARP-1 are dispensable for human immunodeficiency virus type 1 integration. J. Virol. 79:2973-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baekelandt, V., A. Claeys, P. Cherepanov, E. De Clercq, B. De Strooper, B. Nuttin, and Z. Debyser. 2000. DNA-dependent protein kinase is not required for efficient lentivirus integration. J. Virol. 74:11278-11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barlev, N. A., V. Poltoratsky, T. Owen-Hughes, C. Ying, L. Liu, J. L. Workman, and S. L. Berger. 1998. Repression of GCN5 histone acetyltransferase activity via bromodomain-mediated binding and phosphorylation by the Ku-DNA-dependent protein kinase complex. Mol. Cell. Biol. 18:1349-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beall, E. L., A. Admon, and D. C. Rio. 1994. A Drosophila protein homologous to the human p70 Ku autoimmune antigen interacts with the P transposable element inverted repeats. Proc. Natl. Acad. Sci. USA 91:12681-12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blier, P. R., A. J. Griffith, J. Craft, and J. A. Hardin. 1993. Binding of Ku protein to DNA: measurement of affinity for ends and demonstration of binding to nicks. J. Biol. Chem. 268:7594-7601. [PubMed] [Google Scholar]
- 7.Brown, P. O. 1990. Integration of retroviral DNA. Curr. Top. Microbiol. Immunol. 48:19-48. [DOI] [PubMed] [Google Scholar]
- 8.Brussel, A., O. Delelis, and P. Sonigo. 2005. Alu-LTR real-time nested PCR assay for quantifying integrated HIV-1 DNA. Methods Mol. Biol. 304:139-154. [DOI] [PubMed] [Google Scholar]
- 9.Busschots, K., J. Vercammen, S. Emiliani, R. Benarous, Y. Engelborghs, F. Christ, and Z. Debyser. 2005. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J. Biol. Chem. 280:17841-17847. [DOI] [PubMed] [Google Scholar]
- 10.Ciuffi, A., M. Llano, E. Poeschla, C. Hoffmann, J. Leipzig, P. Shinn, J. R. Ecker, and F. Bushman. 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 11:1287-1289. [DOI] [PubMed] [Google Scholar]
- 11.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 12.Daniel, R., G. Kao, K. Taganov, J. G. Greger, O. Favorova, G. Merkel, T. J. Yen, R. A. Katz, and A. M. Skalka. 2003. Evidence that the retroviral DNA integration process triggers an ATR-dependent DNA damage response. Proc. Natl. Acad. Sci. USA 100:4778-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daniel, R., R. A. Katz, and A. M. Skalka. 1999. A role for DNA-PK in retroviral DNA integration. Science 284:644-647. [DOI] [PubMed] [Google Scholar]
- 14.Difilippantonio, M. J., J. Zhu, H. T. Chen, E. Meffre, M. C. Nussenzweig, E. E. Max, T. Ried, and A. Nussenzweig. 2000. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature 404:510-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Downs, J. A., and S. P. Jackson. 1999. Involvement of DNA end-binding protein Ku in Ty element retrotransposition. Mol. Cell. Biol. 19:6260-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emiliani, S., A. Mousnier, K. Busschots, M. Maroun, M. B. Van, D. Tempe, L. Vandekerckhove, F. Moisant, L. Ben-Slama, M. Witvrouw, F. Christ, J. C. Rain, C. Dargemont, Z. Debyser, and R. Benarous. 2005. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J. Biol. Chem. 280:25517-25523. [DOI] [PubMed] [Google Scholar]
- 17.Falzon, M., J. W. Fewell, and E. L. Kuff. 1993. EBP-80, a transcription factor closely resembling the human autoantigen Ku, recognizes single- to double-strand transitions in DNA. J. Biol. Chem. 268:10546-10552. [PubMed] [Google Scholar]
- 18.Gaken, J. A., M. Tavassoli, S. U. Gan, S. Vallian, I. Giddings, D. C. Darling, J. Galea-Lauri, M. G. Thomas, H. Abedi, V. Schreiber, M. J. Menissier-de, M. K. Collins, S. Shall, and F. Farzaneh. 1996. Efficient retroviral infection of mammalian cells is blocked by inhibition of poly(ADP-ribose) polymerase activity. J. Virol. 70:3992-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galande, S., and T. Kohwi-Shigematsu. 1999. Poly(ADP-ribose) polymerase and Ku autoantigen form a complex and synergistically bind to matrix attachment sequences. J. Biol. Chem. 274:20521-20528. [DOI] [PubMed] [Google Scholar]
- 20.Galande, S., and T. Kohwi-Shigematsu. 2000. Caught in the act: binding of Ku and PARP to MARs reveals novel aspects of their functional interaction. Crit. Rev. Eukaryot. Gene Expr. 10:63-72. [PubMed] [Google Scholar]
- 21.Galy, V., J. C. Olivo-Marin, H. Scherthan, V. Doye, N. Rascalou, and U. Nehrbass. 2000. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature 403:108-112. [DOI] [PubMed] [Google Scholar]
- 22.Giampuzzi, M., G. Botti, M. Di Duca, L. Arata, G. Ghiggeri, R. Gusmano, R. Ravazzolo, and A. Di Donato. 2000. Lysyl oxidase activates the transcription activity of human collagene III promoter. Possible involvement of Ku antigen. J. Biol. Chem. 275:36341-36349. [DOI] [PubMed] [Google Scholar]
- 23.Giffin, W., J. Kwast-Welfeld, D. J. Rodda, G. G. Prefontaine, M. Traykova-Andonova, Y. Zhang, N. L. Weigel, Y. A. Lefebvre, and R. J. Hache. 1997. Sequence-specific DNA binding and transcription factor phosphorylation by Ku autoantigen/DNA-dependent protein kinase. Phosphorylation of Ser-527 of the rat glucocorticoid receptor. J. Biol. Chem. 272:5647-5658. [DOI] [PubMed] [Google Scholar]
- 24.Ha, H. C., K. Juluri, Y. Zhou, S. Leung, M. Hermankova, and S. H. Snyder. 2001. Poly(ADP-ribose)polymerase-1 is required for efficient HIV-1 integration. Proc. Natl. Acad. Sci. USA 98:3364-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hazuda, D. J., P. Felock, M. Witmer, A. Wolfe, K. Stillmock, J. A. Grobler, A. Espeseth, L. Gabryelski, W. Schleif, C. Blau, and M. D. Miller. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646-650. [DOI] [PubMed] [Google Scholar]
- 26.Hyrien, O., M. Debatisse, G. Buttin, and V. S. de. 1987. A hotspot for novel amplification joints in a mosaic of Alu-like repeats and palindromic A+T-rich DNA. EMBO J. 6:2401-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacque, J. M., and M. Stevenson. 2006. The inner-nuclear-envelope protein emerin regulates HIV-1 infectivity. Nature 441:641-645. [DOI] [PubMed] [Google Scholar]
- 28.Jeanson, L., and J. F. Mouscadet. 2002. Ku Represses the HIV-1 Transcription. Identification of a putative Ku binding site homologous to the mouse mammary tumor virus NRE1 sequence in the HIV-1 long terminal repeat. J. Biol. Chem. 277:4918-4924. [DOI] [PubMed] [Google Scholar]
- 29.Jeanson, L., F. Subra, S. Vaganay, M. Hervy, E. Marangoni, J. Bourhis, and J. Mouscadet. 2002. Effect of Ku80 depletion on the preintegrative steps of HIV-1 replication in human cells. Virology 300:100. [DOI] [PubMed] [Google Scholar]
- 30.Jeggo, P. A., and L. M. Kemp. 1983. X-ray-sensitive mutants of Chinese hamster ovary cell line: isolation and cross-sensitivity to other DNA-damaging agents. Mutat. Res. 112:313-327. [DOI] [PubMed] [Google Scholar]
- 31.Jeong Br, B. R., D. Wu-Scharf, C. Zhang, and H. Cerutti. 2002. Suppressors of transcriptional transgenic silencing in Chlamydomonas are sensitive to DNA-damaging agents and reactivate transposable elements. Proc. Natl. Acad. Sci. USA 99:1076-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kameoka, M., S. Nukuzuma, A. Itaya, Y. Tanaka, K. Ota, Y. Inada, K. Ikuta, and K. Yoshihara. 2005. Poly(ADP-ribose)polymerase-1 is required for integration of the human immunodeficiency virus type 1 genome near centromeric alphoid DNA in human and murine cells. Biochem. Biophys. Res. Commun. 334:412-417. [DOI] [PubMed] [Google Scholar]
- 33.Lau, A., R. Kanaar, S. P. Jackson, and M. J. O'Connor. 2004. Suppression of retroviral infection by the RAD52 DNA repair protein. EMBO J. 23:3421-3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, G., C. Nelsen, and E. A. Hendrickson. 2002. Ku86 is essential in human somatic cells. Proc. Natl. Acad. Sci. USA 99:832-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, L., J. M. Olvera, K. E. Yoder, R. S. Mitchell, S. L. Butler, M. Lieber, S. L. Martin, and F. D. Bushman. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20:3272-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lieber, M. R., Y. Ma, U. Pannicke, and K. Schwarz. 2003. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell. Biol. 4:712-720. [DOI] [PubMed] [Google Scholar]
- 37.Linares-Cruz, G., H. Bruzzoni-Giovanelli, V. Alvaro, J. P. Roperch, M. Tuynder, D. Schoevaert, M. Nemani, S. Prieur, F. Lethrosne, L. Piouffre, V. Reclar, A. Faille, D. Chassoux, J. Dausset, R. B. Amson, F. Calvo, and A. Telerman. 1998. p21WAF-1 reorganizes the nucleus in tumor suppression. Proc. Natl. Acad. Sci. USA 95:1131-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maertens, G., P. Cherepanov, W. Pluymers, K. Busschots, E. De Clercq, Z. Debyser, and Y. Engelborghs. 2003. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278:33528-33539. [DOI] [PubMed] [Google Scholar]
- 39.Maroun, M., O. Delelis, G. Coadou, T. Bader, E. Segeral, G. Mbemba, C. Petit, P. Sonigo, J. C. Rain, J. F. Mouscadet, R. Benarous, and S. Emiliani. 2006. Inhibition of early steps of HIV-1 replication by SNF5/Ini1. J. Biol. Chem. 281:22736-22743. [DOI] [PubMed] [Google Scholar]
- 40.Mimori, T., M. Akizuki, H. Yamagata, S. Inada, S. Yoshida, and M. Homma. 1981. Characterization of a high molecular weight acidic nuclear protein recognized by autoantibodies in sera from patients with polymyositis-scleroderma overlap. J. Clin. Investig. 68:611-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitchell, R. S., B. F. Beitzel, A. R. Schroder, P. Shinn, H. Chen, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2:E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mulder, L. C., L. A. Chakrabarti, and M. A. Muesing. 2002. Interaction of HIV-1 integrase with DNA repair protein hRad18. J. Biol. Chem. 277:27489-27493. [DOI] [PubMed] [Google Scholar]
- 43.Oh, W. J., E. K. Kim, J. H. Ko, S. H. Yoo, S. H. Hahn, and O. J. Yoo. 2002. Nuclear proteins that bind to metal response element a (MREa) in the Wilson disease gene promoter are Ku autoantigens and the Ku-80 subunit is necessary for basal transcription of the WD gene. Eur. J. Biochem. 269:2151-2161. [DOI] [PubMed] [Google Scholar]
- 44.Reddy, S. V., O. Alcantara, and D. H. Boldt. 1998. Analysis of DNA binding proteins associated with hemin-induced transcriptional inhibition: the hemin response element binding protein is a heterogeneous complex that includes the Ku protein. Blood 91:1793-1801. [PubMed] [Google Scholar]
- 45.Ruiz, M. T., D. Matheos, G. B. Price, and M. Zannis-Hadjopoulos. 1999. OBA/Ku86: DNA binding specificity and involvement in mammalian DNA replication. Mol. Biol. Cell 10:567-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saez-Cirion, A., M.-A. Nicola, G. Pancino, and S. L. Shorte. 2006. Quantitative real-time analysis of HIV-1 gene expression dynamics in single living primary cells. Biotechnology 1:1-8. [DOI] [PubMed] [Google Scholar]
- 47.Schild-Poulter, C., D. Matheos, O. Novac, B. Cui, W. Giffin, M. T. Ruiz, G. B. Price, M. Zannis-Hadjopoulos, and R. J. Hache. 2003. Differential DNA binding of Ku antigen determines its involvement in DNA replication. DNA Cell Biol. 22:65-78. [DOI] [PubMed] [Google Scholar]
- 48.Schroder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 49.Singleton, B. K., A. Priestley, H. Steingrimsdottir, D. Gell, T. Blunt, S. P. Jackson, A. R. Lehmann, and P. A. Jeggo. 1997. Molecular and biochemical characterization of xrs mutants defective in Ku80. Mol. Cell. Biol. 17:1264-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siva, A. C., and F. Bushman. 2002. Poly(ADP-ribose) polymerase 1 is not strictly required for infection of murine cells by retroviruses. J. Virol. 76:11904-11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smider, V., W. K. Rathmell, M. R. Lieber, and G. Chu. 1994. Restoration of X-ray resistance and V(D)J recombination in mutant cells by Ku cDNA. Science 266:288-291. [DOI] [PubMed] [Google Scholar]
- 52.Taccioli, G. E., T. M. Gottlieb, T. Blunt, A. Priestley, J. Demengeot, R. Mizuta, A. R. Lehmann, F. W. Alt, S. P. Jackson, and P. A. Jeggo. 1994. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science 265:1442-1445. [DOI] [PubMed] [Google Scholar]
- 53.Takata, M., M. S. Sasaki, E. Sonoda, C. Morrison, M. Hashimoto, H. Utsumi, Y. Yamaguchi-Iwai, A. Shinohara, and S. Takeda. 1998. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17:5497-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tuteja, R., and N. Tuteja. 2000. Ku autoantigen: a multifunctional DNA-binding protein. Crit. Rev. Biochem. Mol. Biol. 35:1-33. [DOI] [PubMed] [Google Scholar]
- 55.Willis, D. M., A. P. Loewy, N. Charlton-Kachigian, J. S. Shao, D. M. Ornitz, and D. A. Towler. 2002. Regulation of osteocalcin gene expression by a novel Ku antigen transcription factor complex. J. Biol. Chem. 277:37280-37291. [DOI] [PubMed] [Google Scholar]
- 56.Yoder, K., A. Sarasin, K. Kraemer, M. McIlhatton, F. Bushman, and R. Fishel. 2006. The DNA repair genes XPB and XPD defend cells from retroviral infection. Proc. Natl. Acad. Sci. USA 103:4622-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoder, K. E., and F. D. Bushman. 2000. Repair of gaps in retroviral DNA integration intermediates. J. Virol. 74:11191-11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zufferey, R., T. Dull, R. J. Mandel, A. Bukovsky, D. Quiroz, L. Naldini, and D. Trono. 1998. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 72:9873-9880. [DOI] [PMC free article] [PubMed] [Google Scholar]