Abstract
Acute arenavirus disease in primates, like Lassa hemorrhagic fever in humans, begins with flu-like symptoms and leads to death approximately 2 weeks after infection. Our goal was to identify molecular changes in blood that are related to disease progression. Rhesus macaques (Macaca mulatta) infected intravenously with a lethal dose of lymphocytic choriomeningitis virus (LCMV) provide a model for Lassa virus infection of humans. Blood samples taken before and during the course of infection were used to monitor gene expression changes that paralleled disease onset. Changes in blood showed major disruptions in eicosanoid, immune response, and hormone response pathways. Approximately 12% of host genes alter their expression after LCMV infection, and a subset of these genes can discriminate between virulent and nonvirulent LCMV infection. Major transcription changes have been given preliminary confirmation by quantitative PCR and protein studies and will be valuable candidates for future validation as biomarkers for arenavirus disease.
Arenaviruses are occasionally transmitted from rodents to human beings, thereby causing acute lethal disease. One arenavirus, Lassa fever virus, has caused outbreaks in villages and hospitals of West Africa and has been classified as a category A biothreat in the United States. Laboratory studies using a related arenavirus, lymphocytic choriomeningitis virus (LCMV), showed that mice can develop vigorous antiviral cytotoxic-T-lymphocyte responses that may become immunopathological (23). The murine disease is alleviated by immune suppression (31, 44), unlike the acute viral disease in guinea pig and primate models (28, 54, 52, 60). The progression of LCMV-associated hemorrhagic fever in macaques is very similar to that seen for Lassa fever in humans (19, 22, 46, 47, 48, 68) and provides a practical model for disease studies in well-controlled laboratory environments.
Our experiments on rhesus macaques were designed to model human disease following a lethal dose of virus. Needlestick injuries estimated to deliver approximately 102 to 104 infectious particles from patient blood that contains 105 to 107 infectious particles per milliliter have been described previously (62). In both human beings and macaques, intravenous exposure is followed approximately 1 week later by fever (5). In the monkey model, viremia is detectable in blood approximately 4 days after the initial exposure (22, 48). The blood later exhibits elevated liver enzymes and coagulation defects (18, 27). Additional disease signs include petechial rashes, pulmonary edema, cardialgia, renal failure, hemorrhage, and hypoxia, leading to death approximately 2 weeks after virus exposure (46, 62).
Although disease progression in the monkey model is often monitored by liver biopsy and bronchoalveolar lavage (47), venipuncture is less invasive and more practical for monitoring human beings. DNA microarray technology makes it possible to investigate the expression of thousands of genes in a blood sample simultaneously, and profiles of these genes may reveal prognostic biomarkers and insights into the molecular events causing viral hemorrhagic fever.
We characterized changes of gene expression in peripheral blood mononuclear cells (PBMC) isolated at different times from rhesus macaques that were uninfected, infected with the virulent strain LCMV-WE, or infected with the nonvirulent strain LCMV-Armstrong (LCMV-Arm). Although arenaviruses replicate well in periarterial spaces, liver, spleen, and peripheral lymphoid organs soon after inoculation (46, 65), they cannot infect most PBMC (primarily lymphocytes and immature monocytes), and hence viremia does not appear until day 4 in this model. Our goal was to identify molecular events in blood that could (i) serve as markers for disease progression and (ii) discriminate between virulent and nonvirulent infection. We characterized two major stages, previremic and viremic, and compared samples to uninfected samples or samples of blood from monkeys infected with LCMV that did not have disease. This analysis of the primate transcriptome after in vivo LCMV infection reveals critical molecular events that can be related to disease progression.
MATERIALS AND METHODS
Experimental samples.
Twenty-one healthy adult rhesus macaques of 5 to 9 years of age were used for this terminal study as described previously (22). At least three blood samples were obtained before infection: typically they were 1 month, 1 week, and a few minutes before infection. Once the animals were infected, some were bled within 30 min after infection, and some were bled 3 and 5 days after infection, but the majority of samples used for microarray analysis were obtained on the day of euthanasia. Before euthanasia, blood was taken for chemistries and hematology, and then blood was obtained in EDTA tubes and processed at room temperature to obtain PBMC for flow cytometry and RNA extraction. PBMC extraction for RNA occurred within 30 min of drawing from the animal to minimize gene expression changes due to sample processing. PBMC were used instead of whole blood so that results could be compared with studies of PBMC responses to virus in vitro.
LCMV infection of rhesus macaques is uniformly lethal at a dose of 103 PFU of LCMV-WE but shows no pathology after a dose of 103 to 108 PFU of LCMV-Arm (22, 46, 47, 68). Infection in macaques was determined by plaque assay of infectious particles in plasma, by infectious center assay of PBMC, and by reverse transcription-PCR (RT-PCR) to detect viral RNA in tissues as described previously (22, 43, 46, 48).
Flow cytometry analysis.
Fluorescence-activated cell sorter analysis was performed on monkey PBMC samples to identify percentages of cell subsets as described elsewhere (reference 68 and J. D. Rodas, C. Cairo, T. Ruckwardt, M. Djavani, J. C. Zapata, J. Bryant, C. D. Pauza, I. S. Lukashevich, and M. S. Salvato, submitted for publication). Overall, there were no dramatic changes in the circulating blood cell populations of rhesus macaques. Depletion of NK and γδ T cells (less than 5% of blood cells) was detected in both LCMV-WE and LCMV-Arm samples.
RNA preparation and GeneChip hybridization.
Total RNA was prepared from LCMV-infected or uninfected PBMC subjected to TRIzol (GIBCO-BRL) extraction and purification using an RNeasy system (QIAGEN, Valencia, CA) according to the manufacturer's instructions. A QIAGEN RNase-free DNase supplement kit was used to ensure that the RNA had no DNA contamination. RNA quantities were determined by measuring the absorbance at 260 nm. All RNA samples were checked for both quality and quantity on an Agilent 2100 BioAnalyzer system (Agilent Technologies, Palo Alto, CA). RNA that passed this initial quality control screen was then labeled according to the standard target labeling protocols provided by Affymetrix and hybridized to the GeneChip human genome U133 Plus 2.0 array (Affymetrix, Santa Clara, CA) as described by the manufacturer (http://www.affymetrix.com). This chip fully covers the human genome with 54,000 probe sets that represent approximately 22,000 genes.
Microarrays were washed by using a GeneChip fluidics station (Affymetrix). Staining with R-phycoerythrin-streptavidin mix (Molecular Probes) was followed by antibody amplification with a biotinylated streptavidin antibody (Vector Laboratories). Scanning was carried out by using a GeneArray scanner (Hewlett-Packard). The use of the Affymetrix human genome microarrays for monitoring transcriptome changes in rhesus macaque tissues has been validated by other studies (1, 69, 77).
Of the 24 hybridizations used for this analysis, 3 were repeated to determine that the chip-to-chip hybridization mean variation was less than 5%. This agrees with a study comparing different commercially available microarray platforms in which Affymetrix chips had a mean coefficient of variation of less than 5%, whereas the next closest commercial platform had a mean of 10% (71).
Microarray data analysis.
Microarray data analysis was performed using the Array Data Analysis and Management System (ADAMS), currently being developed at VBI (http://pathport.vbi.vt.edu). The system uses publicly available tools for analysis of the data. Briefly, raw probe intensities were normalized and summarized using a robust multichip average of G+C content algorithm (gcRMA algorithm) (79). The detection calls (present, marginal, or absent) for each probe set were obtained using the mas5calls function in the Affy R package (29). For paired comparisons, only genes with at least one present call across the compared samples were included. A total of 24 samples were used to generate the microarray data (http://www.ncbi.nlm.nih.gov/geo/). The data from the 24 samples were grouped into four as follows to perform the analysis of variance (ANOVA): uninfected controls (11 samples), samples infected with the nonvirulent strain LCMV-Arm (5 samples), samples infected with the virulent strain LCMV-WE and showing the previremic response (4 samples), and samples infected with LCMV-WE and showing viremic response (4 samples). The P values were corrected using the Benjamini and Hochberg false-discovery-rate test (6).
Pairwise comparisons were also performed to calculate the changes (n-fold) using single pre- and postinfection samples from the same animals. A total of 6,416 genes with changes (n-fold) greater than or equal to two were used for further functional characterization of the genes. Among the 6,416 genes, we identified a total of 3,520 genes with P values from the ANOVA of ≤0.05 and 1,843 genes with P values of ≤0.01. The microarray data and the related experimental information from this study can be accessed at http://www.ncbi.nlm.nih.gov/geo/, platform number GSE5790.
Quantitative real-time reverse transcriptase PCR.
Selected genes from the microarray analysis were subjected to validation by quantitative real-time RT-PCR to determine the extent to which gene expression was up- or downregulated as a result of infection. Monkey and human gene-specific primers were used to validate expression levels for selected host genes. The PCR primer pair sequences will be supplied upon request. Briefly, total RNA (1 μg) from each sample was used to generate a cDNA template using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). The total reaction volume was 20 μl. The reaction was incubated as follows in a Tetrad thermocycler (MJResearch, Waltham, MA): 5 min at 25°C, 30 min at 42°C, 5 min at 85°C, and a hold at 4°C. cDNA products were diluted 1:10 in diethylpyrocarbonate-treated water. The incubations also were performed on controls with no RNA template and with the reverse transcriptase enzyme omitted. The PCR primer pairs were designed based on previously published sequences (GenBank) and using the Oligo 6 primer design software (Molecular Biology Insights, Cascade, CO).
The PCR used cDNA and Taq DNA polymerase (Invitrogen, Carlsbad, CA) and previously described conditions (21, 22). Each gene amplicon was purified by using a MiniElute PCR purification kit (QIAGEN). The purified amplicon for each gene was quantified on an agarose gel and with a GeneQuant Pro spectrophotometer (Amersham Biosciences, Piscataway, NJ). These purified amplicons were used further to optimize the real-time PCR conditions and to generate the standard curves in the real-time PCR assay. Primer concentrations and annealing temperatures were optimized for the iCycler iQ system (Bio-Rad) for each set of primers by use of the system's gradient protocol. PCR efficiencies were maintained at 100% for each primer set during optimization and also during the real-time PCR of sample cDNA.
RT-PCR products were analyzed by 1% agarose gel electrophoresis. The amplification plots were expressed as threshold cycle (CT) values. The CT is the reaction cycle at which PCR products or amplicons reach a threshold level of detection: the lower the CT value, the more abundant the substrate. CT values were normalized by using GAPDH (glyceraldehyde-3-phosphate dehydrogenase) or rRNA 18S amplicons as standards. Dissociation analysis of the PCR products was used to confirm specificity.
Protein determination using SearchLight proteome arrays.
Plasma samples were collected from uninfected and LCMV-infected rhesus macaques and spun down to separate the debris, and then the supernatants were stored in 0.5-ml aliquots at −70°C and shipped frozen for analysis on sandwich enzyme-linked immunosorbent assays (ELISAs) on SearchLight proteome arrays (Pierce, Boston, MA).
Proteomic array data analysis.
Proteomic array data were calculated as mean (± standard error of the mean) or median (range) values. Statistical significance between groups was determined using a one-way repeated-measures ANOVA. This accounts for individual differences among animals, thus removing animal variability from the analysis. After ANOVA, the Student t test was used to determine statistical significance between groups, with a maximum ANOVA P value of 0.05 considered as significant.
Ingenuity Pathway Analysis.
The Ingenuity Pathway Analysis software tool was used to analyze 400 genes that discriminated between virulent and benign infections with changed P values of <0.05 in order to get a better idea of the most likely downstream events.
RESULTS
Most differentially expressed transcripts were downregulated at the onset of viremia.
Gene expression data from PBMC of animals infected with the virulent strain LCMV-WE were compared to similar data from either uninfected animals (control animals) or animals infected with a nonvirulent strain, LCMV-Arm. Rhesus macaque samples were hybridized to human Affymetrix chips. Genes with changes in expression of at least twofold were designated “differentially expressed” (Fig. 1; also see Table SI in the supplemental material). Out of 54,000 probe sets on the human chip, 6,416 (∼12%) were identified as differentially expressed genes (in Table SI in the supplemental material, these genes are arranged in the order of magnitude of differential expression). We use the word “genes” instead of “probe sets” herein for simplicity, even though each probe set actually identifies a subportion of a gene. We chose to use PBMC rather than whole blood because we wanted our results to be comparable to results from in vitro studies using PBMC.
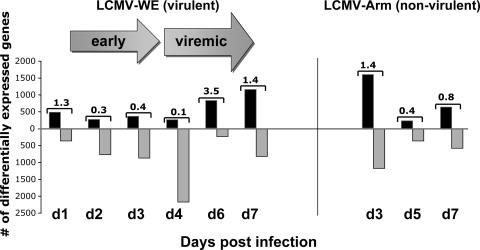
FIG. 1.
Macaque PBMC genes that are differentially modulated in LCMV-infected cells and control uninfected cells. Virulent (LCMV-WE) and nonvirulent (LCMV-Arm) infections are compared in terms of the time courses of up- and downregulated genes. Values on the bar represent the ratios between up- and downregulated genes. There are more differentially expressed genes that are down- rather than upregulated. For these data we used rhesus macaques inoculated with a lethal dose (1 × 103 PFU) of LCMV-WE (day 1 [n = 2], day 2 [n = 1], day 3 [n = 1], day 4 [n = 1], day 6 [n = 2], day 7 [n = 1]), three monkeys inoculated with 1 × 103 PFU LCMV-Arm virus ([day 3 [n = 1], day 5 [n = 1], day 7 [n = 1]), and five uninfected control monkeys. “Early” means in the previremic stage of infection. The microarray data and the related experimental information from this study can be accessed at http://www.ncbi.nlm.nih.gov/geo/, platform number GSE5790.
Virus was detectable in these monkeys at day 4 after infection by plaque assay and by RT-PCR of plasma virion RNA (48). We also used an infectious center assay to measure virus in PBMC from three monkeys per day. Arenaviruses replicate in cells of the reticuloendothelial system, primarily monocyte-derived macrophages, dendritic cells, and endothelial cells, but fail to replicate in the most abundant cells found in PBMC, which are immature monocytes and lymphocytes. Consequently, the infectious centers were very low and not detectable until day 6 for LCMV-WE-infected monkeys (i.e., 0.45 infectious centers per 106 PBMC [mean for three monkeys]). Thus, virus was not detectable in the circulation for the first 3 days, and we therefore defined days 1 to 3 as the early previremic stage and days 4 to 7 as the viremic stage. Although the LCMV-Arm strain replicated in cell culture as well as LCMV-WE, LCMV-Arm-infected monkeys did not experience viremia; that is, viral nucleic acids were detected in tissues, but virus loads were never high enough to be detected in circulation. The LCMV-Arm-infected monkeys resisted a lethal challenge with LCMV-WE, further confirming that they had been productively infected and immunized (68). An overview of gene expression in monkeys shows a trend to downregulate genes at the onset of viremia followed by a sharp increase in the number of upregulated genes. Those monkeys that did not experience viremia (LCMV-Arm-infected monkeys) showed no dramatic downregulation trend for the overall numbers of differentially expressed genes (Fig. 1). The number of downregulated genes in the LCMV-WE-infected macaques was significantly greater than the number seen for LCMV-Arm-infected monkeys (P < 0.05); by a two-way comparison based on the averages over the 7-day study period for each virus.
Changes in circulating blood cell populations did not significantly affect blood gene expression profiles.
Using flow cytometry (Rodas et al., submitted), we analyzed fluctuations of CD8+ (Tc) cells, CD4+ (TH) cells, CD20+ (B) cells, CD3-CD16+ (NK) cells, CD3+ CD16+ (NK T) cells, and CD3− γδ TcR+ cells (γδ T lymphocytes). By day 2, we detected a 20 to 70% decrease in NK and γδ T cells that lasted through day 7, without significant fluctuations in the other cell subsets (Rodas et al., submitted). We examined the current transcriptome data set for fluctuations in CD16 and γδ TcR gene expression and saw that they remained below our twofold cutoff throughout the study (see Table SI in the supplemental material). This is reasonable, since the NK and γδ T-cell populations constitute less than 5% of the total circulating PBMC, and their depletion during infection would not be expected to have a large impact on gene expression.
Gene expression profiling that discriminates between virulent and nonvirulent virus infection.
After all results were normalized to those for uninfected controls, PBMC gene expression profiles showed significant differences between virulent and nonvirulent infections (Fig. 2). We determined that the levels of approximately 478 genes discriminated between nonvirulent and virulent LCMV infections (Fig. 2). Although data for individual animals are shown, statistical significance for the 478 genes was achieved by pooling results from eight animals (LCMV-WE infected) for comparison with results from three animals (LCMV-Arm infected). Genes that were downregulated during virulent LCMV-WE virus replication were relatively unchanged or upregulated with nonvirulent LCMV-Arm infection and vice versa (genes up with LCMV-WE were unchanged or down with LCMV-Arm). The 478 genes that differ most between virulent and nonvirulent infections are listed in Tables 1, 2, and 3 and in Tables SI to SV in the supplemental material. Tables 1, 2, and 3 are grouped temporally as previremic (Table 1), upregulated viremic (Table 2) and downregulated viremic (Table 3). The expression levels for some of these genes were verified by real-time PCR or by SearchLight ELISA (Table 4).
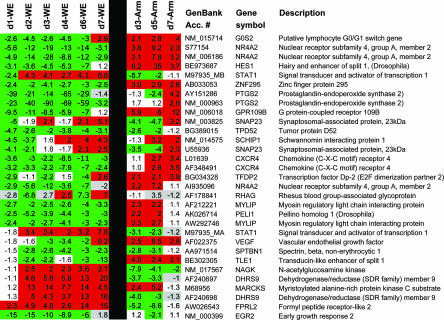
FIG. 2.
Gene expression profiles distinguish between infection with hemorrhagic fever virus and infection with a benign virus. Blood was obtained from rhesus macaques on the days indicated, and the most prominent differentially expressed genes between the two viruses are represented here. Animal numbers are as described for Fig. 1, so for days 1 and 6 the geometric mean of data from two animals is presented. In addition to the 31 genes depicted here, there are approximately 450 genes that distinguish these two infections with a P value confidence level of <0.05 (see Tables SI to SV in the supplemental material). Differential expression, i.e., the level of a gene's expression after infection normalized to its expression level before infection, is shown. The green color represents downregulated gene expression, and the red color represents upregulated gene expression. White values are below the level of ±twofold deviation from baseline, and gray values are below white values in detection quality. Days postinfection (d1, etc.) are indicated.
TABLE 1.
Potential signature genes for early (day 1 to day 3) stage of LCMV-WE infection
| GenBank accession no. | Description | Gene symbol | Fold changea |
|---|---|---|---|
| NM_000647 | Chemokine (C-C motif) receptor 2 | CCR2 | 3.6 |
| NM_000591 | CD14 antigen | CD14 | 2.3 |
| AF130095 | Fibronectin 1 | FN1 | 3.4 |
| AI459194 | Early growth response 1 | EGR1 | −3.2 |
| NM_000399 | Early growth response 2 (Krox-20 homolog, Drosophila) | EGR2 | −13.3 |
| NM_015714 | Putative lymphocyte G0/G1 switch gene | G0S2 | −3.2 |
| NM_006018 | G protein-coupled receptor 109B | GPR109B | −9.5 |
| BE973687 | Hairy and enhancer of split 1 | HES1 | −3.7 |
| BE898861 | Heterogeneous nuclear ribonucleoprotein C (C1/C2) | HNRPC | −3.1 |
| BE256479 | Heat shock 60-kDa protein 1 (chaperonin) | HSPD1 | −3.1 |
| AF073310 | Insulin receptor substrate 2 | IRS2 | −4.0 |
| AA348410 | Inositol 1,4,5-trisphosphate 3-kinase B | ITPKB | −4.4 |
| NM_002258 | Killer cell lectin-like receptor subfamily B, member 1 | KLRB1 | −4.0 |
| NM_005466 | Mediator of RNA polymerase II transcription | MED6 | −4.3 |
| NM_018835 | Membrane-associated DNA binding protein | MNAB | −3.0 |
| AI935096 | Nuclear receptor subfamily 4, group A, member 2 | NR4A2 | −6.8 |
| NM_014059 | Response gene to complement 32 | RGC32 | −4.9 |
| AY151286 | Prostaglandin-endoperoxide synthase 2 | PTGS2 | −24.5 |
| AB051450 | Transducer of ERBB2, 2 | TOB2 | −3.0 |
| AK025862 | Tumor rejection antigen (gp96) 1 | TRA1 | −3.7 |
| Other | Otherb | 71 genes | ≥−2 |
Geometric means of the changes for days 1, 2, and 3 after infection. Mean changes in expression for four LCMV-WE-infected animals were compared to results from three LCMV-Arm-infected animals, and 91 genes had ANOVA P values of <0.05.
The other 71 genes are listed elsewhere (see Table SV in the supplemental material).
TABLE 2.
Potential signature genes for the viremic (day 4 to day 7) stage of infection with LCMV-WE: the upregulated genes (approximately 94 genes)
| GenBank accession no. | Description | Gene symbol | Fold changea |
|---|---|---|---|
| BC000006 | ATPase, Na+/K+ transporting, beta 1 polypeptide | ATP1B1 | 6.6 |
| AF307338 | B aggressive lymphoma gene | BAL | 8.0 |
| NM_004335 | Bone marrow stromal cell antigen 2 | BST2 | 6.8 |
| AI337069 | Viperin | CIG5 | 8.1 |
| NM_006820 | Chromosome 1 open reading frame 29 | C1orf29 | 13.8 |
| NM_001565 | Chemokine (C-X-C motif) ligand 10 | CXCL10 | 82.5 |
| AF240697 | Dehydrogenase/reductase (SDR family) member 9 | DHRS9 | 12.8 |
| AK002064 | DNA polymerase-transactivated protein 6 | DNAPTP6 | 11.6 |
| AA633203 | Epithelial stromal interaction 1 (breast) | EPSTI1 | 8.9 |
| NM_005101 | IFN-α-inducible protein (clone IFI-15K) | G1P2 | 13.2 |
| BC001782 | Growth arrest-specific 2 like 1 | GAS2L1 | 5.8 |
| BC002666 | Guanylate binding protein 1, IFN inducible, 67 kDa | GBP1 | 20.9 |
| NM_004120 | Guanylate binding protein 2, IFN inducible | GBP2 | 8.9 |
| NM_000161 | GTP cyclohydrolase 1 (dopa-responsive dystonia) | GCH1 | 16.4 |
| NM_004751 | Glucosaminyl (N-acetyl) transferase 3, mucin type | GCNT3 | 6.8 |
| NM_000165 | Gap junction protein, alpha 1, 43 kDa (connexin 43) | GJA1 | 6.7 |
| AI719730 | Guanylate cyclase 1, soluble, alpha 3 | GUCY1A3 | 5.5 |
| NM_017912 | Hect domain and RLD 6 | HERC6 | 10.2 |
| NM_018326 | Immunity-associated protein 4 | HIMAP4 | 12.0 |
| NM_016323 | Hect domain and RLD 5 | HERC5 | 8.2 |
| NM_005345 | Heat shock 70-kDa protein 1A | HSPA1A | 9.4 |
| NM_005532 | IFN-α-inducible protein 27 | IFI27 | 92.6 |
| NM_006417 | IFN-induced protein 44 | IFI44 | 28.1 |
| NM_022168 | IFN induced with helicase C domain 1 | IFIH1 | 7.4 |
| NM_001548 | IFN-induced protein with tetratricopeptide repeats 1 | IFIT1 | 52.7 |
| BE888744 | IFN-induced protein with tetratricopeptide repeats 2 | IFIT2 | 47.5 |
| AI075407 | IFN-induced protein with tetratricopeptide repeats 4 | IFIT4 | 14.5 |
| NM_006548 | IGF-II mRNA-binding protein 2 | IMP-2 | 6.8 |
| AF098114 | Integrin, alpha 2b (platelet glycoprotein) | ITGA2B | 5.0 |
| M68956 | Myristoylated alanine-rich protein kinase C substrate | MARCKS | 11.1 |
| NM_017947 | Molybdenum cofactor sulfurase | MOCOS | 7.7 |
| NM_002462 | Myxovirus (influenza virus) resistance 1 | MX1 | 9.6 |
| NM_002463 | Myxovirus (influenza virus) resistance 2 (mouse) | MX2 | 14.6 |
| NM_002534 | 2′,5′-oligoadenylate synthetase 1, 40/46 kDa | OAS1 | 5.0 |
| NM_016817 | 2′,5′-oligoadenylate synthetase 2, 69/71 kDa | OAS2 | 15.4 |
| NM_006187 | 2′,5′-oligoadenylate synthetase 3, 100 kDa | OAS3 | 4.0 |
| NM_003733 | 2′,5′-oligoadenylate synthetase-like | OASL | 18.7 |
| NM_016619 | Placenta-specific 8 | PLAC8 | 10.3 |
| AF178841 | Rhesus blood group-associated glycoprotein | RHAG | 6.3 |
| AI304317 | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide | RIG-I | 7.2 |
| AB028976 | Sterile alpha motif domain containing 4 | SAMD4 | 12.3 |
| X15132 | Superoxide dismutase 2, mitochondrial | SOD2 | 8.4 |
| BC002704 | Signal transducer and activator of transcription 1, 91 kDa | STAT1 | 10.6 |
| U57059 | Tumor necrosis factor (ligand) superfamily, member 10 | TNFSF10 | 13.8 |
| NM_004223 | Ubiquitin-conjugating enzyme E2L 6 | UBE2L6 | 4.3 |
| Other | Other genesb | 49 genes | ≥2.0 |
Geometric mean of the change for days 4, 6, and 7 after infection. Mean changes in expression for four LCMV-WE-infected animals were compared to results from three LCMV-Arm-infected animals and 94 upregulated genes had an ANOVA P value of <0.05.
The other 49 upregulated genes can be found by inspection of Table SI in the supplemental material.
TABLE 3.
Potential signature genes for the viremic (day 4 to day 7) stage of infection with LCMV-WE: the downregulated genes (approximately 293 genes)
| GenBank accession no. | Description | Gene symbol | Fold changea |
|---|---|---|---|
| NM_004038 | Amylase, alpha 2B; pancreatic | AMY2B | −6.3 |
| AI743534 | Rho GTPase activating protein 24 | ARHGAP24 | −8.2 |
| NM_021813 | BTB and CNC homology 1, basic leu zipper transfactor 2 | BACH2 | −6.8 |
| AW303865 | Chromosome 20 open reading frame 22 | C20orf22 | −6.6 |
| NM_001765 | CD1C antigen, c polypeptide | CD1C | −8.4 |
| M74721 | CD79A antigen (immunoglobulin-associated alpha) | CD79A | −7.8 |
| AJ224869 | Chemokine (C-X-C motif) receptor 4 | CXCR4 | −7.0 |
| BG251521 | MRNA; cDNA DKFZp586B211 | DKFZp586B211 | −8.0 |
| AI332397 | Eukaryotic translation initiation factor 4A, isoform 2 | EIF4A2 | −5.6 |
| NM_001975 | Enolase 2 (gamma, neuronal) | ENO2 | −4.7 |
| BC000284 | Golgi associated; g-adaptin ear containing; ARF binding 2 | GGA2 | −5.6 |
| NM_002120 | Major histocompatibility complex, class II, DO beta | HLA-DOB | −9.0 |
| NM_002118 | Major histocompatibility complex, class II, DM beta | HLA-DMB | −4.7 |
| BG397856 | Major histocompatibility complex, class II, DQ alpha 1 | HLA-DQA1 | −4.7 |
| AI638155 | Interleukin 24 | IL-24 | −4.8 |
| AW005982 | Metastasis associated in lung adenocarcinoma transcript 1 | MALAT-1 | −6.4 |
| NM_014048 | MKL/myocardin-like 2 | MKL2 | −4.8 |
| AI808597 | Membrane-spanning 4 domains, subfamily A, member 1 | MS4A1 | −5.5 |
| NM_013262 | Myosin regulatory light chain-interacting protein | MYLIP | −4.7 |
| U85430 | Nuclear factor of activated T cells, c | NFATC3 | −6.3 |
| NM_002628 | Profilin 2 | PFN2 | −6.3 |
| NM_000963 | Prostaglandin-endoperoxide synthase 2 | PTGS2 | −74.5 |
| AV705803 | Splicing factor proline/glu rich | SFPQ | −4.9 |
| AA524053 | Splicing factor, arginine/serine rich 7, 35 kDa | SFRS7 | −5.6 |
| AA971514 | Spectrin, beta, nonerythrocytic 1 | SPTBN1 | −6.5 |
| NM_031214 | TAR DNA binding protein | TARDBP | −5.8 |
| AA812232 | Thioredoxin-interacting protein | TXNIP | −5.4 |
| S80876 | Zinc finger protein, subfamily 1A, 1 (Ikaros) | ZNFN1A1 | −6.7 |
| Other | Otherb | 266 genes | −2 to −4.5 |
Geometric means of transcriptome changes for days 4, 6, and 7 after infection with LCMV-WE. Mean changes in expression for four LCMV-WE-infected animals were compared to results from three LCMV-Arm-infected animals, and 293 downregulated genes had ANOVA P values of <0.05.
The other 266 downregulated genes can be found elsewhere (see Table SI in the supplemental material).
TABLE 4.
Confirmation of expression patterns of selected genes by quantitative RT-PCRa
| GenBank accession no. | Gene symbol | Fold change in gene expression for macaques (DNA array/real-time PCR) onb:
|
|||||
|---|---|---|---|---|---|---|---|
| Day 1 | Day 2 | Day 3 | Day 4 | Day 6 | Day 7 | ||
| NM_004335 | BST2 | 1.0/−1.5 | 2.0/−1.5 | 2.8/2.1 | 3.0/4.1 | 7.9/4.4 | 8.3/3.0 |
| NM_001565 | CXCL10 | 1.1/−1.0 | 1.0/7.5 | 3.9/3.9 | 12.3/21.4 | 134/119.3 | 49.6/4.9 |
| NM_000399 | EGR2 | −4.7/−31.9 | −14.6/−50.1 | −10.2/−5.0 | −8.9/−31.7 | −5.9/−66.3 | 1.8/−127.4 |
| NM_006018 | GPR109B | −4.2/−5.1 | −10.6/−2.0 | −8.5/−11.8 | −5.9/−22.4 | −10.0/−16.4 | 1.2/−7.4 |
| NM_002120 | HLA-DOB | −2.5/−3.8 | −1.2/2.8 | −2.2/−2.9 | −2.8/−2.0 | −6.5/−14.7 | −14.5/−44.9 |
| NM_000576 | IL-1-b | −1.0/−3.6 | −2.4/−4.5 | −3.6/−32.0 | −2.8/−2.0 | −3.7/−81.6 | −1.2/−261.4 |
| NM_002463 | MX2 | 1.0/−2.9 | 15.8/16.9 | 15.3/4.3 | 12.2/9.1 | 16.4/5.2 | 13.2/4.5 |
| NM_016817 | OAS2 | 1.1/−4.0 | 7.6/18.0 | 7.2/5.1 | 12.2/10.7 | 17.2/7.6 | 15.1/4.5 |
| NM_000963 | PTGS2 | −10.5/−75.1 | −39.7/−10.3 | −89.9/−99.7 | −69.1/−125.8 | −77.0/−254.8 | −3.2/−51.3 |
| BC002704 | STAT1 | 1.6/−4.2 | 8.2/8.9 | 4.8/3.8 | 4.5/3.5 | 10.9/2.0 | 16.2/ND |
| AK027071 | TGF-b1I4 | −1.0/−2.0 | −3.3/−3.2 | 1.8/−3.3 | 1.4/−1.1 | 3.9/−3.9 | 5.7/ND |
| D50683 | TGF-BRII | −1.1/1.1 | −1.3/2.2 | −1.3/−2.0 | −3.5/1.3 | −2.0/−2.3 | −1.2/−34.1 |
| NM_014886 | TINP1 | −1.3/3.6 | −1.4/5.3 | −2.0/−2.0 | −3.0/3.4 | −3.4/−4.6 | −2.8/−9.0 |
| AW872374 | TLR8 | 2.4/2.7 | −1.0/4.7 | −1.0/−1.4 | −1.1/−14.3 | 2.7/ND | 6.9/ND |
| U57079 | TNFSF10 | −1.0/2.7 | 2.0/9.5 | 4.9/2.0 | 2.8/7.2 | 14.3/4.0 | 23.6/2.4 |
| AF022375 | VEGF | 1.1/−7.1 | −2.0/1.1 | −3.0/−2.6 | −3.4/−2.0 | −2.1/1.0 | 9.2/ND |
Primer pairs used for real-time RT-PCR will be supplied upon request.
Change (n-fold) was used to evaluate a gene's mRNA expression level by comparing LCMV-WE-infected samples to control samples (from the same animals preinfection) in terms of the numbers of PBMC. P values for the real-time PCR are at <0.05. ND, not determined.
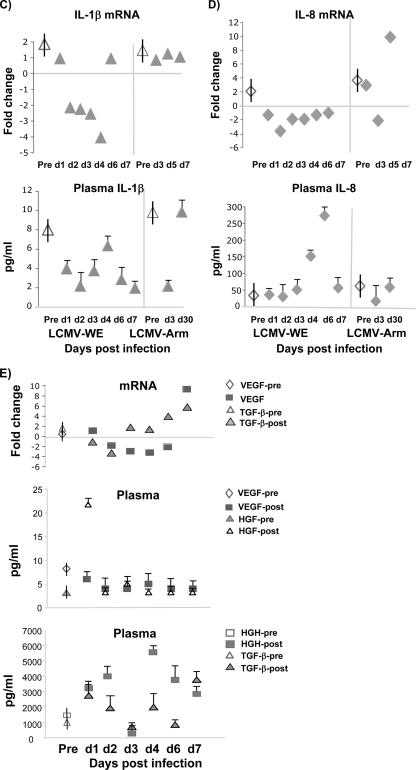
Genes with dramatically different expression in virulent versus nonvirulent infections included the gene for PTGS2 (prostaglandin-endoperoxide synthase 2), also known as cyclooxygenase 2 (COX-2). We confirmed the reduced expression (10- to 250-fold) of PTGS2 during early and viremic stages of LCMV-WE infection by quantitative RT-PCR. In contrast to the LCMV-WE suppression of COX-2, this transcript was upregulated by twofold in the nonvirulent LCMV-Arm infection. The decreased PTGS2 mRNA expression in LCMV-WE infection was correlated with decreases in mRNA levels of vascular endothelial growth factor (VEGF) (7-fold down) and interleukin-1 β (IL-1β) (261-fold down), both of which are known to upregulate PTGS2 (34, 56). Another gene that was dramatically downregulated (127-fold) during the virulent infection was early growth response 2 (EGR2), a gene from a family of genes that function with VEGF to promote lymph node development (8). Thus, some observations from the initial microarray data were confirmed by similar changes in a group of coordinately regulated genes and by quantitative PCR studies (discussed below).
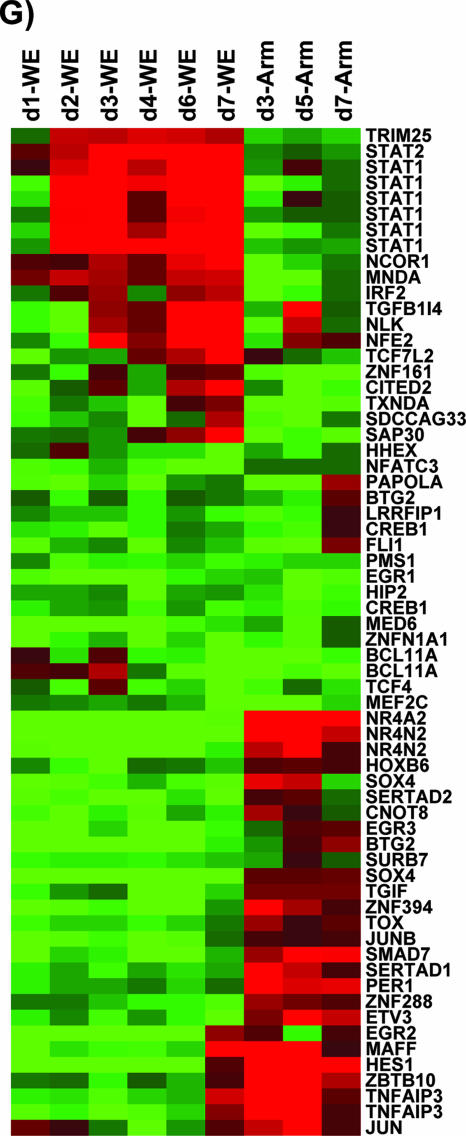
Functional grouping of genes regulated by LCMV infection.
Genes regulated by LCMV were first grouped based on their cellular functions. Over 65% of the 6,416 differentially regulated genes were categorized as immune, proinflammatory, or stress response genes (Fig. 3; also see Tables SI to SV in the supplemental material for additional online data). A number of genes encoding transcription control factors (approximately 50) were downregulated, while 25 of them were upregulated in LCMV-WE-infected PBMC. The transcription factor human homeobox gene (HHEX) (see Table SIV in the supplemental material), also known as proline-rich homeobox (PRH), was not differentially expressed in PBMC, contrary to what we described previously for liver (22).
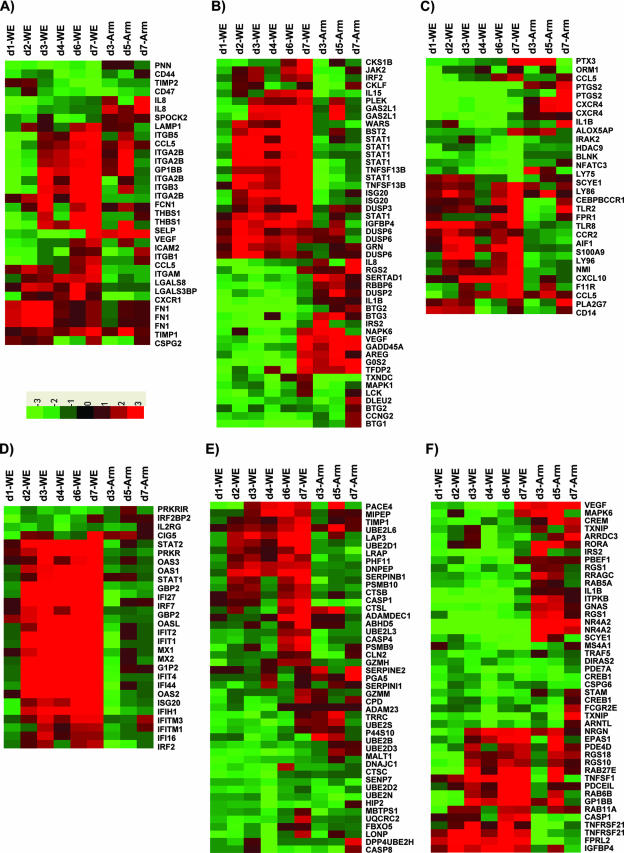
FIG. 3.
Functional categories of transcripts (see Tables SI to SV in the supplemental material). Differentially expressed genes with confidence level P values of <0.05 were grouped into functional categories including cell adhesion (A), cell proliferation (B), inflammatory (C), IFN response (D), proteolysis (E), signal transduction (F), and transcription (G). On the heat map, the green color represents downregulated gene expression and the red color represents upregulated gene expression (the scale at the bottom of panel A indicates relative expression change). Days postinfection (d1, etc.) are indicated.
Remarkable changes were seen in the group of genes controlling the eicosanoid biosynthetic pathway. Prostaglandins and leukotrienes are potent eicosanoid lipid mediators derived from phospholipase-released arachidonic acid and are involved in numerous homeostatic functions and inflammation. Transcription profiling during virulent infection revealed altered gene expression of eicosanoid pathway enzymes as follows: for arachidonate 5-lipoxygenase (ALOX5), expression was up and then down; for ALOX5-activating protein (ALOX5AP), expression was always down; for PTGS1, expression was always up; and for PTGS2, expression was always down (see Table SI in the supplemental material). Most notably, PTGS2 (encoding COX-2), mentioned in the previous section as a gene dramatically suppressed in virulent infections (Fig. 2), also stood out in the functional groupings of inflammatory responses.
Temporal grouping of genes: early host response to the virulent strain of LCMV.
Infection by the virulent LCMV-WE strain at the earliest (previremic [days 1 to 3]) time points revealed a number of downregulated genes greater than that of upregulated genes in macaque PBMC (Fig. 1 and 2; also see Table SI in the supplemental material). Prominent early downregulated genes included PTGS2, G protein-coupled receptor (GPR), EGR1 and -2, nuclear receptor subfamily 4 group A (NR4A2), chemokine (C-X-C motif) receptor 4 (CXCR4), insulin receptor substrate 2 (IRS2), and response gene to complement 32 (RGC32). Prominent genes that were upregulated during the previremic phase of the virulent infection included interferon (IFN)-inducible genes (e.g., CIG5, IFI44, IFIT4, GBP1, OAS2, OAS-L, and MX2), signal transducer and activator of transcription 1 (STAT1), dehydrogenase/reductase 9 (DHRS9), myristoylated alanine-rich protein kinase C substrate (MARCKS), and S100 calcium binding protein P (S100P). Profiling of the virulent infection demonstrated significant differences from the nonvirulent LCMV-Arm infection even before the onset of viremia (Table 1; also see Table SV in the supplemental material). Most of the differentially expressed genes remained unchanged after day 4 of infection (see Table SI in the supplemental material).
Viremic stage of response to the virulent strain of LCMV.
The host transcriptional response to LCMV-WE viremia was dominated by immune response, stress response, and proinflammatory genes. We also observed specific upregulation of genes involved in antiviral responses, including IL-15 (4-fold), as well as in a number of type I and II IFN-responsive genes, such as GBP1 (>37-fold), GBP2 (>12-fold), G1P2 (>15-fold), MX1 (>15-fold), and MX2 (>16-fold) (see Table SI in the supplemental material). Mx antiviral proteins belong to the family of GTPases and are induced preferentially by IFN-α/β (25). Viperin (cig5), encoding an antiviral protein, is also upregulated by LCMV-WE infection but is downregulated with nonvirulent LCMV-Arm infection. Viperin is an IFN-inducible protein and is also upregulated by human cytomegalovirus and vesicular stomatitis virus infections (7, 15). Viperin and Mx proteins have been shown to impair influenza virus, vesicular stomatitis virus, rabies virus, and hepatitis C virus replication (15, 33, 40, 80). Increases in the expression of genes encoding neutrophil and/or monocyte/macrophage-targeted chemokines and chemokine receptors were observed by day 6 of the viremic stage. Interestingly, the gene which proved to be most strongly induced (119-fold) among all 6,416 differentially regulated transcripts was the IFN-stimulated chemokine CXCL10, also known as IP-10, which has a critical role in recruiting T-helper cells and other lymphocytes (see Table SI in the supplemental material). Expression of CCL3 (also known as MIP-1α), CCL4 (also known as MIP-1β), CCL20, and the CCL3/CCL4 receptor CCR5 were all increased by 6 days postinfection. By contrast, the related CCL5 (RANTES) was downregulated in the previremic stage and was then unremarkable in the viremic stage. Cytokines, chemokines, and chemokine receptors taken together represented the strongest induction among all functional subclasses of immune/stress/inflammatory response genes (Tables 2 and 3).
Confirming microarray data by RT-PCR.
In previous publications, we confirmed viral modulation of mRNA levels by use of quantitative real-time RT-PCR as well as measures of protein expression (21, 22). Real-time PCR was carried out using cDNA from LCMV-Arm and -WE-infected animals and from preinfection control animals to determine gene expression levels. Modulation of the specific mRNA for 16 genes relative to the expression of GAPDH was assessed at various times after infection. These genes were selected from different categories that contained the most highly up- or downregulated genes. Microarray gene expression levels were compared to those obtained by real-time PCR, using the same monkey blood samples that were used in the microarray hybridization. The microarray data were correlated with those obtained by quantitative real-time RT-PCR (Table 4), and sustained up- or downregulation was often also seen by PCR. The magnitude of up- or downregulation was usually more extreme in PCR than in microarray, since PCR data are accurate over a broader range.
Plasma chemokine and cytokine protein expression levels in LCMV-infected rhesus macaques.
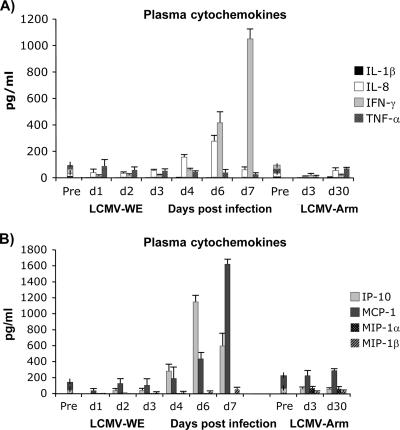
To determine whether plasma chemokine and cytokine concentrations were significantly different between virulent LCMV and nonvirulent virus throughout the experiment, samples from infected rhesus macaques were analyzed by multiplexed sandwich ELISA. Plasma protein levels of LCMV-infected animals were compared to those of the control preinfection animals. The levels of plasma cytokines remained unchanged from the preinfected levels during the previremic stage of infection. After day 4 of infection (viremic stage), IFN-γ, IP-10, and IL-8 levels were markedly upregulated in macaque plasma and remained elevated up to day 7 (Fig. 4A).
FIG. 4.
Plasma protein level was affected by LCMV infection. Cytokine and chemokine concentrations in plasma samples of uninfected control and LCMV-infected monkeys were determined. Cytochemokines (A and B), IL-1β (C), IL-8 (D), and angiogenesis and growth factors (E) were measured using SearchLight protein arrays. Results are expressed as means ± standard deviations (error bars) from at least three monkeys and three replicates per time point. Based on the Student t test, we observed statistically significant differences (P < 0.05) between LCMV-infected plasma samples and control preinfection plasma. Vertical bars superimposed upon symbols signify average plasma protein levels of preinfection control samples (n = 21). Pre and post, preinfection and postinfection, respectively.
IFN-γ-inducible protein 10 (IP-10 or CXCL10) is a CXC chemokine that binds to the CXCR3 receptor on activated T lymphocytes and NK cells (73). We measured plasma levels of IP-10 in animals infected with virulent or nonvirulent LCMV strains. IP-10 was elevated markedly in plasma and remained elevated between day 4 and day 7, and levels were positively correlated with IFN-γ concentrations and plasma viremia beginning 4 days after infection (Fig. 4B). Monocyte chemoattractant protein 1 (MCP-1), known for its ability to loosen endothelial cell junctions (41) and to participate in the breakdown of the blood-brain barrier (75), had a similarly high plasma profile in samples from virulently infected monkeys but not in samples from uninfected or LCMV-Arm-infected monkeys (Fig. 4B). Analysis of secreted proteins in plasma showed decreased levels of VEGF, IL-1β, and tumor necrosis factor alpha (TNF-α) (Fig. 4C to E). Since VEGF is one of the principal factors responsible for inducing blood mononuclear cell COX-2 expression (34), it is reasonable that the downregulation of VEGF (Fig. 4E) should be concomitant with the downregulation of COX-2.
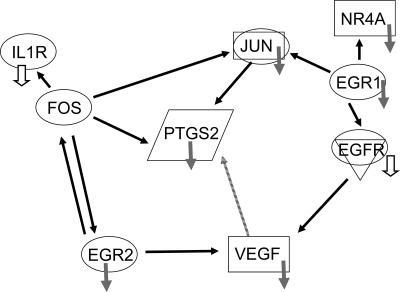
A pathway analysis revealed the connections between a few important gene products, such as the transcription factors EGR1 and -2 and the COX-2 gene PTGS2 (Fig. 5). Additional nodes in this pathway included two gene products (IL-1R and epidermal growth factor receptor [EGFR]) that were prominent in a “kinomics” study by Bowick et al. (10). The severe downregulation of EGR1 and EGR2 during the virulent infection is directly linked to the downregulation of PTGS2, VEGF, IL-1R, and EGFR.
FIG. 5.
Pathway analysis reveals interactions between the major gene products affected by virulent infection. EGFR and IL-1R are included in the diagram to show that our results corroborate those from Bowick et al. (10), in which guinea pigs infected with virulent Pichinde virus P18 have much lower levels of phosphorylated EGFR and IL-1R than guinea pigs infected with benign Pichinde virus P2. Arrows pointing from one gene product to another signify activation. Small gray arrows signify downregulation, with the open arrows derived from kinomic results from the work of Bowick et al. (10). EGR1 and EGR2 are transcription factors, NR4A is a retinoic acid receptor, avian sarcoma virus oncogene homolog JUN is a transcription factor, murine sarcoma virus oncogene homolog FOS is a transcription factor, PTGS2 encodes COX-2, and IL-1R is an IL receptor protein. A connection between PTGS2 and VEGF was published previously (2) but not described in the Ingenuity Pathway Analysis, so it is indicated by a stippled arrow.
DISCUSSION
Transcription, or “transcriptome,” profiling of PBMC was used to identify gene expression changes in macaques during lethal infection with hemorrhagic fever virus. We identified genome-wide expression changes due to infection, characterized genes that were differentially expressed with respect to a nonvirulent arenavirus, and confirmed our most important results by real-time RT-PCR and by ELISA-based protein analysis.
During days 2 to 4, the global impact of LCMV-WE infection was to reduce host gene expression. With the onset of viremia (after day 4), the trend was reversed as host genes were turned on in response to the rising viral burden (Fig. 1). This finding resembles an observation made by the Shenk laboratory about human cytomegalovirus in tissue culture: gene expression was predominantly downregulated in infected cells, but UV-inactivated virus caused upregulation of a large number of mRNAs, many of which contained IFN-responsive genes and encoded proinflammatory cytokines (11). Abundant viral replication, including the production of noninfectious particles, appears to drive the expression of IFN-responsive genes. A study of influenza-exposed cell cultures from the Katze laboratory revealed that a substantial proportion of the transcriptome response to viral infection can be driven by viral antigens in the absence of replication (30). Thus, the broad pattern of gene upregulation during the viremic phase of infection may reflect the host response to viral particles with an ensuing activation of IFN to control the spreading infection.
A recent profiling study with influenza virus-infected macaques demonstrated that gene expression arrays can be used to detect infection after exposure but prior to the onset of symptoms or virus shedding (3). In our study, we identified 91 genes expressed before the onset of viremia that differed significantly between virulent and nonvirulent infections (Table 1). Several of these early host responses have also been noted for murine LCMV infections. For example, CCR2, the MCP-1 receptor that recruits monocytes to inflamed sites, was upregulated in LCMV-WE-infected monkeys and in LCMV-infected mice but not in vesicular stomatitis virus-infected mice (57). Murine brains persistently infected with LCMV upregulated STAT1 and IFN-stimulated genes (39) that are prominent early in the monkey virulent infection; however, the murine tissues upregulated the major histocompatibility complex and β2-microglobulin genes, but these are downregulated in monkeys. The monkey fibronectin gene was another gene upregulated early; similarly, in murine LCMV hepatitis, fibronectin deposits accompany viral antigen in hepatic sinusoids (42). Early growth response genes EGR1 and EGR2 are downregulated early in virulent monkey infections. In acute murine LCMV infections, EGR genes are critical for the development of a vigorous cytotoxic-T-lymphocyte response (72), and early downregulation of EGR1 and -2 in monkeys could be a mechanism for limiting the development of LCMV-specific cell-mediated immunity. Heat shock protein gp96 is involved in the stable antigen presentation of mature LCMV NP (4), and the gp96 gene, TRA1, was downregulated early in infected monkeys, indicating another mechanism for limiting antiviral immune responses. The resemblance between murine responses to LCMV infection and previremic primate responses supports the role of these transcripts as early indicators of infection.
The IFN response pathways constitute an important antiviral response during LCMV infection. Many IFN-inducible genes, including the MX2 gene, were highly induced by day 2 in LCMV-WE-infected PBMC (Fig. 3D). Murine studies showed that LCMV triggers early and high levels of IFN production in nonplasmacytoid dendritic cells (20). However, in our monkey studies plasma levels of type I IFN were unremarkable, though type II IFN levels did rise with viremia. The IFN-inducible genes are notable as a group because they are more readily upregulated by inactivated viral particles than by infectious particles (references 11 and 51 and M. S. Salvato, unpublished data). Reduced expression of IFN-inducible genes in the LCMV-Arm-infected monkeys is due either to lower levels of viral replication or to the successful suppression of the IFN pathway by LCMV-Arm.
TNF-α/NF-κB-responsive genes were suppressed during smallpox infection and strongly upregulated during the viremic phase of Ebola virus infection in a monkey transcriptome study by Rubins et al. (69). In our study, many of these same genes, categorized as IFN responsive or inflammatory, were also upregulated (with a few notable exceptions) during the viremic stage of virulent infection and were relatively diminished in expression during nonvirulent infection. The exceptions include IL-8, VEGF, IL-1β, and CXCR4, which were downregulated, while most inflammatory genes (CXCL10, CCR5, CCR2, and TLR8) were upregulated in a virulent (lethal) infection. We do not know how these genes would behave in an animal that survives virulent disease. In a study of people infected with Lassa fever virus, survivors had higher levels of the proinflammatory cytokines (IL-8 and IP-10) than the fatalities (49), indicating that these cytochemokines could be essential components of host defense.
The notion that activated inflammatory pathways can avert lethal viral disease has been pursued in several animal studies. In a mouse model for Ebola, inoculation routes that elicited earlier and higher-level type I IFN promoted survival (50). Treatment of Marburg virus-infected monkeys with IFN-α prolonged survival but could not save the animals (38). In mice, the use of CpG oligonucleotides to stimulate type I IFN via TLR9 reduced arenavirus choriomeningitis (58). Unfortunately, type I IFNs not only induce antiviral responses but also have deleterious immune-suppressive activities. For example, type I and type III IFN responses synergize in suppressing the CD4 proliferative responses to respiratory syncytial virus infection (14). Initially, cell proliferation is blocked by IFN-mediated delays to cell cycling, and once cells have proliferated, IFN promotes apoptosis of activated lymphocytes (24). Our current hypothesis is that early expression of inflammatory genes stimulates acquired immunity but that high expression of inflammatory genes during the viremic phase contributes to the immune suppression of virus-specific host responses.
In support of this hypothesis, we note that innate effectors such as TLR1, TLR2, TLR4, and TLR8 are upregulated late during the viremic phase. TLR10, which is expressed in B cells, respiratory epithelia, and dendritic cells (32), is downregulated by late viremia along with the majority of HLA genes, which are downregulated in LCMV-WE-infected tissues compared to what is seen for LCMV-Arm-infected tissues. Reduced HLA expression likely causes a defect in antigen presentation that could contribute to pathogenesis.
Cell culture studies have been extremely useful in identifying host responses that are directly connected to the functions of viral gene products. For example, the NS proteins of respiratory syncytial virus and the NP protein of LCMV suppress IRF-3 and block NF-κB-promoted expression of IFN-β, IFN-α, and proinflammatory chemokines like IL-8 and TNF-α (53, 74, 76). However, the IFN-suppressive activity of LCMV NP is found in both virulent and benign LCMV isolates, so its connection to pathogenesis is unclear. Cell culture studies with the LCMV Z gene product also failed to find disease-related mutations but did identify important virus-host interactions (12, 16, 22, 59). Our working hypothesis is that the toxic activities of viral gene products like NP or Z must reach a threshold that is dependent on polymerase activity to cause disease.
At first glance, cell culture studies that compare virulent and nonvirulent viruses do not support our findings about gene expression in vivo. A comparison of Lassa virus and Mopeia virus in cell culture showed that TNF-α and IL-8 are downregulated by the virulent virus in relation to the nonvirulent virus (45). In monkeys, although IL-8 is downregulated, TNF superfamily genes are upregulated, and several other NF-κB-driven responses rise during the viremic stage of the virulent disease. Comparison of virulent and nonvirulent arenavirus isolates, Pichinde virus isolates P18 and P2, respectively, in a monocytic cell line predicted the upregulation of gene products (Nurr1, Fez, Striatin, and PTEN) (9); however, the mRNAs for those products were downregulated in our study. A study using liver cell cultures infected with Marburg and Ebola viruses related virulence to increased suppression of the type I IFN and inflammatory responses, downregulation of coagulation genes, and upregulation of immune response genes (35). In our monkey studies, these three gene categories during the viremic phase of LCMV-WE disease look just the opposite of how they appear during primate infection with virulent Ebola virus (69). Our view is that the cell culture studies accurately depict virus infection in the microcosms of cells that are initially infected in vivo, but those results are drowned out by the responses from uninfected cells. Thus, the previremic stage in vivo reflects a small amount of virus in an overwhelmingly large population of uninfected cells, whereas the viremic stage, similar to what is seen in cell culture studies with high virus input, is driven by virus but dominated by host efforts to suppress viral replication.
A recent paper by Bowick et al. (10) compares guinea pigs infected with virulent and benign arenaviruses, Pichinde virus isolates P18 and P2, respectively. Using “kinomics” to assess kinase activity in peritoneal macrophages, this group showed that virulent infection significantly downregulated kinase activity as well as the activity of several signaling pathways. Their findings at the protein level are corroborated by our study, in which several mRNAs with products involved in signal transduction are downregulated in the virulent infection (Fig. 3F). Their most prominent findings, that virulent infection downregulates epidermal growth factor receptor (EGFR) and IL-1 receptor (IL-1R), are corroborated in a pathway analysis of our most prominent early gene downregulations (Fig. 5). Similar to our study, their study found major early responses that are not obviously related to results from virus-infected cell cultures (9). For example, we saw early downregulations of PTGS2 (encoding COX-2), NR4A2 (encoding nuclear receptor NURR1), and GPR109B (encoding G-protein coupled receptor 109B) that were not seen in PBMC cultures exposed to LCMV-WE or LCMV-Arm (J. C. Zapata, M. M. Djavani, and M. S. Salvato, unpublished observations).
Our observation of decreased PTGS2 (encoding COX-2) is remarkable in these studies. COX-2 is the key enzyme in prostaglandin biosynthesis; it is upregulated by many viruses (13, 17, 36, 37, 66, 67, 78), and cytomegalovirus is so dependent on COX-2 for replication that it encodes a COX-2 homologue (70). COX-2 expression can be upregulated by TNF-α, IL-1β, or a variety of growth factors like VEGF and EGRF (2), but virulent arenaviruses in cell culture usually downregulate such products of NF-κB/RBP-mediated transcription (22, 26, 45). In fact, the genes for IL-1β, VEGF, and IL-8 are all downregulated in virulent monkey infections compared to what is seen for nonvirulent monkey infections. Downregulation of COX-2 occurs during overdose of drugs such as rofecoxib (Vioxx), which are known to deplete prostacyclins, destroy clotting, and cause hemorrhage (63). Our findings with COX-2 corroborate studies by Fisher-Hoch and Cummins et al. that show a decrease in prostacyclin in monkeys with Lassa fever (18, 28). The decrease in PTGS2 expression during virulent disease is not directly attributable to viral gene functions but can provide a mechanism for the coagulation defects (18, 55), cardiac dysfunction (61, 64), and capillary leakage (55) that are hallmarks of arenavirus hemorrhagic fever.
In summary, microarray analysis of blood from rhesus monkeys revealed how infection with a virulent arenavirus regulates gene expression in an animal model. The early stage of the host response to virulent LCMV infection is dominated by innate immune responses based on complement, IFN, and proinflammatory processes. The alteration in the transcriptome during viremia strongly suggests that the higher level of replication of the virulent virus contributes to the pathogenicity of LCMV-WE-associated hemorrhagic fever. Of 54,000 probe sets in a full human genome array, 12% were differentially regulated in blood during primate infection with the virulent LCMV-WE. Roughly 400 of these genes differed in expression between LCMV-WE and LCMV-Arm infections, and of these genes, 91 differed during the early or previremic stage and could potentially be biomarkers of disease that are capable of classifying the disease prior to the onset of viremia. The characterization of gene expression changes provides a dynamic view of the host-pathogen interaction and corroborates a recent publication (10) on the guinea pig model for Lassa fever.
Supplementary Material
Acknowledgments
The animal work carried out at University of Maryland Biotechnology Institute, Institute of Human Virology, was supported by start-up funds to M. S. Salvato from the Institute of Human Virology with some support from NIH grants to M. S. Salvato (AI053619, AI053620, and a subcontract from the Mid-Atlantic Regional Centers of Excellence and Emerging Infectious Disease Research [MARCE; U54 AI057168 to M. Levine] for diagnostics for flu-like diseases). The bioinformatics data analysis at VBI was funded by Department of Defense grant DAAD 13-02-C-0018 to B. Sobral.
We are grateful for Susan Martino-Catt and Clive Evans for helpful comments and technical support.
Footnotes
Published ahead of print on 23 May 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ace, C. I., and W. C. Okulicz. 2004. Microarray profiling of progesterone-regulated endometrial genes during the rhesus monkey secretory phase. Reprod. Biol. Endocrinol. 2:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackerman, W. E., IV, B. H. Rovin, and D. A. Kniss. 2004. Epidermal growth factor and interleukin-1beta utilize divergent signaling pathways to synergistically upregulate cyclooxygenase-2 gene expression in human amnion-derived WISH cells. Biol. Reprod. 71:2079-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baas, T., C. R. Baskin, D. L. Diamond, A. Garcia-Sastre, H. Bielefeldt-Ohmann, T. M. Tumpey, M. J. Thomas, V. S. Carter, T. H. Teal, N. Van Hoeven, S. Proll, J. M. Jacobs, Z. R. Caldwell, M. A. Gritsenko, R. R. Hukkanen, D. G. Camp II, R. D. Smith, and M. G. Katze. 2006. An integrated molecular signature of disease: analysis of influenza virus-infected macaques through functional genomics and proteomics. J. Virol. 80:10813-10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basta, S., R. Stoessel, M. Basler, M. van den Broek, and M. Groettrup. 2005. Cross-presentation of the long-lived lymphocytic choriomeningitis virus nucleoprotein does not require neosynthesis and is enhanced via heat shock proteins. J. Immunol. 175:796-805. [DOI] [PubMed] [Google Scholar]
- 5.Bausch, D. G., S. S. S. Sesay, and B. Oshin. 2004. In memoriam Aniru Conteh: on the front lines of Lassa fever. Emerg. Infect. Dis. 10:1889-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57:289-300. [Google Scholar]
- 7.Boudinot, P., S. Riffault, S. Salhi, C. Carrat, C. Sedlik, N. Mahmoudi, B. Charley, and A. Benmansour. 2000. Vesicular stomatitis virus and pseudorabies virus induce a vig1/cig5 homologue in mouse dendritic cells via different pathways. J. Gen. Virol. 81:2675-2682. [DOI] [PubMed] [Google Scholar]
- 8.Bourbie-Vaudaine, S., N. Blanchard, C. Hivroz, and P. H. Romeo. 2006. Dendritic cells can turn CD4+ T lymphocytes into vascular endothelial growth factor-carrying cells by intercellular neuropilin-1 transfer. J. Immunol. 177:1460-1469. [DOI] [PubMed] [Google Scholar]
- 9.Bowick, G. C., S. M. Fennewald, B. L. Elsom, J. F. Aronson, B. A. Luxon, D. G. Gorenstein, and N. K. Herzog. 2006. Differential signaling networks induced by mild and lethal hemorrhagic fever virus infections. J. Virol. 80:10248-10252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowick, G. C., S. M. Fennewald, E. P. Scott, L. Zhang, B. L. Elsom, J. F. Aronson, H. M. Spratt, B. A. Luxon, D. G. Gorenstein, and N. K. Herzog. 2007. Identification of differentially activated cell-signaling networks associated with Pichinde virus pathogenesis using systems kinomics. J. Virol. 81:1923-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Browne, E. P., B. Wing, D. Coleman, and T. Shenk. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell Dwyer, E. J., H. Lai, R. C. MacDonald, M. S. Salvato, and K. L. Borden. 2000. The lymphocytic choriomeningitis virus RING protein Z associates with eukaryotic initiation factor 4E and selectively represses translation in a RING-dependent manner. J. Virol. 74:3293-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carey, M. A., J. A. Bradbury, J. M. Seubert, R. Langenbach, D. C. Zeldin, and D. R. Germolec. 2005. Contrasting effects of cyclooxygenase-1 (COX-1) and COX-2 deficiency on the host response to influenza A viral infection. J. Immunol. 175:6878-6884. [DOI] [PubMed] [Google Scholar]
- 14.Chi, B., H. L. Dickensheets, K. M. Spann, M. A. Alston, C. Luongo, L. Dumoutier, J. Huang, J. C. Renauld, S. V. Kotenko, M. Roederer, J. A. Beeler, R. P. Donnelly, P. L. Collins, and R. L. Rabin. 2006. Alpha and lambda interferon together mediate suppression of CD4 T cells induced by respiratory syncytial virus. J. Virol. 80:5032-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chin, K.-C., and P. Cresswell. 2001. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. USA 98:15125-15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornu, T. I., and J. C. de la Torre. 2002. Characterization of the arenavirus RING finger Z protein regions required for Z-mediated inhibition of viral RNA synthesis. J. Virol. 76:6678-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crofford, L. J., K. T. McDonagh, S. Guo, H. Mehta, H. Bian, L. M. Petruzelli, and B. J. Roessle. 2005. Adenovirus binding to cultured synoviocytes triggers signaling through MAPK pathways and induces expression of cyclooxygenase-2. J. Gene Med. 7:288-296. [DOI] [PubMed] [Google Scholar]
- 18.Cummins, D., S. P. Fisher-Hoch, K. Walshe, I. J. H. Mackie, D. Bennett, G. Perez, B. Farrar, S. J. Machin, and J. B. McCormick. 1989. A plasma inhibitor of platelet aggregation in patients with Lassa fever. Br. J. Haematol. 72:543-548. [DOI] [PubMed] [Google Scholar]
- 19.Danes, L., R. Benda, and M. Fuchsova. 1963. Experimentalni inhalacni nakaza opic druhu macacus cynomolgus a macacus rhesus virem lymphocitarni choriomeningitidy (kmenem WE). Bratisl. Lek. Listy 43:71-79. [PubMed] [Google Scholar]
- 20.Diebold, S. S., M. Montoya, H. Unger, L. Alexopoulou, P. Roy, L. E. Haswell, A. Al-Shamkhani, R. Flavell, P. Borrow, and C. Reis e Sousa. 2003. Viral infection switches non-plasmacytoid dendritic cells into high interferon producers. Nature 424:324-328. [DOI] [PubMed] [Google Scholar]
- 21.Djavani, M., J. D. Rodas, I. S. Lukashevich, D. Horejsh, P. P. Pandolfi, K. L. B. Borden, and M. S. Salvato. 2001. Role of the promyelocytic leukemia protein PML in the interferon sensitivity of lymphocytic choriomeningitis virus. J. Virol. 75:6204-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Djavani, M., I. Topisirovic, J. C. Zapata, M. Sadowska, Y. Yang, I. S. Lukashevich, K. L. B. Borden, C. D. Pauza, and M. S. Salvato. 2005. The proline-rich homeodomain (PRH/HEX) protein is down-regulated in liver during infection with lymphocytic choriomeningitis virus. J. Virol. 79:2461-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doherty, P. C., and R. M. Zinkernagel. 1974. T-cell mediated immunopathology in viral infections. Transplant. Rev. 19:89-120. [DOI] [PubMed] [Google Scholar]
- 24.Dondi, E., G. Roue, V. J. Yuste, S. A. Susin, and S. Pellegrini. 2004. A dual role of IFN-alpha in the balance between proliferation and death of human CD4+ T lymphocytes during primary response. J. Immunol. 173:3740-3747. [DOI] [PubMed] [Google Scholar]
- 25.Engelhardt, O. G., E. Ullrich, G. Kochs, and O. Haller. 2001. Interferon-induced antiviral Mx1 GTPase is associated with components of the SUMO-1 system and promyelocytic leukemia protein nuclear bodies. Exp. Cell Res. 271:286-295. [DOI] [PubMed] [Google Scholar]
- 26.Fennewald, S. M., J. F. Aronson, L. Zhang, and N. K. Herzog. 2002. Alterations in NF-κB and RBP-Jκ by arenavirus infection of macrophages in vitro and in vivo. J. Virol. 76:1154-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisher-Hoch, S. P., J. B. McCormick, D. Sasso, and R. B. Craven. 1988. Hematologic dysfunction in Lassa fever. J. Med. Virol. 26:127-135. [DOI] [PubMed] [Google Scholar]
- 28.Fisher-Hoch, S. P. 1993. Arenavirus pathophysiology, p. 299-317. In M. Salvato (ed.), The Arenaviridae. Plenum Press, New York, NY.
- 29.Gautier, L., L. Cope, B. M. Bolstad, and R. A. Irizarry. 2004. Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20:307-315. [DOI] [PubMed] [Google Scholar]
- 30.Geiss, G. K., M. C. An, R. E. Bumgarner, E. Hammersmark, D. Cunningham, and M. G. Katze. 2001. Global impact of influenza virus on cellular pathways is mediated by both replication-dependent and -independent events. J. Virol. 75:4321-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilden, D. H., G. A. Cole, A. A. Monjan, and N. Nathanson. 1972. Immunopathogenesis of acute central nervous system disease produced by LCMV. I. Cyclophosphamide-mediated induction of virus-carrier state in adult mice. J. Exp. Med. 135:860-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hasan, U., C. Chaffois, C. Gaillard, V. Saulnier, E. Merck, S. Tancredi, C. Guiet, F. Briere, J. Vlach, S. Lebecque, G. Trinchieri, and E. E. Bates. 2005. Human TLR10 is a functional receptor, expressed by B cells and plasmacytoid dendritic cells, which activates gene transcription through MyD88. J. Immunol. 174:2942-2950. [DOI] [PubMed] [Google Scholar]
- 33.Helbig, K. J., D. T. Lau, L. Semendric, H. A. Harley, and M. R. Beard. 2005. Analysis of ISG expression in chronic hepatitis C identifies viperin as a potential antiviral effector. Hepatology 42:702-710. [DOI] [PubMed] [Google Scholar]
- 34.Hernandez, G. L., O. V. Volpert, M. A. Iniguez, E. Lorenzo, S. Martinez-Martinez, R. Grau, M. Fresno, and J. M. Redondo. 2001. Selective inhibition of vascular endothelial growth factor-mediated angiogenesis by cyclosporin A: roles of the nuclear factor of activated T cells and cyclooxygenase 2. J. Exp. Med. 193:607-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kash, J. C., E. Muhlberger, V. Carter, M. Grosch, O. Perwitasari, S. C. Proll, M. J. Thomas, F. Weber, H. D. Klenk, and M. G. Katze. 2006. Global suppression of the host antiviral response by Ebola- and Marburgviruses: increased antagonism of the type I interferon response is associated with enhanced virulence. J. Virol. 80:3009-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaul, R., S. C. Verma, M. Murakami, K. Lan, T. Choudhuri, and E. S. Robertson. 2006. Epstein-Barr virus protein can upregulate cyclo-oxygenase-2 expression through association with the suppressor of metastasis Nm23-H1. J. Virol. 80:1321-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, H. T., W. Qiang, N. Liu, V. L. Scofield, P. K. Wong, and G. Stoica. 2005. Up-regulation of astrocyte cyclooxygenase-2, CCAAT/enhancer-binding protein-homology protein, glucose-related protein 78, eukaryotic initiation factor 2 alpha, and c-Jun N-terminal kinase by a neurovirulent murine retrovirus. J. Neurovirol. 11:166-179. [DOI] [PubMed] [Google Scholar]
- 38.Kolokol'tsov, A. A., I. A. Davidovich, M. A. Strel'tsova, A. E. Nesterov, O. A. Agafonova, and A. P. Agafonov. 2001. The use of interferon for emergency prophylaxis of Marburg hemorrhagic fever in monkeys. Bull. Exp. Biol. Med. 132:686-688. [DOI] [PubMed] [Google Scholar]
- 39.Kunz, S., J. M. Rojek, A. J. Roberts, D. B. McGavern, M. B. Oldstone, and J. C. de la Torre. 2006. Altered central nervous system gene expression caused by congenitally acquired persistent infection with lymphocytic choriomeningitis virus. J. Virol. 80:9082-9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leroy, M., G. Pire, E. Baise, and D. Desmecht. 2006. Expression of the interferon-alpha/beta-inducible bovine Mx1 dynamin interferes with replication of rabies virus. Neurobiol. Dis. 21:515-521. [DOI] [PubMed] [Google Scholar]
- 41.Lin, C. F., S. C. Chiu, Y. L. Hsiao, S. W. Wan, H. Y. Lei, A. L. Shiau, H. S. Liu, T. M. Yeh, S. H. Chen, C. C. Liu, and Y. S. Lin. 2005. Expression of cytokine, chemokine, and adhesion molecules during endothelial cell activation induced by antibodies against dengue virus nonstructural protein 1. J. Immunol. 174:395-403. [DOI] [PubMed] [Google Scholar]
- 42.Lohler, J., J. Gossmann, T. Kratzberg, and F. Lehmann-Grube. 1994. Murine hepatitis caused by lymphocytic choriomeningitis virus. I. The hepatic lesions. Lab. Investig. 70:263-278. [PubMed] [Google Scholar]
- 43.Lukashevich, I. S., A. D. Vasiuchkov, R. F. Mar'iankova, and V. I. Votyakov. 1982. Factors affecting plaque formation by Lassa virus in Vero cells. Vopr. Virusol. January/February(1):57-61. (In Russian.) [PubMed]
- 44.Lukashevich, I. S., S. V. Orlova, R. F. Mar'iankova, and N. D. Barkar. 1985. Pathogenicity of Lassa virus for laboratory mice. Vopr. Virusol. 30:595-599. (In Russian.) [PubMed] [Google Scholar]
- 45.Lukashevich, I. S., R. Maryankova, A. S. Vladyko, N. Nashkevich, S. Koleda, M. Djavani, D. Horejsh, N. Voitenok, and M. S. Salvato. 1999. Lassa and Mopeia replication in human monocyte/macrophages and in endothelial cells: different effects on IL-8 and TNF-α gene expression. J. Med. Virol. 59:552-560. [PMC free article] [PubMed] [Google Scholar]
- 46.Lukashevich, I. S., M. Djavani, J. D. Rodas, J. C. Zapata, A. Usborne, C. Emerson, J. Mitchen, P. B. Jahrling, and M. S. Salvato. 2002. Hemorrhagic fever occurs after intravenous, but not after intragastric, inoculation of rhesus macaques with lymphocytic choriomeningitis virus. J. Med. Virol. 67:171-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lukashevich, I. S., I. Tikhonov, J. D. Rodas, J. C. Zapata, Y. Yang, M. Djavani, and M. S. Salvato. 2003. Arenavirus-mediated liver pathology: acute lymphocytic choriomeningitis infection of rhesus macaques is characterized by high interleukin-6 expression and hepatocyte proliferation. J. Virol. 77:1727-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lukashevich, I. S., J. D. Rodas, I. Tikhonov, J. C. Zapata, Y. Yang, M. Djavani, and M. S. Salvato. 2004. LCMV-mediated hepatitis in rhesus macaques: WE and not Arm strain activates hepatocytes and induces liver regeneration. Arch. Virol. 385:1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahanty, S., D. G. Bausch, R. L. Thomas, A. Goba, A. Bah, C. J. Peters, and P. E. Rollin. 2001. Low levels of interleukin-8 and interferon-inducible protein-10 in serum are associated with fatal infections in acute Lassa fever. J. Infect. Dis. 183:1713-1721. [DOI] [PubMed] [Google Scholar]
- 50.Mahanty, S., M. Gupta, J. Paragas, M. Bray, R. Ahmed, and P. E. Rollin. 2003. Protection from lethal infection is determined by innate immune responses in a mouse model of Ebola virus infection. Virology 312:415-424. [DOI] [PubMed] [Google Scholar]
- 51.Marcus, P. I., J. M. Rojek, and M. J. Sekellick. 2005. 2005. Interferon induction and/or production and its suppression by influenza A viruses. J. Virol. 79:2880-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martinez Peralta, L. A., M. Laguens, C. Ponzinibbio, and R. P. Laguens. 1990. Infection of guinea pigs with two strains of lymphocytic choriomeningitis virus. Medicina (Buenos Aires) 50:225-229. [PubMed] [Google Scholar]
- 53.Martinez-Sobrido, L., E. I. Zuniga, D. Rosario, A. Garcia-Sastre, and J. C. de la Torre. 2006. Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J. Virol. 80:9192-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCormick, J. B., D. H. Walker, I. J. King, P. A. Webb, L. H. Elliott, S. G. Whitfield, and K. M. Johnson. 1986. Lassa virus hepatitis: a study of fatal Lassa fever in humans. Am. J. Trop. Med. Hyg. 35:401-407. [DOI] [PubMed] [Google Scholar]
- 55.McCormick, J. B., and S. P. Fisher-Hoch. 2002. Lassa fever. Curr. Top. Microbiol. Immunol. 262:75-109. [DOI] [PubMed] [Google Scholar]
- 56.Mrak, R. E., and W. S. Griffin. 2001. Interleukin-1, neuroinflammation, and Alzheimer's disease. Neurobiol. Aging 22:903-908. [DOI] [PubMed] [Google Scholar]
- 57.Nansen, A., O. Marker, C. Bartholdy, and A. R. Thomsen. 2000. CCR2+ and CCR5+ CD8+ T cells increase during viral infection and migrate to sites of infection. Eur. J. Immunol. 30:1797-1806. [DOI] [PubMed] [Google Scholar]
- 58.Pedras-Vasconcelos, J. A., D. Goucher, M. Puig, L. H. Tonelli, V. Wang, S. Ito, and D. Verthelyi. 2006. CpG oligodeoxynucleotides protect newborn mice from a lethal challenge with the neurotropic Tacaribe arenavirus. J. Immunol. 176:4940-4949. [DOI] [PubMed] [Google Scholar]
- 59.Perez, M., D. L. Greenwald, and J. C. de la Torre. 2004. Myristoylation of the RING finger Z protein is essential for arenavirus budding. J. Virol. 78:11443-11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peters, C. J., P. B. Jahrling, C. T. Liu, R. H. Kenyon, K. T. McKee, and J. G. Barrera Oro. 1987. Experimental studies of arenaviral hemorrhagic fevers. Curr. Top. Microbiol. Immunol. 134:5-68. [DOI] [PubMed] [Google Scholar]
- 61.Peters, C. J., C. T. Liu, G. W. Anderson, Jr., J. C. Morrill, and P. B. Jahrling. 1989. Pathogenesis of viral hemorrhagic fevers: Rift Valley fever and Lassa fever contrasted. Rev. Infect. Dis. 11(Suppl. 4):S743-S749. [DOI] [PubMed] [Google Scholar]
- 62.Price, M. E., S. P. Fisher-Hoch, R. B. Craven, and J. B. McCormick. 1988. A prospective study of maternal and fetal outcome in acute Lassa fever infection during pregnancy. BMJ 297:584-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Psaty, B. M., and C. D. Furberg. 2005. COX-2 inhibitors—lessons in drug safety. N. Engl. J. Med. 352:1133-1135. [DOI] [PubMed] [Google Scholar]
- 64.Qian, C., P. B. Jahrling, C. J. Peters, and C. T. Liu. 1994. Cardiovascular and pulmonary responses to Pichinde virus infection in strain 13 guinea pigs. Lab. Anim. Sci. 44:600-607. [PubMed] [Google Scholar]
- 65.Rai, S. K., B. K. Micales, M. S. Wu, D. S. Cheung, T. D. Pugh, G. E. Lyons, and M. S. Salvato. 1997. Timed appearance of lymphocytic choriomeningitis virus after gastric inoculation of mice. Am. J. Pathol. 151:633-639. [PMC free article] [PubMed] [Google Scholar]
- 66.Ray, N., and L. W. Enquist. 2004. Transcriptional response of a common permissive cell type to infection by two diverse alphaherpesviruses. J. Virol. 78:3489-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Richardson, J. Y., M. G. Ottolini, L. Pletneva, M. Boukhvalova, S. Zhang, S. N. Vogel, G. A. Prince, and J. C. Blanco. 2005. Respiratory syncytial virus (RSV) infection induces cyclooxygenase 2: a potential target for RSV therapy. J. Immunol. 174:4356-4364. [DOI] [PubMed] [Google Scholar]
- 68.Rodas, J. D., I. S. Lukashevich, J. C. Zapata, C. Cairo, I. Tikhonov, M. Djavani, C. D. Pauza, and M. S. Salvato. 2004. Mucosal arenavirus infection of primates can protect them from lethal hemorrhagic fever. J. Med. Virol. 72:424-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rubins, K. H., L. E. Hensley, P. B. Jahrling, A. R. Whitney, T. W. Geisbert, J. W. Huggins, A. Owen, J. W. LeDuc, P. O. Brown, and D. A. Relman. 2004. The host response to smallpox: analysis of the gene expression program in peripheral blood cells in a nonhuman primate model. Proc. Natl. Acad. Sci. USA 101:15190-15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rue, C. A., M. A. Jarvis, A. J. Knoche, H. L. Meyers, V. R. DeFilippis, S. G. Hansen, M. Wagner, K. Fruh, D. G. Anders, S. W. Wong, P. A. Barry, and J. A. Nelson. 2004. A cyclooxygenase-2 homologue encoded by rhesus cytomegalovirus is a determinant for endothelial cell tropism. J. Virol. 78:12529-12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shi, L., L. H. Reid, W. D. Jones, R. Shippy, J. A. Warrington, S. C. Baker, P. J. Collins, et al. 2006. The microarray quality control (MACQ) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 24:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singh, A., J. Svaren, J. Grayson, and M. Suresh. 2004. CD8 T cell responses to lymphocytic choriomeningitis virus in early growth response gene 1-deficient mice. J. Immunol. 173:3855-3862. [DOI] [PubMed] [Google Scholar]
- 73.Soejima, K., and B. J. Rollins. 2001. A functional IFN-gamma-inducible protein-10/CXCL10-specific receptor expressed by epithelial and endothelial cells that is neither CXCR3 nor glycosaminoglycan. J. Immunol. 167:6576-6582. [DOI] [PubMed] [Google Scholar]
- 74.Spann, K. M., K. C. Tran, and P. L. Collins. 2005. Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-κB, and proinflammatory cytokines. J. Virol. 79:5353-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Streblow, D. N., S. L. Orloff, and J. A. Nelson. 2001. Do pathogens accelerate atherosclerosis? J. Nutr. 131:27985-28045. [DOI] [PubMed] [Google Scholar]
- 76.Tian, B., Y. Zhang, B. A. Luxon, R. P. Garofalo, A. Casola, M. Sinha, and A. R. Brasier. 2002. Identification of NF-κB-dependent gene networks in respiratory syncytial virus-infected cells. J. Virol. 76:6800-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang, Z., M. G. Lewis, M. E. Nau, A. Arnold, and M. T. Vahey. 2004. Identification and utilization of inter-species conserved (ISC) probesets on Affymetrix human GeneChip platforms for the optimization of the assessment of expression patterns in non human primate (NHP) samples. BMC Bioinformatics 26:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waris, G., and A. Siddiqui. 2005. Hepatitis C virus stimulates the expression of cyclooxygenase-2 via oxidative stress: role of prostaglandin E2 in RNA replication. J. Virol. 79:9725-9734. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 79.Wu, Z., R. A. Irizarry, R. Gentleman, F. Martinez Murillo, and F. Spencer. 2004. A model based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99:909-917. [Google Scholar]
- 80.Zurcher, T., J. Pavlovic, and P. Staeheli. 1992. Nuclear localization of mouse Mx1 protein is necessary for inhibition of influenza virus. J. Virol. 66:5059-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.