Abstract
Hepatitis C virus (HCV) is a major human pathogen that causes serious liver disease, including cirrhosis and hepatocellular carcinoma. The primary target cells of HCV are hepatocytes, and entry is restricted by interactions of the envelope glycoproteins, E1 and E2, with cellular receptors. E1 and E2 form noncovalently linked heterodimers and are heavily glycosylated. Glycans contribute to protein folding and transport as well as protein function. In addition, glycans associated with viral envelopes mask important functional domains from the immune system and attenuate viral immunogenicity. Here, we explored the role of N- and O-linked glycans on E2, which is the receptor binding subunit of the HCV envelope. We identified a number of glycans that are critical for viral entry. Importantly, we showed that the removal of several glycans significantly increased the inhibition of entry by sera from HCV-positive individuals. Only some of the glycans that affected entry and neutralization were also important for CD81 binding. Our results show that HCV envelope-associated glycans play a crucial role in masking functionally important regions of E2 and suggest a new strategy for eliciting highly neutralizing antibodies against this virus.
Hepatitis C virus (HCV) is a single-stranded positive RNA flavivirus that causes serious liver disease in humans (2, 21, 29, 47). Approximately 15% of newly infected individuals clear the virus, and an estimated 170 million people worldwide are persistently infected with HCV (11, 31). Persistence probably results from antigenic variability that allows the virus to escape host immunity (39). It is presently unclear what roles the different arms of the immune system play in the outcome of HCV infection. Several groups have shown that the clearance of HCV infection correlates with a strong, early cytotoxic-T-lymphocyte response (11, 31, 54). Both neutralizing and nonneutralizing antibodies are elicited by natural HCV infection, but their roles in viral clearance remain contested (4, 9, 10, 33). High antibody titers elicited by the vaccination of chimpanzees with recombinant HCV envelope glycoproteins correlate with protection or a delay in disease onset after challenge with live virus (1, 16, 45). Furthermore, individuals negative for HCV RNA but positive for HCV antibodies are 12 times more likely to clear a second infection than individuals infected for the first time (35). These studies support the premise that neutralizing antibodies are an essential component of protective immunity, but its correlates remain to be identified.
The infectious HCV particle is enveloped by a lipid membrane comprising E1-E2 heterodimers that specifically restrict viral tropism to hepatocytes (8, 27, 30, 47, 53). Based on analogies to alphaviral envelope glycoproteins as well as structural modeling, it has been postulated that E1 serves as the fusogenic subunit and that E2 acts as the receptor binding subunit of the HCV envelope (22, 24, 32, 42, 60). Alternatively, HCV E1 may facilitate E1-E2 folding into a metastable conformation and E2 may carry both receptor binding and fusion activities, as is the case for flaviviruses (18, 28, 46, 57). The E1 and E2 envelope glycoproteins are heavily glycosylated. There is one potential O-linked glycosylation site on E1, and four out of five putative N-linked glycosylation sites are occupied (36). E2 comprises four potential O-linked glycosylation sites, and all 11 N-linked glycosylation sites are occupied (25, 52). HCV glycosylation sites are highly conserved, indicating that they are critical for envelope glycoprotein structure and function. For other viruses, glycans have been shown to modulate antigenicity and immunogenicity as well as receptor recognition (56).
The CD81 tetraspanin was identified as the first putative HCV receptor by its ability to bind specifically and with high affinity to a soluble form of HCV envelope glycoprotein E2 (41). Subsequently, it was shown that CD81 expression is necessary but not sufficient for entry into target cells, indicating a requirement for additional hepatocyte-specific molecules (5, 13, 58). Other cell surface molecules that are reported to play a role in HCV entry include low-density lipoprotein receptor, the scavenger receptor class B type I, and glycosaminoglycans (6, 20, 51). Evans et al. recently demonstrated a requirement for the tight-junction protein claudin-1 in HCV entry (17). C-type lectins DC-SIGN (dendritic cell-specific ICAM-3-grabbing nonintegrin) and L-SIGN (liver- and lymph node-specific ICAM-3-grabbing nonintegrin, or DC-SIGNR) mediate trans infection of target cells after the capture of HCV particles (12, 23, 34, 44). The recognition of high-mannose-content oligosaccharides in the viral envelope glycoproteins is critical for binding and trans infection (23, 26, 34). The in vivo capture of HCV by L-SIGN on liver sinusoidal endothelial cells and DC-SIGN on dendritic cells may concentrate virus in the liver (3, 43).
Here, we report that HCV envelope-associated glycans play a major role in entry, CD81 binding, and neutralization. Using site-directed mutagenesis to remove O and N glycosylation sites, we showed that approximately half of all E2-associated glycans significantly modulate the entry of HCV pseudoparticles (HCVpp) into target cells. The removal of individual glycans did not significantly affect HCVpp capture and trans infection mediated by C-type lectins. Importantly, we show that the removal of several glycans significantly increased sensitivity to neutralization by sera from HCV-positive individuals. Glycans that affected entry and neutralization did not necessarily influence the binding of E2 to CD81. We conclude that glycans play a critical role in HCV interactions with CD81 as well as other envelope-mediated processes. The ability to modulate the sensitivity of HCV to neutralization by removing certain glycans points to a novel approach for HCV vaccine development.
MATERIALS AND METHODS
Cells, sera, and antibodies.
Cells were grown under standard conditions in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, 2 mM l-glutamine, 1× minimal essential medium nonessential amino acids, and 1× sodium pyruvate. Human Huh-7 hepatoma cells were provided by R. Chowdhurry (Albert Einstein College of Medicine, Bronx, NY). Human 293T kidney endothelial cells and HeLa cervical epithelial cells were purchased from the American Type Culture Collection. HeLa cells stably expressing L-SIGN or DC-SIGN were generated as previously described (23) and maintained in medium supplemented with 800 μg/ml of G418 (Life Technologies).
HCV-positive and HCV-negative control sera were collected at Montefiore Medical Center, Bronx, NY. Murine anti-E2 monoclonal antibody (MAb) 091b-5 was purchased from Austral Biologicals. Murine anti-CD81 MAb JS81 was purchased from BD Pharmingen. The rat anti-hemagglutinin (anti-HA) MAb 3F10 was purchased from Roche. The phycoerythrin (PE)-conjugated goat anti-rat immunoglobulin G (IgG) antibody and the mouse anti-V5 MAb were purchased from Invitrogen. The horseradish peroxidase (HRP)-conjugated goat anti-rat and sheep anti-mouse IgG antibodies were purchased from GE Healthcare.
E2 mutagenesis.
Insertional PCR-based mutagenesis was used to append a V5 tag (5′GGT AAG CCT ATC CCT AAC CCT CTC CTC GGT CTC GAT TCT ACG3′) to the 3′ end of the E2 coding sequence in the pcDNA3.1-E1E2 expression vector described previously (13). Similarly, an HA tag (5′TAT CCT TAT GAC GTA CCT GAC TAT GCT TCA CTG3′) was added to the 3′ end of the sequence encoding E2 ectodomain residues 384 to 661 in the PPI4 vector (50). Asparagines at N glycosylation sites and threonines at potential O glycosylation sites were replaced with alanine by using the QuikChange kit according to the instructions of the manufacturer (Stratagene). Substitutions were introduced into the E1-E2-V5 coding sequence used for generating pseudoparticles as well as into the sequence coding for the soluble E2-HA ectodomain (7). Nucleotide sequencing was performed to ascertain the presence of the appropriate substitutions.
Production of pseudoparticles and quantification of E2 incorporation.
HCVpp were generated as previously described (7). Briefly, 293T cells were transfected with the NLluc+env− human immunodeficiency virus type 1 (HIV-1)-based reporter vector and a vector expressing wild-type or mutant E1-E2 in a 1:2 ratio with a total of 12 μg of DNA per 10 cm of tissue culture plate. Cell culture supernatants were collected 48 h posttransfection and clarified with 0.45-μm-pore-size filters (Millipore). HCVpp were further purified by layering on 20% sucrose (4:1, vol/vol) and spinning for 2 h at 25,000 rpm in an SW28 rotor. HCVpp pellets were resuspended in phosphate-buffered saline (PBS). Serial dilutions of heat-denatured HCVpp in 0.1% NP-40 in PBS were captured onto Galanthus nivalis lectin-coated Immulon 2 HB microtiter plates (Thermo Electron Corporation). Binding was detected with the anti-V5 MAb (1:1,000) followed by anti-mouse IgG-HRP (1:2,000). All incubations and washes were performed with 0.5% Tween 20 in PBS. The TMB substrate kit (Pierce) was used to reveal antibody binding at a wavelength of 450 nm.
For HIV-1 p24 quantification, MaxiSorp plates (Nalge Nunc) coated with an anti-p24 HIV-1 MAb (ImmunoDiagnostics) were used to capture serial dilutions of HCVpp samples in a mixture of 0.5% Tween 20, 0.1% casein, 1.44 M NaCl, and 250 mM Trizma base, pH 7.5, also used as the incubation and wash buffer. Incubation with a rabbit anti-p24 antibody was followed by labeling with streptavidin-HRP (BD Pharmingen). Sigma FAST OPD substrate was used to reveal antibody binding by measuring specific absorbance at 490 nm using an enzyme-linked immunosorbent assay (ELISA) microplate reader.
Western blotting of E2 proteins.
Similar quantities of purified soluble E2 (sE2) proteins (approximately 100 ng) were separated on 10 to 20% polyacrylamide denaturing gels (Bio-Rad). After transfer onto nitrocellulose, wild-type and mutant sE2 proteins were detected by labeling with anti-HA antibodies (1:1,000) followed by an HRP-conjugated anti-rat secondary antibody (1:10,000). Blocking and incubations with antibodies were performed with 0.5% milk in Tris-buffered saline, whereas 0.1% Tween 20 in Tris-buffered saline was used for washing. The PerkinElmer Western Lightning kit chemiluminescent reagent was used to reveal HRP activity according to the manufacturer's instructions. A similar procedure was used to detect HCVpp-associated E2, but detection was performed with an anti-V5 antibody (1:1,000).
Infection and trans infection assays.
For determining the efficiency of entry mediated by E2 mutant proteins, Huh-7 hepatoma target cells (104) were infected with 50 μl of sucrose cushion-purified HCVpp. For neutralization, serial dilutions of HCV-positive sera (50 μl) were added to cells prior to infection with HCVpp. For trans infection assays, parental, L-SIGN-positive, or DC-SIGN-positive HeLa cells (2 × 104) were spin inoculated with 250 μl of viral supernatant for 1 h at 4°C and then incubated for an additional hour at 37°C. After three washes with serum-free medium to remove unbound virus, cells were cocultured with Huh-7 target cells (4 × 104). For all assays, luciferase activity (expressed as relative light units [RLU]) in cell lysates was measured 48 h after infection by using the luciferase assay system (Promega).
sE2 production and binding to CD81.
293T cells were calcium phosphate transfected with vectors expressing wild-type or mutant sE2. The medium was replaced the next day with fully supplemented Dulbecco's modified Eagle's medium without fetal bovine serum and with 10 mM sodium butyrate to boost transgene expression. Supernatants were collected 48 h posttransfection and concentrated 50-fold by centrifugation through Amicon ultracentrifugal filter devices with a 50-kDa cutoff (Millipore). Serial dilutions of sE2 proteins were captured onto G. nivalis-coated microtiter plates and detected with anti-HA MAb 3F10 (1:1,000) followed by an anti-rat IgG-HRP (1:2,000). The TMB substrate kit (Pierce) was used to reveal antibody binding at a wavelength of 450 nm. In parallel, wild-type sE2 was quantified by detection with anti-E2 MAb 091b-5 (500 ng/ml) by using a concentration curve generated with sE2 purchased from Austral. Concentrations of mutant sE2 proteins were derived by fractional equivalence calculations using 50% optical densities at 450 nm as obtained with the anti-HA MAb.
293T cells (3 × 105) were incubated with serial dilutions of sE2 beginning with 100 μg/ml for 1 h at room temperature in Dulbecco's PBS supplemented with 1% bovine serum albumin and 0.05% sodium azide. Following two washes with PBS, cells were incubated with anti-HA MAb 3F10 (1:100) followed by a secondary, PE-conjugated antibody (1:100). The 50% effective concentration (EC50) was the concentration of sE2 that generated half-maximal mean fluorescence intensity as measured by flow cytometry. For the inhibition of sE2 binding, cells were incubated for 1 h at 4°C with a nonspecific murine IgG or JS81 (10 μg/ml) followed by wild-type or mutant sE2 proteins at the EC50.
Computational analyses of the E2 primary sequence.
ClustalW software (http://www.ebi.ac.uk/clustalw/) was used to generate alignments of all full-length E2 sequences available in the HCV database (http://hcv.lanl.gov/content/hcv-db/index), including those of genotypes 1 through 6. Consensus software (http://www.bork.embl-heidelberg.de/Alignment/consensus.html) was then used to create amino acid consensus sequences of HCV E2 and to identify variable and conserved regions. Consensus sequences were further analyzed with PredictProtein (http://www.predictprotein.org/), which relies on neural networks to predict secondary structures and folding patterns. Similar analyses were used to determine the degree of conservation of putative E2 glycosylation sites. NetOGlyc 3.1 (http://www.cbs.dtu.dk/services/NetOGlyc/) was used to predict O glycosylation sites.
RESULTS
Organization and glycosylation of the HCV envelope glycoprotein E2.
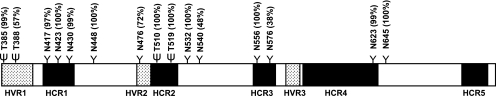
Mature, full-length E2 comprises amino acids 384 to 746 of the HCV polyprotein. We devised a rational model of the E2 ectodomain (residues 384 to 715) based on present knowledge about its structure and function, coupled with computational analyses of its primary structure. Table1 lists the different E2 domains in a 90% consensus sequence as well as the corresponding domains in the subtype 1a H77 isolate used in this study. Our model predicts three solvent-exposed hypervariable regions (HVR1 to HVR3). Despite the high degree of genetic variability of HVR1, this region may be structurally constrained, because amino acid substitutions do not follow a random pattern (40). The E2 ectodomain also comprises five hyperconserved regions (HCR1 to HCR5), which are predicted to be buried within the protein. The central region of E2, including HCR1 through HCR2, comprises the putative CD81 binding site, and we observed that most bulky residues in this tract are essential for viral entry (T. Dragic, unpublished results; 14, 19, 37, 38, 48). HCR2 also comprises the fusion peptide predicted by Yagnik et al. (57), whereas HCR5 constitutes an E1-E2 heterodimerization domain (15).
TABLE 1.
Structural organization of HCV E2 ectodomaina
| 90% Consensus sequence domain | H77 domain | Identification | Exposureb | Secondary structure(s)c | Functiond |
|---|---|---|---|---|---|
| 384-406 | 384-405 | HVR1 | Exposed | α | |
| 413-431 | 412-430 | HCR1 | Buried | β | CD81 binding site |
| 474-485 | 473-482 | HVR2 | Exposed | CD81 binding site | |
| 486-523 | 483-520 | HCR2 | Buried | β | CD81 binding site |
| 552-568 | 549-565 | HCR3 | Buried | ||
| 575-587 | 572-578 | HVR3 | Exposed | ||
| 590-632 | 581-623 | HCR4 | Buried | β | |
| 692-710 | 683-701 | HCR5 | Buried | α/β | Heterodimerization domain |
A model of E2 was generated based on present knowledge of E2 function and computational analyses. Numbers indicate the positions of the E2 residues that delineate the boundaries of hypervariable and hyperconserved structural domains in the 90% consensus sequence and the H77 isolate. The 90% consensus sequence is based on all E2 sequences in the HCV database, regardless of genotype.
Exposure indicates whether these domains are predicted to be exposed on the protein surface or buried.
α or β indicates whether these domains are predicted to be characterized by alpha-helices or beta-sheets, respectively.
Data are based on functional analyses reported in the literature.
Figure 1 illustrates the relative distributions of the different E2 domains. The positions and conservation of putative glycosylation sites in the H77 isolate are also indicated. Altogether, there are four predicted O glycosylation sites and 11 confirmed N glycosylation sites (25). Six of the 15 glycans are grouped in the amino-terminal end of E2, mostly in HVR1 and HCR1. A second cluster of five glycans is located mostly in HVR2 and HCR2. The remaining four N-glycans are paired in HCR3 and HCR4. As indicated in Fig. 1, E2-associated glycans are highly conserved across all HCV subtypes, with some exceptions: T388 is absent in genotype 6 (with the exception of subtype 6d); N476 is absent in some genotype 1 and 2 sequences but is conserved in genotypes 3, 4, and 6; N540 is found in all genotype 1 and 2 sequences but not in the other genotypes; and the frequency of N576 varies among the different subtypes and genotypes. The overall high degree of conservation, however, confirms the structural and functional importance of E2-associated glycans.
FIG. 1.
E2 ectodomain organization and glycosylation. The hypervariable (shaded boxes) and hyperconserved (black boxes) regions are shown, as are the positions of N (Y) and O (ψ) glycosylation sites. The percentages of conservation of the various glycosylation sites are indicated in parentheses.
E2-associated glycans play a role in HCV entry.
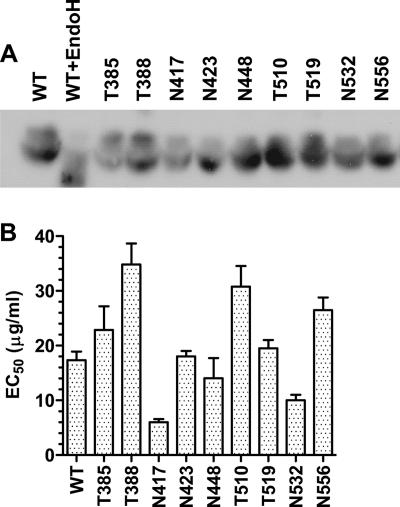
Replacements of single residues with alanine were introduced at all 15 putative N and O glycosylation sites in E2 of the H77 isolate. C-terminal V5 tags were also appended to wild-type and mutant E2 proteins for detection and quantification. Retroviral pseudoparticles (HCVpp) with wild-type and mutant proteins were generated and further purified by sucrose cushioning. We confirmed by Western blotting that the 11 N glycosylation sites were occupied, as revealed by an apparent downshift in the sizes of the mutant protein bands compared to those for wild-type E2 (25; data not shown). Similar results with sE2 proteins that were used to investigate CD81 binding were observed (see Fig. 4A). We were not able to demonstrate a downshift for HCVpp-associated E2 mutated at various O glycosylation sites. The size difference between wild-type E2 and mutant proteins lacking a single O-glycan chain (∼1 to 2 kDa) could not be resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis because E2 migrates as a diffuse band. However, similar analyses of sE2 proteins showed that the four predicted O glycosylation sites were occupied (Fig. 4A).
FIG. 4.
Binding of sE2 to cell surface CD81. (A) sE2 proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by Western blotting using an anti-HA MAb. The diffuse appearance of the E2 bands is expected because the E2 proteins are high-mannose-content proteins. WT indicates wild-type sE2, WT+EndoH indicates sE2 treated with endoglycosidase H, and mutant proteins are indicated by the relevant residue letter code followed by the position number in full-length E1-E2. (B) Serial dilutions of wild-type or mutant HA-tagged sE2 starting at 100 μg/ml were incubated with 293T cells, which naturally express CD81. The binding of sE2 was detected by flow cytometry after labeling with an anti-HA MAb. The EC50 is the concentration of sE2 that generated half the maximum mean fluorescence intensity. All values are means ± standard deviations of results from three independent experiments. The statistical significance of mean EC50s was determined using a two-tailed t test. Differences between wild-type sE2 and T385, N423, N448, and T519 sE2 mutant proteins were not significant (P > 0.2). For all other mutant proteins, P values were <0.03.
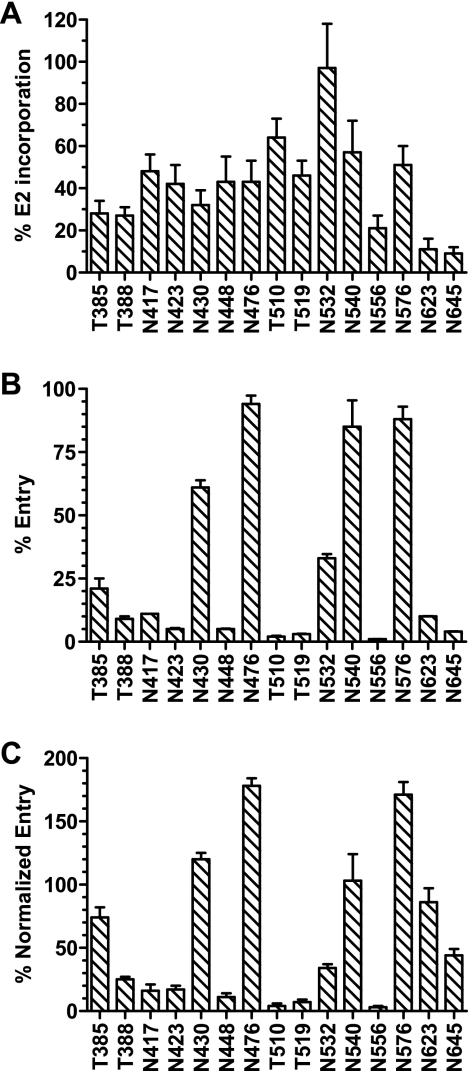
In parallel, E2 incorporation into purified HCVpp was determined by a G. nivalis capture-based ELISA. ELISA quantifications were confirmed by E2 band intensity after Western blotting, indicating that the removal of single glycans does not significantly affect the G. nivalis-mediated capture of pseudoparticles. The HIV-1 core (p24) content was also determined by an ELISA and used to standardize measurements of E2 incorporation into HCVpp (Fig. 2A). Only mutant proteins with alanine at positions N623 and N645 were incorporated at approximately 10% of wild-type levels and therefore considered to be significantly compromised in their ability to exit the endoplasmic reticulum and pseudotype retroviral particles. All of the other E2 glycosylation mutant proteins were incorporated into pseudoparticles at levels ranging from 25 to 100% of that of the wild-type protein.
FIG. 2.
Incorporation of E2 glycosylation mutant proteins into HCVpp and entry mediated by E2 mutant proteins. (A) The incorporation of V5-tagged wild-type and mutant E2 glycoproteins into HCVpp was determined by ELISA. HIV-1 p24 was also quantified (as nanograms per milliliter) by standard ELISA. The percentage of E2 incorporation was calculated using the following formula: [(50% optical density at 450 nm for mutant E2/50% optical density at 450 nm for wild-type E2)/(p24 ng/ml in HCVpp with mutant E2/p24 ng/ml in HCVpp with wild-type E2)] × 100. Mutant proteins are indicated by the relevant residue letter code followed by the position number in full-length E1-E2. (B) The entry of HCVpp bearing wild-type or mutant E2 into Huh-7 cells was assessed by measuring the luciferase activity in cell lysates 48 h postinfection. The percentage of entry was calculated using the following formula: (RLU for mutant E2/RLU for wild-type E2) × 100. (C) Levels of entry into Huh-7 cells mediated by the various E2 glycosylation mutants were standardized with levels of E2 incorporation into HCVpp. All values are means ± standard deviations of results from three independent experiments.
More than half of the glycosylation mutant proteins critically disabled the entry of HCVpp into Huh-7 hepatoma cells (Fig. 2B). When entry levels were normalized for E2 incorporation, alanine residues at N-glycosylation positions N417, N423, N448, and N556 were found to result in entry levels of <20% of that mediated by wild-type E2 (Fig. 2C). T388A, T510A, and T519A O glycosylation mutant proteins also drastically reduced HCVpp entry. Substitutions at positions N532 and N645 moderately impaired entry, whereas changes at glycosylation positions T385, N430, N540, and N623 were inert. Finally, mutations at positions N476 and N576 increased the level of entry by approximately twofold compared to that mediated by wild-type E2. These observations indicate that multiple E2-associated glycans modulate HCVpp entry into target cells. Whereas the removal of numerous glycans had a negative impact on entry, it is interesting that the removal of two E2 glycans actually improved HCVpp entry.
Trans infection is not affected by the removal of single glycans.
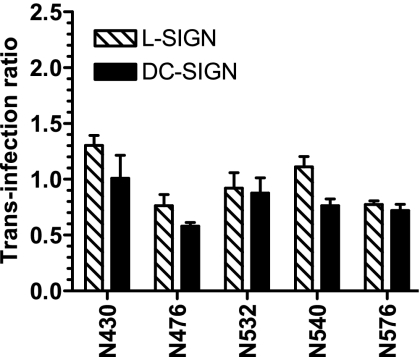
E2 glycosylation mutant proteins were also tested for their abilities to mediate HCVpp transfer to target cells by C-type lectins L-SIGN and DC-SIGN, which recognize high-mannose-content glycans on viral envelopes. Most of the mutant proteins could not be tested because they mediated low levels of entry and therefore did not generate reliable trans infection signals. For all of the other mutant proteins, trans infection levels had to be corrected for variability in entry levels, as well as compared to those mediated by wild-type E2. We expressed this normalized comparison as the trans infection ratio, which is equal to 1 for wild-type E2. Thus, a ratio of >1 indicated an increase in trans infection, whereas a ratio of <1 indicated a decrease in trans infection. We found that the removal by mutagenesis of single N glycosylation sites did not modulate the trans infection ratio in a statistically significant manner (Fig. 3). This result suggests that C-type lectins make contact with multiple glycans on the virion surface and that the removal of single glycans is not sufficient to down-regulate this interaction. Alternatively, it is the glycans that we were unable to investigate in this assay that are important for capture and trans infection.
FIG. 3.
C-type lectin-mediated trans infection mediated by E2 glycosylation mutants. HeLa cells and L-SIGN- or DC-SIGN-positive derivatives were incubated with HCVpp bearing wild-type or mutant envelope glycoproteins, washed, and cocultured with Huh-7 hepatoma cells. Luciferase activity in cell lysates was determined 48 h later. To calculate the ratio of trans infection, the following formula was used: (RLU for trans infection with mutant E2/RLU for trans infection with wild-type E2)/(RLU for entry of mutant E2/RLU for entry of wild-type E2). All values are means ± standard deviations of results from three independent experiments. Means of trans infection ratios for mutants were not significantly different as determined by a two-tailed t test (P > 0.05). Mutant proteins are indicated by the relevant residue letter code followed by the position number in full-length E1-E2.
Envelope-associated glycans modulate neutralization by HCV-positive sera.
Glycans associated with viral envelopes have been shown to decrease the immunogenicity of viral particles by mimicking host proteins and masking important neutralization epitopes (59). In order to test whether HCV E2-associated glycans participate in the latter function, we tested the ability of sera from two HCV subtype 1a-infected individuals (designated J and I) to inhibit entry mediated by the different E2 glycosylation mutant proteins (Table 2). Pooled sera from four HCV-negative individuals did not affect HCVpp entry, whereas positive sera did not inhibit entry mediated by particles pseudotyped with vesicular stomatitis virus G protein (data not shown). Certain mutant proteins could not be analyzed in the neutralization assay because their low levels of entry precluded the generation of statistically reliable data. The levels of sensitivity of most mutant proteins to neutralization varied by less than twofold from that of the wild-type protein. Several mutant proteins, however, were found to be >2-fold more sensitive to neutralization than the wild-type E2 protein: the T385 and N532 mutant proteins were approximately three- and fivefold more sensitive than the wild type to HCV 1a-infected serum from donor J, whereas the T385, T388, N417, and N532 mutant proteins were two- to sixfold more sensitive to HCV 1a-infected serum from donor I.
TABLE 2.
Titers of HCV-positive sera resulting in 50% inhibition of HCVpp entrya
| Form of E2 | 50% Neutralization titer (difference [n-fold] from titer for wild type) of sera positive for HCV subtype:
|
||||||
|---|---|---|---|---|---|---|---|
| 1a (donor J) | 1a (donor I) | 1b | 2b | 3a | 4e | 5a | |
| Wild type | 7.8E−05 (1.0) | 5.8E−05 (1.0) | 1.0E−03 (1.0) | 1.8E−04 (1.0) | 1.9E−02 (1.0) | 9.0E−03 (1.0) | 6.0E−05 (1.0) |
| T385 | 3.0E−05 (2.6) | 2.7E−05 (2.2) | 4.7E−04 (2.1) | 2.6E−05 (6.9) | 3.2E−03 (6.0) | 1.0E−03 (9.0) | 7.3E−05 (0.8) |
| T388 | 7.5E−05 (1.0) | 2.0E−05 (2.9) | 1.6E−03 (0.6) | 1.6E−04 (1.1) | 2.3E−02 (0.8) | 1.0E−03 (9.0) | 1.0E−05 (6.0) |
| N417 | 5.5E−05 (1.4) | 1.0E−05 (5.8) | 3.0E−04 (3.3) | 1.4E−04 (1.2) | 2.1E−02 (0.9) | 1.0E−03 (9.0) | 1.0E−05 (6.0) |
| N430 | 2.0E−04 (0.4) | 1.9E−04 (0.3) | 7.0E−03 (0.1) | 2.4E−04 (0.7) | 2.3E−02 (0.8) | 5.5E−03 (1.6) | 5.0E−05 (1.2) |
| N476 | 1.1E−04 (0.7) | 5.7E−05 (1.0) | 1.0E−03 (1.0) | 2.6E−04 (0.7) | 2.3E−02 (0.8) | 2.5E−03 (3.6) | 1.3E−04 (0.5) |
| N532 | 1.7E−05 (4.6) | 1.0E−05 (5.8) | 2.8E−04 (3.5) | 6.8E−05 (2.6) | 1.6E−02 (1.2) | 1.0E−03 (9.0) | 3.8E−05 (1.6) |
| N540 | 1.1E−04 (0.7) | 5.5E−05 (1.0) | 1.5E−03 (0.7) | 2.0E−04 (0.9) | 2.3E−02 (0.8) | 1.3E−03 (6.8) | 4.9E−05 (1.2) |
| N576 | 6.7E−05 (1.2) | 5.0E−05 (1.2) | 1.0E−03 (1.0) | 1.1E−04 (1.6) | 1.9E−02 (1.0) | 2.5E−03 (3.6) | 3.5E−05 (1.7) |
| N645 | 5.5E−05 (1.4) | 4.8E−05 (1.2) | 1.0E−03 (1.0) | 3.3E−04 (0.5) | 2.3E−02 (0.8) | 1.0E−03 (9.0) | 5.5E−05 (1.1) |
Sera from individuals infected with the indicated HCV subtypes were tested by serial dilution for their abilities to inhibit the entry of HCVpp bearing wild-type and mutant E2 proteins of the H77 subtype 1a isolate. Mutant proteins are indicated by the relevant residue letter code followed by the position number in full-length E1-E2. To calculate the percentage of inhibition of entry, the following formula was used: (RLU with serum sample/RLU without serum sample) × 100. The 50% neutralization titers are the serum dilutions at which 50% inhibition of entry was achieved. The difference (n-fold) from the 50% neutralization titer for the wild type was calculated by using the following formula: (50% neutralization titer for wild-type E2)/(50% neutralization titer for indicated form of E2). All values are means of results from three independent experiments. The standard deviations were all within 20% of the mean 50% neutralization titer. All mean 50% neutralization titers deviating by more than 2 standard deviations from the mean 50% neutralization titer for the wild type were considered statistically significant.
To determine whether T385, T388, N417, and N532 contribute to cross-clade epitopes, we also tested the ability of sera from patients infected with other HCV subtypes, including 1b, 2b, 3a, 4e, and 5a, to inhibit entry mediated by E2 glycosylation mutant proteins (Table 2). For all five sera tested, we found that the removal of some combination of residues at positions T385, T388, N417, and N532 conferred neutralization sensitivity 2- to 10-fold higher than that of the wild-type envelope glycoprotein. Only serum from the 4e-infected individual was also highly neutralizing for HCVpp carrying mutant proteins with substitutions at several other positions, including N476, N540, and N576. We note that certain substitutions, i.e., those at N430 and N476, decreased sensitivity to neutralization, although their effects were not as consistent as those of the mutations that increased sensitivity.
E2-associated glycans modulate interactions with the CD81 entry receptor.
E2 proteins mutated at glycosylation sites that affected HCVpp entry or neutralization were tested for binding to the CD81 entry receptor. A C-terminal HA tag was appended to the sequence encoding the soluble ectodomain of E2 (residues 384 to 661) to allow quantification and detection of the protein by flow cytometry independently of E2 epitopes, which might be modified by the removal of glycans. Selected glycosylation sites were replaced with alanines by site-directed mutagenesis, and soluble envelope glycoproteins were generated by transient expression, followed by concentration and quantification. A downshift in the sizes of all mutant proteins was revealed by Western blotting, indicating that the alanine residues we introduced effectively abolished glycosylation at these sites (Fig. 4A). Serial dilutions of wild-type or mutant sE2 were incubated with 293T cells, which express high levels of CD81 but are resistant to HCV entry because they lack putative entry cofactors. The binding of sE2 was detected by staining with an anti-HA MAb followed by a PE-conjugated secondary antibody. Concentrations of sE2 that generated half-maximal binding to cells (EC50s) were determined by flow cytometry analyses (Fig. 4B). Note that the binding of all mutant E2 proteins to 293T cells was mediated by CD81 alone because it could be completely inhibited by an anti-CD81 MAb (data not shown).
The EC50 of wild-type sE2 was 16 μg/ml. The removal of E2 glycosylation sites either decreased, increased, or had no effect on the level of CD81 binding. Mutants that were impaired for entry or sensitive to neutralization were not necessarily impaired for binding to CD81. Thus, E2 proteins carrying mutations at positions N417 and N532 exhibited significantly increased binding to CD81, despite being impaired in their ability to mediate HCVpp entry. Mutations at both of these sites also increased sensitivity to neutralization. In contrast, substitutions at positions T385, N423, N448, and T519 did not significantly affect binding to CD81, yet mutations at N423 and N448 had a strong negative impact on entry and the mutation of T385 increased sensitivity to neutralization. Finally, mutations of T510 and N556 decreased binding to CD81 and also impaired entry. The replacement of residue T388 decreased binding to CD81 and increased neutralization sensitivity.
DISCUSSION
In this report, we investigated the various functional roles of HCV E2-associated glycans, including those in entry, trans infection mediated by C-type lectins, neutralization, and receptor binding. In general, glycosylation affects protein folding as well as protein function. Glycans associated with viral envelopes also play a major role in masking neutralization epitopes and modulating the overall immunogenicity of viral particles. The E2 envelope glycoprotein comprises 11 N-linked glycans and four O-linked glycans (25, 52). We found that the removal of any one of the N glycosylation sites at positions 417, 423, 448, and 556 or the putative O glycosylation sites at positions 388, 510, and 519 significantly impaired the ability of E2 to mediate HCVpp entry into hepatoma cells, thereby confirming and extending the findings by Goffard et al. (25). The replacement of glycans at positions N623 and N645 severely impaired E2 incorporation into pseudoparticles, which suggests that these glycans affect E2 folding. We propose that glycans, other than those at positions 623 and 645, modulate heterodimerization, receptor interactions, or conformational changes leading up to membrane fusion. Note that E2 glycan function was tested using retroviral pseudoparticles generated in 293T cells. Although all human cells are likely to recognize and use the same glycosylation motifs, it is possible that chain branching patterns may not be identical in 293T cells and primary hepatocytes. Nevertheless, the observation that human sera recognize and neutralize HCVpp indicates that pseudoparticle-associated glycans do not dramatically alter the conformation and presentation of the E2 envelope glycoprotein.
We determined the effects of various glycans on E2 binding to the CD81 entry receptor. Substitutions at positions T388, T510, and N556 decreased the apparent affinity of E2 for CD81, whereas substitutions at positions T385, N423, N448, and T519 had no effect on binding to CD81. Finally, substitutions at positions N417 and N532 increased the apparent affinity of E2 for CD81. Goffard et al. previously reported that E1-E2 heterodimerization was significantly reduced by mutations at positions 540, 556, and 623 and that mutations at the two latter positions also impaired CD81 binding (25). Several studies have pointed to the following E2 tracts as being important for binding to CD81: 407 to 424 (38), 480 to 493 (19, 55), 544 to 551 (19), and 613 to 618 (48). A recent study has shown that E2 residues W420, Y527, W529, G530, and D535 are critical for CD81 interactions. The removal of glycans in the vicinity of these residues may therefore overexpose the CD81 binding site, leading to the increased binding of N417 and N532 mutant proteins that we observed. The reasons why this increase in CD81 binding leads to a decrease in viral entry are probably complex. One possibility is that E2 cannot dissociate easily from CD81 in order to progress through subsequent stages of entry. In contrast, glycans that decreased binding to CD81 also decreased the entry of HCVpp into hepatoma cells. Glycans that did not modulate CD81 binding but nonetheless had an impact on entry probably regulate E2 interactions with other receptors or E1-E2 conformational changes leading to membrane fusion.
Because glycans modulate microbial antigenicity and immunogenicity, we also investigated the effects of E2 glycan removal on antibody neutralization. We found that the presence of glycans at positions T385, T388, N417, and N532 attenuates neutralization, probably by masking important functional domains on E2. The removal of any one of these glycans significantly increased sensitivity to neutralization by sera from individuals infected with different HCV subtypes, and each serum sample was responsive to a particular subset of these glycans, the most commonly recognized being N532. Note that all glycans that modulated sensitivity to neutralization were located in the N-terminal half of E2, indicating that this domain is exposed on the virion surface. As mentioned earlier, the removal of glycans at positions N417 and N532 probably exposes the CD81 binding site, rendering it more accessible to CD81 as well as neutralizing antibodies. Substitutions at positions 385 and 388 must affect neutralization through a different mechanism, perhaps by modulating interactions with other putative entry factors. MAbs against HVR1, where T385 and T388 are located, inhibit the binding of soluble E2 and virions to target cells, suggesting that this region regulates E2-receptor interactions (48, 49, 61). HVR1 could be likened to the V3 loop of HIV-1, which is both functionally indispensable for coreceptor interactions and highly variable for escape from neutralization.
The high level of conservation of HCV E2 glycans is indicative of the dual role they play as key functional elements and key neutralization determinants. In evolving with its host, HCV, like HIV-1, has had to compromise between efficient entry and efficient escape from neutralization. Hence, it has evolved glycosylation sites that attenuate both CD81 binding and antibody recognition of this critical functional domain. Unlike HIV-1, however, HCV does not appear to have a shifting glycan shield. Indeed, the degree of conservation of the 15 glycosylation sites in HCV E2 is very high. The HCV envelope, therefore, does not appear to have the plasticity that would allow it to shuffle the positions of its glycans in order to escape neutralization without also severely compromising folding and function. This finding suggests that it may be possible to elicit neutralizing antibodies against HCV by using glycosylation mutants as immunogens.
Acknowledgments
This work was supported by NIH grant AI060390. This work was also supported in part by the NIAID Centers for AIDS Research grant AI051519 to Albert Einstein College of Medicine. F.K. is supported by Progenics Pharmaceuticals, Inc. E.F. is supported in part by Molecular and Cell Biology and Genetics NIH training grant GM07491.
Footnotes
Published ahead of print on 16 May 2007.
REFERENCES
- 1.Abrignani, S., M. Houghton, and H. H. Hsu. 1999. Perspectives for a vaccine against hepatitis C virus. J. Hepatol. 31(Suppl. 1):259-263. [DOI] [PubMed] [Google Scholar]
- 2.Alter, H. J., and L. B. Seef. 1993. Transfusion-associated hepatitis, p. 467-498. In A. J. Zuckerman and H. C. Thomas (ed.), Viral hepatitis. Churchill Livingstone, Edinburgh, United Kingdom.
- 3.Baribaud, F., S. Pohlmann, G. Leslie, F. Mortari, and R. W. Doms. 2002. Quantitative expression and virus transmission analysis of DC-SIGN on monocyte-derived dendritic cells. J. Virol. 76:9135-9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartosch, B., J. Bukh, J. C. Meunier, C. Granier, R. E. Engle, W. C. Blackwelder, S. U. Emerson, F. L. Cosset, and R. H. Purcell. 2003. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc. Natl. Acad. Sci. USA 100:14199-14204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartosch, B., A. Vitelli, C. Granier, C. Goujon, J. Dubuisson, S. Pascale, E. Scarselli, R. Cortese, A. Nicosia, and F. L. Cosset. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624-41630. [DOI] [PubMed] [Google Scholar]
- 7.Bertaux, C., and T. Dragic. 2006. Different domains of CD81 mediate distinct stages of hepatitis C virus pseudoparticle entry. J. Virol. 80:4940-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boisvert, J., X. S. He, R. Cheung, E. B. Keeffe, T. Wright, and H. B. Greenberg. 2001. Quantitative analysis of hepatitis C virus in peripheral blood and liver: replication detected only in liver. J. Infect. Dis. 184:827-835. [DOI] [PubMed] [Google Scholar]
- 9.Burioni, R., N. Mancini, F. Canducci, S. Carletti, A. Grieco, M. Perotti, G. Serafini, E. Berardinelli, S. Bighi, P. E. Varaldo, and M. Clementi. 2003. Humoral immune response against hepatitis C virus. J. Biol. Regul. Homeost. Agents 17:125-127. [PubMed] [Google Scholar]
- 10.Burioni, R., N. Mancini, S. Carletti, M. Perotti, A. Grieco, F. Canducci, P. E. Varaldo, and M. Clementi. 2004. Cross-reactive pseudovirus-neutralizing anti-envelope antibodies coexist with antibodies devoid of such activity in persistent hepatitis C virus infection. Virology 327:242-248. [DOI] [PubMed] [Google Scholar]
- 11.Cooper, S., A. L. Erickson, E. J. Adams, J. Kansopon, A. J. Weiner, D. Y. Chien, M. Houghton, P. Parham, and C. M. Walker. 1999. Analysis of a successful immune response against hepatitis C virus. Immunity 10:439-449. [DOI] [PubMed] [Google Scholar]
- 12.Cormier, E. G., R. J. Durso, F. Tsamis, L. Boussemart, C. Manix, W. C. Olson, J. P. Gardner, and T. Dragic. 2004. L-SIGN (CD209L) and DC-SIGN (CD209) mediate transinfection of liver cells by hepatitis C virus. Proc. Natl. Acad. Sci. USA 101:14067-14072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cormier, E. G., F. Tsamis, F. Kajumo, R. J. Durso, J. P. Gardner, and T. Dragic. 2004. CD81 is an entry coreceptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 101:7270-7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drummer, H. E., I. Boo, A. L. Maerz, and P. Poumbourios. 2006. A conserved Gly436-Trp-Leu-Ala-Gly-Leu-Phe-Tyr motif in hepatitis C virus glycoprotein E2 is a determinant of CD81 binding and viral entry. J. Virol. 80:7844-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drummer, H. E., and P. Poumbourios. 2004. Hepatitis C virus glycoprotein E2 contains a membrane-proximal heptad repeat sequence that is essential for E1E2 glycoprotein heterodimerization and viral entry. J. Biol. Chem. 279:30066-30072. [DOI] [PubMed] [Google Scholar]
- 16.Esumi, M., T. Rikihisa, S. Nishimura, J. Goto, K. Mizuno, Y. H. Zhou, and T. Shikata. 1999. Experimental vaccine activities of recombinant E1 and E2 glycoproteins and hypervariable region 1 peptides of hepatitis C virus in chimpanzees. Arch. Virol. 144:973-980. [DOI] [PubMed] [Google Scholar]
- 17.Evans, M. J., T. von Hahn, D. M. Tscherne, A. J. Syder, M. Panis, B. Wolk, T. Hatziioannou, J. A. McKeating, P. D. Bieniasz, and C. M. Rice. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801-805. [DOI] [PubMed] [Google Scholar]
- 18.Ferlenghi, I., M. Clarke, T. Ruttan, S. L. Allison, J. Schalich, F. X. Heinz, S. C. Harrison, F. A. Rey, and S. D. Fuller. 2001. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell 7:593-602. [DOI] [PubMed] [Google Scholar]
- 19.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flint, M., E. R. Quinn, and S. Levy. 2001. In search of hepatitis C virus receptor(s). Clin. Liver Dis. 5:873-893. [DOI] [PubMed] [Google Scholar]
- 21.Fry, D. E., and L. M. Flint, Jr. 1997. Hepatitis: an overview of important issues. Bull. Am. Coll. Surg. 82:8-13. [PubMed] [Google Scholar]
- 22.Fuller, S. D., J. A. Berriman, S. J. Butcher, and B. E. Gowen. 1995. Low pH induces swiveling of the glycoprotein heterodimers in the Semliki Forest virus spike complex. Cell 81:715-725. [DOI] [PubMed] [Google Scholar]
- 23.Gardner, J., R. J. Durso, R. R. Arrigale, G. P. Donovan, P. J. Maddon, T. Dragic, and W. C. Olson. 2003. L-SIGN is a liver-specific capture receptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 100:4498-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garry, R. F., and S. Dash. 2003. Proteomics computational analyses suggest that hepatitis C virus E1 and pestivirus E2 envelope glycoproteins are truncated class II fusion proteins. Virology 307:255-265. [DOI] [PubMed] [Google Scholar]
- 25.Goffard, A., N. Callens, B. Bartosch, C. Wychowski, F. L. Cosset, C. Montpellier, and J. Dubuisson. 2005. Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins. J. Virol. 79:8400-8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helle, F., C. Wychowski, N. Vu-Dac, K. R. Gustafson, C. Voisset, and J. Dubuisson. 2006. Cyanovirin-N inhibits hepatitis C virus entry by binding to envelope protein glycans. J. Biol. Chem. 281:25177-25183. [DOI] [PubMed] [Google Scholar]
- 27.Kato, N., T. Nakazawa, T. Mizutani, and K. Shimotohno. 1995. Susceptibility of human T-lymphotropic virus type I infected cell line MT-2 to hepatitis C virus infection. Biochem. Biophys. Res. Commun. 206:863-869. [DOI] [PubMed] [Google Scholar]
- 28.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lauer, G. M., and B. D. Walker. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41-52. [DOI] [PubMed] [Google Scholar]
- 30.Lavillette, D., A. W. Tarr, C. Voisset, P. Donot, B. Bartosch, C. Bain, A. H. Patel, J. Dubuisson, J. K. Ball, and F. L. Cosset. 2005. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology 41:265-274. [DOI] [PubMed] [Google Scholar]
- 31.Lechner, F., D. K. Wong, P. R. Dunbar, R. Chapman, R. T. Chung, P. Dohrenwend, G. Robbins, R. Phillips, P. Klenerman, and B. D. Walker. 2000. Analysis of successful immune responses in persons infected with hepatitis C virus. J. Exp. Med. 191:1499-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 33.Logvinoff, C., M. E. Major, D. Oldach, S. Heyward, A. Talal, P. Balfe, S. M. Feinstone, H. Alter, C. M. Rice, and J. A. McKeating. 2004. Neutralizing antibody response during acute and chronic hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 101:10149-10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lozach, P. Y., H. Lortat-Jacob, A. de Lacroix de Lavalette, I. Staropoli, S. Foung, A. Amara, C. Houles, F. Fieschi, O. Schwartz, J. L. Virelizier, F. Arenzana-Seisdedos, and R. Altmeyer. 2003. DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J. Biol. Chem. 278:20358-20366. [DOI] [PubMed] [Google Scholar]
- 35.Mehta, S. H., A. Cox, D. R. Hoover, X. H. Wang, Q. Mao, S. Ray, S. A. Strathdee, D. Vlahov, and D. L. Thomas. 2002. Protection against persistence of hepatitis C. Lancet 359:1478-1483. [DOI] [PubMed] [Google Scholar]
- 36.Meunier, J. C., A. Fournillier, A. Choukhi, A. Cahour, L. Cocquerel, J. Dubuisson, and C. Wychowski. 1999. Analysis of the glycosylation sites of hepatitis C virus (HCV) glycoprotein E1 and the influence of E1 glycans on the formation of the HCV glycoprotein complex. J. Gen. Virol. 80:887-896. [DOI] [PubMed] [Google Scholar]
- 37.Owsianka, A., R. F. Clayton, L. D. Loomis-Price, J. A. McKeating, and A. H. Patel. 2001. Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J. Gen. Virol. 82:1877-1883. [DOI] [PubMed] [Google Scholar]
- 38.Patel, A. H., J. Wood, F. Penin, J. Dubuisson, and J. A. McKeating. 2000. Construction and characterization of chimeric hepatitis C virus E2 glycoproteins: analysis of regions critical for glycoprotein aggregation and CD81 binding. J. Gen. Virol. 81:2873-2883. [DOI] [PubMed] [Google Scholar]
- 39.Pawlotsky, J. M. 1998. Hepatitis C virus infection: virus/host interactions. J. Viral Hepat. 5(Suppl. 1):3-8. [DOI] [PubMed] [Google Scholar]
- 40.Penin, F., C. Combet, G. Germanidis, P. O. Frainais, G. Deleage, and J. M. Pawlotsky. 2001. Conservation of the conformation and positive charges of hepatitis C virus E2 envelope glycoprotein hypervariable region 1 points to a role in cell attachment. J. Virol. 75:5703-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 42.Pletnev, S. V., W. Zhang, S. Mukhopadhyay, B. R. Fisher, R. Hernandez, D. T. Brown, T. S. Baker, M. G. Rossmann, and R. J. Kuhn. 2001. Locations of carbohydrate sites on alphavirus glycoproteins show that E1 forms an icosahedral scaffold. Cell 105:127-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pohlmann, S., E. J. Soilleux, F. Baribaud, G. J. Leslie, L. S. Morris, J. Trowsdale, B. Lee, N. Coleman, and R. W. Doms. 2001. DC-SIGNR, a DC-SIGN homologue expressed in endothelial cells, binds to human and simian immunodeficiency viruses and activates infection in trans. Proc. Natl. Acad. Sci. USA 98:2670-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pohlmann, S., J. Zhang, F. Baribaud, Z. Chen, G. J. Leslie, G. Lin, A. Granelli-Piperno, R. W. Doms, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J. Virol. 77:4070-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puig, M., M. E. Major, K. Mihalik, and S. M. Feinstone. 2004. Immunization of chimpanzees with an envelope protein-based vaccine enhances specific humoral and cellular immune responses that delay hepatitis C virus infection. Vaccine 22:991-1000. [DOI] [PubMed] [Google Scholar]
- 46.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 47.Rice, C. M. 1996. Flaviviridae: the viruses and their replication, p. 931-1034. In B. N. Fields (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 48.Roccasecca, R., H. Ansuini, A. Vitelli, A. Meola, E. Scarselli, S. Acali, M. Pezzanera, B. B. Ercole, J. McKeating, A. Yagnik, A. Lahm, A. Tramontano, R. Cortese, and A. Nicosia. 2003. Binding of the hepatitis C virus E2 glycoprotein to CD81 is strain specific and is modulated by a complex interplay between hypervariable regions 1 and 2. J. Virol. 77:1856-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosa, D., S. Campagnoli, C. Moretto, E. Guenzi, L. Cousens, M. Chin, C. Dong, A. J. Weiner, J. Y. Lau, Q. L. Choo, D. Chien, P. Pileri, M. Houghton, and S. Abrignani. 1996. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc. Natl. Acad. Sci. USA 93:1759-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanders, R. W., L. Schiffner, A. Master, F. Kajumo, Y. Guo, T. Dragic, J. P. Moore, and J. M. Binley. 2000. Variable-loop-deleted variants of the human immunodeficiency virus type 1 envelope glycoprotein can be stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits. J. Virol. 74:5091-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scarselli, E., H. Ansuini, R. Cerino, R. M. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Slater-Handshy, T., D. A. Droll, X. Fan, A. M. Di Bisceglie, and T. J. Chambers. 2004. HCV E2 glycoprotein: mutagenesis of N-linked glycosylation sites and its effects on E2 expression and processing. Virology 319:36-48. [DOI] [PubMed] [Google Scholar]
- 53.Sung, V. M., S. Shimodaira, A. L. Doughty, G. R. Picchio, H. Can, T. S. Yen, K. L. Lindsay, A. M. Levine, and M. M. Lai. 2003. Establishment of B-cell lymphoma cell lines persistently infected with hepatitis C virus in vivo and in vitro: the apoptotic effects of virus infection. J. Virol. 77:2134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thimme, R., J. Bukh, H. C. Spangenberg, S. Wieland, J. Pemberton, C. Steiger, S. Govindarajan, R. H. Purcell, and F. V. Chisari. 2002. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc. Natl. Acad. Sci. USA 99:15661-15668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Triyatni, M., J. Vergalla, A. R. Davis, K. G. Hadlock, S. K. Foung, and T. J. Liang. 2002. Structural features of envelope proteins on hepatitis C virus-like particles as determined by anti-envelope monoclonal antibodies and CD81 binding. Virology 298:124-132. [DOI] [PubMed] [Google Scholar]
- 56.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 57.Yagnik, A. T., A. Lahm, A. Meola, R. M. Roccasecca, B. B. Ercole, A. Nicosia, and A. Tramontano. 2000. A model for the hepatitis C virus envelope glycoprotein E2. Proteins 40:355-366. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, J., G. Randall, A. Higginbottom, P. Monk, C. M. Rice, and J. A. McKeating. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 78:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, M., B. Gaschen, W. Blay, B. Foley, N. Haigwood, C. Kuiken, and B. Korber. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229-1246. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, W., S. Mukhopadhyay, S. V. Pletnev, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2002. Placement of the structural proteins in Sindbis virus. J. Virol. 76:11645-11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zibert, A., E. Schreier, and M. Roggendorf. 1995. Antibodies in human sera specific to hypervariable region 1 of hepatitis C virus can block viral attachment. Virology 208:653-661. [DOI] [PubMed] [Google Scholar]