Abstract
Human immunodeficiency virus type 1 (HIV-1) Vif counteracts the antiviral activity of the human cytidine deaminase APOBEC3G (APO3G) by inhibiting its incorporation into virions. This has been attributed to the Vif-induced degradation of APO3G by cytoplasmic proteasomes. We recently demonstrated that although APO3G has a natural tendency to form RNA-dependent homo-multimers, multimerization was not essential for encapsidation into HIV-1 virions or antiviral activity. We now demonstrate that a multimerization-defective APO3G variant (APO3G C97A) is able to assemble into RNase-sensitive high-molecular-mass (HMM) complexes, suggesting that homo-multimerization of APO3G and assembly into HMM complexes are unrelated RNA-dependent processes. Interestingly, APO3G C97A was highly resistant to Vif-induced degradation even though the two proteins were found to interact in coimmunoprecipitation experiments and exhibited partial colocalization in transfected HeLa cells. Surprisingly, encapsidation and antiviral activity of APO3G C97A were both inhibited by Vif despite resistance to degradation. These results demonstrate that targeting of APO3G to proteasome degradation and interference with viral encapsidation are distinct functional properties of Vif.
The human immunodeficiency virus type 1 (HIV-1) accessory protein Vif plays an important role in regulating virus infectivity (10, 42). It is now well established that HIV-1 Vif can counteract the human cytidine deaminase APOBEC3G (APO3G). The inhibition of APO3G has been attributed to a reduction in cellular expression levels, which is due to Vif-mediated degradation of APO3G by cytoplasmic proteasomes (8, 24, 29, 30, 39, 41, 50). In the absence of Vif, APO3G is efficiency packaged into HIV virions. The presence of APO3G in the virus can result in hypermutation of the viral minus-strand cDNA during reverse transcription (RT) (12, 22, 26, 28, 49, 51), inhibition of RT (11a), or deamination-independent inactivation of the virus (2, 32, 34, 40).
APO3G has the propensity to form multimers, a property that it shares with other members of the APOBEC family of proteins (13, 21, 31, 34, 40, 47). In our previous study, we demonstrated that human APO3G is capable of forming multimeric complexes in transfected HeLa cells (34). The multimerization of APO3G was sensitive to RNase treatment, suggesting an involvement of viral or cellular RNA in this process. Interestingly, multimerization was also exquisitely sensitive to mutation of a single cysteine residue (C97) that is part of an N-terminal zinc finger motif. However, the C97A mutation abolished neither catalytic nor antiviral activity of APO3G (34). Mutation of C288 and C291, on the other hand, did not impair APO3G oligomerization but completely abrogated APO3G deaminase activity. Nevertheless, this deaminase-deficient APO3G mutant was packaged into vif-defective HIV-1 virions and retained partial antiviral activity when transiently expressed in HeLa cells (34).
APO3G assembles into intracellular high-molecular-mass (HMM) ribonucleoprotein complexes in activated CD4+ T cells and transfected cell lines (6, 7, 11, 20). APO3G associated with HMM complexes was reported to be enzymatically inactive and is presumed to lack antiviral activity (6). Since both multimerization of APO3G and assembly into HMM complexes are RNA dependent, we investigated the ability of the multimerization-incompetent APO3G C97A mutant to form HMM complexes in transfected HeLa cells. We found that APO3G C97A retained its ability to form HMM complexes. Assembly into HMM complexes was sensitive to RNase treatment, although the effects of RNase treatment were different for wild-type (wt) and mutant APO3G protein. We also tested the sensitivity of APO3G C97A to inhibition by Vif. Interestingly, we found that APO3G C97A was resistant to degradation by Vif. In addition to the C97A mutation, mutation of the nearby C100 residue in APO3G C100S also rendered the protein resistant to degradation by Vif while mutation of cysteine residues in the C-terminal zinc finger motif did not affect sensitivity to Vif. These results suggest that sensitivity of APO3G to degradation by Vif is dependent on determinants in the N-terminal zinc finger domain. Importantly, we found that despite resistance to degradation, encapsidation and antiviral activity of APO3G C97A were still efficiently controlled by Vif. These results suggest that Vif has evolved distinct mechanisms for excluding APO3G from HIV-1 virions. One involves intracellular degradation of APO3G; the other regulates packaging of APO3G through a degradation-independent pathway.
MATERIALS AND METHODS
Plasmids.
The vif-defective molecular clone pNL4-3ΔVif (17) was used for the production of virus stocks. Construction of pcDNA-APO3Gmyc for the expression of C-terminally epitope-tagged wt human APO3G was reported elsewhere (15). Most experiments were performed using untagged APO3G. For the expression of untagged human APO3G, a stop codon was introduced into the pcDNA-APO3G-MycHis vector by PCR-based mutagenesis. Untagged APO3G was efficiently expressed from the resulting pcDNA-APO3G vector and identified in immunoblots using the ApoC17 polyclonal antibody (see below). Mutation of cysteine residues C97A, C100S, C288S, and C291A in human APO3G either alone or in combination was accomplished by PCR-based mutagenesis of pcDNA-APO3G (34). The presence of the desired mutations and the absence of additional mutations were verified for each construct by sequence analysis. For transient expression of Vif (strain HXB2), the subgenomic expression vector pNL-A1 (42) was employed. This plasmid expresses all HIV-1 proteins except Gag and Pol. A vif-defective variant, pNL-A1vif(−), was constructed by deletion of an NdeI/PflMI fragment (17). Construction of pNL-A1/AgmVif, carrying the vif gene of simian immunodeficiency virus (SIV) strain SIVagm9063 in the backbone of pNL-A1, was described elsewhere (44). Vector pNL-A1/MacVif for the expression of SIVmac239 Vif protein was constructed by replacing the vif gene in pNL-A1/AgmVif using standard PCR techniques. The construction of pcDNA-hVif for the expression of NL4-3 Vif from a codon-optimized vector under the transcriptional control of a cytomegalovirus promoter has been described elsewhere (33). The Cul5 expression vector pCul5-HA was a generous gift from X. F. Yu (50). An Rbx binding mutant of Cul5-hemagglutinin (HA) was constructed by site-directed mutagenesis using an approach similar to that described in Yu et al. (50).
Antisera.
APO3G was identified using a polyclonal rabbit serum against recombinant human APO3G (15) or against a synthetic peptide comprising the 17 C-terminal residues of APO3G (anti-ApoC17; available through the NIH AIDS Research and Reagent Program, catalog no. 10082). Serum from an HIV-positive patient (APS) was used to detect HIV-1-specific capsid (CA) proteins. A monoclonal antibody to HIV-1 Vif (monoclonal antibody no. 319) was a gift from Michael Malim and was used for all immunoblot analyses and for immunocytochemistry. Polyclonal antibodies to SIVagm Vif and SIVmac Vif were raised in rabbits by immunization with purified recombinant proteins. Tubulin was identified using a monoclonal antibody to α-tubulin (Sigma-Aldrich, Inc., St. Louis, MO). For immunoprecipitation of myc-tagged APO3G, a rabbit polyclonal anti-myc antibody was employed (Sigma-Aldrich, Inc., St. Louis, MO). For immunostaining of HA-tagged APO3G, an HA-specific mouse monoclonal antibody (Research Diagnostics, Inc., Concord, MA) was used. For immunostaining of APO3G C97Amyc, a Myc-specific polyclonal rabbit antibody was used (Sigma-Aldrich, Inc., St. Louis, MO). Cy2- and Texas Red-conjugated secondary antibodies were obtained from Jackson Laboratory (Jackson Immunoresearch Laboratory, West Grove, PA).
Tissue culture and transfections.
HeLa cells were propagated in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS). For transfection, HeLa cells were grown in 25-cm2 flasks to about 80% confluence. Cells were transfected using LipofectAMINE PLUS (Invitrogen Corp., Carlsbad, CA), following the manufacturer's recommendations. A total of 5 to 6 μg of plasmid DNA per 25-cm2 flask (5 × 106 cells) was used. Total amounts of transfected DNA were kept constant in all samples of any given experiment by adding empty-vector DNA [pcDNA3.1 or pNL-A1vif(−)] as appropriate. Cells were harvested at 24 to 48 h posttransfection.
Preparation of virus stocks.
Virus stocks were prepared by transfecting HeLa cells with appropriate plasmid DNAs. Virus-containing supernatants were harvested 24 to 48 h after transfection. Cellular debris was removed by centrifugation (3 min, 3,000 × g), and clarified supernatants were filtered (0.45 μm) to remove residual cellular contaminants. For determination of viral infectivity, unconcentrated filtered supernatants were used for the infection of LuSIV indicator cells. For immunoblot analysis of viral proteins, virus-containing supernatants (7 ml) were concentrated by ultracentrifugation through 4 ml of 20% sucrose in phosphate-buffered saline (PBS) as described previously (15).
Infectivity assay.
To determine viral infectivity, virus stocks were normalized for equal levels of reverse transcriptase activity and used to infect LuSIV cells (5 × 105) in a 24-well plate in a total volume of 1.2 to 1.4 ml. LuSIV cells are derived from CEMx174 cells and contain a luciferase indicator gene under the control of the SIVmac239 long terminal repeat (35). These cells were obtained through the NIH AIDS Research and Reference Reagent Program (catalog no. 5460) and were maintained in complete RPMI 1640 medium supplemented with 10% FBS and hygromycin B (300 μg/ml). Cells were infected for 24 h at 37°C. Cells were then harvested and lysed in 150 μl of Promega 1× reporter lysis buffer (Promega Corp., Madison, WI). To determine the luciferase activities in the lysates, 50 μl of each lysate was combined with luciferase substrate (Promega Corp., Madison, WI) by automatic injection and light emission was measured for 10 seconds at room temperature in a luminometer (Optocomp II; MGM Instruments, Hamden, CT).
Immunoblotting.
For immunoblot analysis of intracellular proteins, whole-cell lysates were prepared as follows. Cells were washed once with PBS, suspended in PBS (400 μl/107 cells), and mixed with an equal volume of sample buffer (4% sodium dodecyl sulfate [SDS], 125 mM Tris-HCl, pH 6.8, 10% 2-mercaptoethanol, 10% glycerol, and 0.002% bromphenol blue). Proteins were solubilized by boiling them for 10 to 15 min at 95°C, with occasional vortexing of the samples to shear cellular DNA. Residual insoluble material was removed by centrifugation (2 min, 15,000 rpm, in an Eppendorf Minifuge). For immunoblot analysis of virus-associated proteins, concentrated viral pellets were suspended in a 1:1 mixture of PBS and sample buffer and boiled. Cell lysates and viral extracts were subjected to SDS-polyacrylamide gel electrophoresis; proteins were transferred to polyvinylidene difluoride membranes and reacted with appropriate antibodies as described in the text. Membranes were then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Amersham Biosciences, Piscataway, NJ) and visualized by enhanced chemiluminescence (Amersham Biosciences).
Immunoprecipitation analysis.
For coimmunoprecipitation analysis of APO3G and Vif, cell lysates were prepared as follows. Cells were washed once with PBS and lysed in 300 μl of lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.5% Triton X-100). The cell extracts were clarified at 13,000 × g for 3 min, and the supernatant was incubated on a rotating wheel for 1 h at 4°C with protein A-Sepharose coupled with anti-myc rabbit polyclonal antibodies (Sigma-Aldrich, Inc., St. Louis, MO). Immune complexes were washed three times with 50 mM Tris, 300 mM NaCl, and 0.1% Triton X-100, pH 7.4. Bound proteins were eluted from beads by heating them in sample buffer for 5 min at 96°C and analyzed by immunoblotting.
FPLC analysis.
HeLa cells were transfected with appropriate plasmid DNAs. After 24 h, cells were washed once with PBS and lysed for 30 min at 4°C in 1 ml of lysis buffer (50 mM HEPES, 125 mM NaCl, 0.2% NP-40, 0.1 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail [complete mini; Roche Diagnostics, Indianapolis, IN]). Cell extracts were clarified by centrifugation at 13,000 × g for 15 min at 4°C before injection for fast-performance liquid chromatography (FPLC). Samples were loaded on a Superose 6HR 10-30 column and separated in running buffer (10% glycerol, 50 mM HEPES, pH 7.4, 125 mM NaCl, 0.1% NP-40, 1 mM dithiothreitol). Twenty-five fractions of 1 ml each were collected at a flow rate of 0.2 ml/min. Samples were concentrated at 4,000 × g for 10 min using Amicon Ultra 10K tubes (Millipore) and mixed 1:1 with sample buffer. Samples were boiled for 5 min at 96°C and analyzed by immunoblotting. For RNase treatment, cell lysates were incubated for 1 h at 37°C with 100 μg/ml of RNase A. The cell extracts were then subjected to FPLC analysis as described above.
Immunofluorescence and confocal microscopy.
HeLa cells were transfected as indicated in the text. Transfected cells were trypsinized, and single-cell suspensions were distributed into 12-well plates containing 0.13-mm coverslips. Cells were grown for 15 to 24 h at 37°C in Dulbecco's modified Eagle's medium containing 10% FBS. Cells were fixed at −20°C in precooled methanol (−20°C) for 10 min, followed by two washes in PBS. Coverslips were stored in PBS at 4°C until use. For antibody staining, coverslips were incubated in a humid chamber at 37°C for 1 h with primary antibodies at appropriate dilutions in 1% bovine serum albumin in PBS. Coverslips were washed once in PBS (5 min, room temperature) and incubated with Cy2- or Texas Red-conjugated secondary antibodies (diluted in 1% bovine serum albumin in PBS) for 30 min at 37°C in a humid chamber. Coverslips were then washed twice with PBS and mounted onto microscope slides with glycerol gelatin (Sigma-Aldrich, Inc., St. Louis, MO) containing 0.1 M N-propyl gallate (Sigma) to prevent bleaching. For confocal microscopy, a Zeiss LSM 410 inverted laser scanning microscope (LSM) equipped with a krypton-argon mixed-gas laser was employed. Images were acquired with a Plan-Apochromat 63×/1.4-oil-immersion objective (Zeiss). For two-color analysis, objects were excited using 488/568-nm laser lines. Green and red emissions were simultaneously recorded through appropriate filters (515- to 540-nm band pass filter for Cy2 and 590-nm long pass filter for Texas Red) and stored in separate (red and green) image channels. At the same time, bright-field images (Nomarski optics) were collected and stored in a third (blue) channel. Image quality was enhanced during data acquisition using the LSM line average feature (8 or 16×). Postacquisition digital image enhancement was performed using the LSM software.
RESULTS
APO3G C97A is resistant to degradation by HIV-1 Vif.
We and others previously reported that APO3G forms homo-multimers (34, 40). Multimerization of APO3G was RNase sensitive, suggesting that APO3G multimerization was mediated or facilitated by an RNA bridge. We also found that mutation of cysteine 97 (C97A) in human APO3G, which is part of an N-terminal zinc finger motif, inhibited homo-multimerization (34). However, APO3G C97A, despite its inability to form homo-multimers, was efficiently packaged into HIV-1 virions and exhibited antiviral activity (34). The main goal of the current study was to examine the sensitivity of APO3G C97A to inhibition by Vif.
We first compared the effects of Vif on the stability of wt APO3G and APO3G C97A. Expression vectors for wt and mutant APO3G were cotransfected into HeLa cells together with increasing amounts of the Vif expression vector pNL-A1. The ratios of APO3G/Vif vectors were 1:0.5 (Fig. 1A, lanes 3 and 6) and 1:1.5 (Fig. 1A, lanes 4 and 7), respectively. The vif-defective pNL-A1Vif(−) plasmid was included as a control (Fig. 1A, lanes 2 and 5). A control expressing Vif in the absence of APO3G was also included (Fig. 1A, lane 1). The total amounts of transfected DNA were kept constant in all samples by using empty pcDNA3.1 or pNL-A1vif(−) DNA as appropriate. Cells were harvested at 24 h posttransfection. Whole-cell lysates were subjected to immunoblot analysis using antibodies to APO3G (Fig. 1A, top). The same blot was subsequently reprobed with monoclonal antibodies to Vif (Fig. 1A, middle) or α-tubulin as a loading control (Fig. 1A, bottom). APO3G-specific bands were quantified by densitometric scanning, and results are presented as percentages of the value for APO3G in the absence of Vif, which was defined as 100% (Fig. 1A, right). As expected, the expression of wt APO3G decreased with increasing amounts of Vif (Fig. 1A, lanes 2 to 4). Interestingly, expression of increasing amounts of Vif did not result in a decrease in APO3G C97A levels (Fig. 1A, lanes 5 to 7). These results suggest that APO3G C97A has acquired resistance to Vif-induced degradation.
FIG. 1.
APO3G C97A is resistant to Vif-induced degradation. (A) HeLa cells were transfected with vectors expressing untagged wt APO3G (lanes 2 to 4) or APO3G C97A (lanes 5 to 7) together with vif-deficient pNL-A1 (lanes 2 and 5) or increasing amounts of the Vif-expressing pNL-A1 vector as described in the text (lanes 3, 4, 6, and 7). Lane 1 is a control that expresses Vif in the absence of APO3G. The total amount of transfected DNA in each sample was adjusted to 5 μg using empty pcDNA3.1 vector DNA (lane 1) or pNL-A1vif(−) DNA (lanes 2 to 7). Cells were harvested 24 h after transfection, and whole-cell lysates were analyzed by immunoblotting using an APO3G-specific rabbit polyclonal antibody (ApoC17) followed by incubation with an HRP-conjugated anti-rabbit antibody (APO3G). The same blot was subsequently reblotted with a Vif-specific monoclonal antibody (Vif), followed by probing with an antibody to α-tubulin (tubulin). Proteins are identified on the right. APO3G-specific protein bands were quantified by densitometric scanning of the gel, and signal intensities were calculated as percentages of the signal observed in the absence of Vif (right side). (B) HeLa cells were transfected and analyzed as for panel A, except that pNL-A1 was replaced by the pcDNA-hVif vector carrying a codon-optimized vif gene. DNA amounts were adjusted to 5 μg using empty pcDNA3.1 vector DNA. (C) HeLa cells (5 × 106) were transfected with constant amounts of myc-tagged wt pcDNA-APO3Gmyc and untagged pcDNA-APO3G C97A together with increasing amounts of pcDNA-hVif. Plasmid ratios of hVif/APO3G vectors were 0 (lane 2), 0.25 (lane 3), and 0.5 (lane 4). Total amounts of DNA were adjusted to 5 μg by addition of empty-vector DNA (pcDNA3.1) as appropriate. A mock-transfected sample (no DNA) was included as a control (lane 1). Whole-cell lysates were prepared 24 h after transfection and subjected to immunoblotting using an APO3G-specific peptide antibody (top) or a Vif monoclonal antibody (bottom). Proteins are identified on the right. Quantitation of APO3G-specific bands for panels B and C was done as for panel A.
We have previously reported that Vif expressed from pNL-A1 only modestly reduces the steady-state expression of APO3G (16). It is therefore possible that the lack of effect of Vif on APO3G C97A in Fig. 1A was due in part to the inefficiency of Vif rather than an inherent resistance of APO3G to Vif. To test this possibility, we used a codon-optimized vector for the expression of hVif (33), which in other experiments had a more pronounced effect on the stability of wt APO3G than Vif expressed from pNL-A1 (unpublished data). The experimental setup was the same as that for Fig. 1A except for the different Vif expression vector. The results are shown in Fig. 1B. APO3G-specific bands were quantified as for Fig. 1A, and the results are shown on the right side in Fig. 1B. Expression of hVif resulted indeed in more efficient degradation of wt APO3G than that seen with pNL-A1-based Vif (Fig. 1B, lanes 2 to 4). However, despite the more pronounced effect of hVif on wt APO3G, the levels of APO3G C97A remained constant or even increased slightly at the highest concentration of hVif (Fig. 1B, lanes 5 to 7). These results support the conclusion that APO3G C97A has indeed acquired resistance to Vif-induced proteasomal degradation.
To further confirm the resistance of APO3G C97A to Vif-induced degradation, we studied the effects of hVif on APO3G C97A in cells cotransfected with wt APO3G. HeLa cells were transfected with a mixture of wt APO3G and APO3G C97A vectors together with increasing amounts of hVif plasmid DNA (Fig. 1C, lanes 3 and 4). To allow discrimination between wt and mutant protein, wt APO3G was expressed as an epitope-tagged form carrying a 26-residue myc-his epitope tag (34). The inherent sensitivity of myc-tagged APO3G to degradation by Vif is similar to that of untagged APO3G (data not shown). A mock-transfected sample was included as a background control (Fig. 1C, lane 1). Cells were harvested 24 h after transfection, and whole-cell lysates were analyzed by immunoblotting for expression of APO3G (Fig. 1C, top) or Vif (Fig. 1C, middle). The APO3G blot was subsequently reprobed with a tubulin-specific antibody (Fig. 1C, bottom). APO3G-specific bands were quantified as for Fig. 1A and B, and the results are shown on the right of Fig. 1C. In the absence of Vif, wt APO3G and APO3G C97A were easily detectable in the immunoblot although levels of APO3G C97A were somewhat lower than those of wt APO3G in this experiment (Fig. 1C, lane 2). Importantly, the presence of hVif almost completely eliminated wt APO3G while expression of APO3G C97A was again slightly increased in the presence of hVif (Fig. 1C, lanes 3 and 4). The fact that hVif selectively induces degradation of wt APO3G when both wt and mutant protein are coexpressed in the same cell confirms the resistance of APO3G C97A to Vif-induced degradation and demonstrates that the lack of degradation is not due to an inherent deficiency in our experimental setup.
HIV-1 Vif is able to bind APO3G C97A.
Previous data showed that Vif-induced degradation of APO3G requires the physical interaction of the two proteins. Indeed, a single amino acid change in APO3G at residue 128 was sufficient to severely inhibit the ability of HIV-1 Vif to interact with human APO3G (3, 27, 38). We therefore wanted to test the ability of Vif to coimmunoprecipitate with APO3G C97A to see if resistance to degradation was caused by a loss of Vif/APO3G interaction. For this experiment, myc epitope-tagged variants of wt APO3G (Fig. 2A, lanes 2 and 5) and APO3G C97A (Fig. 2A, lanes 3 and 6) were cotransfected into HeLa cells together with pcDNA-hVif. To prevent degradation of wt APO3G, samples were also cotransfected with pCul5-RbxHA expressing a dominant-negative form of Cul5 that was previously shown to inhibit Vif-dependent degradation of APO3G (50). A sample expressing hVif in the absence of APO3G was included as a negative control (Fig. 2A, lanes 1 and 4). Cells were harvested 24 h after transfection, and cell lysates were prepared as described in Materials and Methods. An aliquot of each cell lysate was directly analyzed by immunoblotting for the presence of APO3G (Fig. 2A, top, lanes 1 to 3) or Vif (Fig. 2A, bottom, lanes 1 to 3). The remaining lysates were immunoprecipitated with a polyclonal myc-specific antibody, and the immunoprecipitated samples were then subjected to immunoblotting using antibodies to APO3G (Fig. 2A, top, lanes 4 to 6) or Vif (Fig. 2A, bottom, lanes 4 to 6). APO C97A was expressed at somewhat lower levels in this experiment than wt APO3G (Fig. 2A, top, compare lanes 2 and 3), and as a result, the amounts of APO3G C97A precipitated by the myc-specific polyclonal antibody were lower as well (Fig. 2A, top, compare lanes 5 and 6). Vif was found to coprecipitate with both wt APO3G and APO3G C97A (Fig. 2A, bottom, lanes 5 and 6). Vif was not immunoprecipitated in the absence of APO3G (Fig. 2A, bottom, lane 4), demonstrating the specificity of the interaction of Vif with wt APO3G and APO3G C97A. For a more quantitative analysis of the Vif-APO3G interactions, Vif coimmunoprecipitation by APO3G was quantified from three independent experiments. Vif- and APO3G-specific bands were densitometrically scanned, and signal intensities were corrected for the differences in the amounts of APO3G immunoprecipitated. The results are shown in Fig. 2B and suggest that APO3G and APO3G C97A have very similar affinities for Vif in the in vitro pull-down experiment. Thus, mutation of C97A did not significantly affect the interaction of Vif and APO3G C97A, suggesting that resistance of APO3G C97A to degradation by Vif is not due to a lack of protein-protein interaction.
FIG. 2.
APO3G C97A binds to Vif. (A) HeLa cells were cotransfected with pcDNA-hVif and myc-tagged wt pcDNA-APO3Gmyc (lanes 2 and 5) or myc-tagged pcDNA-APO3G C97A-myc (lane 3 and 6). To prevent degradation of wt APO3G by Vif, the dominant-negative Cul5 mutant pCul5-RbxHA was cotransfected in all samples. Total DNA amounts were adjusted to 5 μg in each sample by using pcDNA3.1 vector DNA. As a control, HeLa cells were transfected with pcDNA-hVif and pCul5-RbxHA in the absence of APO3G (lanes 1 and 3). Samples were analyzed either directly (left panels) or following immunoprecipitation (IP) with a myc-specific polyclonal antibody (right panels). Cell lysates and immunoprecipitates were analyzed by immunoblotting using an APO3G-specific rabbit polyclonal antibody, followed by incubation with an HRP-conjugated anti-rabbit antibody (top panels), or a monoclonal antibody to Vif, followed by incubation with an HRP-conjugated anti-mouse antibody (bottom panels). Proteins are identified on the right. In the immunoprecipitated samples, the HRP-conjugated second antibody reacted with both the APO3G-specific antibodies and the myc-specific antibody used for the immunoprecipitation (immunoglobulin G [IgG]). (B) The relative binding efficiencies of Vif and APO3G were determined by quantifying the efficiency of Vif coimmunoprecipitation with APO3G. APO3G- and Vif-specific protein bands in the right panels of panel A (IP) were quantified by densitometric scanning. Signals were corrected for differences in the amounts of immunoprecipitated APO3G protein. The signal intensity of Vif coprecipitated with wt APO3G was defined as 100%. Error bars in the APO3G C97A sample (C97A) reflect the standard deviation calculated from three independent co-IP studies.
APO3G C97A is insensitive to degradation by different Vif species.
It was recently reported that the change of a single aspartic acid residue at position 128 in human APO3G to lysine rendered the protein insensitive to HIV-1 Vif but sensitive to SIVagm Vif (3, 27, 38, 48). To determine if mutation of C97 caused a similar switch in species-specific sensitivity to Vif, we analyzed the sensitivity of APO3G C97A to degradation by SIVagm9063 and SIVmac239 Vif. HeLa cells were cotransfected with vif-defective pNL-A1 (Fig. 3, lane 1) or pNL-A1 variants expressing HIV-1 Vif (Fig. 3, lane 2), SIVagm Vif (Fig. 3, lane 3), or SIVmac Vif (Fig. 3, lane 4) together with vectors expressing APO3G C97A (Fig. 3A and B) or wt APO3G (Fig. 3C and D). Cells were harvested 24 h later, and whole-cell lysates were prepared. APO3G proteins were identified by immunoblotting using the ApoC17 peptide antibody. Vif expression was determined by immunoblotting using a mixture of three different Vif-specific antibodies (Fig. 3A and D). HIV-1 Vif was identified using a high-titer monoclonal antibody (no. 319), while SIVagm and SIVmac Vif proteins were identified by polyclonal antibodies raised against recombinant Vif proteins. The SIV Vif-specific antisera exhibited lower specific titers than the monoclonal antibody to HIV-1 Vif, which could explain the stronger signal observed for HIV-1 Vif than for SIVagm and SIVmac Vif in this experiment. The relative mobilities of Vif proteins varied according to the sizes of the Vif proteins (HIV-1, 192 residues; SIVagm, 231 residues; SIVmac, 214 residues). As expected, wt APO3G was sensitive to HIV-1 and SIVmac Vif (Fig. 3D, lanes 2 and 4) but was resistant to SIVagm Vif (Fig. 3D, lane 3), evident from the reduced protein levels compared to those for the Vif-negative control (Fig. 3D, lane 1). In contrast, APO3G C97A was resistant to all of the Vif isolates tested (Fig. 3B), suggesting that mutation of residue C97 induced a general resistance to Vif-induced degradation.
FIG. 3.
APO3G C97A is resistant to Vif from HIV-1, SIVagm, and SIVmac. HeLa cells were transfected with vectors expressing wt APO3G (C and D) or APO3G C97A (A and B) together with vif-deficient pNL-A1 (lane 1), pNL-A1 (lane 2), pNL-A1/AgmVif (lane 3), or pNL-A1/MacVif (lane 4). Whole-cell lysates were analyzed for the expression of APO3G by using a rabbit polyclonal peptide antibody (B and D) or a mixture of antibodies to Vif consisting of a monoclonal antibody to HIV-1 Vif plus polyclonal rabbit antibodies to SIVagm Vif and SIVmac Vif (A and C). Vif-specific antibodies were subsequently identified with a mixture of HRP-conjugated anti-mouse and anti-rabbit antibodies. Proteins are identified on the right.
Changes in the N-terminal but not the C-terminal zinc finger domain of APO3G confer resistance to Vif-induced degradation.
The results from Fig. 1 and 3 demonstrate that Vif is unable to cause degradation of APO3G C97A. In our previous study, we had analyzed additional APO3G mutants and found that APO3G C100S, although able to multimerize, was very inefficiently packaged into HIV particles and, as a result, had largely lost its antiviral activity. Furthermore, a double mutant, APO3G C97A/C100S, did not multimerize, was inefficiently packaged, and had no detectable antiviral activity. Finally, APO3G C288S/C291A, carrying mutations in the C-terminal zinc finger motif, was found to multimerize, was efficiently encapsidated into HIV-1 virions, and exhibited residual antiviral activity despite a lack of catalytic activity (34).
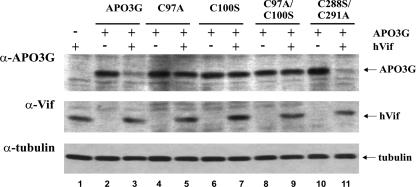
We employed all of these mutants to determine whether the loss of sensitivity of APO3G C97A to Vif-induced degradation was correlated with the inability of APO3G to multimerize. HeLa cells were transfected with vectors encoding wt APO3G (Fig. 4, lanes 2 and 3) or the APO3G mutants C97A (Fig. 4, lanes 4 and 5), C100S (Fig. 4, lanes 6 and 7), C97A/C100S (Fig. 4, lanes 8 and 9), and C288S/C291A (Fig. 4, lanes 10 and 11) in the presence (Fig. 4, odd-numbered lanes) or absence (Fig. 4, even-numbered lanes) of hVif. Whole-cell lysates were prepared 24 h after transfection, and cell lysates were analyzed by immunoblotting using an APO3G-specific antibody (Fig. 4, top). The same blot was then sequentially reblotted with monoclonal antibodies to Vif (Fig. 4, middle) and α-tubulin (Fig. 4, bottom). As expected, wt APO3G was exquisitely sensitive to degradation (Fig. 4, lanes 2 and 3) while APO3G C97A was resistant to Vif (Fig. 4, lanes 4 and 5). Interestingly, APO3G C100S (Fig. 4, lanes 6 and 7) and the C97A/C100S double mutant (Fig. 4, lanes 8 and 9) also exhibited resistance to Vif. In contrast, mutation of the C-terminal zinc finger domain had no effect on Vif sensitivity and APO3G C288S/C291A was degraded with efficiency similar to that for wt APO3G in hVif-expressing cells (Fig. 4, lanes 10 and 11). These results indicate that the C-terminal zinc finger domain, which is critical for catalytic activity, is not important for sensitivity to Vif-induced degradation. In contrast, mutation of either cysteine residue in the N-terminal zinc finger domain of APO3G induced resistance to degradation by Vif irrespective of the protein's ability to multimerize. As noted before, the experiments shown here involved untagged APO3G proteins. Similar results were obtained in experiments using C-terminally epitope-tagged APO3G proteins (data not shown). These results suggest that sensitivity to degradation is not dependent on APO3G homo-multimerization but is determined by sequences mapping to or near the N-terminal zinc finger domain.
FIG. 4.
Mutation of cysteine residues in the N-terminal but not the C-terminal zinc finger motif of APO3G confers resistance to Vif. HeLa cells were transfected with pcDNA-APO3G (lanes 2 and 3), pcDNA-APO3G C97A (lanes 4 and 5), pcDNA-APO3G C100S (lanes 6 and 7), pcDNA-APO3G C97A/C100S (lanes 8 and 9), or pcDNA-APO3G C288S/C291A (lanes 10 and 11) in the absence (even-numbered lanes) or presence (odd-numbered lanes) of pcDNA-hVif plasmid DNA at a 1:1 ratio. All APO3G vectors encoded untagged APO3G proteins. A sample transfected with pcDNA-hVif in the absence of APO3G was included as a control (lane 1). Total DNA amounts were adjusted to 5 μg in each sample by using empty pcDNA3.1 vector DNA. Cells were harvested 24 h after transfection and subjected to immunoblotting as described for Fig. 2.
Colocalization of APO3G C97A and HIV-1 Vif in HeLa cells.
Previous immunofluorescence analyses showed that APO3G is a cytoplasmic protein (1, 16, 29, 45). Interestingly, the cytoplasmic distribution of APO3G was not uniform; instead, APO3G was found to accumulate in cytoplasmic compartments characterized as P bodies or stress granules (11, 20, 46). Although the significance of this observation remains under investigation, it may be related to the assembly of APO3G into HMM complexes (6, 7, 11, 20). We have previously found that HIV-1 Vif when expressed from pNL-A1 partially colocalized with APO3G in transfected HeLa cells (16). Even though APO3G C97A retained its ability to physically interact with HIV-1 Vif in in vitro pull-down experiments (Fig. 2), this interaction could have occurred after cell lysis (29) and it is conceivable that resistance of APO3G C97A is due to intracellular mislocalization.
To analyze the intracellular distribution of APO3G C97A, we performed two experiments. In the first experiment (Fig. 5A to D), we compared the subcellular distribution of wt APO3G and APO3G C97A by employing differentially tagged APO3G proteins that allowed recognition by epitope tag-specific antibodies. HeLa cells were transfected with equimolar amounts of APOBEC3G-HA (41) and APO3G C97A-myc (34) expression vectors. Cells were then stained with HA- and Myc-specific antibodies and analyzed by confocal microscopy as described in Materials and Methods. Both wt APO3G (Fig. 5A) and APO3G C97A (Fig. 5B) were found in cytoplasmic compartments including cytoplasmic bodies. Interestingly, the distribution of APO3G C97A was more diffuse than that of wt APO3G and included some nuclear staining. Overlay of wt and mutant APO3G panels revealed colocalization of wt and mutant APO3G in the cytoplasmic bodies and partial colocalization throughout the cytoplasm. In the second experiment (Fig. 5E to H), we investigated the effect of the C97A mutation on colocalization of APO3G and Vif. HeLa cells were transfected with pcDNA-APO3G C97A and pNL-A1 at 1:0.5 molar ratios. Samples were stained for Vif (Fig. 5E) and APO3G C97A (Fig. 5F) and processed for confocal microscopy as described in Materials and Methods. Aside from double-positive cells, we also found some cells expressing only APO3G C97A. Figure 5E to H shows such an example of single- and double-positive cells. Although not typical, this image was chosen because it confirms on a single-cell level that expression of Vif had no effect on the expression of APO3G C97A. As in Fig. 5B, staining of APO3G C97A in Fig. 5F was more generally cytoplasmic than that of wt APO3G (Fig. 5A), supporting the notion that the cellular distribution of APO3G C97A was not affected by the presence or absence of an epitope tag or the use of different primary antibodies. Importantly, APO3G C97A exhibited partial cytoplasmic colocalization with Vif similar to that previously observed for wt APO3G (16). These results suggest that resistance of APO3G C97A to Vif-induced degradation is not due to a dramatic change in its subcellular distribution.
FIG. 5.
Intracellular distribution of APO3G C97A. (A to D) HeLa cells were transfected with equal amounts of APOBEC3G-HA and pcDNA-APO3G C97Amyc vector DNA. Transfected cells were grown in 12-well plates on coverslips. Cells were fixed with methanol 20 h after being transfected and reacted with an HA-specific mouse monoclonal antibody and a myc-specific polyclonal rabbit antibody, followed by incubation with Cy2-conjugated donkey anti-mouse and Texas Red-conjugated donkey anti-rabbit antibodies. All antibodies were used at 1:200 dilutions. Images of Cy2-stained wt APO3G (A) and Texas Red-stained APO3G C97A (B) were acquired simultaneously and stored in separate image channels. Bright-field (Nomarski) images were stored in a third image channel (C). Colocalization of wt and mutant APO3G was determined by combining the green and red image channels (D). (E to H) HeLa cells were transfected with untagged pcDNA-APO3G C97A and pNL-A1 plasmid DNAs at 1:0.5 molar ratios. Cells were processed for image analysis as described for panels A to D. Cells were stained with a Vif-specific mouse monoclonal antibody and an APO3G-specific rabbit polyclonal antibody. Cy2 and Texas Red-conjugated secondary antibodies were as for panels A to D. Images of Cy2-stained Vif (E) and Texas Red-stained APO3G C97A (F) were acquired simultaneously and stored in separate image channels. Bright-field (Nomarski) images were stored in a third image channel (G). Colocalization of Vif and APO3G C97A was determined by combining the green and red image channels (H).
APO3G C97A forms HMM complexes in HeLa cells.
The immunocytochemical analysis whose results are shown in Fig. 5B revealed the presence of APO3G C97A in cytoplasmic bodies. This suggested that APO3G C97A retained its ability to assemble into HMM complexes despite its inability to form homo-multimers. To compare the assembly of wt APO3G and APO3G C97A into HMM complexes, we subjected wt and mutant APO3G to FPLC (Fig. 6). HeLa cells were separately transfected with plasmids for the expression of wt APO3G or APO3G C97A. Cells were harvested 24 h later and prepared for FPLC analysis as described in Materials and Methods. Samples were either left untreated (Fig. 6A) or treated with RNase A prior to FPLC analysis (Fig. 6B). Individual FPLC fractions were analyzed by immunoblotting for the presence of APO3G. Consistent with a previous report (6), wt APO3G was found enriched in HMM fractions (>400 kDa) of the HeLa extracts (Fig. 6A, APO3G). RNase treatment shifted APO3G to low-molecular-mass (LMM) fractions (Fig. 6B, APO3G), confirming the involvement of RNA in the formation of APO3G HMM complexes. Interestingly, APO3G C97A was also found enriched in HMM fractions (>400 kDa). Unlike for wt APO3G, however, a significant amount of APO3G C97A accumulated in complexes of intermediate mass (Fig. 6A, C97A, fractions 13 to 15). RNase treatment effectively dissociated the HMM complexes of APO3G C97A but did not dissociate the intermediate-mass complexes (Fig. 6B, C97A, fractions 14 and 15). These results demonstrate that mutation of C97A in APO3G, which abolishes multimerization of the protein, did not prohibit its assembly into HMM complexes. However, there was a qualitative difference between wt and mutant APO3G in the sense that the C97A mutant appeared to form additional RNase-insensitive intermediate-mass complexes. APO3G-reactive bands with slower mobilities were evident in the LMM fractions of both wt and mutant APO3G (Fig. 6A, fractions 16 to 18). These bands were not evident in RNase-treated samples (Fig. 6B), and their nature is not clear.
FIG. 6.
APO3G C97A forms RNase-sensitive HMM complexes. (A) HeLa cells (5 × 106) were transfected in a 25-cm2 flask with 5 μg of pcDNA-APO3G, encoding untagged wt APO3G (APO3G), or pcDNA-APO3G C97A, encoding the multimerization-defective untagged APO3G C97A mutant (C97A). Cells were harvested 24 h after transfection, washed in PBS, and lysed for 30 min at 4°C in 1 ml of lysis buffer (50 mM HEPES, 125 mM NaCl, 0.2% NP-40, 0.1 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail). Cell extracts were clarified by centrifugation at 13,000 × g for 15 min at 4°C before injection for FPLC. A Superose 6HR 10-30 column was used with running buffer (10% glycerol, 50 mM HEPES, pH 7.4, 125 mM NaCl, 0.1% NP-40, 1 mM dithiothreitol). Twenty-five 1-ml fractions were collected, and individual samples were concentrated at 4,000 × g for 10 min using Amicon Ultra 10K tubes (Millipore). Concentrated samples were mixed with equal volumes of 2× loading buffer (4% SDS, 125 mM Tris-HCl, pH 6.8, 10% 2-mercaptoethanol, 10% glycerol, and 0.002% bromphenol blue) and heated for 10 min at 95°C prior to gel electrophoresis. Proteins were separated by SDS-polyacrylamide gel electrophoresis and analyzed by immunoblotting using an APO3G-specific rabbit polyclonal peptide antibody (ApoC17). Proteins from a gel filtration calibration kit (Amersham Biosciences, Piscataway, NJ) were used to calibrate the column. The relative positions of albumin (67 kDa), catalase (232 kDa), ferritin (440 kDa), and thyroglobulin (669 kDa) are indicated. (B) Samples were prepared and treated as for panel A except that cell lysates were incubated for 1 h at 37°C with RNase A (100 μg/ml) prior to FPLC analysis.
Vif inhibits encapsidation of APO3G C97A and restores viral infectivity.
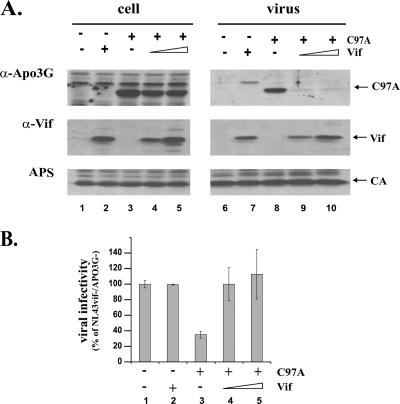
HIV-1 Vif can counteract the antiviral activity of APO3G. This inhibition is generally believed to be due to a reduction in cellular APO3G expression, which has been attributed to Vif-mediated degradation of APO3G by cytoplasmic proteasomes. We wanted to verify the correlation between the cellular steady-state level and antiviral activity of APO3G in the presence of Vif. For that purpose, we determined both the packaging efficiency and the antiviral activity of APO3G C97A for HIV-1 virions (Fig. 7). HeLa cells were transfected with vif-defective pNL4-3 together with pcDNA-APO3G C97A and pNL-A1 for the expression of Vif. Two ratios of APO3G/Vif vectors were tested: 1:0.3 (Fig. 7A, lanes 4 and 9) and 1:1 (Fig. 7A, lanes 5 and 10). For comparison, packaging of APO3G C97A was determined in the absence of Vif (Fig. 7A, lanes 3 and 8). Virus production in the absence of both APO3G and Vif (Fig. 7A, lanes 1 and 6) or in the presence of Vif only (Fig. 7A, lanes 2 and 7) was analyzed in parallel. Cell lysates (Fig. 7A, lanes 1 to 5) and concentrated cell-free virus preparations (Fig. 7A, lanes 6 to 10) were analyzed 48 h after transfection by immunoblotting using an APO3G-specific antibody (Fig. 7A, APO3G). The same blot was then reblotted first with a monoclonal antibody to Vif (Fig. 7A, Vif) followed by an HIV-positive human patient serum (Fig. 7A, APS). Consistent with our previous results (34), APO3G C97A was efficiently packaged into vif-defective HIV-1 virions (Fig. 7A, APO3G, lane 8). Surprisingly, expression of Vif effectively blocked encapsidation of APO3G C97A (Fig. 6A, lanes 9 and 10). The intracellular level of APO3G C97A in the absence of Vif (Fig. 6A, lane 3) appeared slightly elevated in this experiment compared to that in the Vif-expressing samples (Fig. 6A, lanes 4 and 5). However, this effect was not linked to Vif expression levels (Fig. 6A, lanes 4 and 5) and was not observed in repeat experiments and thus reflects experimental error. Importantly, the difference was minuscule compared to that for the effect of Vif on APO3G C97A encapsidation.
FIG. 7.
Vif inhibits the packaging and antiviral activity of APO3G C97A. HeLa cells were transfected with vif-deficient pNL4-3 (lanes 1 to 10) together with pcDNA-APO3G C97A (lanes 3 to 5 and 8 to 10) and vif-defective pNL-A1vif(−) (lanes 1, 3, 6, and 8) or increasing amounts of Vif-expressing pNL-A1 (lanes 4, 5, 9, and 10) vector DNA. Total amounts of transfected DNA were adjusted to 6 μg in each sample by using pcDNA3.1 vector DNA. (A) Cell lysates (lanes 1 to 5) and virus-containing supernatants (lanes 6 to 10) were analyzed by immunoblotting as described for Fig. 2 except that tubulin antibody was replaced by an HIV-positive patient serum (APS) for the identification of capsid protein (CA). Proteins are identified on the right. (B) Virus-containing supernatants from panel A were normalized for equivalent amounts of reverse transcriptase activity and used to infect LuSIV indicator cells (35) for determination of viral infectivity as described in Materials and Methods. The luciferase activity induced by virus lacking Vif and APO3G was defined as 100% (lane 1). The infectivities of the remaining viruses were calculated relative to that of the control virus. Error bars reflect standard deviations for triplicate independent infections.
The infectivities of the viruses produced in Fig. 6A were analyzed in a single-cycle infectivity assay as described in Materials and Methods. The infectivity of virus produced in the absence of APO3G C97A and Vif (Fig. 7B, lane 1) was defined as 100% and used to calculate the relative infectivities of the remaining virus samples. As expected, expression of Vif in the absence of APO3G did not affect viral infectivity, due to the permissive nature of the HeLa producer cell line (Fig. 7B, lane 2). Consistent with our previous report (34), expression of APO3G C97A in the absence of Vif inhibited viral infectivity (Fig. 7B, lane 3). In contrast, expression of Vif resulted in the production of fully infectious viruses (Fig. 7B, lanes 4 and 5). Thus, Vif is able to inhibit packaging and to counteract the antiviral activity of APO3G C97A despite its resistance to Vif-induced intracellular degradation.
DISCUSSION
In this study, we identified for the first time an APO3G mutant that is resistant to degradation by Vif yet remained fully sensitive to Vif with respect to virus encapsidation and antiviral activity. These results support parallel studies, in which we concluded that HIV-1 Vif can inhibit the antiviral activity of wt human APO3G in the absence of degradation (Kao et al., unpublished results). The results from both studies lead us to conclude that the effect of Vif on the intracellular stability of APO3G and inhibition of its antiviral activity are separable functions of Vif.
It remains to be investigated why APO3G C97A is resistant to degradation by Vif. Our data clearly demonstrate that resistance to degradation is unrelated to the inability of APO3G C97A to multimerize since mutation of C100 in the N-terminal zinc finger motif of APO3G, which does not affect multimerization, also rendered the protein resistant to degradation. However, we were unable to test the effect of Vif on the antiviral activity of APO3G C100S since this APO3G mutant is not packaged into HIV-1 virions and therefore does not have antiviral activity (34).
A series of previous studies established that a single amino acid change at the nearby D128 residue induced resistance to degradation by HIV-1 Vif (3, 27, 38, 48). In the case of the D128K mutation, resistance to degradation was explained by an inability of APO3G to interact with HIV-1 Vif (3, 27, 38). Interestingly, the D128K mutation in APO3G did not induce a general resistance to Vif but, in fact, altered its species-specific sensitivity, resulting in sensitivity to degradation by SIVagm Vif (3, 27, 38, 48). Our data on APO3G C97A demonstrate that mutation of human APO3G at position C97 does not cause a similar change in species-specific sensitivity but rather imposes a general resistance to Vif (Fig. 3). The fact that the viral packaging and antiviral activity of APO3G C97A remain sensitive to HIV-1 Vif suggests that resistance to degradation is not caused by an inability of APO3G C97A to interact with Vif. In fact, coimmunoprecipitation analyses demonstrated that the protein retained the ability to interact with Vif in vitro (Fig. 2). These results are consistent with previous reports demonstrating that the binding of HIV-1 Vif to human APO3G is necessary but not sufficient to overcome the inhibitory effects of the deaminase (29, 50). It should be noted, however, that Vif and APO3G expressed in separate cultures were previously found to interact when cell lysates were mixed together (29). Thus, Vif-APO3G complexes can form after cell lysis and the successful coimmunoprecipitation of Vif and APO3G C97A in our experiments does not rule out the possibility that the resistance to degradation is due to a lack of interaction in vivo. While immunocytochemical analyses revealed a broader intracellular distribution of APO3G C97A than of wt APO3G, significant overlap in the intracellular staining of wt and mutant APO3G remained (Fig. 5D). Thus, the resistance to degradation is unlikely due to a gross alteration of the protein's subcellular localization. Furthermore, we observed partial colocalization of Vif and APO3G C97A in transfected HeLa cells (Fig. 5E to H), which resembles that previously observed for wt APO3G (16). It is interesting to note that in some of the experiments the levels of APO3G C97A were slightly increased when hVif was coexpressed (e.g., Fig. 1B and C). The increased levels of APO3G C97A in hVif-expressing cells were not due to loading error, as the levels of tubulin were constant in these experiments. The reason for the increased levels of APO3G C97A in the presence of hVif is unclear and remains under investigation. However, APO3G C97A is inherently less stable than wt APO3G and it is conceivable that the interaction of Vif with APO3G C97A actually stabilizes the protein. Efforts are ongoing to establish an in vitro degradation assay similar to the one developed for Vpu-induced degradation of CD4 that would allow us to address this question (5).
In a recent study, Chiu et al. reported a postentry restriction of HIV-1 by APO3G (6). The authors found that expression of APO3G in an LMM form correlated with high resistance to HIV-1 infection in resting CD4 cells. Activation of CD4+ cells resulted in a shift of APO3G to an enzymatically inactive RNase-sensitive HMM complex that no longer provided protection from HIV-1 infection (6). Because of the inability of APO3G C97A to multimerize (34), we initially hypothesized that APO3G might be unable to form HMM complexes and might therefore represent a constitutively active deaminase with potent antiviral activity. Surprisingly, we found that APO3G C97A retained its ability to form RNase-sensitive HMM complexes (Fig. 6). However, the FPLC profiles were not directly superimposable. Unlike wt APO3G, APO3G C97A associated with intermediate-mass complexes of 200 to 300 kDa. Interestingly, these intermediate complexes appeared to be less RNase sensitive than the HMM complexes. Their nature and composition are under investigation. Nevertheless, we can conclude that formation of HMM complexes is independent of multimerization and neither property is predictive of the sensitivity of APO3G to degradation by Vif.
Our finding that APO3G C97A was resistant to Vif-induced degradation but its antiviral activity remained sensitive to Vif was very surprising. Yet, our data clearly demonstrate that Vif is able to effectively inhibit packaging of APO3G C97A into viral particles and to counteract its antiviral activity (Fig. 7). How Vif inhibits the encapsidation of APO3G C97A remains unclear. The observation that mutations or deletions in various regions of APO3G affect packaging into virus particles (4, 23, 34) argues in favor of a specific packaging mechanism. Moreover, mutations in the viral Gag precursor abolished the packaging of the deaminase enzyme into virus-like particles, suggesting an involvement of Gag, in particular the nucleocapsid NC, in APO3G packaging (1, 25, 37, 43). Finally, packaging of APO3G requires an interaction with RNA. Several studies conclude that viral RNA is not essential for encapsidation of APO3G (1, 4, 9, 25, 31, 43); however, the efficiency of APO3G packaging was clearly enhanced by the presence of viral genomic RNA (19, 43). In our study, we showed that APO3G C97A retained the ability to bind RNA necessary for the formation of HMM complexes (Fig. 6). In fact, RT-PCR analysis confirmed that APO3G C97A retained its ability to interact with viral genomic RNA in transfected HeLa cells (data not shown). It is also well established that HIV-1 Vif is packaged into virus particles (14). It is therefore conceivable that Vif somehow interferes with these APO3G-RNA or APO3G-Gag interactions. Like that of APO3G, packaging of Vif is facilitated by viral genomic RNA (18, 19) and results in the association of Vif with the nucleoprotein complex (18, 19). Experiments are ongoing to test whether Vif can inhibit packaging of APO3G through competitive binding to common packaging signal(s) on Gag or the viral genomic RNA. Such a mechanism could be highly effective, would require only small amounts of Vif, and could explain the degradation-independent inhibition of APO3G encapsidation and antiviral activity.
Acknowledgments
We are grateful to Alicia Buckler-White, Ron Plishka, and Christopher Erb for oligonucleotide synthesis and sequence analysis. We thank Jean-Marie Peloponese for help with the FPLC analyses. We thank Xiao-Fang Yu for the Cul5-HA vector and Warner Greene for APOBEC3G-HA. We further thank Jason Roos and Janice Clements for the LuSIV indicator cell line and Michael Malim for the Vif monoclonal antibody. The latter two reagents were obtained through the NIH Research and Reference Reagent Program.
This work was supported in part by a grant from the NIH Intramural AIDS Targeted Antiviral Program to K.S. and by the Intramural Research Program of the NIH, NIAID.
Footnotes
Published ahead of print on 23 May 2007.
REFERENCES
- 1.Alce, T. M., and W. Popik. 2004. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 279:34083-34086. [DOI] [PubMed] [Google Scholar]
- 2.Bishop, K. N., R. K. Holmes, and M. H. Malim. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 80:8450-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogerd, H. P., B. P. Doehle, H. L. Wiegand, and B. R. Cullen. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. USA 101:3770-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cen, S., F. Guo, M. Niu, J. Saadatmand, J. Deflassieux, and L. Kleiman. 2004. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 279:33177-33184. [DOI] [PubMed] [Google Scholar]
- 5.Chen, M. Y., F. Maldarelli, M. K. Karczewski, R. L. Willey, and K. Strebel. 1993. Human immunodeficiency virus type 1 Vpu protein induces degradation of CD4 in vitro: the cytoplasmic domain of CD4 contributes to Vpu sensitivity. J. Virol. 67:3877-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435:108-114. [DOI] [PubMed] [Google Scholar]
- 7.Chiu, Y. L., H. E. Witkowska, S. C. Hall, M. Santiago, V. B. Soros, C. Esnault, T. Heidmann, and W. C. Greene. 2006. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl. Acad. Sci. USA 103:15588-15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009-2013. [DOI] [PubMed] [Google Scholar]
- 9.Douaisi, M., S. Dussart, M. Courcoul, G. Bessou, R. Vigne, and E. Decroly. 2004. HIV-1 and MLV Gag proteins are sufficient to recruit APOBEC3G into virus-like particles. Biochem. Biophys. Res. Commun. 321:566-573. [DOI] [PubMed] [Google Scholar]
- 10.Fisher, A. G., B. Ensoli, L. Ivanoff, M. Chamberlain, S. Petteway, L. Ratner, R. C. Gallo, and F. Wong-Staal. 1987. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science 237:888-893. [DOI] [PubMed] [Google Scholar]
- 11.Gallois-Montbrun, S., B. Kramer, C. M. Swanson, H. Byers, S. Lynham, M. Ward, and M. H. Malim. 2007. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 81:2165-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Guo, F., S. Cen, M. Niu, J. Saadatmand, and L. Kleiman. 2007. Inhibition of tRNA3Lys-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 80:11710-11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 13.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285-296. [DOI] [PubMed] [Google Scholar]
- 14.Kao, S., H. Akari, M. A. Khan, M. Dettenhofer, X. F. Yu, and K. Strebel. 2003. Human immunodeficiency virus type 1 Vif is efficiently packaged into virions during productive but not chronic infection. J. Virol. 77:1131-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kao, S., M. A. Khan, E. Miyagi, R. Plishka, A. Buckler-White, and K. Strebel. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 77:11398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kao, S., E. Miyagi, M. A. Khan, H. Takeuchi, S. Opi, R. Goila-Gaur, and K. Strebel. 2004. Production of infectious human immunodeficiency virus type 1 does not require depletion of APOBEC3G from virus-producing cells. Retrovirology 1:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karczewski, M. K., and K. Strebel. 1996. Cytoskeleton association and virion incorporation of the human immunodeficiency virus type 1 Vif protein. J. Virol. 70:494-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan, M. A., C. Aberham, S. Kao, H. Akari, R. Gorelick, S. Bour, and K. Strebel. 2001. Human immunodeficiency virus type 1 Vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA. J. Virol. 75:7252-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan, M. A., S. Kao, E. Miyagi, H. Takeuchi, R. Goila-Gaur, S. Opi, C. L. Gipson, T. G. Parslow, H. Ly, and K. Strebel. 2005. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J. Virol. 79:5870-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozak, S. L., M. Marin, K. M. Rose, C. Bystrom, and D. Kabat. 2006. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 281:29105-29119. [DOI] [PubMed] [Google Scholar]
- 21.Lau, P. P., H. J. Zhu, A. Baldini, C. Charnsangavej, and L. Chan. 1994. Dimeric structure of a human apolipoprotein B mRNA editing protein and cloning and chromosomal localization of its gene. Proc. Natl. Acad. Sci. USA 91:8522-8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 23.Li, J., M. J. Potash, and D. J. Volsky. 2004. Functional domains of APOBEC3G required for antiviral activity. J. Cell. Biochem. 92:560-572. [DOI] [PubMed] [Google Scholar]
- 24.Liu, B., X. Yu, K. Luo, Y. Yu, and X. F. Yu. 2004. Influence of primate lentiviral Vif and proteasome inhibitors on human immunodeficiency virus type 1 virion packaging of APOBEC3G. J. Virol. 78:2072-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo, K., B. Liu, Z. Xiao, Y. Yu, X. Yu, R. Gorelick, and X. F. Yu. 2004. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J. Virol. 78:11841-11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 27.Mangeat, B., P. Turelli, S. Liao, and D. Trono. 2004. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 279:14481-14483. [DOI] [PubMed] [Google Scholar]
- 28.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 29.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 30.Mehle, A., B. Strack, P. Ancuta, C. Zhang, M. McPike, and D. Gabuzda. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279:7792-7798. [DOI] [PubMed] [Google Scholar]
- 31.Navarro, F., B. Bollman, H. Chen, R. Konig, Q. Yu, K. Chiles, and N. R. Landau. 2005. Complementary function of the two catalytic domains of APOBEC3G. Virology 333:374-386. [DOI] [PubMed] [Google Scholar]
- 32.Newman, E. N., R. K. Holmes, H. M. Craig, K. C. Klein, J. R. Lingappa, M. H. Malim, and A. M. Sheehy. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166-170. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen, K. L., M. llano, H. Akari, E. Miyagi, E. M. Poeschla, K. Strebel, and S. Bour. 2004. Codon optimization of the HIV-1 vpu and vif genes stabilizes their mRNA and allows for highly efficient Rev-independent expression. Virology 319:163-175. [DOI] [PubMed] [Google Scholar]
- 34.Opi, S., H. Takeuchi, S. Kao, M. A. Khan, E. Miyagi, R. Goila-Gaur, Y. Iwatani, J. G. Levin, and K. Strebel. 2006. Monomeric APOBEC3G is catalytically active and has antiviral activity. J. Virol. 80:4673-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roos, J. W., M. F. Maughan, Z. Liao, J. E. Hildreth, and J. E. Clements. 2000. LuSIV cells: a reporter cell line for the detection and quantitation of a single cycle of HIV and SIV replication. Virology 273:307-315. [DOI] [PubMed] [Google Scholar]
- 36.Roy, B. B., J. Hu, X. Guo, R. S. Russell, F. Guo, L. Kleiman, and C. Liang. 2006. Association of RNA helicase A with human immunodeficiency virus type 1 particles. J. Biol. Chem. 281:12625-12635. [DOI] [PubMed] [Google Scholar]
- 37.Schafer, A., H. P. Bogerd, and B. R. Cullen. 2004. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology 328:163-168. [DOI] [PubMed] [Google Scholar]
- 38.Schrofelbauer, B., D. Chen, and N. R. Landau. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. USA 101:3927-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 40.Shindo, K., A. Takaori-Kondo, M. Kobayashi, A. Abudu, K. Fukunaga, and T. Uchiyama. 2003. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J. Biol. Chem. 278:44412-44416. [DOI] [PubMed] [Google Scholar]
- 41.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 42.Strebel, K., D. Daugherty, K. Clouse, D. Cohen, T. Folks, and M. A. Martin. 1987. The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature 328:728-730. [DOI] [PubMed] [Google Scholar]
- 43.Svarovskaia, E. S., H. Xu, J. L. Mbisa, R. Barr, R. J. Gorelick, A. Ono, E. O. Freed, W. S. Hu, and V. K. Pathak. 2004. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 279:35822-35828. [DOI] [PubMed] [Google Scholar]
- 44.Takeuchi, H., S. Kao, E. Miyagi, M. A. Khan, A. Buckler-White, R. Plishka, and K. Strebel. 2005. Production of infectious SIVagm from human cells requires functional inactivation but not viral exclusion of human APOBEC3G. J. Biol. Chem. 280:375-382. [DOI] [PubMed] [Google Scholar]
- 45.Wichroski, M. J., K. Ichiyama, and T. M. Rana. 2005. Analysis of HIV-1 viral infectivity factor-mediated proteasome-dependent depletion of APOBEC3G: correlating function and subcellular localization. J. Biol. Chem. 280:8387-8396. [DOI] [PubMed] [Google Scholar]
- 46.Wichroski, M. J., G. B. Robb, and T. M. Rana. 2006. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathogens 2:e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu, H., E. S. Svarovskaia, R. Barr, Y. Zhang, M. A. Khan, K. Strebel, and V. K. Pathak. 2004. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl. Acad. Sci. USA 101:5652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu, Q., R. Konig, S. Pillai, K. Chiles, M. Kearney, S. Palmer, D. Richman, J. M. Coffin, and N. R. Landau. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11:435-442. [DOI] [PubMed] [Google Scholar]
- 50.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]