Abstract
We investigated the binding of human parainfluenza virus types 1 and 3 (hPIV1 and hPIV3, respectively) to the glycan array of the Consortium for Functional Glycomics and binding and their release from erythrocytes under conditions where neuraminidase is inactive or active. hPIV1 and hPIV3 bind modifications of Neu5Acα2-3Galβ1-4GlcNAc, including the sialyl-Lewisx motif and structures containing 6-sulfogalactose. hPIV1 and hPIV3 thus bind typical N-linked glycans, in contrast to avian influenza virus H5 hemagglutinin (J. Stevens, O. Blixt, T. M. Tumpey, J. K. Taubenberger, J. C. Paulson, and I. A. Wilson, Science 312:404-410, 2006), which binds less-common motifs. While the receptor is not the sole determinant of tropism, hPIV or H5 influenza virus infection of specific cells that express receptors may contribute to their different pathologies.
Parainfluenza viruses (PIVs) initiate infection by binding to sialic acid receptors on cell surfaces and are released at the end of the replication cycle by the viral neuraminidase (NA) activity. The same viral protein is used for binding and release, the membrane-anchored hemagglutinin-neuraminidase (HN). Human PIVs (hPIVs) bind predominantly to receptors containing α2-3-linked sialic acid (16), in contrast to human influenza viruses, which bind α2-6 sialic acids. Avian influenza viruses bind α2-3 sialic acids, and this specificity is commonly assumed to impede infection of humans, but hPIV readily infects humans. The apparent conundrum may be resolved by differences in binding beyond the sialic acid, so we investigated the specificity of hPIV HN binding using red cell binding assays and the glycoarray of the Consortium for Functional Genomics.
Red blood cell binding and elution.
Agglutination of red cells provides an informative screen of binding and hydrolysis of viral receptors. Agglutination at 4°C measures binding, warming to room temperature shows if the virus can be eluted from red cells, and cooling back to 4°C shows if this elution was by thermal motion (reversible) or by enzymatic removal of receptors (irreversible) (3, 5). We carried out hemagglutination and elution studies using freshly grown hPIV type 1 (hPIV1) or hPIV3 concentrated from tissue culture supernatants. Assays were done at pH 7, where NA is inactive, and at pH 5, where the NA is active as long as the NaCl concentration is ≤150 mM (10, 18).
The results are shown in Table 1. hPIV1 did not elute from guinea pig red cells at pH 7. It eluted at pH 5 but bound back at 4°C, showing that receptors had not been removed. The lower avidity at pH 5 suggests involvement of side chains with a pKa of between 7 and 5 in the interaction. hPIV1 eluted from chicken cells at pH 7 or at pH 5 but rebound at 4°C, showing that elution was due to low avidity, not the NA activity. hPIV1, therefore, binds to receptors on red cells that are resistant to the NA activity, but additional receptors seem to be exposed on guinea pig cells after incubation at room temperature. At pH 5, hPIV3 bound at 4°C to human or guinea pig red cells, and there was no rebinding after elution at room temperature, indicating that the receptors had been removed by the NA activity. At pH 7 there was very low binding to human cells but the hemagglutinin (HA) titer was increased after elution and rebinding, suggesting exposure of a previously hidden receptor, while guinea pig red cells eluted at room temperature and bound back at 4°C. We conclude from these results that the HN of PIV3 destroys its receptors at pH 5, but at pH 7 an activity (which cannot be detected using 4-methylumbelliferyl-N-acetylneuraminic acid [MUN] or sialyllactose substrate) exposes additional receptors.
TABLE 1.
Red cell binding and elution of hPIV1 and hPIV3
| Virus | Source of red cellsa | pH | HA titer (log2)b
|
||
|---|---|---|---|---|---|
| 4°C | After elution at 20°C | After rebinding at 4°C | |||
| hPIV1c | Chicken | 7 | 6 | 0 | 6 |
| 5 | 6 | 0d | 6 | ||
| Guinea pig | 7 | 2.5 | 2.5 | >8 | |
| 5 | 5.5 | 0 | >8 | ||
| hPIV3 | Human | 7 | ≤1 | 0 | 4 |
| 5 | 3 | 0 | 0 | ||
| Guinea pig | 7 | 3 | 0 | 4 | |
| 5 | 3 | 0 | 0 | ||
hPIV1 bound poorly to human red blood cells; hPIV3 did not bind chicken red cells.
Results are shown for one experiment, but the results with different HA amounts and different batches of red cells showed the same pattern.
The NA activity of hPIV1 was ∼1 nmol 3′-sialyllactose cleaved in 30 min per 50 μl virus. hPIV3 activity is too low to measure unless the virus is concentrated ∼100-fold.
In some experiments no elution was observed, but after mixing and resettling there was no rebinding at 22°C, indicating reduced avidity.
Fluorescent labeling of viruses.
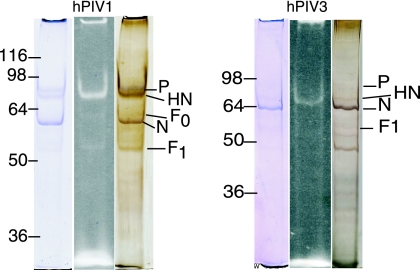
hPIV1 strain C-35 and hPIV3 strain C-243 (ATCC) were propagated in LLC-MK2 cells (ATCC). Virus was adsorbed for 30 minutes, infection medium (Dulbecco modified Eagle medium-F-12 medium [1:1] with ITS+ [BD Biosciences]) was added, and the plates were incubated at 37°C for 4 to 6 days. For PIV1, trypsin (1 μg/ml) was added to the infection medium. Viruses were purified by centrifugation through a 10% to 60% sucrose gradient, pelleted, and resuspended to about 105 HA U/ml. To 100 μl virus was added 10 μl 1 M sodium bicarbonate (pH 9.0) and then 50 micrograms Alexa Fluor 488 succinimidyl ester (Molecular Probes catalog no. A20000). The mixture was gently stirred for 1 hour at room temperature in the dark, transferred to Slide-A-Lyzer minidialysis units (7,000-molecular-weight cutoff; Pierce), and dialyzed at 4°C overnight in the dark. The HA activity was measured to make sure that binding sites had not been inactivated by the reagent. Figure 1 shows that after treatment with Alexa Fluor 488 succinimidyl ester there was fluorescent labeling of a band that corresponds to the known molecular weight and migration of the HN glycoprotein for each virus (4) with much less labeling of F0 or F1.
FIG. 1.
Sodium dodecyl sulfate gel electrophoresis of Alexa-labeled hPIV1 and hPIV3. The fluorescent band (center lane) corresponding to HN is compared to lanes stained with Coomassie blue (left) or Bio-Rad silver reagent (right). Numbers to the left of each panel are molecular masses in kilodaltons.
Glycoarray analysis.
The Alexa 488-labeled virus was diluted in binding buffer at pH 5.5 or 7.0 with the additions of 1% bovine serum albumin and 0.05% Tween 20. Seventy microliters of sample was incubated on the Consortium for Functional Glycomics printed array (v2; 285 oligosaccharides; www.functionalglycomics.org) for 1 hour at room temperature or at 4°C in a humidified chamber. The slide was washed and then dried under a stream of nitrogen. The binding image was read in a Perkin-Elmer Microarray XL4000 scanner and analyzed using Imagene (V.6) image analysis software.
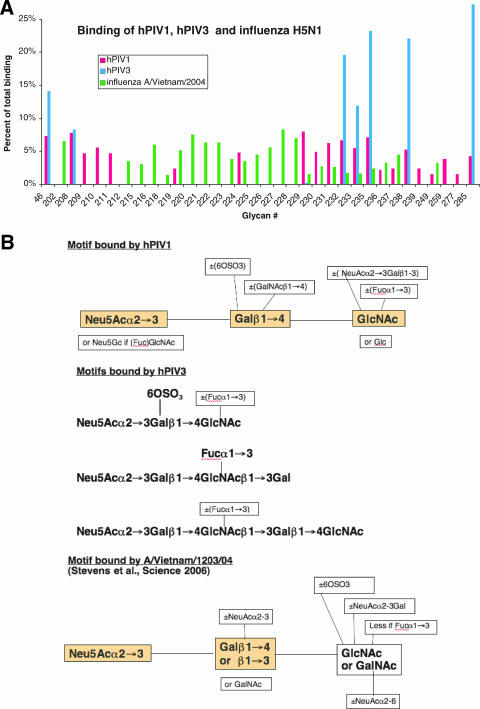
Sample profiles are shown in Fig. 2 and comparative results in Fig. 3A. The structures of the oligosaccharides are shown at www.functionalglycomics.org. Both hPIV1 and hPIV3 bind to oligosaccharides that are modified from NeuAcα2-3Galβ1-4GlcNAc, including the sialyl-Lewisx epitope and its 6′-sulfo derivative. hPIV1 shows an overall decrease in binding at 4°C compared to that at room temperature but no significant qualitative differences. hPIV3 was more restricted in binding than PIV1 and showed somewhat higher selectivity at 4°C than at 22°C. The binding motifs of hPIV1 and hPIV3 deduced from these results are shown in Fig. 3B.
FIG. 2.
Glycoarray analysis of binding specificities of hPIV1 and hPIV2. Each of the 285 glycans is represented six times on the version 2 printed array. The highest and lowest fluorescence readings were discarded, and the remaining four were averaged. The average binding of virus to each glycan is shown as relative fluorescence units (RFU) ± standard error of the mean.
FIG. 3.
Structures bound by hPIV1 and hPIV3 compared to those bound by H5 HA of A/Vietnam/1203/04. The H5 data and motif were calculated from the work of Stevens et al. (15). (A) Binding calculated as a percentage of total binding; the total includes all glycans that bind >1% of the maximum, excluding glycoproteins. (B) The minimum motifs bound by hPIV1 (top panel), hPIV3 (middle panel), and H5 HA (bottom panel). For hPIV1 and H5 HA the minimal binding units are shaded. For hPIV3 there are three minimal motifs as shown.
We looked for evidence of NA activity of hPIV1. There was a general decrease in binding at pH 5.5, which could be due to sialidase activity or to ionization differences, but in most cases we could not correlate sensitivity or resistance with features of the oligosaccharides that might indicate substrate specificity (Fig. 2). Overall, the NeuAcα2-3Gal glycosidic bond is surprisingly resistant, at least when printed on the slide (unpublished data). However, some of the NeuAcα2-8NeuAcα2-3 sialylated sugars bound at pH 5.5 but not at pH 7, suggesting that hPIV1 HN cleaves α2-8-linked sialic acid from α2-3, as does Newcastle disease virus (2), but does not bind to it. hPIV3 sialidase activity is very low, about 1% of that of hPIV1 measured with MUN substrate (our results) or sialyllactose (1, 18), and so a low-pH experiment was not attempted.
Receptor specificity of hPIV1 and hPIV3.
The most detailed studies of receptor binding preferences of hPIVs were from Suzuki et al. (16), who measured virus bound to gangliosides separated by thin-layer chromatography using an overlay technique or gangliosides and oligosaccharides bound to microplate wells. The results showed specific binding of hPIV1 and hPIV3 to long and branched polylactosamine chains with terminal α2-3-linked sialic acid (16). Other studies include human pseudostratified mucociliary airway epithelium, where hPIV3 appeared to bind more strongly to α2-6-linked sialic acid than to α2-3-linked sialic acid based on elimination of infection by sialidase from Arthrobacter ureafaciens (19). However, this result could alternatively be due to release of branched or modified α2-3-linked sialic acid.
The glycoarray experiments encompass a much wider variety of oligosaccharides than previously available, and the results show that both hPIV1 and hPIV3 bind to only a subset of the glycans that contain α2-3-linked sialic acids. The minimum structure bound by hPIV1 is Neu5Acα2-3Galβ1-4GlcNAc, while hPIV3 requires a longer oligosaccharide unless the Gal is sulfated (Fig. 3). Neither binds if the Gal-GlcNAc linkage is β1-3 or if the second sugar is GalNAc. Both bind if the Gal is sulfated and/or the GlcNAc is fucosylated (e.g., 6′-sulfosialyl-Lewisx). There are, however, some interesting differences. hPIV1 can bind if there is a branched GalNAc on the Gal or NeuAc-Gal on the GlcNAc. There was significant binding for N-glycolyl-sialyl-Lewisx (glycan 259) although humans lack the CMP-N-acetylneuraminic acid hydroxylase and the only source of Neu5Gc is dietary (17). hPIV3 is more restricted in its binding (Fig. 2). It can bind the trisaccharide only if the Gal is sulfated, and it binds the tetrasaccharide Neu5Acα2-3Galβ1-4GlcNAcβ1-3Gal only when GlcNAc is fucosylated (sialyl-Lewisx). The addition of more polylactosamine units allows binding even when there is no sulfate or fucose (glycans 235, 238, and 285). Two gangliosides containing NeuAca2-6 have been reported to bind hPIV3 (16), but related structures on the glycan array did not bind.
Comparison of hPIV receptors with avian influenza virus H5N1 receptors.
Avian influenza virus H5N1 does not readily infect humans, nor is it readily transmitted between humans. Shinya et al. demonstrated abundant α2-3 sialic acid in human lungs using Maackia amurensis lectin, but none was detectable in the upper respiratory tract (12). They proposed that since H5N1 viruses bind to 2-3 sialic acid, they can infect only deep in the lungs, thus impeding both infection and transmission. However, primary bronchial epithelial cells from human trachea were shown to stain with both 2-3 and 2-6 sialic acid lectins and to support both human influenza virus and hPIV1 infection (7). Lectin staining of cultured, differentiated human tracheal tissue showed the presence of both 2-3 and 2-6 sialic acid, with goblet cells expressing more 2-3 sialic acid and ciliated cells expressing more 2-6 sialic acid (9). Human and avian virus infections were not restricted to cells showing their preferred receptor, suggesting that the 2-3 and 2-6 distributions were not as separate as indicated by the lectin staining. Nicholls et al. resolved apparent discrepancies by showing that the two lectins from Maackia amurensis bind to different tissues (11). MAH (MAA-2) stained alveolar cells and not upper respiratory tract tissues, in agreement with the previous results. However, MAL (MAA-1) strongly bound to upper respiratory tract cells (11). Crystal structures of MAL and MAH complexed with oligosaccharides confirmed their different specificities: MAL bound strongly to Neu5Acα2-3Galβ1-4GlcNAc, while MAH preferred Neu5Acα2-3Galβ1-3(Neu5Acα2-6)- GalNAc (6).
Glycoarray analysis of the HA of A/Vietnam/1203/2004 (H5N1) showed strong binding to oligosaccharides that would be predicted to bind MAH (15). A sulfate group can replace the branched sialic acid attached to GalNAc at position 3, but Neu5Acα2-3Galβ1-3GalNAc structures dominate the list of those binding the strongest to H5 HA (15). NeuAc2-3Galβ1-4GlcNAc binds about 50% of the maximum, so H5N1 virus might also be expected to infect upper respiratory tract cells, as was observed elsewhere (11). The H5 glycan array experiments used soluble expressed HA cross-linked with antibodies to provide the necessary multivalency, while we used whole hPIV virions. Stevens et al. compared Alexa-labeled whole virus versus baculovirus-expressed HA in the glycan-binding protein database (www.functionalglycomics.org/) and concluded that there is no significant difference in binding (14), and the baculovirus-expressed HA of A/Moscow/10/99 (H3N2) (13) shows the same binding glycans as does Alexa-labeled whole H3N2 virus A/Wyoming/3/03 in our analysis (8).
Cells in the upper respiratory tract, which are rich in ligands for MAL, would be expected to support infection by hPIV1 and hPIV3. Influenza virus H5N1 can weakly bind a few of the same ligands, but its stronger binding specificity is similar to that of MAH, and 13 glycans that are bound by H5 HA show no binding to hPIV. Thus, the different binding specificities of hPIV compared to those of H5N1 are in good agreement with the concentration of MAL ligands in the upper respiratory tract and that of MAH ligands in the lungs and with the reported sites of infection.
Acknowledgments
This work was supported in part by NIAID (AI18203). The glycoarray analysis was carried out by Core H of the Consortium for Functional Glycomics, funded by NIGMS (GM62116).
We thank Helga Veeraprame and Julie Linden for excellent technical assistance.
Footnotes
Published ahead of print on 23 May 2007.
REFERENCES
- 1.Ah-Tye, C., S. Schwartz, K. Huberman, E. Carlin, and A. Moscona. 1999. Virus-receptor interactions of human parainfluenza viruses types 1, 2 and 3. Microb. Pathog. 27:329-336. [DOI] [PubMed] [Google Scholar]
- 2.Corfield, A. P., M. Wember, R. Schauer, and R. Rott. 1982. The specificity of viral sialidases. The use of oligosaccharide substrates to probe enzymic characteristics and strain-specific differences. Eur. J. Biochem. 124:521-525. [PubMed] [Google Scholar]
- 3.Gulati, U., W. Wu, S. Gulati, K. Kumari, J. L. Waner, and G. M. Air. 2005. Mismatched hemagglutinin and neuraminidase specificities in recent human H3N2 influenza viruses. Virology 339:12-20. [DOI] [PubMed] [Google Scholar]
- 4.Henrickson, K. J. 2003. Parainfluenza viruses. Clin. Microbiol. Rev. 16:242-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirst, G. K. 1950. Receptor destruction by viruses of the mumps-NDV-influenza group. J. Exp. Med. 91:161-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imberty, A., C. Gautier, J. Lescar, S. Perez, L. Wyns, and R. Loris. 2000. An unusual carbohydrate binding site revealed by the structures of two Maackia amurensis lectins complexed with sialic acid-containing oligosaccharides. J. Biol. Chem. 275:17541-17548. [DOI] [PubMed] [Google Scholar]
- 7.Kogure, T., T. Suzuki, T. Takahashi, D. Miyamoto, K. I. Hidari, C. T. Guo, T. Ito, Y. Kawaoka, and Y. Suzuki. 2006. Human trachea primary epithelial cells express both sialyl(α2-3)Gal receptor for human parainfluenza virus type 1 and avian influenza viruses, and sialyl(α2-6)Gal receptor for human influenza viruses. Glycoconj. J. 23:101-106. [DOI] [PubMed] [Google Scholar]
- 8.Kumari, K., S. Gulati, D. F. Smith, U. Gulati, R. D. Cummings, and G. M. Air. 2007. Receptor binding specificity of recent human H3N2 viruses. Virol. J. 4:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matrosovich, M. N., T. Y. Matrosovich, T. Gray, N. A. Roberts, and H. D. Klenk. 2004. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. USA 101:4620-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merz, D. C., P. Prehm, A. Scheid, and P. W. Choppin. 1981. Inhibition of the neuraminidase of paramyxoviruses by halide ions: a possible means of modulating the two activities of the HN protein. Virology 112:296-305. [DOI] [PubMed] [Google Scholar]
- 11.Nicholls, J. M., M. C. Chan, W. Y. Chan, H. K. Wong, C. Y. Cheung, D. L. Kwong, M. P. Wong, W. H. Chui, L. L. Poon, S. W. Tsao, Y. Guan, and J. S. Peiris. 2007. Tropism of avian influenza A (H5N1) in the upper and lower respiratory tract. Nat. Med. 13:147-149. [DOI] [PubMed] [Google Scholar]
- 12.Shinya, K., M. Ebina, S. Yamada, M. Ono, N. Kasai, and Y. Kawaoka. 2006. Avian flu: influenza virus receptors in the human airway. Nature 440:435-436. [DOI] [PubMed] [Google Scholar]
- 13.Stevens, J., O. Blixt, L. Glaser, J. K. Taubenberger, P. Palese, J. C. Paulson, and I. A. Wilson. 2006. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J. Mol. Biol. 355:1143-1155. [DOI] [PubMed] [Google Scholar]
- 14.Stevens, J., O. Blixt, J. C. Paulson, and I. A. Wilson. 2006. Glycan microarray technologies: tools to survey host specificity of influenza viruses. Nat. Rev. Microbiol. 4:857-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stevens, J., O. Blixt, T. M. Tumpey, J. K. Taubenberger, J. C. Paulson, and I. A. Wilson. 2006. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312:404-410. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki, T., A. Portner, R. A. Scroggs, M. Uchikawa, N. Koyama, K. Matsuo, Y. Suzuki, and T. Takimoto. 2001. Receptor specificities of human respiroviruses. J. Virol. 75:4604-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varki, A. 2001. Loss of N-glycolylneuraminic acid in humans: mechanisms, consequences, and implications for hominid evolution. Am. J. Phys. Anthropol. Suppl. 33:54-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, Z. M., L. L. Tong, D. Grant, and T. Cihlar. 2001. Expression and characterization of soluble human parainfluenza virus type 1 hemagglutinin-neuraminidase glycoprotein. J. Virol. Methods 98:53-61. [DOI] [PubMed] [Google Scholar]
- 19.Zhang, L., A. Bukreyev, C. I. Thompson, B. Watson, M. E. Peeples, P. L. Collins, and R. J. Pickles. 2005. Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J. Virol. 79:1113-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]