Abstract
Natural polymorphisms in the heterogeneous human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein may have an impact on both sensitivity to entry inhibitors and viral replicative fitness. Of significant interest is variation in the V3 crown due to its involvement in direct engagement with the coreceptor. Two positions in the crown (318 and 319) appear to be important in determining intrinsic susceptibility to multiple entry inhibitors. Thus, we evaluated a series of natural polymorphisms at positions 318 and 319 in three distinct CCR5-tropic envelope genetic backgrounds to address their role in replicative fitness and sensitivity to entry inhibitors. Change at position 319 to each of the three major consensus amino acids (A, T, and R) resulted in variation in sensitivity to entry inhibitors and altered replicative fitness, but the effects of any one amino acid depended on the envelope context. Change of the nearly invariant tyrosine at position 318 to a rare arginine resulted in increased sensitivity to entry inhibitors and decreased replicative fitness independent of envelope context. Polymorphisms in the V3 crown that showed increased susceptibility to entry inhibitors also exhibited decreased entry efficiency, replicative fitness in primary peripheral blood mononuclear cells, and ability to replicate in primary macrophages. These findings suggest that differences in coreceptor affinity and/or avidity may underlie these phenotypic characteristics.
The human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein mediates entry of virus into host cells through sequential interaction with CD4, a coreceptor (either CCR5 or CXCR4), and subsequent membrane fusion. Entry inhibitors can disrupt this process by preventing any one of these critical events (6, 17, 56). Primary HIV-1 isolates display a wide range of susceptibilities to entry inhibitors, with 50% inhibitory concentrations (IC50) varying by as much as 1,000-fold. This is in significant contrast to inhibitors of reverse transcription and protease cleavage, which exhibit modest differences in intrinsic sensitivity across diverse HIV-1 isolates. Large susceptibility variations in primary HIV-1 isolates have been documented for the chemokine derivative AOP-RANTES (50), the fusion inhibitor enfuvirtide (ENF; T-20) (23, 39, 40), and many small molecule coreceptor antagonists (TAK-779 [39, 44], maraviroc [14], SCH-C [44], SCH-D [vicriviroc] [48], and AMD-3100 [23]). Due to the high degree of diversity among HIV-1 env genes of the same or different HIV-1 subtypes, it is difficult to identify specific sequence variations that may be associated with variable sensitivity to entry inhibitors.
Evaluation of intrinsic sensitivity differences to ENF and TAK-779 revealed that kinetic factors of fusion were largely responsible for variations in IC50 (39). Sensitivity to ENF mapped to the V3 loop of env (11), but mutations in the bridging sheet are also sufficient to modulate intrinsic susceptibility to these inhibitors (39, 40). Multiple factors are involved in the efficiency of host cell entry. Upon CD4 binding, structural rearrangements within the envelope occur that reveal the coreceptor binding site. The current model of ternary complex formation favors multiple interaction sites between HIV-1 envelope and CCR5. The bridging sheet and V3 stem interact with the CCR5 N terminus, while the V3 crown interacts with the second extracellular loop of CCR5 (9, 10, 13, 18, 19, 24, 35, 42). The hypervariable V3 loop must evolve by balancing attempts to escape host humoral response with the need to engage the CCR5 coreceptor for host cell entry. The affinity relationship between CCR5 and envelope, which can be modulated by the density of CCR5 on the cell surface, may be important in influencing the efficiency of entry. Some views hold that the major rate-limiting process in host cell entry is the formation of six-helix bundles (34), but other data suggest that ternary complex formation is the major rate-limiting step (31). The affinity relationship between CCR5 and V3 may be important in influencing the efficiency of entry through either of these pathways.
Mechanisms involved in variable susceptibility to chemokines such as CCL5 (RANTES) or their derivatives have not been evaluated. These inhibitors differ from small-molecule CCR5 antagonists in their ability to occupy surface receptor, as well as trigger internalization of CCR5 (33). We have previously determined the sensitivity of a panel of primary HIV-1 isolates from all subtypes to the CCL5 analog AOP-RANTES (50). We found a >30-fold difference in intrinsic susceptibility and suggested that this variability in AOP-RANTES sensitivity may be related to sequence differences in the V3 crown, specifically at positions 318 and 319 (HXB2 numbering). Similar variations at position 319 were observed when the intrinsic sensitivities of a different panel of primary isolate viruses were compared to inhibition by TAK-779 and SCH-C (44), further underscoring the potential relevance of these sites in intrinsic entry inhibitor sensitivity. Treatment of HIV-infected hu-PBL-SCID mice with the CCL5 analog NNY-RANTES selected for mutation at position 318 in the challenge virus, suggesting a potential role of this polymorphism in escape (32).
In the present study, we assessed the effect of natural polymorphisms at position 318 and 319 in the V3 crown on entry inhibitor sensitivity and overall replicative fitness. Previous studies have suggested that replicative fitness of primary HIV-1 isolates is associated with the env gene and overall efficiency of host cell entry (5, 30, 38). Given the central role of the V3 region in coreceptor interactions, it is possible that variation within this region may have a profound impact on the replicative fitness of the whole virus. To assess this, we inserted a series of observed polymorphisms into positions 318 and 319 in three distinct envelope backgrounds: the C2-V3 region of a subtype A primary isolate (A1-92RW009), the gp120 region of a subtype B primary isolate (B5-91US056), and the gp160 region of the Yu-2 strain. These envelopes were inserted into the NL4-3 backbone, and the viruses were tested for sensitivity to a variety of entry inhibitors and other antiretroviral agents. It was determined that polymorphisms at these sites can have a dramatic impact on entry inhibitor sensitivity in a context-dependent and -independent manner. Competitive replication assays further showed that these polymorphisms can significantly impact replicative fitness. In general, entry inhibitor sensitivity in each background correlated with replication capacity, suggesting the involvement of a common factor, such as the affinity and/or avidity of HIV-1 envelope and CCR5.
MATERIALS AND METHODS
Cells and Viruses.
U87 human glioma cells expressing CD4 and either CCR5 or CXCR4 were obtained through the AIDS Research and Reference Reagent Program. These cells were maintained in Dulbecco modified Eagle medium (Mediatech, Inc., Herndon, VA) supplemented with 15% fetal bovine serum (FBS; Mediatech, Inc.), penicillin (100 U/ml), streptomycin (100 μg/ml), Geneticin (G418; 300 μg/ml), and puromycin (1 μg/ml). 293T cells were grown in Dulbecco modified Eagle medium containing 10% FBS and penicillin-streptomycin. Peripheral blood mononuclear cells (PBMC) were extracted from HIV-seronegative donors by Ficoll-Hypaque density gradient centrifugation of heparin-treated venous blood. PBMC were maintained in RPMI 1640 (Mediatech, Inc.) supplemented with 10% FBS and penicillin-streptomycin, and stimulated with phytohemagglutinin (PHA; 2 μg/ml; Sigma-Aldrich, St. Louis, MO) and interleukin-2 (IL-2; 1 ng/ml; BD Pharmingen, San Diego, CA) for 3 days prior to infection. CD4+ T cells were isolated from PHA-blasted PBMC cultures by negative selection using a Miltenyi CD4+ T-cell magnetic isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany) and maintained in RPMI supplemented with 10% FBS and IL-2. Monocytes were isolated from unstimulated PBMC by positive selection for CD14 positive cells (Miltenyi Biotec), and macrophages were matured by adherence to plastic and growth in the presence of granulocyte-macrophage colony-stimulating factor for 7 days. Macrophages were maintained in RPMI supplemented with 10% new human serum (Cellgro) at 37°C in 5% CO2.
Primary HIV-1 isolates were obtained from the AIDS Research and Reference Reagent Program. For all strains used, the letter and number before the dash indicate the subtype of the virus envelope and the laboratory strain number, which precedes the year of isolation, country of origin, and repository strain number (e.g., A1-92RW009 denotes a subtype A virus isolated in Rwanda in 1992). NL4-3-envelope chimeric viruses were generated by using a yeast recombination-gap repair technique (27). Briefly, the C2-V3 region of the subtype A primary isolate A1-92RW009 was PCR amplified using the forward primer E80-RECOM and the reverse primer E105-RECOM and cotransformed into yeast with the SacII-linearized vector pREC-envΔV3/URA3. Recombined plasmids were selected on leucine dropout media containing 5-fluoroorotic acid. The EcoRI-XhoI fragment, which includes the entire HXB2-derived coding region of gp160, as well as the first and second exons of tat and rev, was shuttled into the infectious molecular clone pNL4-3. Cloning of the gp120 region of the subtype B primary isolate B5-91US056 has been previously described (27, 30). Cloning of the Yu-2 gp160 was performed essentially as described by Marozsan and Arts (27). Site-directed mutagenesis was performed on the envelope genes by using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). Infectious chimeric virus was produced by transfecting 293T cells using the Effectene transfection reagent (QIAGEN, Valencia, CA). Supernatant containing virus was collected 2 days posttransfection, clarified by centrifugation at 2,500 rpm for 15 min, and purified through a 0.45-μm-pore-size filter (Millipore, Billerica, MA). Viruses were subsequently passaged through U87-CD4/CCR5 cells to remove debris produced in transfection. Sequence analysis of the C2-V3 region was performed in triplicate on virus stocks to confirm that reversion or mutation did not occur. Virus titers were calculated by using a limiting dilution 50% tissue culture infective dose(s) (TCID50) method on PHA- and IL-2-activated PBMC, CD4+ T cells, macrophages, or U87-CD4/CCR5 cells as previously described (5).
Western detection of viral proteins.
Viral incorporation of gp120 was assessed by Western blotting. Equal TCID50 values of virus stock were first clarified of cell debris by retaining the supernatant after centrifugation at 3,000 rpm for 10 min and then pelleting the virus in supernatant by centrifugation at 38,000 rpm for 45 min. Virus pellets were lysed with sodium dodecyl sulfate lysis buffer, which was then separated on a 10% polyacrylamide electrophoresis gel and transferred to nitrocellulose. Blots were blocked with 5% gelatin solution and probed for gp41 with the Chessie 8 antibody obtained through the AIDS Research and Reference Reagent Program. The presence of gp120 was assessed by using the B13 antibody (kindly provided by George Lewis, The Institute of Human Virology, Baltimore, MD, and Bruce Chesebro, NIAID, Hamilton, MT) (2). Envelope detection was controlled by p24 quantification, assessed using the 24-4 monoclonal antibody (MAb), obtained through the AIDS Research and Reference Reagent Program. Primary blots were incubated with horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Pierce Biotechnology, Inc., Rockford, IL) and revealed by using the ECL Western detection kit (Amersham, Piscataway, NJ).
Single-cycle infection assays.
Envelope expression vectors were generated from the pREC-env shuttle vectors used in the yeast recombination cloning step. Generation of envelope expression vectors was previously described (27). Plasmids were digested with XbaI and religated to align the poly(A) signal sequence with the 3′ end of the envelope open reading frame. Envelope pseudotypes were generated by cotransfection of 293T cells with the 1 μg of the luciferase-encoding pseudotyping vector NLLuc.AM (37) and 1 μg of envelope expression vector. Cells were washed after 24 h, and pseudoviruses were collected after a subsequent 48 h. Relative particle numbers were determined by limiting dilution reverse transcriptase (RT) assay (28). Limiting-dilution infectivity assays were performed to determine a linear range for each pseudovirus for infection of U87-CD4/CCR5 cells. Cells were incubated with 1:5 dilutions of pseudovirus for 48 h, washed with phosphate-buffered saline (PBS), and lysed with 100 μl of Glo lysis buffer (Promega, Madison, WI). Samples were assessed for luciferase activity on the Bio-Rad Lumimark-Plus (Bio-Rad).
Entry inhibitor sensitivity.
Most drug sensitivity assays (excluding sensitivity to 2D7) were performed in multiple cycle replication assays on U87.CD4.CCR5 cells. Cells were added to 96-well plates (104 cells/well) and allowed to adhere overnight. Cells were incubated with serial 10-fold dilutions of drug (CCL3/CCL4/CCL5 [100 nM to 0.1 pM], PSC-RANTES [10 nM to 0.1 pM], ENF [10 μM to 0.01 nM], TAK-779 [10 μM to 0.1 nM], 3TC [10 μM to 0.1 nM], nevirapine [10 μM to 0.1 nM], ritonavir [10 μg/ml to 0.1 ng/ml], 118-d-24 [100 μM to 1 nM], and 2D7 [25 to 0.25 μg/ml]) for 1 h prior to the addition of virus (multiplicity of infection of 0.005). Cells were incubated with virus for 24 h and washed with PBS, and fresh medium that contained the appropriate concentration of drug was replaced. Cell-free supernatant samples were taken each at day 4 to 8 postinfection and monitored for virus production by radioactive RT assay protocol previously described (50). Plots of RT activity versus drug concentration were constructed and analyzed to determine the 50% inhibitory concentration (IC50) for each drug. For the MAb 2D7, cells were incubated with serial 10-fold dilutions of antibody for 1 h prior to the addition of pseudovirus. Cells were incubated for 48 h, washed with PBS, and lysed, and the luciferase activity determined. Plots of luciferase activity versus drug concentration were used to determine IC50 values for each pseudovirus.
Replicative fitness from competition assays.
The replicative fitness of each chimeric virus was determined by using a dual-infection competition assay. Chimeric viruses containing V3 crown polymorphisms were competed against a panel of CCR5-tropic primary isolate viruses (B2-92BR017, Yu-2, B5-91US056, C5-97ZA003, A1-92RW009, and B10-92BR003). Virus was added alone or in pairs to PHA/IL-2-stimulated PBMC at a multiplicity of infection of 0.0005 IU/cell to 5 × 105 cells per well in a 48-well plate (100 IU/well). Fresh IL-2 was added 4 days postinfection. All assays were performed using one donor from a single blood draw in duplicate. Cell-free supernatants were assessed for RT activity at days 4, 8, and 12 postinfection. Cells and cell-free supernatant were harvested 12 days postinfection and stored at −80°C for subsequent analysis. Proviral DNA was extracted from infected PBMC by using a QIAamp DNA blood kit (QIAGEN). Proviral DNA was PCR amplified over the C2-V3 region as previously described (5). For competitions that include viruses with identical C2-V3 regions, PCR amplification was performed over the vif gene region. Nested PCR products from the competitions were analyzed by heteroduplex tracking assay. A radioactive DNA probe was generated by PCR amplification from a plasmid template containing the C2-V3 region of a heterologous virus. The forward primer was labeled with 2 μCi of [γ-32P]ATP using T4 polynucleotide kinase (Invitrogen, Carlsbad, CA). Nested PCRs from competition DNA were annealed to probe by mixing PCR product with radioactive probe, followed by denaturing at 95°C for 3 min and annealing at 37°C for 5 min. Reactions contained DNA annealing buffer (100 mM NaCl, 10 mM Tris-HCl [pH 7.8], and 2 mM EDTA), 10 μl of unlabeled PCR-amplified DNA from the competition, and approximately 400 cpm of probe per reaction. The DNA heteroduplexes were resolved on Tris-borate-EDTA buffer-containing 10% nondenaturing polyacrylamide gels for 2 h at 200 V. Gels were dried and exposed to X-ray film (Eastman Kodak Co., Rochester, NY). Quantification of heteroduplex bands was performed on a Bio-Rad phosphorimager by using QuantityOne software (Bio-Rad, Hercules, CA). Replicative fitness of each chimeric virus was estimated from the relative abundance of each virus quantified from the heteroduplex tracking assay relative to the probe binding of the monoinfection. Production of each virus in the dual infection (f0) was divided by the initial proportion in the inoculum (i0) and is referred to as the relative fitness of that virus (w = f0/i0). The fitness difference (WD) between the two viruses in competition is representative of the relative fitness of the more-fit virus (wM) versus that of the less-fit virus (wL): WD = wM/wL.
RESULTS
Natural variation at HIV-1 V3 positions 318 and 319.
Multiple studies have demonstrated a potential role for positions 318 and 319 (HXB2 numbering) in the V3 region in affecting sensitivity to entry inhibitors. Previous work described a 30-fold variation in sensitivity to the CCL5 analog AOP-RANTES in a panel of diverse HIV-1 isolates (n = 14). Sequence analysis of the V3 region suggested a possible association between AOP-RANTES sensitivity and the identity of the amino acid at position 319 being either a threonine or an alanine (50). A more recent study on the CCR5 MAb PA14 showed a distinct pattern of sensitivity to the CCR5 antagonists SCH-C and TAK-779. This sensitivity was mapped to the V3 crown and included the polymorphisms alanine, threonine, and arginine at position 319 (44). Furthermore, treatment of hu-PBL-SCID mice infected with the HIV-1 strain 242 with NNY-RANTES resulted in mutation in the V3 crown position 318 (H→R), which may have been associated with escape from drug pressure (32). We analyzed the frequency of the amino acids alanine, threonine, and arginine at position 319 and of tyrosine and arginine at position 318 from 31,408 HIV-1 V3 sequences from all subtypes deposited in the Los Alamos database (Table 1) . Alanine, threonine, and arginine are the major consensus amino acids at position 319, but their frequency varies by subtype. Alanine is the consensus amino acid for subtypes A, C, F, G, and H, while threonine is the consensus amino acid for subtypes B and D. In subtype A, the alanine polymorphism is present in the large majority of sequences (84%), while the threonine is relatively rare (7%). In subtype B, both threonine (53%) and alanine (42%) are present in the majority of sequences. The 319 consensus amino acid for subtype CRF01_AE is the more atypical arginine, and alanine and threonine are present at very low abundance. Position 318, on the other hand, is very well conserved across subtypes. The consensus amino acid for all subtypes is tyrosine, and few variations at this site have been recorded. Arginine at position 318 is present in conjunction with either alanine or threonine at position 319 in very low frequencies across subtypes.
TABLE 1.
V3 (318/319) amino acid frequency by subtype
| Subtype | No. of sequences | Consensus (318/319)a | % of HIV-1 sequences with amino acid residues at 318 and 319
|
|||||
|---|---|---|---|---|---|---|---|---|
| 318Y/319A | 318Y/319T | 318Y/319R | 318R/319A | 318R/319T | Other | |||
| A | 3,900 | YA | 84 | 6.8 | 0.1 | 0.02 | 0.2 | 8.6 |
| B | 20,000 | YT | 42 | 53 | 0.09 | 0.01 | 0.3 | 4.4 |
| C | 3,064 | YA | 84 | 3.1 | 0.4 | 0.1 | 0 | 12 |
| D | 1,021 | YT | 18 | 55 | 1.4 | 0.2 | 0 | 24 |
| F | 290 | YA | 66 | 24 | 0.3 | 0 | 0 | 9.3 |
| F1 | 217 | YA | 64 | 24 | 0.5 | 0 | 0 | 11 |
| G | 890 | YA | 81 | 6.0 | 0.3 | 0.1 | 0 | 13 |
| H | 101 | YA | 71 | 11 | 4.0 | 0 | 0 | 14 |
| J | 140 | YA | 84 | 2.9 | 1.4 | 0 | 0 | 12 |
| K | 108 | YA | 63 | 7.4 | 1.9 | 0 | 0 | 28 |
| AE | 1,677 | YR | 0 | 1.3 | 80 | 0 | 0.1 | 19 |
Consensus amino acid frequencies are indicated in boldface under the corresponding genotype heading.
To assess polymorphisms at position 318 and 319 in a systematic fashion, it was essential to use envelope chimeric viruses. To rule out replicative fitness effects caused by other regions of the HIV-1 genome, we used NL4-3 as a neutral backbone for all chimeric viruses. We generated replication-competent chimeric viruses with three different envelope genes in the NL4-3 backbone: (i) an envelope chimeric virus containing the C2-V3 region of a subtype A primary isolate, A1-92RW009, in the HXB2 envelope background; (ii) the gp120 coding region of a subtype B primary isolate, B5-91US056, with the gp41 derived from HXB2; and (iii) the gp160 region of the Yu-2 strain (Fig. 1A). The HXB2 env in the NL4-3 backbone is preferred over the complete NL4-3 sequence because HXB2 lacks the G36D mutation present in the NL4-3 gp41, which confers reduced susceptibility to ENF. Site-directed mutagenesis was performed on these envelopes to introduce the relevant natural polymorphisms at positions 318 and 319 (Fig. 1A). The wild-type sequence for A1-92RW009 and B5-91US056 is Y318A319, and the wild-type Yu-2 sequence is Y318T319 (Fig. 1A). These positions are four amino acids downstream of the conserved GPGX tip of the V3 crown (Fig. 1B).
FIG. 1.
Generation of V3 mutant chimeric viruses. (A) Three different chimeric viruses were generated: NL4-3-V3A1-92RW009 contains the V3 region of the primary, CCR5-tropic isolate A1-92RW009 in the NL4-3 background. NL4-3-gp120B5-91US056 contains the gp120 region derived from the primary, CCR5-tropic isolate B5-91US056 in the NL4-3 background, and NL4-3-gp160Yu-2 is an NL4-3 construct with the full-length envelope coding sequence of the CCR5-tropic isolate Yu-2. Site-directed mutagenesis was performed at positions 318 and 319 to change the wild-type amino acids (A1-92RW009, YA; B5-91US056, YA; Yu-2, YT) into the relevant polymorphisms (318Y/319A, 318Y/319T, 318Y/319R, 318R/319A, and 318R/319T). Sequence of the 35-amino-acid V3 loop is shown, and the crown sequence is boxed. The amino acids at 318 and 319 are indicated in boldface. Viruses containing wild-type sequences are named in boldface. (B) The X-ray crystal structure of the V3 loop derived from Huang et al. (19) is presented as a ribbon and a space-filling model. The crown GPGR of the subtype B JR-FL strain is depicted in this structure (purple). In JR-FL, position 319 is a threonine. Since a tyrosine or arginine at position 318 and an alanine or threonine at position 319 are natural polymorphisms in both subtype A and B, energy minimizations were performed with the T319A substitutions in the JR-FL structure and using the GROMOS96 algorithm within the DeepView/Swiss-PdbViewer v3.7 (assuming a uniform dielectric constant inside the protein). The alanine was easily accommodated without perturbing the minimal energy predicted by the model.
Infectious chimeric virus was produced and analyzed to quantify envelope incorporation. Equal TCID50 values of viral supernatants (103.5 [3,162] infectious units) were pelleted and analyzed by Western blotting to detect HIV-1 gp120 and p24. All chimeric viruses incorporated similar amounts of gp120 with respect to p24 antigen content (Fig. 2A). Processing of the gp160 precursor into gp120 and gp41 occurred with similar efficiencies in all envelopes (Fig. 2A). Coreceptor tropism for each of the V3 mutant envelopes was determined by using a luciferase-based envelope pseudotype assay. Pseudoviruses were produced bearing each mutant envelope, and equivalent particle numbers were used to infect U87 cells expressing either CD4 alone, CD4 and CXCR4, or CD4 and CCR5. All envelopes effectively mediated infection of U87-CD4/CCR5 cells, but not cells expressing CXCR4 or no coreceptor (Fig. 3).
FIG. 2.
Analysis of envelope incorporation by V3 mutant chimeric viruses. To ensure equivalent incorporation of the envelope protein in the chimeric viruses, equal TCID50 of replication competent virus was pelleted at 38,000 rpm and subjected to Western blot. (A) 103.5 infectious units of each virus were pelleted and resuspended in 100 μl of SDS lysis buffer. A 10-μl portion of this sample, or 316 infectious units, was diluted serially 1:5 for each NL4-3-V3A1-92RW009 chimeric virus and probed for gp120 (B13 MAb), gp41 (Chessie 8 MAb), and p24 (24-4 MAb). (B) 316 infectious units of each NL4-3-gp120B5-91US056 and NL4-3-gp160Yu-2 V3 crown mutant chimeric virus were probed for p24 and gp120 content.
FIG. 3.
Coreceptor tropism and pseudovirus infection efficiency. Equivalent particle numbers of pseudovirus, as determined by limiting-dilution RT assay, were used to infect U87-CD4, U87-CD4/CXCR4, or U87-CD4/CCR5 cells. Cells were incubated with pseudovirus for 48 h, the cells were lysed, and the luciferase activity was determined. Pseudoviruses produced without envelope served as a negative control for infection, while treatment of cells with 100 μM 3TC served as a specificity control for luciferase activity. Values are presented as relative light units, and error bars represent the standard deviations from two independent experiments performed in triplicate.
Using luciferase-expressing pseudoviruses, luciferase activity is proportional to the number of particles that infect the cells (37). Thus, efficiency of virus entry can be roughly assessed from luciferase activity. The A-to-T change at position 319 in the V3A1-92RW009 envelope modestly increased entry efficiency, while the A-to-R change modestly decreased efficiency. The Y-to-R change at position 318 resulted in a 100-fold decrease in luciferase activity in conjunction with either an A or T at position 319 of the V3A1-92RW009 envelope (Fig. 3A). In the B5-91US056 envelope a change between A and T had no effect on entry efficiency, while in the Yu-2 gp160 context a change from the wild-type T to alanine at 319 resulted in a 10-fold increase in entry efficiency (Fig. 3B and C). However, insertion of arginine at position 318 or 319 resulted in significant decreases in entry efficiency (>100-fold) in both subtype B envelope contexts (Fig. 3B and C). In the Yu-2 context, insertion of arginine at either position 318 or 319 severely attenuated transduction of U87-CD4/CCR5 cells by pseudovirus, with >1,000-fold decreases in luciferase activity detected (Fig. 3C).
Replicative fitness of V3 mutant chimeric viruses in PBMC.
Competitive replicative fitness of a virus in ex vivo culture assays describes the relative ability of one virus versus another to expand in a cell population. Previous studies have suggested that the entry process is an important determinant of replicative fitness of the wild-type primary HIV-1 isolates (5, 30, 38, 38). We anticipated that polymorphisms within the V3 region, which mediated significant differences in transduction of U87-CD4/CCR5 by pseudoviruses, would have a significant impact on replicative fitness. Thus, we assessed viral replicative fitness in PHA/IL-2 activated PBMC cultures by competing each V3 mutant chimeric virus against each of six primary HIV-1 reference isolates: B2-92BR017, Yu-2, B5-91US056, C5-97ZA003, A1-92RW009, and B10-92BR003 (Fig. 4). This set of reference HIV-1 isolates was selected based on having a range of relative fitness values common to wild-type primary HIV-1 isolates (Fig. 4A) (5). A subset of V3 mutant chimeric viruses was used in pairwise competitions to confirm the fitness order derived from competitions with the reference virus panel (see Fig. S1 at http://www.case.edu/med/microbio/artslab/publications.htm). Relative virus production was monitored at 12 days postinfection by using heteroduplex tracking assay (see Materials and Methods) (3, 5).
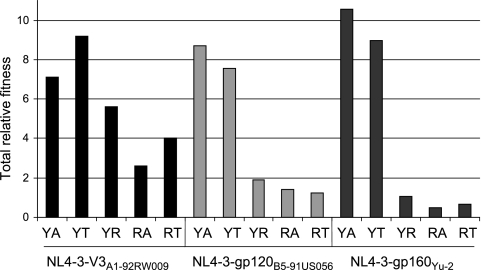
FIG. 4.
Replicative fitness of V3 polymorphisms in three envelope contexts. Each chimeric virus was competed in pairwise dual infection in PHA/IL-2-activated PBMC against a panel of CCR5-tropic primary isolate viruses (B2-92BR017, Yu-2, B5-91US056, C5-97ZA003, A1-92RW009, and B10-92BR003). Relative fitness values were calculated as described in Materials and Methods. Plots are of fitness difference values and are indicative of fold difference replication capacity. Fitness differences are plotted on a logarithmic scale, and bars that fall above the midline (WD = 1) indicate competitions in which the chimeric virus was the winner, whereas bars falling below the midline indicate competitions in which the primary reference isolate was the winner. Bars that lie near the midline (WD = 1) indicate competitions in which both viruses were of nearly equivalent replicative fitness. All competitions were performed in duplicate. (A) Fitness order of full-length viruses used in the reference panel. (B) Fitness difference values of NL4-3-V3A1-92RW009 chimeric viruses with V3 polymorphisms against the reference panel. (C) Fitness difference values of NL4-3-gp120B5-91US056 chimeric viruses with V3 polymorphisms against the reference panel. (D) Fitness difference values of NL4-3-gp160Yu-2 chimeric viruses with V3 polymorphisms against the reference panel.
Insertion of the A1-92RW009 V3 region into the NL4-3 backbone increased the fitness of this chimeric virus over the parental A1-92RW009 primary isolate (Fig. 4B). Comparison of A1-92RW009 to NL4-3-V3A1-92RW009(YA) showed an ∼5-fold increase in replicative efficiency by the chimeric virus. This increased fitness in the NL4-3 backbone may be related to optimization of this laboratory strain through in vitro selection. We consistently observed increases in replicative fitness of the NL4-3/env chimeric virus over the parental primary HIV-1 isolate (Fig. 4B to D). Considering the slight increase in fitness of the chimeric viruses versus the parental strain, the fitness impact of 318 and 319 substitutions were always compared among themselves rather than directly to their parental primary isolate. A change from alanine to threonine at position 319 resulted in a modest increase in replicative fitness consistent with the entry efficiency observed with pseudoviruses (compare Fig. 3A and Fig. 4B). The A→R change at position 319 of NL4-3-V3A1-92RW009(YA) resulted in a significant but still modest decrease in fitness compared to the wild-type chimeric virus (Fig. 4B). However, insertion of arginine at position 318 significantly reduced replicative fitness (Fig. 4B). A threonine as opposed to alanine at position 319 provided some compensation for the decreased fitness conferred by R318 in the NL4-3-V3A1-92RW009(YA) virus (Fig. 4B). A total relative fitness value was derived for the addition of the relative fitness values derived from competitions against the six reference strains (Fig. 5). This collective analysis revealed a fitness order for the V3 polymorphisms in NL4-3-V3A1-92RW009 context: Y318T319 > YA > YR > RT > RA. However, all of these chimeric viruses had fitness values in the range of wild type, CCR5-tropic primary HIV-1 isolates circulating in the infected population.
FIG. 5.
Total relative fitness values for each chimeric virus. Total relative fitness was calculated by adding the relative fitness value (w) of the chimeric virus from each competition with a reference panel virus.
In the subtype A population, alanine is significantly more frequent than threonine at position 319 (84% versus 7%, Table 1), while in the subtype B population, alanine and threonine are present at nearly equal frequency (42% versus 53%, Table 1). A change between A and T at position 319 in both subtype B contexts had modest fitness impact. The wild-type NL4-3-gp120B5-91US056(YA) and NL4-3-gp120B5-91US056(YT) variants had nearly equivalent fitness, and the wild-type YA was slightly more fit that the parental primary B5-91US056 isolate (Fig. 4C). However, the wild-type NL4-3-gp160Yu-2(YT) virus was significantly less fit than the NL4-3-gp160Yu-2(YA), but both chimeric viruses could outcompete the parental Yu-2 virus (Fig. 4D). Insertion of arginine into position V3 position 318 or 319 of NL4-3-gp120B5-91US056 or NL4-3-gp160Yu-2 yielded viruses with significantly reduced replication capacities, as evidenced by their poor ability to compete against the six reference HIV-1 isolates (Fig. 4C and D). Evaluation of total relative fitness values revealed that natural variations at sites 318 and 319 had a greater impact on replicative fitness in the context of the B5-91US056 or Yu-2 envelope than in the A1-92RW009 C2-V3 context (Fig. 5). This decrease in the fitness impact in the NL4-3-V3A1-92RW009 chimeric viruses may again be related to having more of the HXB2 env gene associated with the A1-92RW009 V3 sequence.
To confirm the relative fitness order derived from competitions with reference strains, we competed the NL4-3-V3A1-92RW009 V3 mutant chimeric viruses against each other in pairwise competition. All of the V3 mutant NL4-3-V3A1-92RW009 chimeric viruses contained one of three synonymous mutation vif sequence tags (i.e., vifA [wild-type NL4-3 sequence], vifB, or vifC; see Fig. S1 at http://www.case.edu/med/microbio/artslab/publications.htm). Each tag is a region of altered nucleotide sequence in vif that codes for the same amino acid sequence. These sequence tags were used to distinguish and quantify two NL4-3-V3A1-92RW009 chimeric viruses by heteroduplex tracking assay in a dual infection. Competition between two viruses that are isogenic but differ only in their vif sequence tag showed no fitness difference, indicating that the tag sequences did not affect replicative fitness. A perfect transitive relationship was evident in this fitness matrix, that is, YT had a fitness greater than YA in direct competition, YA > YR, YR > RA, RA > RT, YT > RA, YT > RT, etc (see Fig. S2 at http://www.case.edu/med/microbio/artslab/publications.htm). Fitness differences using pairwise comparison were identical to those found by comparison to the reference panel (see Fig. 5). Thus, all other chimeric viruses were competed solely against the reference panel to determine a replicative fitness value.
Replication of V3 mutant chimeric viruses in macrophage cultures.
The levels of CD4 and CCR5 expressed on the surface of macrophages is significantly lower than the levels of each receptor on CD4+ T cells (12, 25, 26, 54). It is commonly observed that viruses with expanded tropism for macrophages have evolved higher affinity for one or more receptors (16, 36, 49). Thus, we examined whether the chimeric viruses with natural V3 polymorphisms displayed different replication kinetics in macrophage cultures and whether the ability to infect macrophages correlated with replicative fitness in PBMC. Limiting-dilution TCID50 assays were performed side-by-side in macrophage cultures and in CD4+ T-cell cultures from the same donor and blood draw. In the subtype B contexts, the V3 polymorphisms with an arginine at position 318 or 319 that induced the lowest replicative fitness in PBMC cultures failed to grow to detectable levels in macrophage cultures (Table 2). More replication of the NL4-3-V3A1-92RW009 viruses compared to the other chimeric viruses was observed in the macrophage cultures (relative to CD4+ T-cell cultures). NL4-3-V3A1-92RW009 viruses containing arginine at position 318 did replicate in macrophages, but with reduced kinetics compared to the YA and YT polymorphisms in these chimeric viruses (Table 2).
TABLE 2.
Replication of V3 mutant viruses in macrophage cultures
| Day | Log(TCID50 macrophages)/log(TCID50 CD4+ T cells)
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NL4-3-V3A1-92RW009
|
NL4-3-gp120B5-91US056
|
NL4-3-gp160Yu-2
|
|||||||||||||
| YA | YT | YR | RA | RT | YA | YT | YR | RA | RT | YA | YT | YR | RA | RT | |
| 7 | 0.43 | 0.29 | 0.00 | 0.00 | 0.00 | 0.36 | 0.36 | 0.00 | 0.00 | 0.00 | 0.50 | 0.29 | 0.00 | 0.00 | 0.00 |
| 14 | 0.43 | 0.29 | 0.27 | 0.43 | 0.60 | 0.36 | 0.43 | 0.00 | 0.00 | 0.00 | 0.50 | 0.38 | 0.00 | 0.00 | 0.00 |
| 21 | 0.50 | 0.43 | 0.27 | 0.43 | 0.60 | 0.43 | 0.43 | 0.00 | 0.00 | 0.00 | 0.50 | 0.38 | 0.00 | 0.00 | 0.00 |
Antiretroviral susceptibility of conferred by V3 polymorphisms.
Replication-competent chimeric viruses harboring the various V3 polymorphisms were used in drug susceptibility assays with a broad range of antiretroviral compounds in U87-CD4/CCR5 cell cultures. Virus production was measured by determining the RT activity in the supernatant. An exception to these methods was related to the use of single-cycle pseudovirus assays when analyzing sensitivity to the CCR5 MAb, 2D7 (Table 3). Changes at position 318 and 319 mediated differential sensitivity to entry inhibitors in all envelope contexts (Table 3 and Fig. 6). Variable inhibition of the V3 mutant chimeric viruses was observed with (i) native β-chemokines and their analogs (CCL4, CCL5, PSC-RANTES), (ii) monoclonal CCR5 antibodies (2D7), (iii) noncompetitive allosteric inhibitors of CCR5 (TAK-779), and (iv) inhibitors of virus fusion (ENF) (Fig. 6A to C and Table 3). No differences in IC50 values were observed for the V3 mutant chimeric viruses with compounds that inhibit reverse transcription (3TC, nevirapine), protease cleavage (ritonavir), or integrase function (118-d-24) (Fig. 6D and Table 3). Drug sensitivity assays performed with the primary isolate viruses A1-92RW009, B5-91US056, and Yu-2 indicated that the envelope chimeric viruses recapitulate the intrinsic sensitivity of the parental viruses to PSC-RANTES, ENF, TAK-779, and 3TC (Table 4).
TABLE 3.
Sensitivity of V3 mutant viruses to antiretroviral compounds
| Compound | Classa | Sensitivity (IC50)b
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NL4-3-V3A1-92RW009
|
NL4-3-gp120B5-91US056
|
NL4-3-gp160Yu-2
|
||||||||||||||
| YA | YT | YR | RA | RT | YA | YT | YR | RA | RT | YA | YT | YR | RA | RT | ||
| CCL5 (nM) | Chemokine | >100 | >100 | >100 | 3.5 | 0.33 | >100 | >100 | >100 | 10 | 7.4 | >100 | >100 | ND | ND | ND |
| CCL3 (nM) | Chemokine | >100 | >100 | >100 | >100 | >100 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CCL4 (nM) | Chemokine | >100 | >100 | >100 | 33 | 19 | >100 | >100 | >100 | 53 | 31 | >100 | >100 | ND | ND | ND |
| PSC-RANTES (nM) | Chemokine derivative | 0.29 | 0.27 | 0.05 | 0.02 | 0.24 | 0.30 | 0.03 | 0.01 | 0.007 | 0.4 | 0.05 | 0.005 | 0.004 | 0.004 | 0.27 |
| TAK-779 (nM) | CCR5 small molecule | 95 | 24 | 17 | 23 | 2.2 | 2.1 | 45 | 40 | 0.3 | 0.1 | 24 | 18 | 0.48 | 0.11 | 0.08 |
| 2D7 (μg/ml) | CCR5 MAb | 1.3 | 4.7 | 0.81 | 0.013 | 0.007 | 8.4 | 1.2 | 0.01 | 0.002 | 0.003 | 3.5 | 0.07 | ND | ND | ND |
| ENF (nM) | Fusion inhibitor | 21.9 | 24.3 | 0.33 | 1.2 | 0.33 | 3.4 | 16 | 3.1 | 0.38 | 0.11 | 33 | 2.4 | 0.08 | 0.09 | 0.07 |
| 3TC (nM) | NRTI 62 | 50 | 25 | 68 | 20 | 32 | 18 | 44 | 84 | 21 | 77 | 40 | 41 | 22 | 24 | |
| Nevirapine (nM) | NNRTI | 17 | 28 | 20 | 19 | 20 | 25 | 19 | 19 | 22 | 31 | 20 | 23 | ND | ND | ND |
| Ritonavir (ng/ml) | PI | 0.21 | 0.18 | 0.14 | 0.06 | 0.06 | 0.16 | 0.21 | 0.08 | 0.10 | 0.08 | 0.22 | 0.11 | ND | ND | ND |
| 118-D-24 (μM) | Integrase inhibitor | 5.1 | 4.2 | 5.2 | 1.8 | 8.6 | 6.4 | 3.4 | 6.7 | 12.1 | 4.2 | 2.2 | 10.0 | ND | ND | ND |
NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, non-nucleoside reverse transcriptase inhibitor; PI, protease inhibitor.
Concentrations are as indicated in column 1. ND, not determined.
FIG. 6.
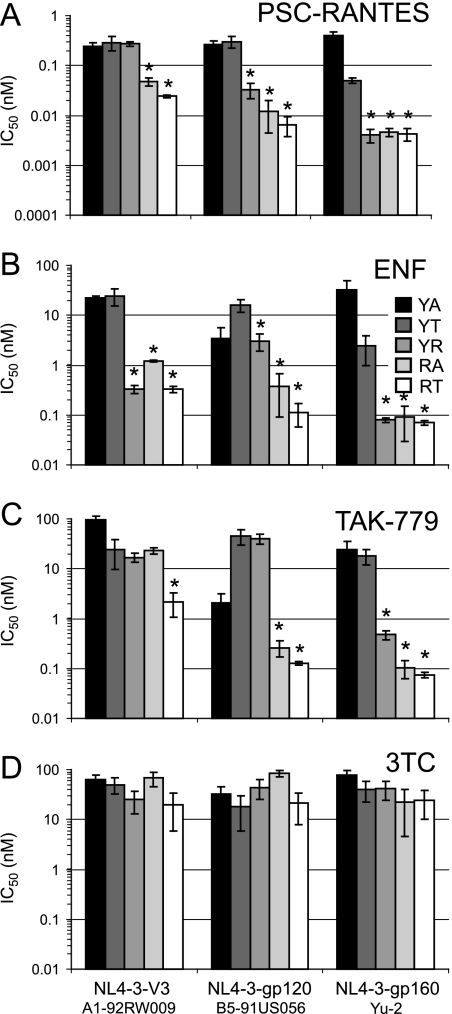
Sensitivity of 318/319 polymorphisms to entry inhibitors. U87-CD4/CCR5 cells were incubated with 10-fold dilutions of PSC-RANTES (A), ENF (B), TAK-779 (C), or 3TC (D) and then exposed to virus for 24 h. After the input virus was washed out, the supernatant samples were analyzed for RT activity at 4, 6, and 8 days postinfection. Drug susceptibility curves (data not shown) were plotted to determine IC50 values using the probit algorithm. IC50 values and standard deviations for each respective drug and virus are plotted in the panels. Asterisks indicate cutoff for significance of P < 0.01 (paired comparison t test).
TABLE 4.
Sensitivity of full-length primary isolate viruses to antiretroviral compounds
| Compound | Sensitivity (IC50 [nM])
|
||
|---|---|---|---|
| A1-92RW009 | B5-91US056 | Yu-2 | |
| PSC-RANTES | 0.17 | 0.14 | 0.02 |
| ENF | 77.0 | 44.3 | 73.1 |
| TAK-779 | 105 | 12.1 | 18.4 |
| 3TC | 60.8 | 42.2 | 54.6 |
In the NL4-3-V3A1-92RW009 virus context, change at position 319 among T, A, and R polymorphisms had only modest effects on sensitivity to most entry inhibitors (Fig. 6A to C). Arginine at 319 in the NL4-3-V3A1-92RW009 viruses resulted in increased susceptibility exclusively to ENF (Fig. 6B). Arginine at position 318 increased sensitivity to nearly all entry inhibitors in conjunction with both alanine and threonine at position 319 (Table 3 and Fig. 6A to C). Only the NL4-3-V3A1-92RW009 viruses containing arginine at position 318 were susceptible to inhibition by CCL5 (RANTES) and CCL4 (MIP-1β), but no viruses were inhibited by CCL3 (MIP-1α) (Table 3).
The 318 and 319 V3 polymorphisms in the subtype B env contexts had similar impacts on sensitivity to entry inhibitors as observed with NL4-3-V3A1-92RW009 viruses. In the NL4-3-gp120B5-91US056 virus context, the T and A polymorphisms at position 319 did not significantly alter sensitivity to most entry inhibitors (Fig. 6A to C; see Fig. S3 at http://www.case.edu/med/microbio/artslab/publications.htm). Arginine at position 319 in this envelope context resulted in increased susceptibility to PSC-RANTES, ENF, and 2D7 (Fig. 6A and B and Table 3). This variant did not exhibit increased susceptibility to TAK-779 (Fig. 6C). Arginine at position 318 in conjunction with either alanine or threonine at position 319 significantly increased sensitivity to all entry inhibitors (Fig. 6A-C and Table 3). The susceptibility of NL4-3-gp120B5-91US056(YT) differed from NL4-3-gp120B5-91US056(RT) by 50-fold to PSC-RANTES and by 100-fold for ENF and TAK-779. As seen in the NL4-3-V3A1-92RW009 context, an arginine at position 318 in the NL4-3-gp120B5-91US056 context resulted in susceptibility to CCL5 and CCL4 (Table 3).
V3 polymorphisms in the NL4-3-gp160Yu-2 context had dramatic effects on sensitivity to entry inhibitors. A change of the wild-type 319T to alanine yielded a NL4-3-gp160Yu-2 virus with reduced susceptibility to PSC-RANTES, ENF, and 2D7 (Fig. 6A to C; Table 3). Insertion of arginine at either position 318 or 319 resulted in NL4-3-gp160Yu-2 viruses with exquisite sensitivity to inhibition by PSC-RANTES, ENF, and TAK-779 (Fig. 6A to C). As described above, the NL4-3-gp160Yu-2 viruses with the V3 polymorphisms were equally sensitive to a protease inhibitor (ritonavir), nucleoside RT inhibitor (3TC), non-nucleoside RT inhibitor (nevirapine), and integrase inhibitor (118-d-24) (Table 3 and Fig. 6D).
Comparing replicative fitness with sensitivity to entry inhibitors.
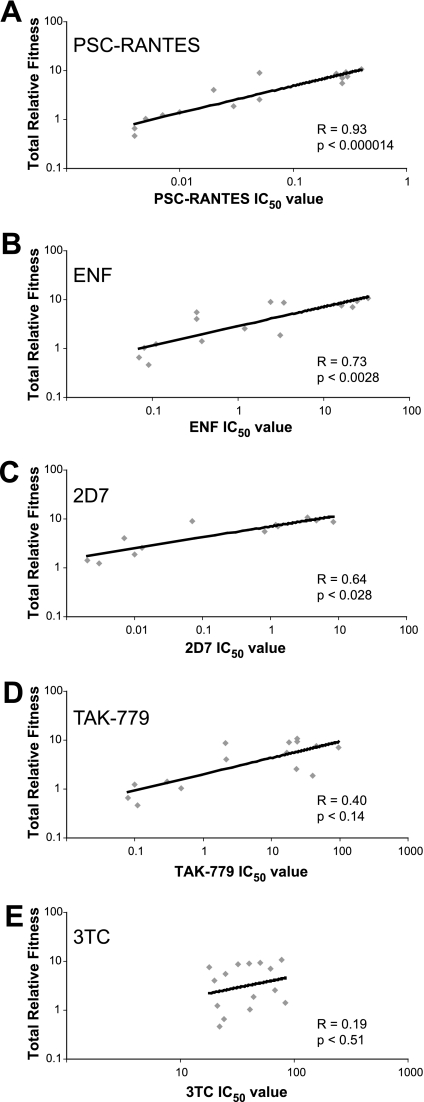
The data described above suggest that replicative fitness in PBMC conferred by variation in the V3 crown is directly related to the efficiency of entry by pseudotyped virus in U87-CD4/CCR5 cells and inversely related to sensitivity to entry inhibitors. Using regression analysis, we observed a highly significant correlation between sensitivity to PSC-RANTES inhibition and total relative fitness (r = 0.93, P < 0.000014, Pearson product-moment correlation [PPMC]) (Fig. 7). The relationships between total relative fitness and IC50 values with ENF (r = 0.73, P < 0.0028, PPMC) and 2D7 (r = 0.64, P < 0.028, PPMC) were weaker but still significant (Fig. 7). In contrast, there was no significant correlation between IC50 values to TAK-779 (r = 0.40, P < 0.14) or 3TC (r = 0.19, P < 0.51) and total relative fitness of these V3 mutant chimeric viruses (Fig. 7). These results suggest that reduced susceptibility to entry inhibitors (as indicated by the increasing IC50 value) may be associated with substitutions in the V3 crown that increase replicative fitness.
FIG. 7.
Correlation between entry inhibitor sensitivity and total relative fitness for V3 crown mutant chimeric viruses. Coefficients were determined by using the PPMC.
DISCUSSION
Changes at position 318 and 319 in the V3 crown, immediately downstream of the GPGX motif, have been implicated in variable susceptibility to entry inhibitors in three separate studies (32, 44, 50). Within the HIV-1 sequence database, position 319 toggles between amino acids alanine and threonine, and to a lesser extent arginine, while the tyrosine at position 318 is less variable (Table 1). In the present study, these polymorphisms were inserted alone or in combination into three distinct envelope contexts. Clonal, chimeric viruses expressing the mutant envelopes in the background of the laboratory strain NL4-3/HXB2 were assessed in drug sensitivity and growth competition assays. In summary, virus containing the generally more frequent polymorphisms at positions 318 and 319 (318, Y; 319, A or T) in the HIV-1 subtype A or B genetic backbone had higher replicative fitness and were less sensitive to entry inhibitors than were viruses containing rare polymorphisms. The impact of polymorphisms at position 319 depended on the env sequence context, while change at position 318 altered replication in a uniform manner, independent of sequence context. Increased sensitivity to entry inhibitors was observed for a wide variety of entry inhibitors, including a CCR5 antagonist (TAK-779), chemokine analog (PSC-RANTES), anti-CCR5 antibody (2D7), native chemokines (CCL5 and CCL4), and fusion inhibitor (ENF).
The broad cross-resistance to entry inhibitors was surprising considering these mutations were present specifically in the V3 crown. The V3 region in env codes for an exposed and immunogenic loop that mediates coreceptor binding and virus entry. X-ray crystallography data on gp120 suggest that the 318 and 319 residues are in a flexible cavity just below the GPGX crown. Mutations in this V3 loop region may affect the gp120 binding affinity to and positioning on the second extracellular loop of CCR5. Weak interactions with CCR5 may enhance the antiviral activity of coreceptor inhibitors, as well as slow the HIV-1 entry process to promote ENF binding and inhibition of fusion. Even though a decrease in overall viral fitness was associated with greater sensitivity to entry inhibitors, viruses harboring these V3 polymorphisms were equivalently sensitive to inhibitors blocking the steps of reverse transcription, DNA integration, and proteolytic processing.
Although the existence of three major consensus amino acids at position 319 of all subtypes suggests flexibility in the V3 crown, natural substitutions at position 319 still altered susceptibility to entry inhibitors and modulated replication capacity (Fig. 5 and 6 and Table 3). The relative impact of the 319-amino-acid change was, however, dependent on the envelope sequence context. Threonine at position 319 in the NL4-3-V3A1-92RW009 had a higher replicative fitness than the more frequent, wild-type alanine (Fig. 5 and Table 1). The frequency of alanine in subtype A is not reflected in subtype B, where threonine is slightly more frequent (53% T versus 42% A). Change from the wild-type T to A at position 319 in Yu-2 caused an increase in replicative fitness (Fig. 5) and increased resistance to PSC-RANTES, ENF, and 2D7 (Fig. 6A and B and Table 3). On the other hand, a single amino acid change (A to T) in an HIV-1 sequence (B5-91US056) of the same subtype (B) and at the same position (i.e., position 319) resulted in no significant changes in replicative fitness (Fig. 4C) or entry inhibitor sensitivity (Fig. 6). The effect of an arginine at position 319 was also context dependent. The impact of this polymorphism in the subtype A V3 background was minimal (Fig. 4B), while this change in either subtype B background drastically impacted replication (Fig. 4C and D) and entry inhibitor sensitivity (Fig. 6).
The 318 position is highly conserved across all HIV-1 isolates. Change of the nearly invariable tyrosine to an arginine resulted in increased sensitivity to entry inhibitors and decreased replicative fitness (Fig. 5 and 6). The magnitude of the effect, however, was again dependent upon the envelope context. In the subtype A V3 background, the fitness of both the R318A319 and R318T319 variant was within the normal range of primary HIV-1 isolates (Fig. 4B), whereas an arginine at position 318 of Yu-2 resulted in severely debilitated viruses (Fig. 4D). Only one other study has described the potential impact of position 318 on susceptibility to entry inhibitors and provided further rationale for our analyses. An H318R substitution was selected in an HIV-1 isolate during treatment of infected hu-PBL-SCID mice with the chemokine analog NNY-RANTES (32). The H318R change appears to have emerged during NNY-RANTES treatment while the wild-type H318 was retained in control treatment. Even though our studies have indicated that the rare R318 enhances sensitivity to entry inhibitors, the R318 in the virus strain used (strain 242) could have still conferred NNY-RANTES resistance relative to the wild-type but highly unique H318. We have shown that the rare R318 virus is less fit than the common Y318 variant (represented in >99.9% of sequences) in three genetic contexts (Fig. 5). We were unable to propagate a Y318H virus, suggesting a highly defective env glycoprotein in these contexts.
We determined ex vivo replicative fitness of these V3 polymorphisms by competing chimeric HIV-1 viruses in human CD8-depleted PBMC. The relative replicative fitness values of these V3 mutant chimeric viruses, derived from competitions with a set of primary CCR5-tropic reference strains, closely matched the efficiency of host cell entry derived from single-round entry assays using envelope pseudotyped viruses (compare Fig. 3 and 5). As an example, we found that in the Yu-2 context, an R at position 318 or 319 resulted in a 1,000-fold reduction in entry efficiency (Fig. 3C) and 100-fold reduction in replicative fitness (Fig. 4C). Given the isogenic background of each virus, it appears that the natural polymorphisms at positions 318 and 319, within a specific env sequence context were specifically responsible for the differences in entry efficiency and replicative fitness. We observed a general correlation between increasing replicative fitness and decreasing sensitivity to most entry inhibitors. This observation extended to a chemokine analog (PSC-RANTES), anti-CCR5 antibody (2D7), and a fusion inhibitor (ENF) (Fig. 7). This broad increase in sensitivity to entry inhibitors was surprising considering we introduced modest changes in a variable region of the V3 crown. However, previous reports have suggested that HIV-1 strains can exhibit a broad cross-resistance to entry inhibitors (20, 41, 43). The strongest and most significant correlate of fitness was sensitivity to PSC-RANTES (r = 0.93), followed by ENF (r = 0.73) and 2D7 (r = 0.64). Correlation between PSC-RANTES sensitivity and replicative fitness with these V3 polymorphisms was somewhat surprising considering the dual inhibitory mechanism of PSC-RANTES (33). However, we hypothesize that differences in susceptibility to PSC-RANTES are more closely related to competitive CCR5 binding between drug and virus than to receptor downregulation (M. A. Lobritz and E. J. Arts, unpublished results). Increased CCR5 affinity conferred by polymorphisms in the V3 loop may increase the ability of envelope to compete with PSC-RANTES for CCR5 docking, which would be reflected in higher IC50 values. Reduced correlation between 2D7 sensitivity and fitness may be a reflection of competition between envelope and 2D7 occurring in an environment of high CCR5 expression, while envelope competes with PSC-RANTES in an environment of low CCR5 expression. We may anticipate a stronger relationship between sensitivity to 2D7 and replicative fitness if drug sensitivity assays are performed in a cell type with lower expression levels of CCR5. However, we also had fewer datum points on the V3 mutant chimeric viruses to test this relationship. Future experiments are exploring the relationship between levels of CCR5 on the cell surface versus inhibition by competitive inhibitors (e.g., 2D7) of viruses with varying fitness.
We also observed a significant relationship between replicative fitness of V3 crown polymorphisms and sensitivity to ENF (Fig. 7). This finding was predicted by previous studies indicating that mutations in the gp120 bridging sheet that affected CCR5 binding and decreased entry efficiency also resulted in increased ENF sensitivity (8, 39, 40). Susceptibility to ENF inhibition exists during a window of time beginning with assembly of hairpin intermediates of C- and N-terminal heptad repeats during CD4 binding and ending with the formation of stable six-helix bundles (39, 47). Differential coreceptor affinities, mediated by the V3 polymorphisms, likely alter the length of this ENF inhibitory window (31, 34). This slower rate of entry also reduces replicative fitness.
Although TAK-779 also inhibits HIV-1 envelope binding to CCR5, there was only a trend relating TAK-779 inhibition and replicative fitness (Fig. 7). We hypothesize that viruses with greater replicative fitness and entry efficiency (related to higher affinity for CCR5) can compete with PSC-RANTES and 2D7 for occupancy of CCR5. In the case of small allosteric molecules such as TAK-779, the inhibitor-bound CCR5 is functionally masked from viral envelope regardless of its coreceptor affinity (7, 45). Differential sensitivity of these mutant V3 chimeric viruses to allosteric compounds may then be more related to the ability of the virus to scavenge drug-free receptor at subsaturating concentrations. However, unlike other small molecule antagonists, TAK-779 binding to CCR5 does not disrupt the 2D7 epitope on the CCR5 second extracellular loop, which is the putative interaction site for the V3 crown (15), and it remains unclear how V3 crown binding to CCR5 is affected by TAK-779.
Studies examining the emergence of HIV-1 resistance to small molecule CCR5 antagonists suggest that a different evolutionary pathway for resistance may be selected in divergent env backgrounds and may be sequence dependent (1, 4, 22, 29, 55). Early in this research, it was clear that primary HIV-1 isolates displayed a wide range of sensitivities to entry inhibitors and to native β-chemokines (11, 23, 48, 50, 53). We have been unsuccessful in our attempts to select for resistance to AOP-RANTES and PSC-RANTES utilizing protocols similar to those successfully used for generating AD101, maraviroc, SCH-D, and TAK-652 resistance in vitro (22, 29, 51, 55). Resistance to small molecule CCR5 inhibitors may be related to usage of both inhibitor-bound CCR5 and enhanced binding to inhibitor-free forms of CCR5 (37, 51, 55). In the present study, we substituted in vitro selection of resistance to entry inhibitors with the careful identification of key residues in the V3 loop associated with intrinsic sensitivity to PSC-RANTES (>50-fold variation in IC50 values). “Resistance” to PSC-RANTES may have emerged in untreated primary HIV-1 isolates as an indirect consequence of virus adaptation to innate (e.g., RANTES) and acquired host selection pressures (e.g., neutralizing antibodies) (21, 46, 52). Changes in the V3 crown that reduced susceptibility to entry inhibitors also increased entry efficiency and fitness, possibly by altering the affinity of the V3 crown for CCR5. This adaptation may also provide a selective advantage for the infecting HIV-1 isolate. Although resistance to most antiretroviral drugs results in decreased fitness, these “resistant” variants may have higher replicative fitness in vitro and in vivo. Increases in CCR5 affinity and as a result, enhanced HIV-1 fitness, may be a key mechanism for HIV-1 resistance to coreceptor inhibitors.
Acknowledgments
This study was supported by NIH grants AI436402, AI49170, AI46283, and T32 GM07250. All virus work was performed in the biosafety level 2 and 3 facilities of the Case Western Reserve/University Hospitals Center for AIDS Research (AI36219).
We are grateful to the AIDS Research and Reference Reagent Program for cell lines, supplies of TAK-779, 118-d-24, and monoclonal antibodies Chessie 8 and 24-4. We further appreciate the supplies of PSC-RANTES from Oliver Hartley and thank Jonathan Karn for equipment for luciferase measurements.
Footnotes
Published ahead of print on 23 May 2007.
REFERENCES
- 1.Aarons, E. J., S. Beddows, T. Willingham, L. Wu, and R. A. Koup. 2001. Adaptation to blockade of human immunodeficiency virus type 1 entry imposed by the anti-CCR5 monoclonal antibody 2D7. Virology 287:382-390. [DOI] [PubMed] [Google Scholar]
- 2.Abacioglu, Y. H., T. R. Fouts, J. D. Laman, E. Claassen, S. H. Pincus, J. P. Moore, C. A. Roby, R. Kamin-Lewis, and G. K. Lewis. 1994. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res. Hum. Retrovir. 10:371-381. [DOI] [PubMed] [Google Scholar]
- 3.Arien, K. K., A. Abraha, M. E. Quinones-Mateu, L. Kestens, G. Vanham, and E. J. Arts. 2005. The replicative fitness of primary human immunodeficiency virus type 1 (HIV-1) group M, HIV-1 group O, and HIV-2 isolates. J. Virol. 79:8979-8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba, M., H. Miyake, X. Wang, M. Okamoto, and K. Takashima. 2007. Isolation and characterization of human immunodeficiency virus type 1 resistant to the small-molecule CCR5 antagonist TAK-652. Antimicrob. Agents Chemother. 51:707-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ball, S. C., A. Abraha, K. R. Collins, A. J. Marozsan, H. Baird, M. E. Quinones-Mateu, A. Penn-Nicholson, M. Murray, N. Richard, M. Lobritz, P. A. Zimmerman, T. Kawamura, A. Blauvelt, and E. J. Arts. 2003. Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C. J. Virol. 77:1021-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 7.Billick, E., C. Seibert, P. Pugach, T. Ketas, A. Trkola, M. J. Endres, N. J. Murgolo, E. Coates, G. R. Reyes, B. M. Baroudy, T. P. Sakmar, J. P. Moore, and S. E. Kuhmann. 2004. The differential sensitivity of human and rhesus macaque CCR5 to small-molecule inhibitors of human immunodeficiency virus type 1 entry is explained by a single amino acid difference and suggests a mechanism of action for these inhibitors. J. Virol. 78:4134-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biscone, M. J., J. L. Miamidian, J. M. Muchiri, S. S. Baik, F. H. Lee, R. W. Doms, and J. D. Reeves. 2006. Functional impact of HIV coreceptor-binding site mutations. Virology 351:226-236. [DOI] [PubMed] [Google Scholar]
- 9.Cormier, E. G., and T. Dragic. 2002. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J. Virol. 76:8953-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cormier, E. G., D. N. Tran, L. Yukhayeva, W. C. Olson, and T. Dragic. 2001. Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J. Virol. 75:5541-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Marzio, P., J. Tse, and N. R. Landau. 1998. Chemokine receptor regulation and HIV type 1 tropism in monocyte-macrophages. AIDS Res. Hum. Retrovir. 14:129-138. [DOI] [PubMed] [Google Scholar]
- 13.Doranz, B. J., Z. H. Lu, J. Rucker, T. Y. Zhang, M. Sharron, Y. H. Cen, Z. X. Wang, H. H. Guo, J. G. Du, M. A. Accavitti, R. W. Doms, and S. C. Peiper. 1997. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J. Virol. 71:6305-6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorr, P., M. Westby, S. Dobbs, P. Griffin, B. Irvine, M. Macartney, J. Mori, G. Rickett, C. Smith-Burchnell, C. Napier, R. Webster, D. Armour, D. Price, B. Stammen, A. Wood, and M. Perros. 2005. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob. Agents Chemother. 49:4721-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dragic, T., A. Trkola, D. A. Thompson, E. G. Cormier, F. A. Kajumo, E. Maxwell, S. W. Lin, W. Ying, S. O. Smith, T. P. Sakmar, and J. P. Moore. 2000. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc. Natl. Acad. Sci. USA 97:5639-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunfee, R. L., E. R. Thomas, P. R. Gorry, J. Wang, J. Taylor, K. Kunstman, S. M. Wolinsky, and D. Gabuzda. 2006. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc. Natl. Acad. Sci. USA 103:15160-15165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 18.Hu, Q., J. O. Trent, G. D. Tomaras, Z. Wang, J. L. Murray, S. M. Conolly, J. M. Navenot, A. P. Barry, M. L. Greenberg, and S. C. Peiper. 2000. Identification of ENV determinants in V3 that influence the molecular anatomy of CCR5 utilization. J. Mol. Biol. 302:359-375. [DOI] [PubMed] [Google Scholar]
- 19.Huang, C. C., M. Tang, M. Y. Zhang, S. Majeed, E. Montabana, R. L. Stanfield, D. S. Dimitrov, B. Korber, J. Sodroski, I. A. Wilson, R. Wyatt, and P. D. Kwong. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koning, F. A., C. Koevoets, T. J. van der Vorst, and H. Schuitemaker. 2005. Sensitivity of primary R5 HTV-1 to inhibition by RANTES correlates with sensitivity to small-molecule R5 inhibitors. Antivir. Ther. 10:231-237. [PubMed] [Google Scholar]
- 21.Koning, F. A., D. Kwa, B. Boeser-Nunnink, J. Dekker, J. Vingerhoed, H. Hiemstra, and H. Schuitemaker. 2003. Decreasing sensitivity to RANTES (regulated on activation, normally T cell-expressed and -secreted) neutralization of CC chemokine receptor 5-using, non-syncytium-inducing virus variants in the course of human immunodeficiency virus type 1 infection. J. Infect. Dis. 188:864-872. [DOI] [PubMed] [Google Scholar]
- 22.Kuhmann, S. E., P. Pugach, K. J. Kunstman, J. Taylor, R. L. Stanfield, A. Snyder, J. M. Strizki, J. Riley, B. M. Baroudy, I. A. Wilson, B. T. Korber, S. M. Wolinsky, and J. P. Moore. 2004. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J. Virol. 78:2790-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labrosse, B., J. L. Labernardiere, E. Dam, V. Trouplin, K. Skrabal, F. Clavel, and F. Mammano. 2003. Baseline susceptibility of primary human immunodeficiency virus type 1 to entry inhibitors. J. Virol. 77:1610-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, B., M. Sharron, C. Blanpain, B. J. Doranz, J. Vakili, P. Setoh, E. Berg, G. Liu, H. R. Guy, S. R. Durell, M. Parmentier, C. N. Chang, K. Price, M. Tsang, and R. W. Doms. 1999. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J. Biol. Chem. 274:9617-9626. [DOI] [PubMed] [Google Scholar]
- 25.Lee, B., M. Sharron, L. J. Montaner, D. Weissman, and R. W. Doms. 1999. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc. Natl. Acad. Sci. USA 96:5215-5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewin, S. R., S. Sonza, L. B. Irving, C. F. McDonald, J. Mills, and S. M. Crowe. 1996. Surface CD4 is critical to in vitro HIV infection of human alveolar macrophages. AIDS Res. Hum. Retrovir. 12:877-883. [DOI] [PubMed] [Google Scholar]
- 27.Marozsan, A. J., and E. J. Arts. 2003. Development of a yeast-based recombination cloning/system for the analysis of gene products from diverse human immunodeficiency virus type 1 isolates. J. Virol. Methods 111:111-120. [DOI] [PubMed] [Google Scholar]
- 28.Marozsan, A. J., E. Fraundorf, A. Abraha, H. Baird, D. Moore, R. Troyer, I. Nankja, and E. J. Arts. 2004. Relationships between infectious titer, capsid protein levels, and reverse transcriptase activities of diverse human immunodeficiency virus type 1 isolates. J. Virol. 78:11130-11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marozsan, A. J., S. E. Kuhmann, T. Morgan, C. Herrera, E. Rivera-Troche, S. Xu, B. M. Baroudy, J. Strizki, and J. P. Moore. 2005. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D). Virology 338:182-199. [DOI] [PubMed] [Google Scholar]
- 30.Marozsan, A. J., D. M. Moore, M. A. Lobritz, E. Fraundorf, A. Abraha, J. D. Reeves, and E. J. Arts. 2005. Differences in the fitness of two diverse wild-type human immunodeficiency virus type 1 isolates are related to the efficiency of cell binding and entry. J. Virol. 79:7121-7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mkrtchyan, S. R., R. M. Markosyan, M. T. Eadon, J. P. Moore, G. B. Melikyan, and F. S. Cohen. 2005. Ternary complex formation of human immunodeficiency virus type 1 Env, CD4, and chemokine receptor captured as an intermediate of membrane fusion. J. Virol. 79:11161-11169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mosier, D. E., G. R. Picchio, R. J. Gulizia, R. Sabbe, P. Poignard, L. Picard, R. E. Offord, D. A. Thompson, and J. Wilken. 1999. Highly potent RANTES analogues either prevent CCR5-using human immunodeficiency virus type 1 infection in vivo or rapidly select for CXCR4-using variants. J. Virol. 73:3544-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pastore, C., G. R. Picchio, F. Galimi, R. Fish, O. Hartley, R. E. Offord, and D. E. Mosier. 2003. Two mechanisms for human immunodeficiency virus type 1 inhibition by N-terminal modifications of RANTES. Antimicrob. Agents Chemother. 47:509-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Platt, E. J., J. P. Durnin, and D. Kabat. 2005. Kinetic factors control efficiencies of cell entry, efficacies of entry inhibitors, and mechanisms of adaptation of human immunodeficiency virus. J. Virol. 79:4347-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Platt, E. J., S. E. Kuhmann, P. P. Rose, and D. Kabat. 2001. Adaptive mutations in the V3 loop of gp120 enhance fusogenicity of human immunodeficiency virus type 1 and enable use of a CCR5 coreceptor that lacks the amino-terminal sulfated region. J. Virol. 75:12266-12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pugach, P., S. E. Kuhmann, J. Taylor, A. J. Marozsan, A. Snyder, T. Ketas, S. M. Wolinsky, B. T. Korber, and J. P. Moore. 2004. The prolonged culture of human immunodeficiency virus type 1 in primary lymphocytes increases its sensitivity to neutralization by soluble CD4. Virology 321:8-22. [DOI] [PubMed] [Google Scholar]
- 37.Pugach, P., A. J. Marozsan, T. J. Ketas, E. L. Landes, J. P. Moore, and S. E. Kuhmann. 2007. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology 361:212-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rangel, H. R., J. Weber, B. Chakraborty, A. Gutierrez, M. L. Marotta, M. Mirza, P. Kiser, M. A. Martinez, J. A. Este, and M. E. Quinones-Mateu. 2003. Role of the human immunodeficiency virus type 1 envelope gene in viral fitness. J. Virol. 77:9069-9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reeves, J. D., J. L. Miamidian, M. J. Biscone, F. H. Lee, N. Ahmad, T. C. Pierson, and R. W. Doms. 2004. Impact of mutations in the coreceptor binding site on human immunodeficiency virus type 1 fusion, infection, and entry inhibitor sensitivity. J. Virol. 78:5476-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Repits, J., M. Oberg, J. Esbjornsson, P. Medstrand, A. Karlsson, J. Albert, E. M. Fenyo, and M. Jansson. 2005. Selection of human immunodeficiency virus type 1 R5 variants with augmented replicative capacity and reduced sensitivity to entry inhibitors during severe immunodeficiency. J. Gen. Virol. 86:2859-2869. [DOI] [PubMed] [Google Scholar]
- 42.Rizzuto, C., and J. Sodroski. 2000. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res. Hum. Retrovir. 16:741-749. [DOI] [PubMed] [Google Scholar]
- 43.Rusert, P., H. Kuster, B. Joos, B. Misselwitz, C. Gujer, C. Leemann, M. Fischer, G. Stiegler, H. Katinger, W. C. Olson, R. Weber, L. Aceto, H. F. Gunthard, and A. Trkola. 2005. Virus isolates during acute and chronic human immunodeficiency virus type 1 infection show distinct patterns of sensitivity to entry inhibitors. J. Virol. 79:8454-8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Safarian, D., X. Carnec, F. Tsamis, F. Kajumo, and T. Dragic. 2006. An anti-CCR5 monoclonal antibody and small molecule CCR5 antagonists synergize by inhibiting different stages of human immunodeficiency virus type 1 entry. Virology 352:477-484. [DOI] [PubMed] [Google Scholar]
- 45.Seibert, C., W. Ying, S. Gavrilov, F. Tsamis, S. E. Kuhmann, A. Palani, J. R. Tagat, J. W. Clader, S. W. McCombie, B. M. Baroudy, S. O. Smith, T. Dragic, J. P. Moore, and T. P. Sakmar. 2006. Interaction of small molecule inhibitors of HIV-1 entry with CCR5. Virology 349:41-54. [DOI] [PubMed] [Google Scholar]
- 46.Stalmeijer, E. H., R. P. Van Rij, B. Boeser-Nunnink, J. A. Visser, M. A. Naarding, D. Schols, and H. Schuitemaker. 2004. In vivo evolution of X4 human immunodeficiency virus type 1 variants in the natural course of infection coincides with decreasing sensitivity to CXCR4 antagonists. J. Virol. 78:2722-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steger, H. K., and M. J. Root. 2006. Kinetic dependence to HIV-1 entry inhibition. J. Biol. Chem. 281:25813-25821. [DOI] [PubMed] [Google Scholar]
- 48.Strizki, J. M., S. Xu, N. E. Wagner, L. Wojcik, J. Liu, Y. Hou, M. Endres, A. Palani, S. Shapiro, J. W. Clader, W. J. Greenlee, J. R. Tagat, S. McCombie, K. Cox, A. B. Fawzi, C. C. Chou, C. Pugliese-Sivo, L. Davies, M. E. Moreno, D. D. Ho, A. Trkola, C. A. Stoddart, J. P. Moore, G. R. Reyes, and B. M. Baroudy. 2001. SCH-C (SCH 351125), an orally bioavailable, small molecule antagonist of the chemokine receptor CCR5, is a potent inhibitor of HIV-1 infection in vitro and in vivo. Proc. Natl. Acad. Sci. USA 98:12718-12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas, E. R., R. L. Dunfee, J. Stanton, D. Bogdan, J. Taylor, K. Kunstman, J. E. Bell, S. M. Wolinsky, and D. Gabuzda. 2007. Macrophage entry mediated by HIV Envs from brain and lymphoid tissues is determined by the capacity to use low CD4 levels and overall efficiency of fusion. Virology 360:105-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torre, V. S., A. J. Marozsan, J. L. Albright, K. R. Collins, O. Hartley, R. E. Offord, M. E. Quinones-Mateu, and E. J. Arts. 2000. Variable sensitivity of CCR5-tropic human immunodeficiency virus type 1 isolates to inhibition by RANTES analogs. J. Virol. 74:4868-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trkola, A., S. E. Kuhmann, J. M. Strizki, E. Maxwell, T. Ketas, T. Morgan, P. Pugach, S. Xu, L. Wojcik, J. Tagat, A. Palani, S. Shapiro, J. W. Clader, S. McCombie, G. R. Reyes, B. M. Baroudy, and J. P. Moore. 2002. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. USA 99:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trkola, A., H. Kuster, C. Leemann, C. Ruprecht, B. Joos, A. Telenti, B. Hirschel, R. Weber, S. Bonhoeffer, and H. F. Gunthard. 2003. Human immunodeficiency virus type 1 fitness is a determining factor in viral rebound and set point in chronic infection. J. Virol. 77:13146-13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trkola, A., W. A. Paxton, S. P. Monard, J. A. Hoxie, M. A. Siani, D. A. Thompson, L. Wu, C. R. Mackay, R. Horuk, and J. P. Moore. 1998. Genetic subtype-independent inhibition of human immunodeficiency virus type 1 replication by CC and CXC chemokines. J. Virol. 72:396-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tuttle, D. L., J. K. Harrison, C. Anders, J. W. Sleasman, and M. M. Goodenow. 1998. Expression of CCR5 increases during monocyte differentiation and directly mediates macrophage susceptibility to infection by human immunodeficiency virus type 1. J. Virol. 72:4962-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Westby, M., C. Smith-Burchnell, J. Mori, M. Lewis, M. Mosley, M. Stockdale, P. Dorr, G. Ciaramella, and M. Perros. 2007. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J. Virol. 81:2359-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]