Abstract
Simian immunodeficiency virus (SIV) SIVsmm naturally infects sooty mangabeys (SMs) and is the source virus of pathogenic infections with human immunodeficiency virus type 2 (HIV-2) and SIVmac of humans and macaques, respectively. In previous studies we characterized SIVsmm diversity in naturally SIV-infected SMs and identified nine different phylogenetic subtypes whose genetic distances are similar to those reported for the different HIV-1 group M subtypes. Here we report that, within the colony of SMs housed at the Yerkes National Primate Research Center, at least four SIVsmm subtypes cocirculate, with the vast majority of animals infected with SIVsmm subtype 1, 2, or 3, resulting in the emergence of occasional recombinant forms. While SIVsmm-infected SMs show a typically nonpathogenic course of infection, we have observed that different SIVsmm subtypes are in fact associated with specific immunologic features. Notably, while subtypes 1, 2, and 3 are associated with a very benign course of infection and preservation of normal CD4+ T-cell counts, three out of four SMs infected with subtype 5 show a significant depletion of CD4+ T cells. The fact that virus replication in SMs infected with subtype 5 is similar to that in SMs infected with other SIVsmm subtypes suggests that the subtype 5-associated CD4+ T-cell depletion is unlikely to simply reflect higher levels of virus-mediated direct killing of CD4+ T-cells. Taken together, this systematic analysis of the subtype-specific features of SIVsmm infection in natural SM hosts identifies subtype-specific differences in the pathogenicity of SIVsmm infection.
The AIDS pandemic originated from zoonotic transmission to humans of simian CD4+ T-cell-tropic lentiviruses that infect African monkey species and are collectively defined as simian immunodeficiency viruses (SIV) (12, 24, 26). In marked contrast to human immunodeficiency virus (HIV)-infected humans, who, if untreated, almost invariably develop progressive CD4+ T-cell depletion and AIDS, the African monkey species naturally infected with SIV are generally spared from any signs of disease (reviewed in references 28, 33, 54, and 64). Conversely, experimental SIV infection of nonnatural host Asian macaque species causes symptoms similar to those described for AIDS patients (simian AIDS) (reviewed in reference 32). At present, it is still unclear why SIV infection is relatively nonpathogenic in natural host monkey species but induces immunodeficiency in nonnatural or recent hosts (including humans). A better understanding of the mechanisms underlying the lack of disease in natural hosts for SIV infection will likely provide important clues as to the pathogenesis of AIDS in HIV-infected individuals (58, 59).
Among the natural hosts for SIV infection, the sooty mangabeys (SMs) (Cercocebus atys) are particularly relevant for two reasons. First, SIVsmm, the virus infecting SMs, is the root cause of the HIV type 2 (HIV-2) epidemic in humans (12, 25). Second, SIVsmm was used to generate the various rhesus macaque (Rh)-adapted SIVmac/SIVsmm viruses (i.e., SIVmac239 and SIVsmm543-3) that are commonly used for studies of AIDS pathogenesis and vaccines (reviewed in reference 32). SIVsmm-infected SMs typically maintain normal CD4+ T-cell counts and do not develop AIDS despite intensive virus replication, with levels of plasma viremia that are as high as, or even higher than, those observed in HIV-infected individuals (11, 51, 56). Therefore, unlike both HIV-infected individuals and experimentally SIV-infected Rhs, in which high levels of virus replication predict faster disease progression (31, 35, 39, 40, 47, 66), in SMs the levels of SIVsmm viral loads (VLs) do not seem to have a predictive value for the outcome of infection (11, 51, 56).
Natural SIV infection of SMs is characterized by near-normal CD4+ T-cell counts in both blood and lymph nodes, normal T-cell turnover, preserved bone marrow and thymic function, and absence of the generalized immune activation and bystander T-cell apoptosis that are typical of HIV-infected individuals (11, 56, 60). Similarly, no signs of disease progression and relatively low levels of immune activation were observed in SMs experimentally infected with uncloned SIVsmm or molecularly cloned SIVmac239 (34, 55). Taken together, these results led to the hypothesis that the attenuated immune activation in SIV-infected SMs is a mechanism that favors the preservation of CD4+ T-cell homeostasis and, thus, the nonpathogenic state of this chronic infection.
Interestingly, the SIVsmm cross-species transmission to both humans and macaques has specific epidemiological and pathogenic features, with some of the strains showing better fitness in terms of transmission to and replication in the new hosts. Only groups A and B of HIV-2 resulted in epidemics (19, 25), whereas the remaining HIV-2 groups act as epidemiological dead ends in humans, with lower pathogenicity and no documented human-to-human transmission (12, 25, 67). Similarly, SIVmac strains clearly have higher pathogenic potential than other cross-species-transmitted SIVsmm strains both in Rhs and in other macaque species, such as pig-tailed macaques or stump-tailed macaques (32, 42, 46). We and others suggested that a key factor beyond these differences in pathogenicity of cross-species-transmitted SIVsmm in the macaque host may be the serial passage in vitro and/or in vivo (4, 6, 8, 10, 30). However, the alternative hypothesis that different SIVsmm strains show intrinsic differences in pathogenicity cannot be dismissed.
The recent discovery of a significant SIVsmm diversity in naturally infected SIVs from primate centers in the United States (3) offered us the possibility to directly test this later hypothesis. In this study we have characterized the SIVsmm diversity within the large colony of naturally SIV-infected SMs housed at the Yerkes National Primate Research Center (YNPRC) and correlated the prognostic markers of disease progression (levels of CD4+ T cells, VLs, and T-cell activation) with SIVsmm lineage. We have also performed a longitudinal retrospective study of the dynamics of virus replication in naturally SIV-infected SMs from the Tulane National Primate Research Center (TNPRC) over a 20-year period. While these studies failed to reveal any significant differences between the known SIVsmm lineages in terms of virus replication or progression to AIDS, a significant association between subtype 5 infection and CD4+ T-cell depletion was observed.
MATERIALS AND METHODS
Animals.
The cross-sectional study included 110 naturally SIV-infected SMs from YNPRC and 29 from TNPRC. At least one blood sample was collected from all animals. The retrospective study involved serum samples collected over a 20-year period from 20 naturally infected SMs from the TNPRC, which were infected with six different subtypes as follows: eight with subtype 1 (AO24, EO38, EO39, EO41, GO79, M923, M927, and M939), three with subtype 2 (D177, GO80, and M946), three with subtype 3 (AO23, F102, and M942), three with subtype 4 (G930, G931, and G932), two with subtype 5 (D175 and FO98), and one with subtype 6 (D215). At least five serum samples and up to 18 serum samples were available from each SM included in this retrospective study. Moreover, for the SMs in the TNPRC colony, a prospective study of virus replication in plasma samples was carried out for 6 years to observe the dynamics of SIVsmm VLs. All SMs and Rhs housed at the YNPRC and TNPRC were maintained in accordance with NIH guidelines (44).
Nucleic acid extractions.
RNA was extracted from plasma and serum of all SIVsmm-infected SMs from TNPRC and YNPRC using the QIAamp viral RNA kit (QIAGEN, Valencia, CA), as described previously (3).
PCR and sequencing.
Nested PCR was performed to obtain amplified fragments from the gag, env, and pol regions, as described previously (3). A 793-bp gag fragment was obtained by a nested PCR protocol using GagA/GagB and GagC/GagF primers, as described previously (25). Alternatively, primers GF1/GR1 and GF2A/GR3 (13) were used in a nested PCR to generate a 909-bp fragment in the gag region. These two fragments completely overlapped. Nested primers were used for sequencing. A 438-bp fragment in the gp36 env region was obtained using a nested PCR protocol with the primers EF4/ER1 and EF5A/ER2A (12). A 602-bp pol integrase fragment was obtained using a slight variation of previously described primers used to amplify divergent SIVs (14-17). Polis4B (5′-CCA GCH CAY AAA GGW ATA GGW GGA AA-3′) and PolORB (5′-ACT GCH CCT TCH CCT TTC CA-3′) were used in the first round of amplification, and Polis4B was again used in a seminested reaction with Unipol2B (5′-CCC CTA TTC CTC CCY TTC TTT TAA-3′). PCR conditions were as previously described (3).
PCR products were purified using the QIAquick gel extraction kit or the PCR purification kit (QIAGEN, Valencia, CA) and sequenced by using direct sequencing and dye terminator methodologies (ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction kit with AmpliTaq FS DNA polymerase [Applied Biosystems, Foster City, CA]) on an automated sequencer (ABI 373, stretch model; Applied Biosystems). Sequencing was performed using the inner primers of each reaction.
Phylogenetic analysis.
The gag, pol, and env nucleotide sequence alignments were obtained from the Los Alamos National Laboratory HIV Sequence Database (http://hiv-web.lanl.gov). Newly derived SIVsmm sequences were aligned using the CLUSTALW (63) profile alignment option. The resulting alignments were adjusted manually where necessary. Regions of ambiguous alignment and all gap-containing sites were excluded.
Phylogenetic trees were inferred from the nucleotide sequence alignments by the neighbor-joining method (52) using PAUP* (62) and based upon the HKY85 model of nucleotide substitution (29). The reliability of branching order was assessed by performing 1,000 bootstrap replicates, again utilizing neighbor joining and the HKY85 model. Phylogenetic trees were also inferred by maximum likelihood using PAUP* with models inferred from the alignment using Modeltest (50). The neighbor-joining tree topology was used as the starting tree in a heuristic search employing tree-bisection-reconnection branch swapping.
Determination of plasma viral RNA.
For the cross-sectional study, SIVsmm RNA quantitation was performed by real-time PCR as follows. RNA was extracted and reverse transcribed as described previously (55). Real-time PCR was performed by amplification of 20 μl cDNA in a 50-μl reaction mixture containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 4 mM MgCl2, 0.2 μM forward primer, 0.3 μM reverse primer, 0.1 μM probe, and 5 U of AmpliTaq Gold DNA polymerase (reagents from Applied Biosystems, Foster City, CA). Primer and probe sequences were targeted to the 5′ untranslated region of the SIVsmm genome: forward primer, 5′-GGCAGG AAAATCCCTAGCAG-3′; reverse primer, 5′-GCCCTTACTGCCTTCACTCA-3′; probe, 5′-(6-carboxyfluorescein)-AGTCCCTGTTCRGGCGCCAA-(6-carboxyltetramethylrhodamine). Amplicon accumulation was monitored with an ABI PRISM 7700 sequence detection system (Applied Biosystems) with the following cycling conditions: 50°C for 2 min, 95°C for 10 min, 40 cycles at 93°C for 30 s, and 59.5°C for 1 min. RNA copy number was determined by comparison to an external standard curve consisting of in vitro transcripts representing bases 216 to 2106 of the SIVmac239 genome.
For the retrospective study of SIVsmm virus replication dynamics in naturally infected SMs, VL was measured on serum stored at −70°C and thawed only once before testing. For the prospective study, VLs were quantified on fresh plasma samples using the branched DNA (bDNA) assay (Bayer Reference Testing Laboratory, Emeryville, CA) that we have previously reported to properly detect different SIVsmm lineages (2, 3).
Lymphocyte studies and flow cytometry.
Whole blood was stained using a whole-blood lysis technique as previously described (48). Monoclonal antibodies for flow cytometry which were originally designed to detect human molecules have been shown by us and others to be cross-reactive with SMs (55, 56, 65). The antibodies used were anti-CD4-phycoerythrin (clone SK3), anti-CD4-allophycocyanin (clone SK3), and anti-CD8-allophycocyanin (clone SK1) (all from Becton Dickinson, San Jose, CA) and anti-CD3-phycoerythrin (clone SP34-2) (from BD Pharmingen, San Diego, CA). Flow cytometric acquisition and analysis of samples were performed on at least 100,000 events on a FACSCalibur flow cytometer driven by the CellQuest software package (Becton Dickinson). Analysis of the acquired data was performed using FlowJo software (Tree Star, Inc., Ashland, OR). Cell blood counts were performed for each animal and each time point and were used to determine the absolute numbers of CD4+ T cells.
Statistical analyses.
VL data and CD4+ T-cell levels were displayed using the box-and-whisker plot method. In all figures displaying box plots, the box extends from the 25th percentile to the 75th percentile, with a horizontal line at the median, and the whiskers extend down to the 10th percentile and up to the 90th percentile. Each outlier is shown as an individual point outside the plots. SigmaPlot 5.0 software (SPSS, Inc., Richmond, CA) was used for box plot construction.
The Kruskal-Wallis one-way analysis of variance with the post hoc Tukey test was used to analyze the statistically significant differences in VLs and CD4+ T-cell counts between SMs infected with different SIVsmm subtypes or uninfected SMs, with P < 0.05 being considered significant.
Nucleotide sequence accession numbers.
The nucleotide sequences of the gag, pol, and env sequences from SIVsmm-infected SMs were deposited in GenBank (accession numbers EF569684 to EF569963).
RESULTS
Four distinct SIVsmm subtypes cocirculate at the YNPRC.
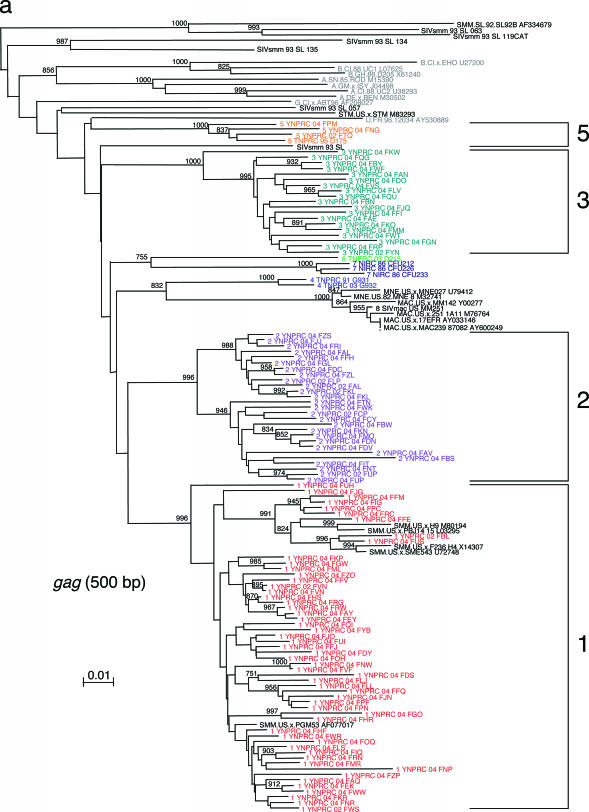
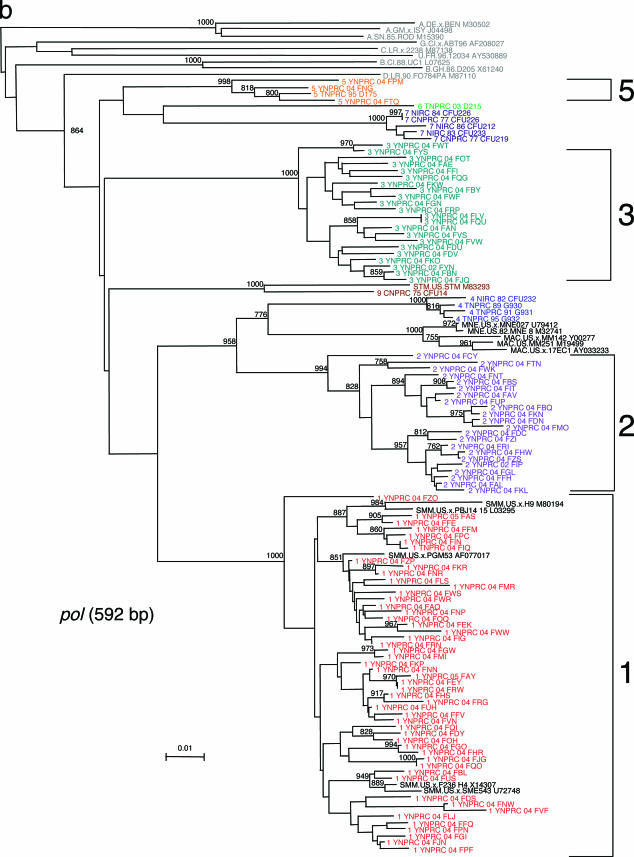
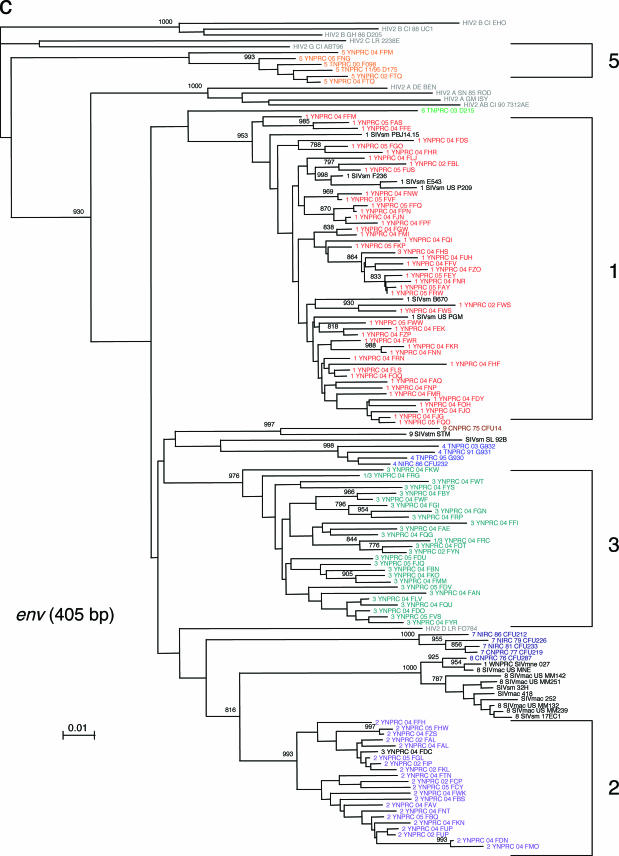
This extensive study of SIVsmm diversity in the colony of SMs housed at the YNPRC was prompted by our recent finding that SIVsmm diversity at the TNPRC is much higher than was initially believed, with six different phylogenetic lineages resulting from independent introductions of SIVsmm and now cocirculating at the TNPRC (3). Since part of the TNPRC colony was established with animals from YNPRC, in a previous study we investigated the SIVsmm diversity in the YNPRC colony and confirmed the presence of subtypes 1, 2, 3, and 5 in a small group of samples (3). To better characterize the extent of virus diversity and to evaluate its biological consequences, we have now extended this investigation to the entire colony of SIVsmm-infected SMs from YNPRC. To the best of our knowledge, this assessment of SIV diversity is the largest ever performed in a natural host species. We included in our study plasma samples from 110 SIVsmm-infected SMs from the YNPRC colony. At least one fragment was obtained in 105 SIVsmm-infected SMs. For the remaining ones, low VL levels precluded a positive PCR amplification, in spite of repeated attempts. Consistent with our previous report (3), the sequence analysis of SIVsmm samples identified subtypes 1, 2, 3, and 5 (Table 1; Fig. 1a to c). Similar to the distribution at the TNPRC, the distribution of SIVsmm subtypes at the YNPRC is uneven, with subtype 1 strains being the major viral form at YNPRC with 51% (54/105). Subtypes 2 and 3 have relatively similar prevalences, accounting for 20% (21/105) and 23% (24/105) of cases, respectively. Subtype 5 was detected in only three monkeys (4%). Interestingly, three SMs at the YNPRC showed recombinant profile patterns, as follows: FDv (2gag/3pol/3pol), FRc (1gag/3env), and FPc (1gag/1pol/2env).
TABLE 1.
SIVsmm subtype distribution in different age groups of SIVsmm-infected SMs from YNPRC
| Age (yr) | No. (%) of SMs
|
|||||
|---|---|---|---|---|---|---|
| Total | With subtype:
|
Recombinant | ||||
| 1 | 2 | 3 | 5 | |||
| <10 | 19 (18) | 9 (48) | 4 (21) | 5 (26) | 1 (5) | |
| 11-15 | 50 (48) | 26 (52) | 10 (20) | 13 (26) | 1 (2) | |
| 16-20 | 22 (21) | 12 (54) | 7 (32) | 2 (9) | 1 (5) | |
| 21-25 | 11 (10) | 6 (55) | 4 (36) | 1 (9) | ||
| >25 | 3 (3) | 1 (33) | 2 (66) | |||
| Total | 105 (100) | 54 (51) | 21 (20) | 24 (23) | 3 (3) | 3 (3) |
FIG. 1.
SIVsmm diversity in SMs from YNPRC. Subtypes are clusters of SIVs that are highly related and branch together. SIVsmm strains belonging to different subtypes are color coded: red, subtype 1; violet, subtype 2; turquoise, subtype 3; blue, subtype 4; orange, subtype 5; light green, subtype 6. Reference SIVsmm strains and strains from wild-caught SMs are shown in black, whereas HIV-2 strains are shown in light gray. The trees are based on gag (500-bp) (a), pol (592-bp) (b), and env (405-bp) (c) fragments after gap-containing sites have been removed. The phylogenetic trees were estimated by the neighbor-joining method on amino acid sequences. The reliability was estimated from 1,000 bootstrap replicates; only relevant bootstrap values are shown. The scale bar indicates amino acid replacements per site. Strain nomenclature includes the assigned lineage, the primate center of origin, the year of strain collection, and the monkey identification.
We then compared the relative prevalences of different SIVsmm subtypes among age groups (Table 1). We did not observe any significant correlation for the four subtypes cocirculating at the YNPRC, suggesting that there is no significant difference in strain transmissibility over time. Interestingly, the recombinant strains are present only in a young animal and in two very old animals. This result is not entirely unexpected, as the chances of superinfection and recombination may increase with the duration of infection.
Similar levels of in vivo viral replication between SIVsmm strains belonging to different subtypes.
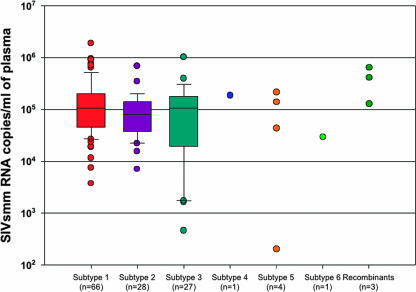
We then pooled data relative to all SIVsmm-infected SMs for which sequence data were available (i.e., both YNPRC and TNPRC) and compared the in vivo viral replication levels for the SIVsmm strains belonging to different subtypes (Fig. 2). The cross-sectional analysis showed a slight, albeit statistically significant, higher level of viral replication for subtype 1 strains (average, 203,162 SIVsmm RNA copies/ml; limits, 3,730 to 1,909,700 SIVsmm RNA copies/ml) than for subtype 2 strains (average, 109,858 SIVsmm RNA copies/ml; limits, 6,980 to 678,000 SIVsmm RNA copies/ml; P = 0.025), subtype 3 strains (average, 141,884 SIVsmm RNA copies/ml; limits, 460 to 1,010,000 SIVsmm RNA copies/ml; P = 0.025), and subtype 5 strains (average, 72,901 SIVsmm RNA copies/ml; limits, 301 to 187,000 SIVsmm RNA copies/ml; P = 0.0322). No statistically significant difference was observed for the recombinant strains (average, 185,613 SIVsmm RNA copies/ml; limits, 1,330 to 453,000 SIVsmm RNA copies/ml; P > 0.5) (Fig. 2). The slightly higher levels of VL observed in subtype 1-infected SMs might suggest either that subtype 1 strains are associated with replicative fitness in vivo (thus also explaining the higher prevalence of this subtype in both colonies) or that the VL quantification assay used is more sensitive for subtype 1 strains, i.e., the most numerous known at the time of assay design and optimization.
FIG. 2.
Cross-sectional analysis of plasma VL levels in 137 SMs infected with different SIVsmm subtypes. Subtypes are color coded as follows: red, subtype 1; violet, subtype 2; turquoise, subtype 3; blue, subtype 4; orange, subtype 5; light green, subtype 6. VL was quantified by bDNA (detection limit, 125 copies/ml) and real-time PCR (detection limit, 100 copies/ml). Box-and-whisker plots show the boxes that extend from the 25th percentile to the 75th percentile, with a horizontal line at the median, and the whiskers extend down to the 10th percentile and up to the 90th percentile. Each outlier is shown as an individual point outside the plots.
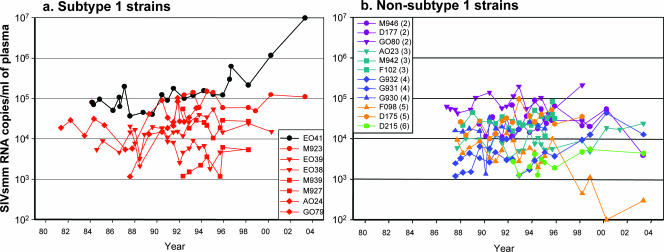
To further address these issues, we performed a retrospective study of the dynamics of VL in a subset of SIVsmm-infected SMs housed at TNPRC by using stored serum samples that were collected over a 20-year period. Due to the nature of samples, the VL levels quantified on sera were lower than those obtained by quantification in plasma. Importantly, this retrospective study failed to reveal any statistically significant difference in viral replication between subtype 1 (Fig. 3a) strains and non-subtype 1 strains (Fig. 3b). This analysis also revealed that the set point levels of virus replication are very stable over long periods of time in chronically SIVsmm-infected SMs, thus identifying another notable difference between nonpathogenic SIV infection of natural hosts and HIV infection of humans, where virus replication tends to increase during the course of infection.
FIG. 3.
Retrospective analysis of the dynamics of VL in SMs infected with different SIVsmm subtypes over a 20-year period showed no statistically significant difference in viral replication between subtype 1 (a) and non-subtype 1 (b) infections. Subtypes are color coded as follows: red, subtype 1; violet, subtype 2; turquoise, subtype 3; blue, subtype 4; orange, subtype 5; light green, subtype 6. Black lines show the dynamics of VLs in an SM that progressed to AIDS (36). VL was quantified by bDNA (detection limit, 125 copies/ml) on serum samples stored at −80°C.
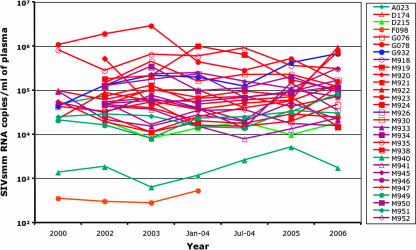
As quantification of SIVsmm VL in serum samples may not accurately reflect the actual level of in vivo viral replication, we also collected and analyzed a set of longitudinal data in SIVsmm-infected SMs from the TNPRC in which the level of viral replication was measured in freshly collected plasma samples over a 6-year period. As shown in Fig. 4, VL levels quantified on fresh plasma exhibited remarkably well-conserved patterns and confirmed the lack of any significant difference between the different SIVsmm subtypes with respect to trends of viral replication over time.
FIG. 4.
Prospective analysis of the dynamics of viral replication of different SIVsmm subtypes over 6 years. No statistical difference in VLs between subtype 1 (red), subtype 2 (violet); subtype 3 (turquoise), subtype 4 (blue), subtype 5 (orange), and subtype 6 (light green) was seen. VL was quantified by bDNA (detection limit, 125 copies/ml).
SIVsmm subtype 5 infection is associated with significant in vivo CD4+ T-cell depletion.
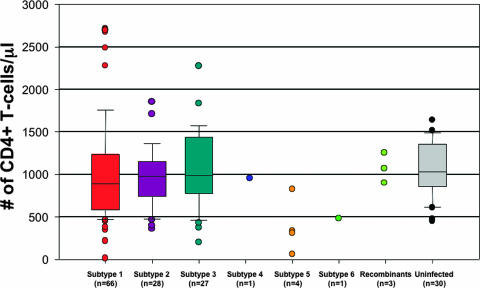
To then determine whether infection with different SIVsmm subtypes is associated with specific immunological features, we next compared the levels of CD4+ T cells in the SMs infected with different SIVsmm phylogenetic subtypes to those measured in non-SIVsmm-infected SMs. As shown in Fig. 5, no statistically significant difference was observed between uninfected SMs (average, 1,061 ± 55 CD4+ T cells/μl; limits, 453 to 1,640 CD4+ T cells/μl) and those infected with subtype 1 (average, 1,016 ± 77 CD4+ T cells/μl; limits, 3 to 2,717 CD4+ T cells/μl), subtype 2 (average, 982 ± 74 CD4+ T cells/μl; limits, 357 to 1,854 CD4+ T cells/μl), and subtype 3 (average, 1,062 ± 95 CD4+ T cells/μl; limits, 200 to 2,273 CD4+ T cells/μl). Interestingly, we found a significant difference in the CD4+ T-cell counts between uninfected SMs and those infected with subtype 5 strains (average, 407 ± 210 CD4+ T cells/μl; limits, 24 to 890 CD4+ T cells/μl) (P < 0.02). Similarly, when the CD4+ T-cell counts of subtype 5-infected SMs were compared to those of SMs infected with subtype 1, subtype 2, and subtype 3, we observed statistically significant differences (P < 0.02). Consistent with this clear trend towards lower CD4+ T-cell counts in subtype 5-infected SMs is the intriguing observation that three out of four (75%) of these animals exhibited a CD4+ T-cell count of <500/μl, while only 11.2% of the non-subtype-5-infected SMs showed a decline of CD4+ T cells below 500/μl. Note that one subtype 5-infected SM that harbored the lowest VL (<500 copies/ml) showed the most severe CD4+ T-cell depletion, with only three CD4+ T cells/μl being detected in this animal. As none of the subtype 5-infected SMs with low CD4+ T-cell counts have hitherto shown any signs of illness, the actual clinical significance of the CD4+ T-cell depletion associated with SIVsmm subtype 5 infection is still unknown.
FIG. 5.
Cross-sectional analysis of CD4+ T-cell levels in SMs infected with different SIVsmm subtypes. Subtypes are color coded: subtype 1, red; subtype 2, violet; subtype 3, turquoise; subtype 4, blue; subtype 5, orange; subtype 6, light green. Statistically significant lower CD4+ T-cell counts were observed in subtype 5-infected SMs. Box-and-whisker plots show the boxes that extend from the 25th percentile to the 75th percentile, with a horizontal line at the median, and the whiskers extend down to the 10th percentile and up to the 90th percentile. Each outlier is shown as an individual point outside the plots.
DISCUSSION
In this study we have characterized the SIVsmm diversity in the YNPRC SM colony. Based on a large set of data, we have investigated if viral diversity has any significant impact on SIVsmm pathogenesis in its natural hosts, the SMs. We did not observe any significant difference in virulence between different SIVsmm subtypes in regard to viral replication or disease progression. However, we found a significant correlation between infection with SIVsmm subtype 5 and lower CD4+ T-cell counts, suggesting that at least this virus subtype may influence the immunological course of infection in the natural SIVsmm hosts.
There are two major reasons to study SIVsmm diversity in naturally infected SMs. The first reason is that the magnitude of diversity of different SIVsmm subtypes is similar to that of HIV-1 group M subtypes (3), thus indicating that SIVsmm subtypes can be used as a model of the effect of virus diversity in the setting of studies of candidate HIV/AIDS vaccines. To this end it should be remembered that the problem of viral diversity is arguably the most formidable obstacle for the development of an effective HIV vaccine (22, 27). The second reason is to characterize the virus and host factors underlying the lack of pathogenicity of SIVsmm in SMs. As recent studies (53) indicated that viral factors, such as nef, may be involved in determining the nonpathogenic phenotype of natural SIV infection, an extensive investigation of the relationship between different SIVsmm strains and the outcome of infection may help elucidate the role of virus-related factors in SMs.
In humans, the study of HIV subtype distribution allowed worldwide viral spread to be monitored: numerous studies have shown that subtype prevalence can vary significantly from country to country and within population groups and can change with time (7, 49). Numerous studies have reported that in HIV-1-infected patients, viral diversity potentially impacts the pathogenesis of infection, although such data were not always confirmed (1, 5, 7, 18, 20, 21, 57). Finally, we and others have shown that HIV diversity may have a significant impact on the efficacy of currently available diagnostic and monitoring tests as well as of antiretrovirals (5, 7, 18, 37, 38, 41, 49, 61).
In this study, we used a very large set of samples to systematically investigate the impact of viral diversity on SIV pathogenesis in its natural SM host. Our present data confirmed a preliminary study (3) in demonstrating the cocirculation of four different SIVsmm clusters at the YNPRC. We found that subtype 1, which includes reference strains B670, H9/PBj, and H4/660/543-3 (23, 30, 32, 43, 45), is the most prevalent, being responsible for approximately half of the cases (Table 1). Subtypes 2 and 3 are each found in approximately 20% of animals, thus accounting for the majority of the remaining samples. Not unexpectedly, the cocirculation of different viral subtypes resulted in the emergence of recombinant strains that we found in three SIVsmm-infected SMs. It should be noted that the current analysis relies on limited genomic information (i.e., amplification of relatively short fragments of different genomic regions); therefore, the actual number of recombinants could have been underestimated because of strains harboring the recombinant sequences in regions that were not analyzed.
To assess the circulation of SIVsmm subtypes over time, we first used the age of infected SMs as a surrogate of the time of infection; however, we found no significant differences in the relative prevalences of the different subtypes in animals of specific age groups, suggesting that the transmission of different SIVsmm subtypes was evenly distributed over time. In this perspective, the higher prevalence of recombinant strains in older animals is not surprising, as the probability of superinfection and recombination increases with the length of infection (and thus with age). The infection of a young animal with a recombinant strain is probably the result of the transmission of the recombinant form and may indicate that recombinant forms started to circulate in the colony, similar to what was described elsewhere for HIV-1 strains isolated from regions in which different subtypes cocirculate (9).
Our large cross-sectional and longitudinal analyses failed to reveal any significant difference in the in vivo replication of SIVsmm strains belonging to different subtypes. One strength of this study was the availability of a unique set of longitudinal samples that allowed us to perform long-term (i.e., up to 20 years) follow-up of the virus replication dynamics. We thus determined that, although the cross-sectional study suggested that the VLs in subtype 1-infected SMs are higher than in those infected with non-subtype 1 strains, both retrospective and prospective longitudinal studies failed to identify any subtype 1-specific features and showed similar levels of viral replication for the different strains belonging to subtypes 1 to 6 (Fig. 3 and 4). Interestingly, the retrospective analysis of VLs over a 20-year period revealed that the set point VL was peculiarly high in the SM that progressed to AIDS after an incubation period of 18 years (36), suggesting that a threshold of viral replication might predict disease progression in those rare cases of AIDS in natural hosts of SIVs.
Intriguingly, we found that SIVsmm subtype 5-infected SMs consistently displayed lower levels of CD4+ T cells than did SMs infected with other SIVsmm subtypes or uninfected animals. A recent survey of the majority of SIV-infected SMs housed at the YNPRC colony revealed that while 85 to 90% of these animals exhibit the healthy levels of CD4+ T cells previously described (56), a subset (10 to 15%) of SIV-infected SMs showed significant CD4+ T-cell depletion (60). Irrespective of these low CD4+ T-cell counts, these animals are still asymptomatic, although the recent description of a case of AIDS in a CD4-depleted, SIV-infected SM at the TNPRC (36) suggests that this asymptomatic phase may be temporary. In any event, the discovery of these “CD4-low” SMs identified a group of animals whose infection may represent an intermediate step between the nonprogressing infection of most SIV-infected SMs and the typical disease progression of HIV-infected individuals or SIV-infected Rhs (60). In this study we showed that, although “CD4-low” SMs were found among animals infected with diverse SIVsmm subtypes (suggesting that the CD4+ T-cell depletion is due to specific host factors), there was a strong association between infection with subtype 5 of SIVsmm and low CD4+ T-cell counts, with three out of four subtype 5-infected SMs showing levels of CD4+ T cells below 500/μl. These low CD4+ T-cell counts were not due to a down-modulation of the CD4 molecule from the cell surface, as no CD3+ cells with intracellular expression of CD4 were found (data not shown). The significance of this “CD4-low” phenotype is unknown. It should be noted, however, that in a group of experimentally infected SMs, a severe CD4+ T-cell depletion occurred in a subset of SMs, as a consequence of an expanded coreceptor usage, even though these animals remained asymptomatic after >5 years of follow-up (J. M. Milush, J. D. Reeves, S. Gordon, D. Zhou, A. Muthukumar, D. Kosub, E. Chacko, L. Giavedoni, C. Ibegbu, K. Stefano-Cole, J. Miamidian, M. Paiardini, A. P. Barry, S. Staprans, G. Silvestri, and D. L. Sodora, submitted for publication). Understanding why SIV-infected SMs only rarely develop CD4+ T-cell depletion and, perhaps even more puzzlingly, tend to avoid progression to AIDS even when their CD4+ T-cell counts reach AIDS-defining levels could be helpful to delineate the mechanisms underlying the highly variable time to AIDS in CD4+ T-cell-depleted HIV-infected humans.
In conclusion, this study of the subtype-specific features of SIVsmm infection in natural SM hosts identified, for the first time, subtype-specific differences in the pathogenicity of SIVsmm infection. It is hoped that the identification of these differences may provide guidance for further studies of HIV pathogenesis and vaccines where pathogenic SIVsmm infection of nonnatural hosts (i.e., Asian macaques) is used as an experimental model.
Acknowledgments
We thank Andrew Lackner and Preston Marx for helpful discussion. We thank Benton Lawson, Meredith Hunter, Nathalia Katz, and the veterinary and animal care staff of TNPRC and YNPRC for their service and expertise.
This work was supported by funds from grants RO1 AI065325 and P20 RR020159 (C.A.), RO1 AI52755 and RO1 AI66998 (G.S.), RO1 AI064066 and R21 AI069935 (I.P.), and P51 RR000164 (TNPRC) and RR00165 (YNPRC) from the National Institute of Allergy and Infectious Diseases and from the National Center for Research Resources.
Footnotes
Published ahead of print on 16 May 2007.
REFERENCES
- 1.Apetrei, C., D. Descamps, G. Collin, I. Loussert-Ajaka, F. Damond, M. Duca, F. Simon, and F. Brun-Vezinet. 1998. Human immunodeficiency virus type 1 subtype F reverse transcriptase sequence and drug susceptibility. J. Virol. 72:3534-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apetrei, C., B. Gormus, I. Pandrea, M. Metzger, P. ten Haaft, L. N. Martin, R. Bohm, X. Alvarez, G. Koopman, M. Murphey-Corb, R. S. Veazey, A. A. Lackner, G. Baskin, J. Heeney, and P. A. Marx. 2004. Direct inoculation of simian immunodeficiency virus from sooty mangabeys in black mangabeys (Lophocebus aterrimus): first evidence of AIDS in a heterologous African species and different pathologic outcomes of experimental infection. J. Virol. 78:11506-11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apetrei, C., A. Kaur, N. W. Lerche, M. Metzger, I. Pandrea, J. Hardcastle, S. Fakelstein, R. Bohm, J. Kohler, V. Traina-Dorge, T. Williams, S. Staprans, G. Plauche, R. S. Veazey, H. McClure, A. A. Lackner, B. Gormus, D. L. Robertson, and P. A. Marx. 2005. Molecular epidemiology of SIVsm in U.S. primate centers unravels the origin of SIVmac and SIVstm. J. Virol. 79:8991-9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apetrei, C., N. W. Lerche, I. Pandrea, B. Gormus, M. Metzger, G. Silvestri, A. Kaur, R. Bohm, D. L. Robertson, J. Hardcastle, A. A. Lackner, and P. A. Marx. 2006. Kuru experiments triggered the emergence of pathogenic SIVmac. AIDS 20:317-321. [DOI] [PubMed] [Google Scholar]
- 5.Apetrei, C., I. Loussert-Ajaka, D. Descamps, F. Damond, S. Saragosti, F. Brun-Vezinet, and F. Simon. 1996. Lack of screening test sensitivity during HIV-1 non-subtype B seroconversions. AIDS 10:F57-F60. [DOI] [PubMed] [Google Scholar]
- 6.Apetrei, C., and P. A. Marx. 2004. Simian retroviral infections in human beings. Lancet 364:137-138. [DOI] [PubMed] [Google Scholar]
- 7.Apetrei, C., P. A. Marx, and S. M. Smith. 2004. The evolution of HIV and its consequences. Infect. Dis. Clin. N. Am. 18:369-394. [DOI] [PubMed] [Google Scholar]
- 8.Apetrei, C., D. L. Robertson, and P. A. Marx. 2004. The history of SIVs and AIDS: epidemiology, phylogeny and biology of isolates from naturally SIV infected non-human primates (NHP) in Africa. Front. Biosci. 9:225-254. [DOI] [PubMed] [Google Scholar]
- 9.Butler, I. F., I. Pandrea, P. A. Marx, and C. Apetrei. 2007. HIV genetic diversity: biological and public health consequences. Curr. HIV Res. 5:23-45. [DOI] [PubMed] [Google Scholar]
- 10.Cayabyab, M., G. B. Karlsson, B. A. Etemad-Moghadam, W. Hofmann, T. Steenbeke, M. Halloran, J. W. Fanton, M. K. Axthelm, N. L. Letvin, and J. G. Sodroski. 1999. Changes in human immunodeficiency virus type 1 envelope glycoproteins responsible for the pathogenicity of a multiply passaged simian-human immunodeficiency virus (SHIV-HXBc2). J. Virol. 73:976-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakrabarti, L. A., S. R. Lewin, L. Zhang, A. Gettie, A. Luckay, L. N. Martin, E. Skulsky, D. D. Ho, C. Cheng-Mayer, and P. A. Marx. 2000. Normal T-cell turnover in sooty mangabeys harboring active simian immunodeficiency virus infection. J. Virol. 74:1209-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, Z., A. Luckay, D. L. Sodora, P. Telfer, P. Reed, A. Gettie, J. M. Kanu, R. F. Sadek, J. Yee, D. D. Ho, L. Zhang, and P. A. Marx. 1997. Human immunodeficiency virus type 2 (HIV-2) seroprevalence and characterization of a distinct HIV-2 genetic subtype from the natural range of simian immunodeficiency virus-infected sooty mangabeys. J. Virol. 71:3953-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, Z., P. Telfer, A. Gettie, P. Reed, L. Zhang, D. D. Ho, and P. A. Marx. 1996. Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J. Virol. 70:3617-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Courgnaud, V., B. Abela, X. Pourrut, E. Mpoudi-Ngole, S. Loul, E. Delaporte, and M. Peeters. 2003. Identification of a new simian immunodeficiency virus lineage with a vpu gene present among different Cercopithecus monkeys (C. mona, C. cephus, and C. nictitans) from Cameroon. J. Virol. 77:12523-12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Courgnaud, V., P. Formenty, C. Akoua-Koffi, R. Noe, C. Boesch, E. Delaporte, and M. Peeters. 2003. Partial molecular characterization of two simian immunodeficiency viruses (SIV) from African colobids: SIVwrc from Western red colobus (Piliocolobus badius) and SIVolc from olive colobus (Procolobus verus). J. Virol. 77:744-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Courgnaud, V., X. Pourrut, F. Bibollet-Ruche, E. Mpoudi-Ngole, A. Bourgeois, E. Delaporte, and M. Peeters. 2001. Characterization of a novel simian immunodeficiency virus from guereza colobus monkeys (Colobus guereza) in Cameroon: a new lineage in the nonhuman primate lentivirus family. J. Virol. 75:857-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Courgnaud, V., M. Salemi, X. Pourrut, E. Mpoudi-Ngole, B. Abela, P. Auzel, F. Bibollet-Ruche, B. Hahn, A.-M. Vandamme, E. Delaporte, and M. Peeters. 2002. Characterization of a novel simian immunodeficiency virus with a vpu gene from greater spot-nosed monkeys (Cercopithecus nictitans) provides new insights into simian/human immunodeficiency virus phylogeny. J. Virol. 76:8298-8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Damond, F., C. Apetrei, D. Descamps, F. Brun-Vezinet, and F. Simon. 1999. HIV-1 subtypes and plasma RNA quantification. AIDS 13:286-288. [DOI] [PubMed] [Google Scholar]
- 19.Damond, F., C. Apetrei, D. L. Robertson, S. Souquiere, A. Lepretre, S. Matheron, J. C. Plantier, F. Brun-Vezinet, and F. Simon. 2001. Variability of human immunodeficiency virus type 2 (HIV-2) infecting patients living in France. Virology 280:19-30. [DOI] [PubMed] [Google Scholar]
- 20.Descamps, D., C. Apetrei, G. Collin, F. Damond, F. Simon, and F. Brun-Vezinet. 1998. Naturally occurring decreased susceptibility of HIV-1 subtype G to protease inhibitors. AIDS 12:1109-1111. [PubMed] [Google Scholar]
- 21.Descamps, D., G. Collin, F. Letourneur, C. Apetrei, F. Damond, I. Loussert-Ajaka, F. Simon, S. Saragosti, and F. Brun-Vezinet. 1997. Susceptibility of human immunodeficiency virus type 1 group O isolates to antiretroviral agents: in vitro phenotypic and genotypic analyses. J. Virol. 71:8893-8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desrosiers, R. C. 2004. Prospects for an AIDS vaccine. Nat. Med. 10:221-223. [DOI] [PubMed] [Google Scholar]
- 23.Fultz, P. N. 1994. SIVsmmPBj14: an atypical lentivirus. Curr. Top. Microbiol. Immunol. 188:65-76. [DOI] [PubMed] [Google Scholar]
- 24.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 25.Gao, F., L. Yue, D. L. Robertson, S. C. Hill, H. Hui, R. J. Biggar, A. E. Neequaye, T. M. Whelan, D. D. Ho, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1994. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J. Virol. 68:7433-7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao, F., L. Yue, A. T. White, P. G. Pappas, J. Barchue, A. P. Hanson, B. M. Greene, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 1992. Human infection by genetically diverse SIVSM-related HIV-2 in West Africa. Nature 358:495-499. [DOI] [PubMed] [Google Scholar]
- 27.Garber, D. A., G. Silvestri, and M. B. Feinberg. 2004. Prospects for an AIDS vaccine: three big questions, no easy answers. Lancet Infect. Dis. 4:397-413. [DOI] [PubMed] [Google Scholar]
- 28.Gordon, S., I. Pandrea, R. Dunham, C. Apetrei, and G. Silvestri. 2005. The call of the wild: what can be learned from studies of SIV infection of natural hosts?, p. 2-29. In T. Leitner, B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, S. Wolinsky, and B. Korber (ed.), HIV sequence compendium 2004. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM.
- 29.Hasegawa, M., H. Kishino, and T. Yano. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160-174. [DOI] [PubMed] [Google Scholar]
- 30.Hirsch, V., D. Adger-Johnson, B. Campbell, S. Goldstein, C. Brown, W. R. Elkins, and D. C. Montefiori. 1997. A molecularly cloned, pathogenic, neutralization-resistant simian immunodeficiency virus, SIVsmE543-3. J. Virol. 71:1608-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirsch, V. M., T. R. Fuerst, G. Sutter, M. W. Carroll, L. C. Yang, S. Goldstein, M. Piatak, Jr., W. R. Elkins, W. G. Alvord, D. C. Montefiori, B. Moss, and J. D. Lifson. 1996. Patterns of viral replication correlate with outcome in simian immunodeficiency virus (SIV)-infected macaques: effect of prior immunization with a trivalent SIV vaccine in modified vaccinia virus Ankara. J. Virol. 70:3741-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirsch, V. M., and P. R. Johnson. 1994. Pathogenic diversity of simian immunodeficiency viruses. Virus Res. 32:183-203. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, P. R., and V. M. Hirsch. 1991. Pathogenesis of AIDS: the non-human primate model. AIDS 5(Suppl. 2):S43-S48. [PubMed] [Google Scholar]
- 34.Kaur, A., R. M. Grant, R. E. Means, H. McClure, M. Feinberg, and R. P. Johnson. 1998. Diverse host responses and outcomes following simian immunodeficiency virus SIVmac239 infection in sooty mangabeys and rhesus macaques. J. Virol. 72:9597-9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lifson, J. D., M. A. Nowak, S. Goldstein, J. L. Rossio, A. Kinter, G. Vasquez, T. A. Wiltrout, C. Brown, D. Schneider, L. Wahl, A. L. Lloyd, J. Williams, W. R. Elkins, A. S. Fauci, and V. M. Hirsch. 1997. The extent of early viral replication is a critical determinant of the natural history of simian immunodeficiency virus infection. J. Virol. 71:9508-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ling, B., C. Apetrei, I. Pandrea, R. S. Veazey, A. A. Lackner, B. Gormus, and P. A. Marx. 2004. Classic AIDS in a sooty mangabey after an 18-year natural infection. J. Virol. 78:8902-8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loussert-Ajaka, I., T. D. Ly, M. L. Chaix, D. Ingrand, S. Saragosti, A. M. Courouce, F. Brun-Vezinet, and F. Simon. 1994. HIV-1/HIV-2 seronegativity in HIV-1 subtype O infected patients. Lancet 343:1393-1394. [DOI] [PubMed] [Google Scholar]
- 38.Mauclere, P., F. Damond, C. Apetrei, I. Loussert-Ajaka, S. Souquiere, L. Buzelay, P. Dalbon, M. Jolivet, M. Mony Lobe, F. Brun-Vezinet, F. Simon, and F. Barin. 1997. Synthetic peptide ELISAs for detection of and discrimination between group M and group O HIV type 1 infection. AIDS Res. Hum. Retrovir. 13:987-993. [DOI] [PubMed] [Google Scholar]
- 39.Mellors, J. W., A. Munoz, J. V. Giorgi, J. B. Margolick, C. J. Tassoni, P. Gupta, L. A. Kingsley, J. A. Todd, A. J. Saah, R. Detels, J. P. Phair, and C. R. Rinaldo, Jr. 1997. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann. Intern. Med. 126:946-954. [DOI] [PubMed] [Google Scholar]
- 40.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 41.Michael, N. L., S. A. Herman, S. Kwok, K. Dreyer, J. Wang, C. Christopherson, J. P. Spadoro, K. K. Young, V. Polonis, F. E. McCutchan, J. Carr, J. R. Mascola, L. L. Jagodzinski, and M. L. Robb. 1999. Development of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 and performance of an improved AMPLICOR HIV-1 MONITOR test with isolates of diverse subtypes. J. Clin. Microbiol. 37:2557-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morton, W. R., L. Kuller, R. E. Benveniste, E. A. Clark, C. C. Tsai, M. J. Gale, M. E. Thouless, J. Overbaugh, and M. G. Katze. 1989. Transmission of the simian immunodeficiency virus SIVmne in macaques and baboons. J. Med. Primatol. 18:237-245. [PubMed] [Google Scholar]
- 43.Murphey-Corb, M., L. N. Martin, S. R. Rangan, G. B. Baskin, B. J. Gormus, R. H. Wolf, W. A. Andes, M. West, and R. C. Montelaro. 1986. Isolation of an HTLV-III-related retrovirus from macaques with simian AIDS and its possible origin in asymptomatic mangabeys. Nature 321:435-437. [DOI] [PubMed] [Google Scholar]
- 44.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC.
- 45.Novembre, F. J., J. De Rosayro, S. P. O'Neil, D. C. Anderson, S. A. Klumpp, and H. M. McClure. 1998. Isolation and characterization of a neuropathogenic simian immunodeficiency virus derived from a sooty mangabey. J. Virol. 72:8841-8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Novembre, F. J., V. M. Hirsch, H. M. McClure, P. N. Fultz, and P. R. Johnson. 1992. SIV from stump-tailed macaques: molecular characterization of a highly transmissible primate lentivirus. Virology 186:783-787. [DOI] [PubMed] [Google Scholar]
- 47.Nowak, M. A., A. L. Lloyd, G. M. Vasquez, T. A. Wiltrout, L. M. Wahl, N. Bischofberger, J. Williams, A. Kinter, A. S. Fauci, V. M. Hirsch, and J. D. Lifson. 1997. Viral dynamics of primary viremia and antiretroviral therapy in simian immunodeficiency virus infection. J. Virol. 71:7518-7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pandrea, I., C. Apetrei, J. Dufour, N. Dillon, J. Barbercheck, M. Metzger, B. Jacquelin, R. Bohm, P. A. Marx, F. Barre-Sinoussi, V. M. Hirsch, M. C. Müller-Trutwin, A. A. Lackner, and R. S. Veazey. 2006. Simian immunodeficiency virus SIVagm.sab infection of Caribbean African green monkeys: new model of the study of SIV pathogenesis in natural hosts. J. Virol. 80:4858-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peeters, M., and P. M. Sharp. 2000. Genetic diversity of HIV-1: the moving target. AIDS 14(Suppl. 3):S129-S140. [PubMed] [Google Scholar]
- 50.Posada, D., and K. A. Crandall. 2001. Selecting the best-fit model of nucleotide substitution. Syst. Biol. 50:580-601. [PubMed] [Google Scholar]
- 51.Rey-Cuille, M. A., J. L. Berthier, M. C. Bomsel-Demontoy, Y. Chaduc, L. Montagnier, A. G. Hovanessian, and L. A. Chakrabarti. 1998. Simian immunodeficiency virus replicates to high levels in sooty mangabeys without inducing disease. J. Virol. 72:3872-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 53.Schindler, M., J. Munch, O. Kutsch, H. Li, M. L. Santiago, F. Bibollet-Ruche, M. C. Muller-Trutwin, F. J. Novembre, M. Peeters, V. Courgnaud, E. Bailes, P. Roques, D. L. Sodora, G. Silvestri, P. M. Sharp, B. H. Hahn, and F. Kirchhoff. 2006. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell 125:1055-1067. [DOI] [PubMed] [Google Scholar]
- 54.Silvestri, G. 2005. Naturally SIV-infected sooty mangabeys: are we closer to understanding why they do not develop AIDS? J. Med. Primatol. 34:243-252. [DOI] [PubMed] [Google Scholar]
- 55.Silvestri, G., A. Fedanov, S. Germon, N. Kozyr, W. J. Kaiser, D. A. Garber, H. McClure, M. B. Feinberg, and S. I. Staprans. 2005. Divergent host responses during primary simian immunodeficiency virus SIVsm infection of natural sooty mangabey and nonnatural rhesus macaque hosts. J. Virol. 79:4043-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silvestri, G., D. L. Sodora, R. A. Koup, M. Paiardini, S. P. O'Neil, H. M. McClure, S. I. Staprans, and M. B. Feinberg. 2003. Nonpathogenic SIV infection of sooty mangabeys is characterized by limited bystander immunopathology despite chronic high-level viremia. Immunity 18:441-452. [DOI] [PubMed] [Google Scholar]
- 57.Simon, F., S. Souquiere, F. Damond, A. Kfutwah, M. Makuwa, E. Leroy, P. Rouquet, J. L. Berthier, J. Rigoulet, A. Lecu, P. T. Telfer, I. Pandrea, J. C. Plantier, F. Barre-Sinoussi, P. Roques, M. C. Muller-Trutwin, and C. Apetrei. 2001. Synthetic peptide strategy for the detection of and discrimination among highly divergent primate lentiviruses. AIDS Res. Hum. Retrovir. 17:937-952. [DOI] [PubMed] [Google Scholar]
- 58.Stebbing, J., B. Gazzard, and D. C. Douek. 2004. Where does HIV live? N. Engl. J. Med. 350:1872-1880. [DOI] [PubMed] [Google Scholar]
- 59.Stevenson, M. 2003. HIV-1 pathogenesis. Nat. Med. 9:853-860. [DOI] [PubMed] [Google Scholar]
- 60.Sumpter, B., R. Dunham, S. Gordon, J. Engram, M. Hennessy, A. Kinter, M. Paiardini, B. Cervasi, N. Klatt, H. McClure, J. M. Milush, S. Staprans, D. L. Sodora, and G. Silvestri. 2007. Correlates of preserved CD4+ T cell homeostasis during natural, nonpathogenic simian immunodeficiency virus infection of sooty mangabeys: implications for AIDS pathogenesis. J. Immunol. 178:1680-1691. [DOI] [PubMed] [Google Scholar]
- 61.Swanson, P., V. Soriano, S. G. Devare, and J. Hackett, Jr. 2001. Comparative performance of three viral load assays on human immunodeficiency virus type 1 (HIV-1) isolates representing group M (subtypes A to G) and group O: LCx HIV RNA quantitative, AMPLICOR HIV-1 MONITOR version 1.5, and Quantiplex HIV-1 RNA version 3.0. J. Clin. Microbiol. 39:862-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swofford, D. 1999. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 63.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.VandeWoude, S., and C. Apetrei. 2006. Going wild: lessons from T-lymphotropic naturally occurring lentiviruses. Clin. Microbiol. Rev. 19:728-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Veazey, R., B. Ling, I. Pandrea, H. McClure, A. Lackner, and P. Marx. 2003. Decreased CCR5 expression on CD4+ T cells of SIV-infected sooty mangabeys. AIDS Res. Hum. Retrovir. 19:227-233. [DOI] [PubMed] [Google Scholar]
- 66.Watson, A., J. Ranchalis, B. Travis, J. McClure, W. Sutton, P. R. Johnson, S. L. Hu, and N. L. Haigwood. 1997. Plasma viremia in macaques infected with simian immunodeficiency virus: plasma viral load early in infection predicts survival. J. Virol. 71:284-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamaguchi, J., S. G. Devare, and C. A. Brennan. 2000. Identification of a new HIV-2 subtype based on phylogenetic analysis of full-length genomic sequence. AIDS Res. Hum. Retrovir. 16:925-930. [DOI] [PubMed] [Google Scholar]