Abstract
The penton base is a major capsid protein of human adenoviruses (HAdV) which forms the vertices of the capsid and interacts with hexon and fiber protein. Two hypervariable loops of the penton are exposed on the capsid surface. Sequences of these and 300 adjacent amino acid residues of all 51 HAdV and closely related simian adenoviruses were studied. Adjacent sequences and predicted overall secondary structure were conserved. Phylogenetic analysis revealed clustering corresponding to the HAdV species and recombination events in the origin of HAdV prototypes. All HAdV except serotypes 40 and 41 of species F exhibited an integrin binding RGD motif in the second loop. The lengths of the loops (HVR1 and RGD loops) varied significantly between HAdV species with the longest RGD loop observed in species C and the longest HVR1 in species B. Long loops may permit the insertion of motifs that modify tissue tropism. Genetic analysis of HAdV prime strain p17′H30, a neutralization variant of HAdV-D17, indicated the significance of nonhexon neutralization epitopes for HAdV immune escape. Fourteen highly conserved motifs of the penton base were analyzed by site-directed mutagenesis of HAdV-D8 and tested for sustained induction of early cytopathic effects. Thus, three new motifs essential for penton base function were identified additionally to the RGD site, which interacts with a secondary cellular receptor responsible for internalization. Therefore, our penton primary structure data and secondary structure modeling in combination with the recently published fiber knob sequences may permit the rational design of tissue-specific adenoviral vectors.
Adenoviridae are nonenveloped, double-stranded DNA viruses with icosahedral capsids (49). Human adenoviruses (HAdV) belong to the genus Mastadenovirus of the family Adenoviridae and are classified into six species, A to F (HAdV-A to HAdV-F). Vertex structures of the capsid of all adenoviruses are formed by the penton base protein pentameres, which are noncovalently bound to each other and to the other major structural capsid proteins hexon and fiber (44, 56). Several other minor structural proteins are involved in stabilizing the capsid, whereas major proteins carry receptor or neutralizing antibody binding sites. The fiber and penton base capsid proteins are the major players in adenovirus cell entry. Fiber knob structures interact with primary cellular receptors, which are CAR, CD46, CD86, sialic acid, and p50, depending on HAdV type (4, 45, 46). Cell entry is gained by interaction with secondary receptors followed by endocytosis (31). In most serotypes, RGD loops extending from the penton base bind to the αvβ3 or αvβ5 integrins (29, 52). The structure of the RGD loops may influence the specificity and affinity of integrin binding, thereby influencing the tropism for different cell types and probably also virulence (56). Moreover, binding of high concentrations of virus capsid or free penton proteins to the secondary receptors causes cells to detach from a monolayer in vitro. This phenomenon can be observed as early as 6 h after exposing cells to highly concentrated virus suspension and is therefore termed as “early CPE” (early cytopathic effects) (43). The early CPE may correlate well to the function of the secondary receptor and therefore be related to tropism (6).
A wide range of different diseases is caused by HAdV depending on their tissue tropism and virulence. For example, species HAdV-A and -F share the same tissue tropism and both cause gastroenteritis. HAdV-C1, -C2, and -C5 are occasionally found in gastroenteritis but infect mainly the upper respiratory tract, as is true of HAdV-B3, -B7, -B16, and -B21. By contrast, HAdV-B11, -B14, -B34, and -B35 are associated with kidney and urinary tract infections (49). HAdV have a high capability for intraspecies recombination and occasional interspecies recombination. Recombinant HAdV isolates detected by serological methods were called “intermediate strains.” Intermediate strains are recombinant viruses between two serotypes presenting hexon of one serotype and fiber knob of another serotype. Hence, the classical typing methods neutralization and hemagglutination inhibition (HI) give contradictory results. Another phenomenon of adenoviral evolution detected by neutralization testing was “prime strains” (p), which are serologically atypical HAdV strains. These strains cannot be neutralized by the reference immune sera against their respective prototype, but they induce neutralizing antibodies against their prototype strain.
The tissue tropism of HAdV types can be related partially to their phylogenetic relationships in the fiber knob region (primary receptor binding site) which was analyzed in detail recently (28). Additional insights into the tropism of adenovirus types were achieved by in vitro studies with newly developed adenovirus vectors for gene therapy which were developed for targeted therapy (18, 55). The penton base protein also plays an important role in the effectivity and specificity of human adenoviral vectors (26). However, only 21 penton base sequences were so far available. Penton phylogenetic data and structure data may help to predict the usage of secondary receptors and facilitate the design of more specific adenoviral gene therapy vectors. Recently, structural studies on HAdV-C2 penton protein and HAdV-B3 were performed (14, 56). A crystallographic model of the HAdV-C2 penton base shows the core of the structure to be a jellyroll motif, an eight-stranded antiparallel β-barrel with an extensive insertion of 308 residues (54% of the protein) between two β-strands of the jellyroll, as well as a second insertion of 54 residues between another pair of strands. The first insertion includes two external loops, one of which (the “RGD loop”) contains the integrin-binding RGD domain and is highly variable between serotypes in sequence and length. It is also highly flexible, and none of the 74 residues of the loop were modeled in the crystal structure. The other hypervariable region 1 (HVR1) loop is shorter and also projects from the outer surface of the penton base into the solvent.
Here in this study, we present penton base and deduced protein sequence data of all 51 human adenoviruses, phylogenetic analysis of the HVR1 and RGD loops, and structure predictions for the penton base proteins of all species. Moreover, the function of conserved motifs in the penton protein of HAdV-D8, which has a clear tropism for cornea cells and causes epidemic keratoconjunctivitis, was analyzed by site-directed mutagenesis. This new data set of all 51 penton base sequences will improve the specific design of adenoviral vectors.
MATERIALS AND METHODS
HAdV strains and cells.
HAdV prototype strains were obtained from the American Type Culture Collection (ATCC, Manassas, VA), except for the HAdV-B14, -D10, -D13, and -D30 prototype strains, which were obtained from our collection at the German National Reference Laboratory for Adenoviruses, Hannover Medical School, Germany, and had been previously typed with cross-neutralization. In addition, prime strain p17′H30 (1) and intermediate strain HAdV-D44H38 were included in this study and were also obtained from our collection. All viruses were propagated on A549 cells (ATCC, CCL-185) on 75-cm2 cell culture flasks. When the cytopathic effect was above 50%, the cells were frozen at −70°C, and DNA was extracted with the QIAGEN blood kit (QIAGEN, Hilden, Germany).
PCR amplification and sequencing of penton loops.
Both hypervariable loops and adjacent and interjacent conserved sequences representing approximately 900 nucleotides (nt) of the penton base gene were amplified by PCR and sequenced. The following serotypes were sequenced: HAdV-A18, -A31, -B14, -B16, -B21, -B34, -B35, -D10, -D15, -D19, -D20, -D22, -D23, -D24, -D25, -D26, -D27, -D28, -D29, -D32, -D33, -D36, -D38, -D39, -D42, -D43, -D44, -D45, -D46, -D47, -D48, -D49, and -D51. HAdV-B34, -B35, and -D46 revealed sequences identical to those already deposited in GenBank. Penton primers (Table 1) were developed based on a multiple alignment with GenBank sequences flanking both loops. PCR was performed in a total volume of 100 μl consisting of HotStar Mix (QIAGEN), 1 μM of each primer, and 3 μl of the purified HAdV DNA. The PCR program starts with activation of the “hot start” DNA polymerase for 15 min at 95°C, followed by 40 cycles consisting of denaturation at 94°C for 20 s, a primer annealing temperature depending on the species for 20 s (Table 1), and elongation at 72°C for 40 s, followed by a final extension step of 72°C for 5 min. Both strands of PCR amplicons were cycle sequenced with rhodamine-labeled dideoxynucleotide chain terminators (DNA sequencing kit; ABI, Warrington, England) and analyzed on an ABI Prism 310 automatic sequencer (Applied Biosystems). PCR primers were used to prime sequencing reactions.
TABLE 1.
Primer pairs used for amplification and sequencing of the central part of the penton base gene due to species HAdV-A, -B, -C, and -D
| HAdV sp. | Primer name: primer positions (bp) | Primer sequence (5′-3′) | Annealing temp (°C) |
|---|---|---|---|
| HAdV-A (primer positions related to HAdV-A12) | P1AL: 13493-13511 | CGCTATTTGGCTCCTACGG | 59 |
| P1AR: 14064-14045 | TCGTTAGTATACACTCCCGG | ||
| P2AL: 13994-14015 | GACACCAGAAACTTTCGTCTGG | 57 | |
| P2AR: 14515-14496 | CGGATCTTGCATCATATCGG | ||
| HAdV-B (primer positions related to HAdV-B35) | P1BL: 13747-13765 | TACGAGAGCGTGATGCAGC | 57 |
| P1BR: 14350-14330 | CGAACTTAACACCAATGTCAC | ||
| P2BL: 13978-13989 | GTGCAGAACAATGACTTTACC | 57 | |
| P2BR: 15222-15203 | AGGAACGTTTTCACTGACGG | ||
| HAdV-C (primer positions related to HAdV-C1) | P1CL: 14181-14200 | ATGTATGAGGAAGGTCCTCC | 60 |
| P1CR: 15180-15161 | TGGCATGATCGTTCATGTCC | ||
| P2CL: 14966-14985 | CCAGGAGGGCTTTAGGATCA | 60 | |
| P2CR: 15848-15829 | CAGGGCCTTGTAAACGTAGG | ||
| HAdV-D (primer positions related to HAdV-D9) | P1DL: 13724-13743 | ACTACCAAAACGACCACAGC | 58 |
| P1DR: 14303-14323 | CCTCATACATGATTCTGAAGC | ||
| P2DL: 14276-14294 | TTCGCAAGAAGCAACCTTT | 54 | |
| P2DR: 14699-14683 | TCTTGCATGAGGTCCGG |
Sequencing of the complete HAdV-D8 penton gene and site-directed mutagenesis.
The HAdV-D8 SalI restriction fragment that contains the penton gene was cloned in the pBluescript II KS(+) vector and partially sequenced to obtain HAdV-D8 penton 5′ and 3′ sequences. HAdV-D8 DNA was amplified by PCR using HAdV-D8 penton-specific primers Pent8B (AGG CGC GCG GTG GTG TCC TCT CCT CC) and Pent8E (AAA GGT GCG ACT AGA GAG CAC GCG CG), cloned in pUC18 plasmid and sequenced by primer walking. The HAdV-D8 BamHI-SalI restriction fragment was cloned in the pQE bacterial expression vector. Highly conserved motifs of the HAdV-D8 penton (Fig. 1) were mutagenized by using the QuikChange site-directed mutagenesis kit (Stratagene, Heidelberg).
FIG. 1.
Multiple amino acid sequence alignment of the whole penton gene including one HAdV member of each species (HAdV-D8 [AJ249343], HAdV-A12 [AC_000005], HAdV-B3 [Z29487], HAdV-C2 [AC_000007], HAdV-E4 [AY594254], HAdV-F40 [M86665]) and one SAdV (SAdV-E25 [AC_000011]), indicating the boundaries of the HVR1 and the RGD loops (boxes). The dots represent identical amino acid residues and dashes represent deletions compared to the query sequence HAdV-D8. In addition, the highly conserved motifs, which were chosen for direct mutagenesis, where marked in bold and underlined in the multiple alignment. The mutagenized amino acid sequence is shown in black above the conserved motif. Furthermore, dodecahedron binding sites were shaded gray, whereas fiber or interpenton binding sites were marked with black.
Expression of HAdV-D8 penton construct.
For expression of the recombinant penton base constructs in Escherichia coli (strain M15), bacteria were grown in 2× YT (yeast extract-tryptone) medium and induced with 0.05 mM isopropyl-β-d-thiogalactopyranoside for 4 h at 37°C. Cells from a 500-ml culture were harvested by centrifugation at 3,000 × g for 15 min. After being frozen and thawed, the pellets were resuspended in 4 ml 0.25% Tween 20-0.1 mM EGTA and sonicated for 6 × 30 s and cooled on ice (Ultrasonic processor XL; Heatsystems, Farmingdale, NY). The lysate was centrifuged at 13,000 × g for 10 min to pellet cell debris. The penton protein-containing supernatant was harvested. All recombinant penton proteins were soluble and concentrated sixfold with microconcentrators (Centricon-30; Amicon). The expression of each penton protein was controlled both by denaturing and native sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 12% polyacrylamide gels. For denatured sample preparation, the lysates were suspended in sample buffer containing 7.5% SDS, 25% glycerine, 7.5% β-mercaptoethanol, and 0.75% bromphenol blue (final concentrations) and boiled prior to being loaded onto SDS-PAGE gels. For native sample preparation, the buffer contained no β-mercaptoethanol, and the samples were not boiled. The Western blotting procedure was performed by standard techniques. The gels were blotted onto nitrocellulose membranes (Gibco BRL). An antibody (RGS-His antibody; QIAGEN) directed against the RGS(H)6 epitope served as the primary antibody, and an anti-mouse antibody conjugated to alkaline phosphatase (Boehringer GmbH, Mannheim, Germany) served as the secondary antibody. The color reaction was developed with 5-bromo-4-chloro-3-indolylphosphate toluidinium (Boehringer).
Detection of early CPE.
Confluent monolayers of HeLa cells in 96-well dishes were washed with phosphate-buffered saline and overlaid with 200 μl of minimum essential medium without calf serum. Twenty microliters of phosphate buffer containing 200 ng penton base proteins was added to each well. Plates were incubated for 4 h at 37°C in a 5% CO2 atmosphere and then examined microscopically to detect early CPE.
Phylogenetic analysis.
Multiple alignment was performed by using the ClustalW algorithm implemented in the BioEdit package (version 7.0.4.1). Phylogenetic analysis was performed by using the MEGA software package (version 3.1). The phylogenetic trees were constructed with the maximum parsimony method because of the high identity between HAdV in the penton gene. Similarity plots were calculated based on a PAM250 matrix with a window size of 10 amino acids with the software Plotcon included in the EMBOSS package (38).
Modeling and structure analysis.
Models of the penton base proteins were initially produced and visualized using the Swiss-Pdb Viewer protein modeling environment (version 3.7) and Visual Molecular Dynamics VMD (version 1.8.5). Based on the crystallography penton data of HAdV-C2 (Pdb accession number, 1X9T), secondary structure (56) predictions were performed by using a structural alignment to guide the threading of model sequences onto the known molecular structures. These raw molecular models were then improved manually by moving gaps to exposed variable regions and optimizing side chain packing. The conformation of each model was further refined by energy minimization using the molecular mechanics program CHARMM (7). The Dynamite webserver (5) was used to perform a CONCOORD based simulation and principle component analysis on the resulting ensemble. The results of this were plotted as covariance web diagrams and porcupine plots. Graphic images prepared with the Swiss-Pdb Viewer protein modeling environment or Visual Molecular Dynamics VMD were exported as a POV 3.5 scene file and rendered with the Persistence of Vision Ray Tracer program (POV-Ray 2000, version 3.6).
Accession numbers.
The new generated partial penton sequences were deposited in the GenBank under the accession numbers DQ375458 to DQ375487. The complete HAdV-D8 penton sequence was deposited in the GenBank under the accession number AJ249343. The following HAdV penton sequences already available in GenBank were used for multiple alignment and phylogenetic analysis: HAdV-A12 (AC_000005), -B3 (Z29487), -B7 (AC_000018), -B11 (AC_000015), -B34 (AY737797), -B35 (AC_000019), -B50 (AY737798), -C1 (AC_000017), -C2 (AC_000007), -C5 (AC_000008), -D9 (AF217407), -D13 (AJ296009), -D17 (AF108105), -D30 (AJ296010), -D37 (AF118437), -D46 (AY875648), -E4 (AY594254), -F40 (M86665), -F41 (AF105145), and -G52 (DQ923122). The following sequences for simian adenoviruses (SAdV) already available in GenBank were also used: SAdV-1 (NC_006879), -B21 (AC_000010), -E22 (AY530876), -E23 (AY530877), -E24 (AY530878), and -E25 (AC_000011).
RESULTS
Primary structure analysis of the penton base.
A central fragment of the penton gene representing the penton base (amino acid positions 106 to 466) (Fig. 1) of all HAdV was sequenced. The deduced amino acid sequences represent the central part of the sequence including the main insertion domain and two adjacent β-strands (B and D) of the jellyroll (56). This region comprises the capsid exterior exposed parts of the penton protein including the HVR1 and RGD loops. The complete data set of the RGD loop sequences confirmed that all human adenoviruses except for the only two members of species F (HAdV-F40 and -F41) contain an integrin binding RGD motif (3, 9). The analyzed penton fragments of all 51 HAdV varied significantly in their lengths (282 to 350 amino acid residues). Amino acid insertions were observed only in the hypervariable loops, with a single exception. All members of species C and F had a single amino acid insertion at position 412 close to the RGD loop (position according to multiple sequence alignment) (Fig. 1), N in the case of species C and HAdV-F41 and G in the case of HAdV-F40.
Phylogenetic analysis of the central penton base fragment.
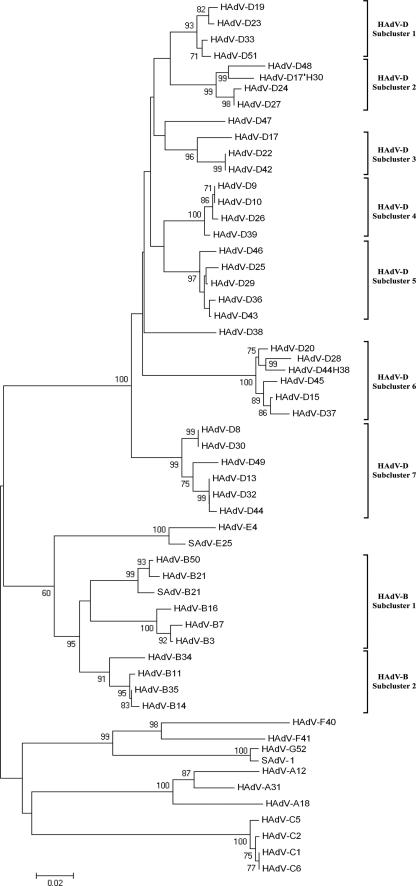
Phylogenetic analysis of the central penton base fragment (amino acid positions 106 to 466) (Fig. 1) demonstrated clustering of all HAdV in major groups corresponding to their species (Fig. 2). HAdV-E4 clustered with SAdV-E22, -E23, -E24, and -E25, with a high bootstrap value (100%; only SAdV-E25 was depicted in Fig. 2) and these together as closest neighbors to the species HAdV-B. Clustering of HAdV-B was in congruence with the subspecies B1/B2 concept supported by a 95% bootstrap value. SAdV-B21 clustered together with HAdV from the subspecies B1 (99% bootstrap value). SAdV-1 was included in our phylogenetic analysis together with the closely related new prototype HAdV-G52 (23), which is claimed to be a new HAdV species (“G”) of its own. As expected, SAdV-1 and HAdV-G52 clustered together and were very closely related to the species HAdV-F (bootstrap values 99 to 100%).
FIG. 2.
Phylogenetic analysis of the central 300 amino acid penton fragment (106 to 466; compare Fig. 1) including SAdV-1, SAdV-B21, and SAdV-E25, as well as prime strain HAdV-D17′H30 and the intermediate strain HAdV-D44H38. Seven subclusters of species HAdV-D and two subclusters of species HAdV-B were deduced from the phylogenetic analysis of the penton base protein.
Interspecies variability in the penton protein was in the range of 23% to 42% divergence (as a comparison fiber knob region: 50% to 59%). By comparing phylogenetic trees of the penton base and hexon ɛ determinant regions (28), several recombination events in the phylogeny of HAdV prototypes were discovered. For example, in the hexon protein HAdV-B14, HAdV-B34 and HAdV-B50 form one cluster, whereas these viruses do not cluster in the penton base protein. HAdV-D39 and HAdV-D43 were closely related in the hexon but 11% divergent in the penton protein.
Furthermore, we included HAdV intermediate strains and prime strains in the phylogenetic analysis. For example, the penton sequence of prime strain p17′H30, which has both intermediate and prime strain properties, is not closely related to any other HAdV prototype (4.6% divergence to HAdV-D48). Its hexon sequence (EF195772) revealed a HAdV-D17 type, whereas its fiber sequence (EF195773) revealed a HAdV-D49 type sequence. Phenotypic characterization in the HI assay was already performed when the strain was discovered (1). Then, HAdV-D49 HI sera were not available, and a reaction of p17′H30 with HAdV-D30 HI sera is not surprising, as cross HI between these types has been previously described (42). Sequencing results indicated at least a double recombination between HAdV-D17, HAdV-D49, and a so-far-unknown HAdV prototype coding for the penton base as ancestors of p17′H30. Another example is the intermediate adenovirus type HAdV-D44H38. Its hexon revealed a HAdV-D44 sequence (EF151417) with an identity of 98% to the prototype strain; the fiber gene (EF151418) had a HAdV-D38 sequence with 99% identity to the prototype strain, but the penton gene (AJ296008) was derived from a so-far-unknown HAdV, with the closest neighbor HAdV-D28 (3.6% divergence). Overall, these results indicate multiple recombination events in the origins of HAdV serotypes, with a breakpoint between the penton gene and the hexon gene.
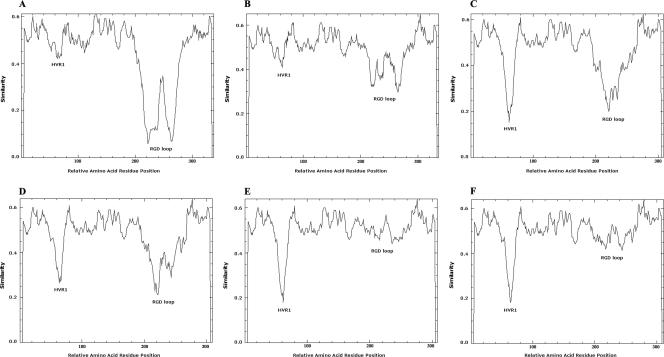
Subclusters formed by the species were examined by similarity plots in order to elucidate hot spots of evolution. In the case of species HAdV-B, subspecies B2 had only limited variability in the penton gene (Fig. 3B). The subspecies B1 cluster exhibited hypervariability in the RGD loop but showed only a limited variability of HVR1 (Fig. 3A). By contrast, an average higher genetic variability was found in HVR1 than in the RGD loop in five of the seven subclusters of species HAdV-D (Fig. 3C, E, and F) but not in subclusters 4 and 5 (Fig. 3D). Moreover, similarity plots of two subclusters, HAdV-D6 and HAdV-D7, of species D revealed an RGD loop, which was not as hypervariable as assumed (Fig. 3E and F). In species HAdV-F, HVR1 and RGD loops were hypervariable, whereas in species C, both loops had only limited variability (data not shown). Species HAdV-A showed a more variable HVR1 than RGD loop. Interestingly, HAdV-A12 was genetically distant from the other members of species A in the HVR1 and had a unique HVR1 sequence. These results showed different patterns of evolution probably caused by different immune selection pressures on the penton surface.
FIG. 3.
Similarity plots of two subclusters from species HAdV-B (A, B) and from species HAdV-D (C through F). Because subclusters HAdV-D1, HAdV-D2, and HAdV-D3 had the same pattern of similarity plots, panel C represents all viruses of these three subclusters. For the same reason, subclusters HAdV-D4 and HAdV-D5 were represented by panel D. All similarity plots were calculated with a window size of 10 amino acids. The members of each subcluster were derived from phylogenetic analysis. In the case of HAdV-D, subclusters 1, 2, and 3, comprise HAdV-D17, -D19, -D22, -D23, -D24, -D27, -D33, -D42, -D47, -D48, and -D51 (C); subclusters 4 and 5 include HAdV-D9, -D10, -D25, -D26, -D29, -D36, -D39, -D43, and -D46 (D); subcluster 6 includes HAdV-D15, -D20, -D28, -D37, and -D45 (E); and subcluster 7 includes HAdV-D8, -D13, -D30, -D32, -D44, and -D49 (F). In the case of species HAdV-B, subcluster 1 comprises HAdV-B3, -B7, -B16, -B21, and -B50 (A) and subcluster 2 comprises HAdV-B11, -B14, -B34, and -B35 (B). The HVR1 and RGD loops were labeled in the similarity plots.
Primary structure of hypervariable loops.
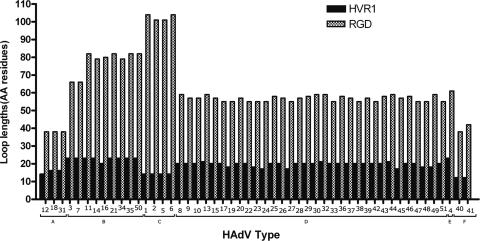
A multiple sequence alignment of all 51 penton fragments confirmed that two hypervariable loops exist in the HAdV penton base protein. Boundaries of both hypervariable loops were deduced from a similarity plot of this multiple sequence alignment (data not shown). Thus, the hypervariable RGD loop was slightly longer than the unresolved RGD loop structure in a crystallographic study of the HAdV-C2 penton base (56). Figure 4 depicts different loop lengths correlating with HAdV species. In general, the loop length of HVR1 varied between 12 and 23 amino acid residues, and the RGD loop length between 38 and 104 amino acid residues. HAdV-E4 and closely related members of the species HAdV-B (except HAdV-B16) had the longest HVR1. The shortest HVR1 loop was detected in the enterotropic species HAdV-F, which also had a short “RGD” loop (even without an RGD motif). The closely related enterotropic species HAdV-A presented also with a small RGD loop, but containing an RGD motif. By contrast, species HAdV-C had the longest RGD loop (101 to 104 residues), whereas the HVR1 of species HAdV-C had a loop length of only 14 amino acid residues (Fig. 4). Because of the growing interest in using SAdV as a backbone for adenoviral therapy vectors, GenBank sequences of SAdV-B21, a member of the species HAdV-B, and SAdV-E22 to SAdV-E25, which are members of the species HAdV-E, were included in the analysis. These SAdV have an RGD motif. The lengths of the loops corresponded to their species: SAdV-B21 HVR1 had a length of 20 amino acid residues and an RGD loop with 83 amino acid residues, both in the same range as the other members of the species HAdV-B. SAdV-E22, -E23, -E24, and -E25 had on average the same loop lengths as HAdV-E4 (HVR1 was an average of 22 amino acid residues in length, and the RGD loop was an average of 58 amino acid residues in length).
FIG. 4.
Lengths of RGD and HVR1 loops for each HAdV type. Boundaries of loops were derived from a multiple sequence alignment containing penton base sequences of all 51 HAdV types (data not shown) and are indicated in Fig. 1.
Overall structure.
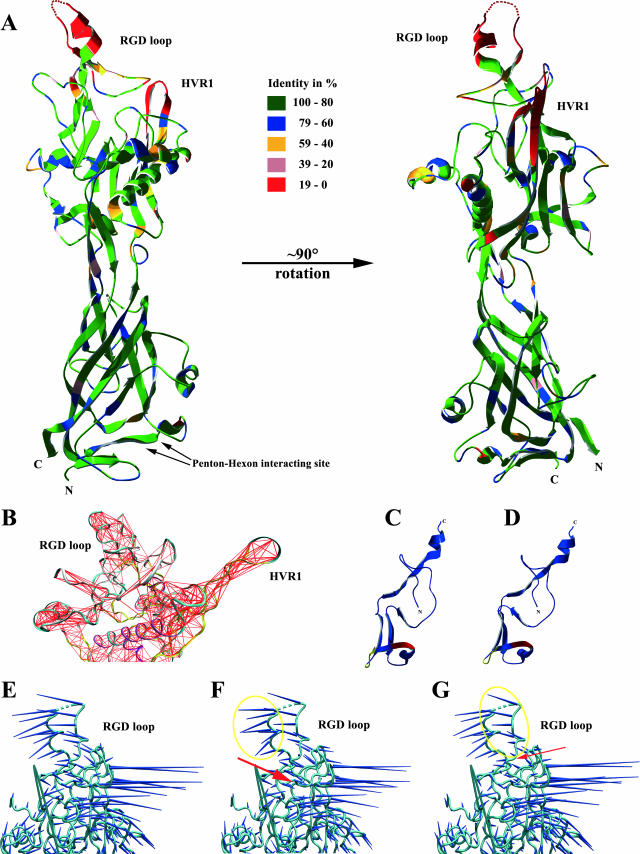
Penton protein alignments consisting of the barrel motif were predicted for all six HAdV species with the help of previously published crystallographic data for HAdV-C2 (56). Divergences from HAdV-C2 were calculated for each species. The barrel structure was predicted to be only 9.8% to 15.5% divergent from the HAdV-C2 barrel motif (Fig. 5A). These predictions should be valid, due to the conserved primary structure of the barrel motif between species. By contrast, the outer part of the remaining penton protein (insertion regions) had an amino acid sequence divergence in a range of 37.3 to 38.8% between the different species, but about half of this divergence is formed by the HVR1 and RGD loops. Secondary structures of the insertion regions excluding these HVRs were predicted for all species by structural alignment and were found to be conserved with slight differences on the outer surface of the capsid.
FIG. 5.
(A) Predicted molecular structure as ribbon diagram of HAdV-D8 penton base protein as a monomer viewed from the side and 90 degrees rotated. The colors represent the sequence identity between the 51 HAdV derived from multiple sequence alignment of the penton base gene. The arrows point to the hexon-penton interaction sites, which are highly conserved between interspecies. HVR1 and RGD loops as well as the N and C termini are labeled. (B) Covariance analysis of the penton base protein excluding the jellyroll motif. Red lines represent connections of two atoms that move together in the penton base protein. HVR1 and RGD loops have the same number of interatom connections relative to the different lengths of these two loops. Panels C and D are a ribbon representation of a part of the penton base protein, which shows amino acid residues from 240 aa to 311 aa (compare Fig. 1). Panel C represents the original HAdV-D8 sequence, and panel D represents the mutation 251 LGI→QES. The mutated site is colored red and the detergent binding site is depicted in green. (E through G) Porcupine molecular dynamics representation of the penton base excluding the highly conserved jellyroll motif. Dark blue spikes indicate relative motion. Panel E shows the original HAdV-D8 penton base, panel F shows the HAdV-D8 penton with the mutation 228 FHPDVVLL→SHPDNNQQ, and panel G shows that with the mutation 377 PDV→QNE. Spikes enclosed entirely by the yellow circle are affected by the mutation site, which is pointed to by red arrows.
In this study, we focused our analysis mainly on the penton base protein of HAdV-D8, which is clearly associated with clinically important epidemic keratoconjunctivitis and causes significant early CPE in cell cultures. First of all, the HAdV-D8 amino acid sequence was superimposed on the crystallographic structure of the HAdV-C2. Because of its high amino acid identity (68%) to HAdV-C2, the calculated three-dimensional structure of the HAdV-D8 penton protein should be an overall valid prediction (34). Like HAdV-C2, the center of the predicted HAdV-D8 penton structure, which is facing the capsid interior, is also formed by a jellyroll motif domain, an eight-stranded antiparallel β-barrel (Fig. 5A). The sequence divergence between these two jellyroll motifs is just 12.6%. By comparison, the part of the penton protein that is located on the outer surface is 36.5% divergent from HAdV-C2. Most of the structure of the loops could not be predicted due to the different length of HVR1 compared to HAdV-C2 and the lack of crystallographic data for most of the RGD loop. By examining the regions of high (>0.9) correlation identified from the molecular dynamics (Dynamite) analysis, the predicted parts of the HVR1 and RGD loops appear with high connectivity (Fig. 5B), indicative of their undergoing concerted motions. It is tempting to speculate on the basis of the results that the internal rigidity reflects the importance of these two neighboring regions. Given this and the relatively unknown role of the HVR1 loop, it is plausible that it might be involved in integrin binding in combination with the RGD loop, or alternatively, in intracellular trafficking.
Relation of conserved motifs to early CPE induction.
Induction of early CPE by the penton base protein is an indicator of functional binding of penton proteins to integrins. Site-directed mutagenesis of the HAdV-D8 penton protein was performed. E. coli-expressed penton proteins had the appropriate size of about 59 kDa on Western blotting using an antibody specific for the His tag (data not shown). Three mutations of the RGD triplet were carried out. All the mutations were chosen in such a way that they changed the biochemical properties of the selected motif to the opposite (polar to nonpolar or hydrophilic to hydrophobic). A mutation of the RGD triplet to RAE led to the loss of induction of early CPE, whereas mutagenesis of the integrin binding RGD motif to alternative integrin binding motifs (309 RGD→EILDV [22, 51] and 308 RGD→DGEA [48]) caused the retention of early CPE induction (Fig. 1 and Fig. 6).
FIG. 6.
Early CPE was induced by E. coli-expressed HAdV-D8 penton base protein (A), and mutated protein 309 RGD→RAE did not cause early CPE (B), whereas mutated protein 309 RGD→DGEA retained early CPE induction (C). Mock-incubated control cell culture (D).
Furthermore, we analyzed whether the site-directed mutagenesis of 13 highly conserved motifs that are not part of the RGD loop (Fig. 1) may influence penton base binding to integrins. Three of these highly conserved motifs (228 FHPDVVLL, 251 LGI, and 377 PDV) were essential for the induction of the early CPE. Mutation of one of these motifs (251 LGI→QES) influenced the secondary structure of a nearby detergent binding site (56) (Fig. 5C and D), probably affecting attachment to cells and also internalization. Interestingly, the other two mutations (228 FHPDVVLL→SHPDNNQQ and 377 PDV→QNE) both substituted a proline residue and were flanking the RGD loop (Fig. 1). The secondary structure of the penton, including the RGD flanking region, was not significantly influenced by these mutations, as studied by computer simulations. Analysis of concerted motions performed with Dynamite did not indicate that the alternative flanking regions have an effect on the bulk dynamics. However, analysis of molecular dynamics represented as porcupine molecular dynamics diagrams of the whole penton base protein indicated slight changes of the molecular motion of the RGD loop flanking regions (Fig. 5E through G), which might indicate that the motion of the RGD loop was reduced by mutations. Consequently, these mutations may influence the unknown molecular dynamics of the RGD loop, although this has not yet been resolved crystallographically and therefore is not available for molecular modeling.
Mutations of the remaining nine highly conserved motifs did not show any change of the early CPE. The functions of four of these highly conserved motifs (164 EGN, 188 GRQ, 340 SYN, and 447 RF) seem to be essential for fiber binding or interpenton binding, which was shown recently (56). The functions of the other four conserved regions (9 PPP, 74 HSN, 212 DPVT, and 410 PV) were recently described as essential motifs for dodecahedron penton particle structure (56). For one region (198 IGVK), it was not possible to find an explanation for why this motif is so highly conserved.
DISCUSSION
The molecular evolution of all 51 HAdV prototypes was recently studied by systematic analysis for two of the three main immunogenic determinants ɛ (hexon) and γ (fiber knob) (28). Phylogenetic analysis of the ɛ determinant revealed the positive selection of immune escape mutations resulting in a higher genetic divergence in this region than in the γ determinant (fiber knob) (28). Phylogeny of the γ determinant compared to the ɛ determinant phylogeny indicated multiple intraspecies recombination events in the origin of HAdV prototypes.
This view has now been supported by the phylogenetic analysis of the hypervariable parts of the penton base protein. The penton gene is located closer to the 5′ end of HAdV (nt 14,151 to 15,866 referring to HAdV-C2) compared to the hexon gene, which is located in the middle of the HAdV genome (nt 18,838 to 21,744 referring to HAdV-C2). Subclustering of species HAdV-D in the penton gene did not coincide with hexon subclusters, indicating multiple recombination events in the origin of HAdV with recombination crossing points between the hexon and penton base genes. Hence, new insights into complex evolution on the type and species levels including intraspecies and interspecies recombination events occurring in the 5′ half of the genome were gained. For example, the penton sequences of HAdV-D15 and HAdV-D29 were unique, whereas their ɛ determinant of the hexon was identical. Interestingly, these two prototypes were also quite different in the γ determinant (located on the fiber gene, nt 31,030 to 32,778 referring to HAdV-C2). Consequently, we derived from these findings that HAdV-D15 and HAdV-D29 have the same ancestor regarding the middle part (hexon region) of their genome. However, due to their divergent penton base and fiber knob sequences, these two prototypes are at least double recombinant viruses (Fig. 2).
Another example for recombination events between the penton and hexon coding regions in the phylogeny of human adenoviruses was demonstrated by sequencing the penton gene (AJ296013) of the HAdV prime strain p17′H30 (1, 2). This isolate was not neutralized by an antiserum against the prototype HAdV-D17, although an antiserum raised against this strain neutralized the HAdV-D17 prototype (1). Surprisingly, sequencing of the p17′H30 hexon gene revealed a high identity (99%) to the HAdV-D17 prototype, although it contains the main neutralization epitope. Therefore, the failure of the anti-HAdV-D17 prototype serum to neutralize p17′H30 suggested that this immune serum reacted mainly with neutralization epitopes other than those on the hexon epitope ɛ, probably with epitopes on the penton base of HAdV-D17 (21). These were highly divergent in the p17′H30 sequence and therefore not recognized by this immune serum. Interestingly, the penton sequence of p17′H30 was not closely related to any other penton sequence. By contrast, the antiserum raised against p17′H30 contained probably neutralizing antibodies against the HAdV-D17 hexon (ɛ) epitope, thus neutralizing the prototype HAdV-D17 (1). Moreover, immune sera raised against HAdV-D30 did not neutralize p17′H30, although p17′H30 is almost sequence identical to HAdV-D30 in the fiber knob region and also cross-reacting in heamgglutination inhibition assays (1, 2). In conclusion, these results suggest the significance of neutralization epitopes on the penton base (15).
Based on the variability and phylogenetic analysis of the penton base gene, the precise definition of the hypervariable regions of the penton gene (HVR1 and RGD loops) was feasible. The variability plots of the different phylogenetic clusters (Fig. 3) revealed different evolutionary pressure on the HVRs. On average, the variability in the short HVR1 was even higher than in the RGD loop. This may indicate that HVR1 is a target of the immune system, putatively of neutralizing antibodies and therefore under evolutionary pressure. In contrast, the RGD loop shows less variability and contains a conserved integrin binding site, but it is also variable, as it is exposed on the capsid exterior and putatively under selective pressure of the immune system. However, these hypervariable regions were flanked by highly conserved regions also exposed on the capsid exterior surface (Fig. 5A). This fact could be of interest for the induction of lymphocytes that target these regions and thus may be cross-reactive for many or even all HAdV types. This may be a promising approach for adoptive immunotherapy in case of highly immunosuppressed stem cell transplant recipients (8).
Recombinant adenoviruses are frequently used as vectors for gene delivery for therapeutic applications (18). In spite of intensive development, their utility has been limited by promiscuous tropism and the host immune response (11, 35, 54). For example, dendritic and hematopoietic cells only express low CAR or even lack CAR and thus do not sufficiently bind primary receptors on the fiber knobs of species C gene therapy vectors (19, 25). The most frequently used approach to modify the tropism of the species C HAdV vectors is the manipulation of the fiber knob domain, which is the primary receptor binding site (36). However, success of fiber modification was limited (36). Therefore, several research groups changed their directions to investigate other adenovirus species that can be used as new backbones for adenoviral vectors. Sirena et al. focused their research on adenoviral vectors based on species B human adenovirus (HAdV-B3) (47), and Ruszics et al. developed a new adenoviral vector based on a species D virus (HAdV-D19a) (40). HAdV-B3 vectors efficiently transduce human hematopoietic cell lines, and HAdV-D19a had a very promising tropism for dendritic and muscle cells (40, 50). These results support the concept that the design of adenoviral vectors should not only be focused on the fiber knob, but also include the penton base and possibly other minor structural proteins.
Penton base pseudotyping could be a feasible and effective approach. This can be derived by the analysis of the genomes from naturally occurring recombinant adenoviruses. Recently, a new intermediate HAdV isolate was described that was a result of a natural recombination from at least three HAdV prototypes (12). Although the neutralization properties of the HAdV-D22 type were retained due to its hexon gene, it gained the capability to cause epidemic keratoconjunctivitis by means of its HAdV-D8 fiber knob and its HAdV-D37 penton base. Interestingly, these ancestors are both EKC-associated HAdV types (12). Moreover, our data about prime strain p17′H30 and intermediate strain HAdV-D44H38 presented in this study demonstrated that naturally occurring recombinant adenovirus capsids consisting of hexon and penton base proteins of two different types of the same species are viable. This approach may be as advantageous for vector retargeting as using a nonspecies HAdV-C backbone if it is performed in combination with the already established fiber knob pseudotyping (17, 24, 41). However, it has not yet been demonstrated that interspecies recombinants containing HAdV-C hexon proteins and, for example, species HAdV-B or HAdV-D penton base protein are stable and viable. So far, our structure predictions for penton base proteins showed an excellent conservation of hexon-penton contact regions (Fig. 5A), suggesting the feasibility of this approach.
Another approach may be to swap only penton HVRs (HVR1 and RGD loops) between types or species. A proof of principle that modification of the penton base is a feasible strategy in the retargeting of HAdV-C5 backbone vectors was the insertion of a hemagglutinin epitope (10). Thus, a retargeted vector with a species HAdV-C backbone, which provides features like the capability of causing persistent infections in immunocompetent patients by its early genes, may be achieved (27, 30). Considering our modeling, swapping of penton base HVRs between HAdV types seems to be feasible, since the structures adjacent to the HVRs are very well conserved between the different HAdV species (Fig. 5A). However, the results of our mutagenesis experiments with HAdV-D8 (228 FHPDVVLL→SHPDNNQQ, 251 LGI→QES, and 377 PDV→QNE) (Fig. 1) indicate that minimal changes of the positions adjacent to the RGD loop may hinder the function of RGD binding.
In addition to structural data on the penton base, systematic retargeting of HAdV vectors by penton base (and fiber) swapping requires detailed knowledge of the tropism of different adenovirus types. Correlation between the disease caused by a HAdV and its cellular tropism is unfortunately not striking. For example, HAdV-D19a is well known as the causative agent of epidemic keratoconjunctivitis, which may suggest a highly specialized tropism for cornea cells. However, extensive in vitro studies revealed a high capability to transfect dendritic cells and muscle cells (40, 50). Therefore, we suggest a systematic study of the cell tropism of all 51 adenovirus types in vitro to predict their tissue tropism as gene therapy vectors. Such an approach should yield an appropriate data set for selecting penton base loop sequences and fiber knob sequences for retargeted vectors. Recently, 20 HAdV types were tested for their capability to infect soft tissue sarcoma cells (20), and HAdV-B35 was found most efficient in transfecting these cells.
Another drawback of the widely used adenoviral species C vectors is the preexisting immunity of the human population, which may be one of the main factors for their low efficacy (16, 33). Therefore, exchange of the main neutralization determinant in the hexon gene with a sequence from a nonhuman adenovirus may be more advantageous in order to circumvent problems of existing immunity. This could be achieved by merely exchanging the ɛ determinant of the hexon gene of current human adenovirus vectors. Alternatively, the backbone for a new adenoviral vector could be a SAdV (13, 37, 39) or bovine adenovirus (53) pseudotyped with human penton and fiber sequences to increase the efficacy of transfection of human cells. Recently developed bacterial artificial chromosome cloning strategies are a promising tool for constructing these vectors (32, 40).
In conclusion, this complete data set of all 51 HAdV penton base sequences in combination with the analysis of conserved motifs and structural predictions may permit the rational design of tissue-specific viral vectors in the future.
Acknowledgments
We thank Guy Schoehn for help with interpreting structural data, Benjamin A. Hall for discussing the molecular dynamics part, and Penelope Kay-Jackson and Peter Fagan for critical readings of the manuscript.
Footnotes
Published ahead of print on 23 May 2007.
REFERENCES
- 1.Adrian, T., B. Best, and R. Wigand. 1987. Serologically atypical adenovirus strains of subgenus D are different in their genome from the respective prototypes. Med. Microbiol. Immunol. 176:217-224. [DOI] [PubMed] [Google Scholar]
- 2.Adrian, T., R. Wigand, and G. Wadell. 1987. Serological and biochemical characteristics of intermediate adenovirus strains of subgenus D. Arch. Virol. 97:347-357. [DOI] [PubMed] [Google Scholar]
- 3.Albinsson, B., and A. H. Kidd. 1999. Adenovirus type 41 lacks an RGD alpha(v)-integrin binding motif on the penton base and undergoes delayed uptake in A549 cells. Virus Res. 64:125-136. [DOI] [PubMed] [Google Scholar]
- 4.Arnberg, N., P. Pring-Akerblom, and G. Wadell. 2002. Adenovirus type 37 uses sialic acid as a cellular receptor on Chang C cells. J. Virol. 76:8834-8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett, C. P., B. A. Hall, and M. E. Noble. 2004. Dynamite: a simple way to gain insight into protein motions. Acta Crystallogr. Sect. D 60:2280-2287. [DOI] [PubMed] [Google Scholar]
- 6.Boudin, M. L., M. Moncany, J. C. D'Halluin, and P. A. Boulanger. 1979. Isolation and characterization of adenovirus type 2 vertex capsomer (penton base). Virology 92:125-138. [DOI] [PubMed] [Google Scholar]
- 7.Brooks, B. R., R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comp. Chem. 4:187-217. [Google Scholar]
- 8.Chatziandreou, I., K. C. Gilmour, A. M. McNicol, M. Costabile, J. Sinclair, D. Cubitt, J. D. Campbell, C. Kinnon, W. Qasim, and H. B. Gaspar. 2007. Capture and generation of adenovirus specific T cells for adoptive immunotherapy. Br. J. Haematol. 136:117-126. [DOI] [PubMed] [Google Scholar]
- 9.Davison, A. J., E. A. Telford, M. S. Watson, K. McBride, and V. Mautner. 1993. The DNA sequence of adenovirus type 40. J. Mol. Biol. 234:1308-1316. [DOI] [PubMed] [Google Scholar]
- 10.Einfeld, D. A., D. E. Brough, P. W. Roelvink, I. Kovesdi, and T. J. Wickham. 1999. Construction of a pseudoreceptor that mediates transduction by adenoviruses expressing a ligand in fiber or penton base. J. Virol. 73:9130-9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Einfeld, D. A., and P. W. Roelvink. 2002. Advances towards targetable adenovirus vectors for gene therapy. Curr. Opin. Mol. Ther. 4:444-451. [PubMed] [Google Scholar]
- 12.Engelmann, I., I. Madisch, H. Pommer, and A. Heim. 2006. An outbreak of epidemic keratoconjunctivitis caused by a new intermediate adenovirus 22/H8 identified by molecular typing. Clin. Infect. Dis. 43:e64-e66. [DOI] [PubMed] [Google Scholar]
- 13.Farina, S. F., G. P. Gao, Z. Q. Xiang, J. J. Rux, R. M. Burnett, M. R. Alvira, J. Marsh, H. C. Ertl, and J. M. Wilson. 2001. Replication-defective vector based on a chimpanzee adenovirus. J. Virol. 75:11603-11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuschiotti, P., G. Schoehn, P. Fender, C. M. Fabry, E. A. Hewat, J. Chroboczek, R. W. Ruigrok, and J. F. Conway. 2006. Structure of the dodecahedral penton particle from human adenovirus type 3. J. Mol. Biol. 356:510-520. [DOI] [PubMed] [Google Scholar]
- 15.Gahery-Segard, H., F. Farace, D. Godfrin, J. Gaston, R. Lengagne, T. Tursz, P. Boulanger, and J. G. Guillet. 1998. Immune response to recombinant capsid proteins of adenovirus in humans: antifiber and anti-penton base antibodies have a synergistic effect on neutralizing activity. J. Virol. 72:2388-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gahery-Segard, H., V. Molinier-Frenkel, C. Le Boulaire, P. Saulnier, P. Opolon, R. Lengagne, E. Gautier, A. Le Cesne, L. Zitvogel, A. Venet, C. Schatz, M. Courtney, T. Le Chevalier, T. Tursz, J. G. Guillet, and F. Farace. 1997. Phase I trial of recombinant adenovirus gene transfer in lung cancer. Longitudinal study of the immune responses to transgene and viral products. J. Clin. Investig. 100:2218-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gall, J., A. Kass-Eisler, L. Leinwand, and E. Falck-Pedersen. 1996. Adenovirus type 5 and 7 capsid chimera: fiber replacement alters receptor tropism without affecting primary immune neutralization epitopes. J. Virol. 70:2116-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh, S. S., P. Gopinath, and A. Ramesh. 2006. Adenoviral vectors: a promising tool for gene therapy. Appl. Biochem. Biotechnol. 133:9-29. [DOI] [PubMed] [Google Scholar]
- 19.Harui, A., M. D. Roth, D. Vira, M. Sanghvi, H. Mizuguchi, and S. K. Basak. 2006. Adenoviral-encoded antigens are presented efficiently by a subset of dendritic cells expressing high levels of alpha(v)beta3 integrins. J. Leukoc. Biol. 79:1271-1278. [DOI] [PubMed] [Google Scholar]
- 20.Hoffmann, D., A. Heim, D. M. Nettelbeck, L. Steinstraesser, and O. Wildner. 2007. Evaluation of twenty human adenovirus types and one infectivity-enhanced adenovirus for the therapy of soft-tissue sarcoma. Hum. Gene Ther. 18:51-62. [DOI] [PubMed] [Google Scholar]
- 21.Hong, S. S., N. A. Habib, L. Franqueville, S. Jensen, and P. A. Boulanger. 2003. Identification of adenovirus (ad) penton base neutralizing epitopes by use of sera from patients who had received conditionally replicative ad (addl1520) for treatment of liver tumors. J. Virol. 77:10366-10375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hynes, R. O. 1992. Integrins: versatility, modulation, and signaling in cell adhesion. Cell 69:11-25. [DOI] [PubMed] [Google Scholar]
- 23.Jones, M. S., II, B. Harrach, R. D. Ganac, M. M. Gozum, W. P. Dela Cruz, B. Riedel, C. Pan, E. L. Delwart, and D. P. Schnurr. 2007. New adenovirus species found in a patient presenting with gastroenteritis. J. Virol. 81:5978-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanerva, A., M. Wang, G. J. Bauerschmitz, J. T. Lam, R. A. Desmond, S. M. Bhoola, M. N. Barnes, R. D. Alvarez, G. P. Siegal, D. T. Curiel, and A. Hemminki. 2002. Gene transfer to ovarian cancer versus normal tissues with fiber-modified adenoviruses. Mol. Ther. 5:695-704. [DOI] [PubMed] [Google Scholar]
- 25.Kim, M., K. R. Zinn, B. G. Barnett, L. A. Sumerel, V. Krasnykh, D. T. Curiel, and J. T. Douglas. 2002. The therapeutic efficacy of adenoviral vectors for cancer gene therapy is limited by a low level of primary adenovirus receptors on tumor cells. Eur. J. Cancer 38:1917-1926. [DOI] [PubMed] [Google Scholar]
- 26.Koizumi, N., K. Kawabata, F. Sakurai, Y. Watanabe, T. Hayakawa, and H. Mizuguchi. 2006. Modified adenoviral vectors ablated for coxsackievirus-adenovirus receptor, alphav integrin, and heparan sulfate binding reduce in vivo tissue transduction and toxicity. Hum. Gene Ther. 17:264-279. [DOI] [PubMed] [Google Scholar]
- 27.Korner, H., U. Fritzsche, and H. G. Burgert. 1992. Tumor necrosis factor alpha stimulates expression of adenovirus early region 3 proteins: implications for viral persistence. Proc. Natl. Acad. Sci. USA 89:11857-11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madisch, I., G. Harste, H. Pommer, and A. Heim. 2005. Phylogenetic analysis of the main neutralization and hemagglutination determinants of all human adenovirus prototypes as a basis for molecular classification and taxonomy. J. Virol. 79:15265-15276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathias, P., T. Wickham, M. Moore, and G. Nemerow. 1994. Multiple adenovirus serotypes use alpha v integrins for infection. J. Virol. 68:6811-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNees, A. L., J. A. Mahr, D. Ornelles, and L. R. Gooding. 2004. Postinternalization inhibition of adenovirus gene expression and infectious virus production in human T-cell lines. J. Virol. 78:6955-6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meier, O., and U. F. Greber. 2004. Adenovirus endocytosis. J. Gene Med. 6(Suppl. 1):S152-S163. [DOI] [PubMed] [Google Scholar]
- 32.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molnar-Kimber, K. L., D. H. Sterman, M. Chang, E. H. Kang, M. ElBash, M. Lanuti, A. Elshami, K. Gelfand, J. M. Wilson, L. R. Kaiser, and S. M. Albelda. 1998. Impact of preexisting and induced humoral and cellular immune responses in an adenovirus-based gene therapy phase I clinical trial for localized mesothelioma. Hum. Gene Ther. 9:2121-2133. [DOI] [PubMed] [Google Scholar]
- 34.Mount, D. W. 2001. Bioinformatics: sequence and genome analysis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Nemerow, G. R. 2000. Adenoviral vectors—new insights. Trends Microbiol. 8:391-394. [DOI] [PubMed] [Google Scholar]
- 36.Noureddini, S. C., and D. T. Curiel. 2005. Genetic targeting strategies for adenovirus. Mol. Pharmacol. 2:341-347. [DOI] [PubMed] [Google Scholar]
- 37.Purkayastha, A., S. E. Ditty, J. Su, J. McGraw, T. L. Hadfield, C. Tibbetts, and D. Seto. 2005. Genomic and bioinformatics analysis of HAdV-4, a human adenovirus causing acute respiratory disease: implications for gene therapy and vaccine vector development. J. Virol. 79:2559-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 39.Roy, S., Y. Zhi, G. P. Kobinger, J. Figueredo, R. Calcedo, J. R. Hiller, H. Feldmann, and J. M. Wilson. 2006. Generation of an adenoviral vaccine vector based on simian adenovirus 21. J. Gen. Virol. 87:2477-2485. [DOI] [PubMed] [Google Scholar]
- 40.Ruzsics, Z., M. Wagner, A. Osterlehner, J. Cook, U. Koszinowski, and H. G. Burgert. 2006. Transposon-assisted cloning and traceless mutagenesis of adenoviruses: development of a novel vector based on species D. J. Virol. 80:8100-8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakurai, F., H. Mizuguchi, and T. Hayakawa. 2003. Efficient gene transfer into human CD34+ cells by an adenovirus type 35 vector. Gene Ther. 10:1041-1048. [DOI] [PubMed] [Google Scholar]
- 42.Schnurr, D., and M. E. Dondero. 1993. Two new candidate adenovirus serotypes. Intervirology 36:79-83. [DOI] [PubMed] [Google Scholar]
- 43.Schrader, E., and R. Wigand. 1981. Neutralization of adenovirus infectivity and cytotoxin in various cell cultures. J. Virol. Methods 2:321-330. [DOI] [PubMed] [Google Scholar]
- 44.Shenk, T. 2001. Adenoviridae: the viruses and their replication. Lippincott-Raven Publishers, New York, NY.
- 45.Short, J. J., A. V. Pereboev, Y. Kawakami, C. Vasu, M. J. Holterman, and D. T. Curiel. 2004. Adenovirus serotype 3 utilizes CD80 (B7.1) and CD86 (B7.2) as cellular attachment receptors. Virology 322:349-359. [DOI] [PubMed] [Google Scholar]
- 46.Sirena, D., B. Lilienfeld, M. Eisenhut, S. Kalin, K. Boucke, R. R. Beerli, L. Vogt, C. Ruedl, M. F. Bachmann, U. F. Greber, and S. Hemmi. 2004. The human membrane cofactor CD46 is a receptor for species B adenovirus serotype 3. J. Virol. 78:4454-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sirena, D., Z. Ruzsics, W. Schaffner, U. F. Greber, and S. Hemmi. 2005. The nucleotide sequence and a first generation gene transfer vector of species B human adenovirus serotype 3. Virology 343:283-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Staatz, W. D., K. F. Fok, M. M. Zutter, S. P. Adams, B. A. Rodriguez, and S. A. Santoro. 1991. Identification of a tetrapeptide recognition sequence for the alpha 2 beta 1 integrin in collagen. J. Biol. Chem. 266:7363-7367. [PubMed] [Google Scholar]
- 49.Swenson, P. D., G. Wadell, A. Allard, and J. C. Hierholzer. 2003. Adenoviruses, p. 1404-1417. In P. R. Murray, E. J. Baron, M. A. Pfaller, J. H. Jorgensen, and R. A. Yolken (ed.), Manual of clinical microbiology, vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 50.Thirion, C., H. Lochmuller, Z. Ruzsics, M. Boelhauve, C. Konig, C. Thedieck, S. Kutik, C. Geiger, S. Kochanek, C. Volpers, and H. G. Burgert. 2006. Adenovirus vectors based on human adenovirus type 19a have high potential for human muscle-directed gene therapy. Hum. Gene Ther. 17:193-205. [DOI] [PubMed] [Google Scholar]
- 51.Wickham, T. J., M. E. Carrion, and I. Kovesdi. 1995. Targeting of adenovirus penton base to new receptors through replacement of its RGD motif with other receptor-specific peptide motifs. Gene Ther. 2:750-756. [PubMed] [Google Scholar]
- 52.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 73:309-319. [DOI] [PubMed] [Google Scholar]
- 53.Wu, Q., and S. K. Tikoo. 2004. Altered tropism of recombinant bovine adenovirus type-3 expressing chimeric fiber. Virus Res. 99:9-15. [DOI] [PubMed] [Google Scholar]
- 54.Young, L. S., and V. Mautner. 2001. The promise and potential hazards of adenovirus gene therapy. Gut 48:733-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young, L. S., P. F. Searle, D. Onion, and V. Mautner. 2006. Viral gene therapy strategies: from basic science to clinical application. J. Pathol. 208:299-318. [DOI] [PubMed] [Google Scholar]
- 56.Zubieta, C., G. Schoehn, J. Chroboczek, and S. Cusack. 2005. The structure of the human adenovirus 2 penton. Mol. Cell 17:121-135. [DOI] [PubMed] [Google Scholar]