Abstract
Vif is a primate lentiviral accessory protein that is crucial for viral infectivity. Vif counteracts the antiviral activity of host deaminases such as APOBEC3G and APOBEC3F. We now report a novel function of African green monkey simian immunodeficiency virus (SIVagm) Vif that promotes replication of SIVagm in human cells lacking detectable deaminase activity. We found that cyclophilin A (CypA) was excluded from wild-type SIV particles but was efficiently packaged into vif-deficient SIVagm virions. The presence of CypA in vif-defective SIVagm was correlated with reduced viral replication. Infection of CypA knockout Jurkat cells or treatment of Jurkat cells with cyclosporine A eliminated the Vif-sensitive inhibition and resulted in replication profiles that were similar for wild-type and vif-deficient SIVagm. Importantly, the inhibitory effect of CypA was restricted to virus-producing cells and was TRIM5α independent. The abilities of SIVagm Vif to inhibit encapsidation of CypA and to increase viral infectivity were shared by rhesus macaque SIV Vif and thus seem to be general properties of SIV Vif proteins. Exclusion of CypA from SIVagm particles was not associated with intracellular degradation, suggesting a mode of Vif action distinct from that proposed for APOBEC3G. This is the first report of a novel vif-sensitive antiviral activity of human CypA that may limit zoonotic transmission of SIV and the first demonstration of CypA encapsidation into a virus other than human immunodeficiency virus type 1.
Replication of primate lentiviruses is cell type specific and is controlled by a number of restriction factors. Host restriction of viral replication can occur at multiple levels, e.g., lack of the appropriate surface receptors required for viral entry or expression of internal host factors with antiviral activity. For instance, Ref1 is expressed in human and other nonmurine cells and imposes a restriction on viral replication similar to that of Fv1 in murine cells (56). Although the precise mechanism of Fv1 restriction remains unclear, the viral determinants for this type of restriction have been mapped to the capsid (CA) protein (21, 37). However, Ref1 was found to restrict retroviral replication at a step prior to reverse transcription, while Fv1 seems to impose a post-reverse transcription block (reviewed in reference 22). Another restriction factor, Lv1, was found to be responsible for restricting human immunodeficiency virus type 1 (HIV-1) but not rhesus macaque simian immunodeficiency virus (SIVmac) replication in Old World monkey cells (40). Both Ref1 and Lv1 were recently identified as tripartite interaction motif 5 alpha (TRIM5α) variants (4, 5, 26, 33, 44, 52). As with Fv1-mediated restriction, viral capsid proteins were found to be the viral determinants of TRIM5α-mediated restriction (14, 24, 26, 42, 43, 56).
Recently, cytidine deaminases were identified as a new class of antiviral factors that target retroviruses such as HIV-1 or SIV (for a review, see references 15 and 23). Most prominent among those factors is APOBEC3G (A3G), a host cytidine deaminase with potent antiviral activity whose function is sensitive to the activity of the HIV-1 Vif protein (46). Unlike TRIM5α or Fv1, A3G does not exert its antiviral activity by targeting the incoming viral capsid protein but instead is packaged into virus particles and inhibits virus replication by targeting single-stranded viral cDNA.
The function of Vif is species specific (39, 48). Accordingly, human A3G is insensitive to African green monkey SIV (SIVagm) Vif, while African green monkey A3G is insensitive to HIV-1 Vif (6, 38, 39, 45, 59). However, such species specificity is not absolute. In fact, we found that SIVagm Vif was able to support replication of SIVagm in the A3G-positive human A3.01 T-cell line. Replication of vif-defective SIVagm in A3.01 cells was severely restricted and resulted in an accumulation of cytidine deaminase-induced G-to-A mutations in the SIVagm genome (54).
In the current study, we extended our analysis of SIVagm replication in human cells. Surprisingly, we found that replication of SIVagm in A3G-negative human Jurkat T cells was still Vif dependent. Yet, vif-defective SIVagm genomes did not accumulate G-to-A mutations, suggesting that the vif-sensitive inhibition in Jurkat cells was not due to the presence of other cytidine deaminases. Interestingly, we found that cyclophilin A (CypA) was efficiently packaged into SIVagm virions in the absence of Vif but was excluded in the presence of Vif. The Vif protein-dependent increase in the infectivity of SIVagm produced from Jurkat cells was directly correlated to the presence or absence of CypA. Accordingly, SIVagm Vif was not required for full viral infectivity in CypA-knockout Jurkat cells or in normal Jurkat cells treated with cyclosporine A (CsA). Silencing of TRIM5α did not overcome the vif-sensitive inhibition of SIVagm in Jurkat cells. Finally, SIVagm Vif did not affect the intracellular stability of CypA. Our data define a novel role for SIVagm Vif in counteracting an APOBEC-independent but SIV-specific antiviral effect of CypA.
MATERIALS AND METHODS
Plasmids.
Full-length molecular clones of HIV-1 NL4-3 (2) and SIVagm9063 (27) were used for the production of wild-type infectious virus. Vif-defective variants of NL4-3 and SIVagm9063 have been described previously (32, 54). A full-length molecular clone of SIVmac239 was generated by ligating SphI- digested p239SpSp5′ and p239SpE3′ fragments of SIVmac239 obtained from Ronald Desrosiers through the NIH AIDS Research and Reference Reagent Program (catalog numbers 829 and 830, respectively). Similarly, a vif-defective variant of the SIVmac carrying a deletion of vif was constructed by ligating SphI-digested p7-21 (Vif defective) and p239SpE3′, obtained from Ronald Desrosiers through the NIH AIDS Research and Reference Reagent Program (catalog numbers 2470 and 830, respectively). The resulting pSIVmac239Vif(−) vector expresses all SIV-encoded proteins except Vif. For transient expression of HIV-1 Vif, the subgenomic expression vector pNL-A1 was used (51). For the expression of SIVmac239 Vif and SIVagm9063 Vif, the vif genes of these isolates were amplified by PCR from the respective full-length molecular clones and subcloned into the BssHII/EcoRI sites of pNL-A1, resulting in pNL-A1/macVif and pNL-A1/agmVif, respectively (54). Plasmid pcDNA-HA-CypA for the expression of N-terminally hemagglutinin (HA)-tagged human cyclophilin A was described previously (44).
Antisera.
A polyclonal antibody to SIVagm Vif was prepared by immunizing rabbits with purified recombinant protein. HIV-1 Vif was detected using a monoclonal antibody (number 319; a gift from Michael Malim). Serum from an HIV-positive patient was used to detect HIV-1-specific CA proteins. A polyclonal antibody to SIVagm CA protein was a gift of Vanessa Hirsch (12). A rabbit polyclonal antibody to CypA was obtained from BIOMOL (BIOMOL Research Laboratories, Inc., Plymouth Meeting, PA). An HA-specific rat monoclonal antibody for immunoprecipitation of HA-CypA was obtained from Roche (Roche Diagnostics, Indianapolis, IN).
Tissue culture and transfections.
HeLa and COS cells were propagated in Dulbecco's modified Eagle medium containing 10% fetal bovine serum (FBS). LuSIV cells are derived from CEMx174 cells and contain a luciferase indicator gene under the control of the SIVmac239 long terminal repeat (LTR). These cells were obtained through the NIH AIDS Research and Reference Reagent Program and were maintained in complete RPMI 1640 medium supplemented with 10% FBS and hygromycin B (300 μg/ml). The human Jurkat T-cell line was cultured in RPMI 1640 medium, 10% FBS. CypA−/− Jurkat cells were reported previously (9). For transfection, HeLa cells and COS cells were grown in 25-cm2 flasks to about 80% confluence. Cells were transfected using LipofectAmine PLUS (Invitrogen Corp., Carlsbad, CA) following the manufacturer's recommendations. A total of 5 μg of plasmid DNA per 25-cm3 flask was used. For transfection of various amounts of Vif expression vectors, all DNA samples were adjusted to equal DNA amounts using a vif-defective pNL-A1 variant. Cells were harvested 48 h posttransfection.
Nucleofection of Jurkat cells.
Jurkat cells (2 × 106 cells) were washed in phosphate-buffered saline (PBS) and suspended in a 100-μl solution of nucleofection V (Amaxa Biosystems, Gaithersburg, MD). A total of 20 μg of plasmid DNA per 2 × 106 cells was used. Nucleofections were carried out using an Amaxa nucleofector device. The nucleofection parameter was A-17. After nucleofection, cells were suspended in complete RPMI 1640 medium supplemented with 10% FBS. After 3 days of culture, both cells and supernatants were collected and analyzed by immunoblotting. A portion of the culture supernatants was used to determine virus production (by reverse transcriptase [RT] assay) and infectivity.
Preparation of virus stocks and immunoblotting.
Virus stocks were prepared by transfecting HeLa cells as previously reported (54). To produce virus stocks from Jurkat cells, Jurkat cells were infected with HeLa cell-derived viruses. Virus production was measured by determining the supernatant RT activity, and virus stocks were harvested at the peak of infection. Virus stocks were normalized for equal RT activity. Immunoblot analyses of cell lysates and viral pellets were performed as previously described (54).
Infection and DNA preparation.
Virus stocks produced from Jurkat cells were treated with 100 U of DNase I (Roche Applied Science, Indianapolis, IN) in the presence of 10 mM MgCl2 for 1 h at 37°C. For heat inactivation, virus (wild-type [WT] strain NL4-3) was incubated at 65°C for 30 min. Virus stocks were quantified by p24 (HIV-1) or p27 (SIV) enzyme-linked immunosorbent assay (ZeptMetrix Corporation, Buffalo, NY). LuSIV cells (5 × 105) were exposed to 100 ng of virus (p24) for 24 h at 37°C. Total DNA was extracted using a DNeasy tissue kit (QIAGEN, Inc., Valencia, CA) following the manufacturer's directions.
Step gradient analysis of virions.
Step gradient analysis of SIV and HIV virions was performed as reported elsewhere (34). Briefly, 2.0 ml of a 60% sucrose solution was placed into the bottom of model SW55 centrifuge tubes and overlaid with 2.1 ml of a 20% sucrose solution. Immediately prior to adding concentrated virus stocks (500 μl), the step gradients were overlaid with 100 μl of a protease inhibitor cocktail (Complete; Boehringer) and 100 μl of either PBS or 1% Triton X-100. Samples were then centrifuged in a model SW55Ti rotor for 75 min at 35,000 rpm at 4°C. Three 1.1-ml fractions were collected from the top, and each fraction was combined with 100 μl of 10× protease inhibitor cocktail. Aliquots of each fraction were processed for immunoblotting.
DNA PCR analysis.
To identify hypermutations of the SIV genome, total DNA from infected Jurkat cells was extracted using a DNeasy tissue kit (QIAGEN, Inc., Valencia, CA) and PCR amplified using a primer pair mapping to the SIVagm 3′ LTR region (54). To identify full-length viral cDNA, total cellular DNA was used for PCR amplification (Expand Long Template PCR system; Roche Diagnostics Corp., Indianapolis, IN) using primers 5′-TTCCTTACTGGGTTCTCTC (nucleotides 677 to 695 in SIVagm) and 3′-TTGTCTCCCTTTTAGTGCT (nucleotides 958 to 976 in SIVagm). PCR products were resolved on 0.8% agarose gels. For coamplification of actin sequences, a human beta-actin primer pair was included in the PCR (RandD Systems, Inc., Minneapolis, MN).
Real-time PCR analysis.
For the detection and quantification of full-length viral DNA by real-time PCR, the sense primer for the SIVagm envelope region was ATCAGAAGAAAAATTATTCAG (nucleotides 6821 to 6841), the antisense primer was AGAGTTAGAGCTAGAGCTGTT (nucleotides 6874 to 6894), and the probe was GTATGGAATGATGCAGAGATCTATTGTAA (nucleotides 6844 to 6872), and the sense primer for the NL4-3 envelope region was CAGGCCTGTCCAAAGGTATCC (nucleotides 6821 to 6841), the antisense primer was TTTAGAATCGCAAAACCAGCC (nucleotides 6894 to 6874), and the probe was TGAGCCAATTCCCATACATTATTGTGCCC (nucleotides 6844 to 6872). PCR was carried out in a spectrofluorometric thermal cycler (ABI PRISM 7700; Applied Biosystems, Inc., Foster City, CA).
Infectivity assay.
LuSIV cells (5 × 105) were infected for 24 h with 100 μl of unconcentrated virus stocks in 24-well plates. Cells were then harvested and lysed in 1× reporter lysis buffer (Promega Corp., Madison, WI). To determine the luciferase activity in the lysates, aliquots of each lysate (50 μl) were combined with luciferase substrate (Promega Corp., Madison, WI) by automatic injection, and light emission was measured in a luminometer (Optocomp II; MGM Instruments, Hamden, CT).
Construction of knockdown cell lines.
Jurkat and LuSIV cells were transduced with an HIV-1-based vector that confers puromycin resistance and delivers a short hairpin RNA (shRNA) expression construct specific either for human TRIM5α (TR5-shRNA) or for luciferase (Luc-shRNA) as previously described (49). Transduced cells were selected with puromycin and tested for the loss of Ref1 restriction activity with vesicular stomatitis virus G protein (VSV-G) pseudotyped N- or B-tropic murine leukemia virus green fluorescent protein (MLVGFP) virions as an indication of effective TRIM5α knockdown.
RESULTS
SIVagm Vif is required for efficient replication in Jurkat cells.
We compared the replication potential of the WT with that of the vif-defective SIVagm in A3G-negative Jurkat cells. As expected, HIV-1 Vif had no effect on the replication efficiency of HIV-1 in these cells (Fig. 1A, right panel). Interestingly, efficient replication of SIVagm9063 and SIVmac239 in Jurkat cells was Vif dependent (Fig. 1A, left and middle panels). Consistent with these observations, analysis of viral infectivity with a single-cycle assay revealed that the infectivity of vif-defective SIV but not HIV-1 was reduced compared to that of their respective WT counterparts (Fig. 1B). Despite the absence of A3G in Jurkat cells (54), the inhibition of vif-defective SIV in Jurkat cells could be caused by cytidine deaminases other than A3G and result in hypermutation of the viral genome in the absence of Vif. While this seemed unlikely given the fact that HIV-1 replication in Jurkat cells was Vif independent, we nevertheless analyzed viral DNA from SIVagm-infected Jurkat cells for evidence of cytidine deamination. Indeed, we found no evidence for cytidine deamination in DNA isolated 2 weeks after infection from Jurkat cells infected with the WT or the vif-defective SIVagm (Fig. 1C). These results suggest that replication of SIV in Jurkat cells is inhibited by a deaminase-independent but Vif-sensitive antiviral host factor.
FIG. 1.
SIVagm Vif is required for efficient replication in Jurkat cells. (A) WT or vif-defective SIVagm, SIVmac239, and HIV-1 NL4-3 stocks were produced with HeLa cells and used to infect Jurkat T cells. Virus production was monitored for 14 days by determining the virus-associated reverse transcriptase activity in the culture supernatants. (B) Culture supernatants from the infections in panel A were collected at peak virus production, adjusted for equal reverse transcriptase activities, and used for the infection of LuSIV cells. Infection was determined 24 h later by measuring the Tat-induced luciferase activity in the target cells. Infectivity of vif-defective viruses was calculated relative to the infectivity of WT viruses, which was defined as 100%. Error bars in panels A and B reflect the standard deviations calculated from triplicate independent infections. (C) Jurkat cells lack cytidine deaminase activity. Total DNA was isolated 14 days after infection from SIVagm-infected cultures shown in panel A. A 323-bp fragment from the 3′ LTR region of the viral genome was PCR amplified, cloned, and sequenced as described previously (54). G-to-A mutations in 9 to 10 independent clones (2,900 to 3,200 total bp each) were analyzed and compared to other nucleotide substitutions. The mutation frequency was calculated as the number of mutations per 100 base pairs. Dots represent results from individual clones.
SIVagm Vif inhibits the packaging of CypA into SIVagm virions and increases viral infectivity.
CypA is a host peptidyl-isomerase that is encapsidated into HIV-1 but not into SIV particles (8, 41, 55). CypA has a positive effect on HIV-1 replication and supports an early step of HIV-1 replication in newly infected target cells (8, 25, 50). However, analysis of the precise function of CypA for HIV-1 replication is still ongoing, and there is no known role for CypA in SIV replication in human or simian cells.
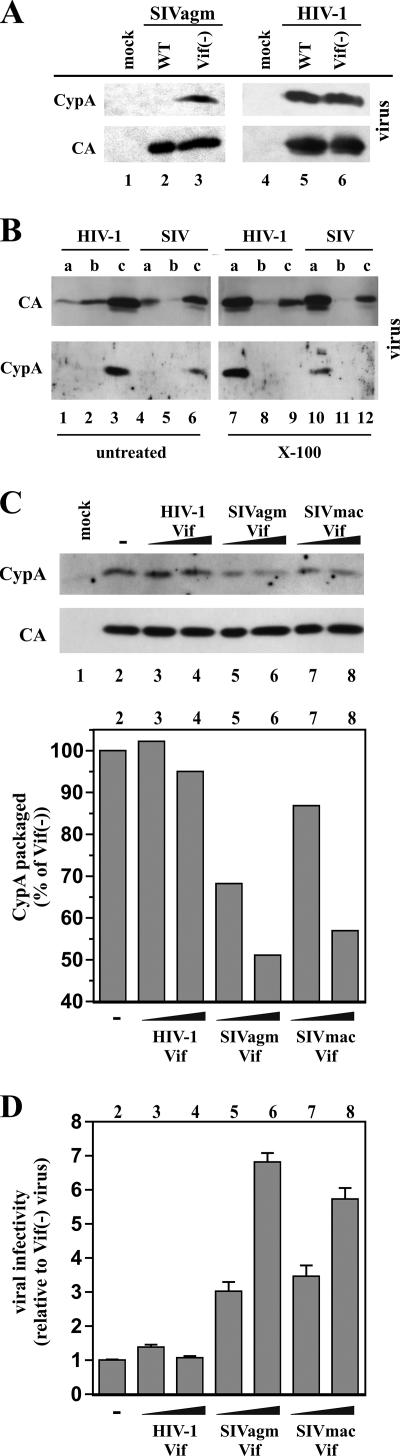
To investigate a possible role for CypA in the inhibition of vif-defective SIVagm in human cells, we compared possible effects of SIVagm Vif and of HIV-1 Vif on the expression or packaging of CypA into SIVagm or HIV-1 particles produced from Jurkat cells. Samples were taken from the cultures shown in Fig. 1A at peak virus production. Concentrated viral pellets were analyzed by immunoblotting (Fig. 2A). HIV-1 Vif had no effect on the packaging of CypA into cell-free HIV-1 particles (Fig. 2A, lanes 5 and 6). Also, consistent with previous reports (16, 18, 41, 55), CypA was largely excluded from the WT SIVagm (Fig. 2A, lane 2). Interestingly, however, CypA was packaged into vif-deficient SIVagm particles (Fig. 2A, lane 3). CypA was absent from culture supernatants from mock-transfected controls (Fig. 2A, lanes 1 and 4). These results demonstrate that packaging of CypA into SIV particles is Vif sensitive and that the reported absence of CypA from SIV virions is due to an activity of Vif.
FIG. 2.
SIVagm Vif inhibits the packaging of CypA in SIVagm virions in Jurkat cells. (A) Virus-containing supernatants from SIVagm (lanes 1 to 3) and HIV-1-infected Jurkat cells (lanes 4 to 6) shown in Fig. 1A were harvested at peak virus production. Virus-containing supernatants were normalized for equal RT activities and concentrated by pelleting through 20% sucrose. Pelleted viruses were then analyzed by immunoblotting using antibodies specific to CypA or SIVagm and HIV-1 CA protein. Proteins are identified on the left. (B) Vif-deficient virions from infected Jurkat cells (as shown in Fig. 1A) were subjected to step gradient analysis in the absence (untreated) or presence (Triton X-100) of detergent. Three fractions (a to c) were collected from each gradient and analyzed by immunoblotting for the presence of SIV or HIV-1 CA protein or CypA. (C) Jurkat cells were nucleofected with pSIVagmVif(−) in the absence of Vif (lane 2) or together with increasing amounts of pNL-A1 (lanes 3 to 4), pNL-A1/agmVif (lanes 5 to 6) or pNL-A1/macVif (lanes 7 to 8). The plasmid ratios (provirus:Vif) were 4:1 (lanes 3, 5, and 7) and 1:1 (lanes 4, 6, and 8). A mock-transfected sample was included as a control (lane 1). Total amounts of transfected plasmid DNA were kept constant by adjusting with appropriate amounts of vif-defective pNL-A1vif(−) DNA. Virus-containing supernatants were harvested 3 days after transfection and processed for immunoblotting as shown in panel A (upper panels). CypA-specific protein bands were quantified by densitometric scanning of the gel and were plotted as a percentage of the Vif-negative control (lane 2) which was defined as 100% (lower panel). Lane numbers correspond to lanes on the immunoblot. (D) Virus-containing supernatants were normalized for equal reverse transcriptase activity and used for the infection of LuSIV indicator cells. Infection was determined 24 h later as described in the legend to Fig. 1B. Error bars reflect the standard deviations calculated from triplicate infections. Lane numbers correspond to lanes on the immunoblot in panel C (top).
We have previously noted that HIV-1 Vif as well as the antiviral host factor APOBEC3G is packaged into the core of HIV-1 virions (34, 35). To investigate a possible core association of CypA with vif-deficient viruses, we performed step gradient analyses of detergent-treated virions, as described previously (34). Proteins not associated or only loosely associated with the viral core are removed by detergent treatment of the particles and will partition to the soluble fraction in this assay (Fig. 2B, fraction a), while core-associated proteins are recovered from the fraction (Fig. 2B, fraction c). A buffer fraction (Fig. 2B, fraction b) separating the fractions (Fig. 2B, fractions a and c) should not contain any viral proteins. Samples of the vif-deficient HIV-1 and SIVagm shown in Fig. 1A were concentrated by pelleting through 20% sucrose and then subjected to step gradient centrifugation in the absence (Fig. 2B, lanes 1 to 6) or presence (Fig. 2B, lanes 7 to 12) of detergent (Fig. 2B, X-100). As expected, the majority of viral CA proteins of HIV-1 and SIVagm as well as virus-associated CypA partitioned with fraction c in the step gradients of untreated viruses (Fig. 2B, lanes 3 and 6). As observed before (34), detergent treatment resulted in the loss of a substantial portion of CA protein to the soluble fraction (Fig. 2B, panel CA, lanes 7 and 10). However, a notable portion of viral CA proteins remained in the detergent-resistant core fraction (Fig. 2B, panel CA, lanes 9 and 12). These results are consistent with the finding that less than half of the virus-associated CA proteins form the mature core (10). Importantly, CypA was highly sensitive to detergent treatment, and virtually all of the virus-associated CypA partitioned with the soluble fraction in the step gradients (Fig. 2B, panel CypA, lanes 7 and 10). These results suggest that the CypA present in HIV-1 and SIVagm virions is not stably associated with the viral core.
Vif proteins encoded by SIVmac and SIVagm are functionally equivalent with respect to their activities toward CypA. This was shown by the electroporation of Jurkat cells with vif-defective SIVagm together with increasing amounts of vectors encoding SIVagm (Fig. 2C, lanes 5 and 6) or SIVmac Vif (Fig. 2C, lanes 7 and 8). The effect of HIV-1 Vif (Fig. 2C, lanes 3 and 4) was analyzed in parallel. All samples were compared to a vif-deficient control (Fig. 2C, lane 2). Both SIVagm Vif and SIVmac Vif reduced packaging of CypA into SIVagm particles in a dose-dependent manner, while HIV-1 Vif had no effect on CypA packaging into SIVagm virions (Fig. 2C, bottom panel). This was paralleled by a dose-dependent increase in the infectivity of viruses produced in the presence of SIV Vif (Fig. 2D, lanes 5 to 8). At the highest levels of transfected Vif, viral infectivity was increased about 6- to 7-fold relative to that of vif-deficient particles. In contrast, HIV-1 Vif did not noticeably affect the packaging of CypA into SIVagm virions (Fig. 2C, bottom panel, lanes 3 and 4) and did not alter SIVagm infectivity (Fig. 2D, lanes 3 and 4). These results suggest that the ability to inhibit encapsidation of CypA is specific to SIV Vif proteins and is conserved among SIV isolates. Similar to SIVagm, vif-defective SIVmac239 virions were found to package CypA (data not shown), suggesting that CypA packaging is a general property of vif-defective SIV virions. Of note, SIV Vif proteins were unable to inhibit CypA packaging into HIV-1 virions (not shown), suggesting that this function of SIV Vif involves a specific interplay with the viral Gag proteins.
The effect of SIVagm Vif on packaging of CypA and viral infectivity is cell type independent.
The results shown in Fig. 2 reveal a correlation between packaging of CypA into SIVagm and reduction in viral infectivity. To see if the effects of SIVagm Vif on CypA packaging are cell type specific or are limited to Jurkat cells, experiments were performed with the HeLa and the African green monkey-derived COS cell lines. HeLa and COS cells were transfected with vif-defective SIVagm plasmid DNA together with increasing amounts of plasmids encoding HIV-1 Vif (Fig. 3A to D, lanes 2 to 3 and 7 to 8) or SIVagm Vif (Fig. 3A to D, lanes 4 to 5 and 9 to 10). Vif expression was monitored by immunoblot analysis of cell lysates (Fig. 3A, upper panels). The differences in mobility of the Vif proteins are explained by the size differences between HIV-1 Vif (192 residues) and SIVagm Vif (231 residues). The presence of equal amounts of CypA in all cell lysates was verified by immunoblotting using a CypA-specific antibody (Fig. 3A, bottom panels). Concentrated cell-free virions were analyzed for the presence of viral CA protein and CypA (Fig. 3B). CypA packaging was quantified by densitometric scanning of the gel. Resulting values were corrected for variations in CA levels and are shown in Fig. 3C. Consistent with the results shown in Fig. 2C, SIVagm Vif but not HIV-1 Vif inhibited CypA packaging into SIVagm particles irrespective of the cellular host. Thus, SIVagm Vif controls the encapsidation of CypA into HeLa- or COS cell-derived SIVagm virions with the same efficiency as that of Jurkat cells, suggesting that packaging of CypA into SIVagm virions is cell type independent.
FIG. 3.
Cell type-independent effect of Vif on the infectivity of SIVagm. HeLa cells (lanes 1 to 5) and COS cells (lanes 6 to 10) were transfected with pSIVagm Vif(−), either in the absence of Vif (lanes 1 and 6) or together with increasing amounts of pNL-A1 (lanes 2 to 3 and 7 to 8) or with pNL-A1/agm Vif (lanes 4 to 5 and 9 to 10). The plasmid ratios (provirus:Vif) were 4:1 (lanes 2, 4, 7, and 9) or 1:1 (lanes 3, 5, 8, and 10). The total amount of plasmid DNA was kept constant by adjusting with appropriate amounts of pNL-A1vif(−) plasmid DNA. (A and B) Cells and virus-containing supernatants were harvested 48 h after transfection. Viruses were concentrated as described in the legend to Fig. 2A, and whole-cell lysates (panel A) and virus samples (panel B) were analyzed by immunoblotting for the presence of Vif, CypA, and CA proteins as indicated. (C) CypA-specific protein bands from the virus fraction were quantified by densitometric scanning of the gel and corrected for variations in CA signals. The results are plotted as percentages of the Vif-deficient controls (lanes 1 and 6), which were defined as 100%. Lane numbers correspond to lanes on the immunoblot in panel B. (D) The infectivity of viruses was determined by infection of LuSIV cells as described in the legend to Fig. 1B. Error bars reflect the standard deviations calculated from triplicate infections. Lane numbers correspond to lanes on the immunoblot in panel B.
The effect of Vif on the infectivity of HeLa or COS cell-derived viruses was analyzed in a single-round infectivity assay. Consistent with the results shown in Fig. 2D, SIVagm Vif increased the infectivity of SIVagm virions from HeLa cells and COS cells in a dose-dependent manner, while HIV-1 Vif had no effect on viral infectivity (Fig. 3D). These results demonstrate that SIVagm Vif acts specifically and in a cell type-independent manner to increase the infectivity of SIVagm virions. The increase in viral infectivity of COS cell-derived viruses in response to increasing expression of SIVagm Vif was more subtle than in HeLa or Jurkat cells (Fig. 3D, compare lanes 4 to 5 and 9 to 10), despite having similar effects on the encapsidation of CypA (Fig. 3C, compare lanes 4 to 5 and 9 to 10). The reason for this is unclear, although the presence of low levels of A3G in COS cells could be a contributing factor.
SIVagm Vif does not induce degradation of CypA.
The analysis of CypA in Fig. 3A provides no indication that the inhibition of CypA encapsidation by SIVagm Vif might be associated with a reduction of cellular CypA levels in HeLa or COS cells. Similarly, Vif had no apparent effect on cellular CypA levels in infected Jurkat cells even at peak virus production when the majority of cells were infected (data not shown). To more directly analyze the possible effect of SIVagm Vif on the stability of human CypA, we performed a pulse-chase analysis of CypA in HeLa cells. HeLa cells were cotransfected with pcDNA-HA-CypA encoding N-terminally HA-tagged human CypA (44) and either pNL-A1vif(−) [Fig. 4A, Vif(−)] or pNL-A1/agmVif (Fig. 4A, Agm Vif). The plasmid ratio of Vif to CypA expression vectors was 10:1. Pulse-chase analysis of the transfected cells was performed 24 h later as described in the legend to Fig. 4. Proteins were immunoprecipitated with an HA-specific rat monoclonal antibody (Fig. 4A, CypA) or an SIVagm Vif-specific rabbit polyclonal antibody (Fig. 4A, Vif). CypA migrated as a doublet in the reducing gel, as reported previously (41). CypA-specific bands (Fig. 4B) and Vif-specific bands (Fig. 4C) were quantified by densitometric scanning of the films, and the results were plotted as percentages of the signal intensities measured at the pulse time points (time zero). CypA was stable over the 2-h observation period in both the presence and the absence of Vif (Fig. 4B). This finding is interesting since the proposed mechanism of A3G exclusion from HIV-1 virions by Vif involves intracellular A3G degradation (reviewed in reference 17) and suggests that inhibition of CypA packaging into SIV particles is regulated by a degradation-independent mechanism. Unlike CypA, SIVagm Vif was unstable and degraded with a half-life of ∼30 min. This is consistent with our previous reports on HIV-1 Vif, which is unstable as well and is degraded by cellular proteasomes in HeLa and H9 cells, with kinetics very similar to that of SIVagm Vif (3, 19).
FIG. 4.
Vif does not induce degradation of CypA. (A) HeLa cells (5 × 106) were transfected with 0.5 μg of pcDNA-HA-CypA and either 4.5 μg of pNL-A1 DNA (Vif+) or 4.5 μg of pNL-A1vif(−) DNA [Vif(−)]. Cells were harvested 24 h later and pulse-labeled for 10 min with [35S]methionine (2 mCi/ml). Unincorporated isotope was removed, and cells were cultured at 37°C in complete RPMI medium. Aliquots were collected at the indicated times and stored on dry ice. Cells were then lysed in 300 μl of lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.5% Triton X-100). The cell extracts were centrifuged at 13,000 × g for 3 min, and half of each supernatant was immunoprecipitated with an HA-specific rat monoclonal antibody (CypA) or a Vif-specific polyclonal rabbit antiserum (Vif). Immunoprecipitated proteins were separated by sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis and visualized by fluorography. (B) CypA-specific bands were quantified by densitometric scanning, and results are plotted as percentages of the CypA signals detected at the pulse time points, which were defined as 100%. (C) Vif-specific bands were quantified as described in the legend to panel B.
Replication of SIVagm in CypA knockout Jurkat cells is Vif independent.
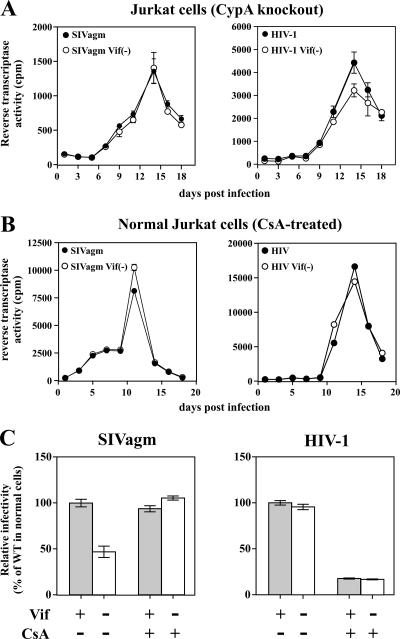
To investigate the role of CypA in HIV-1 or in SIVagm replication in Jurkat cells, we made use of a Jurkat-derived cell line (PPIA−/−) in which both alleles of the PPIA gene encoding CypA have been inactivated (9). Consistent with a previous report (9), replication of HIV-1 in PPIA−/− Jurkat cells was largely vif-independent (Fig. 4A, right panel). However, the overall virus output was reduced about 6-fold compared to that of normal Jurkat cells (cf. with Fig. 1A, right panel). The reduced overall virus replication can be explained at least in part by reduced cell-surface CD4 levels in the CypA knockout line (data not shown). Interestingly, replication of SIVagm in PPIA−/− Jurkat cells was no longer Vif-dependent (Fig. 5A, left panel), and there was no longer a detectable difference in the relative infectivity levels of viruses produced from these cells when they were analyzed with a single-cycle infectivity assay (data not shown). As with HIV-1, overall virus replication was about 5- to 6-fold lower compared to that of normal Jurkat cells (cf. with Fig. 1A, left panel), presumably due to the reduced cell-surface CD4 T-cell level in the knockout line. These results strongly suggest that CypA is responsible for the inhibition of vif-defective SIV in Jurkat cells.
FIG. 5.
Replication of SIVagm in CypA-deficient cells is Vif independent. (A) Virus stocks were produced from transiently transfected HeLa cells and used for the infection of PPIA−/− Jurkat cells. Virus production was monitored for 18 days by determining the virus-associated reverse transcriptase activity in the culture supernatants. (B) CsA eliminates the requirement for Vif for SIVagm replication in Jurkat cells. WT or vif-defective SIVagm (left panel) and HIV-1 (right panel) produced in HeLa cells were used to infect Jurkat cells. Infected cells were cultured in the presence of CsA (2.5 μM). Virus production was monitored for 18 days by determining the virus-associated reverse transcriptase activity in the culture supernatants. (C) The infectivity of the viruses at peak virus production shown in panel B was determined by infection of LuSIV indicator cells, as described in the legend to Fig. 1B. The infectivity of the WT virus in untreated cells was defined as 100%. Error bars reflect the standard deviations calculated from triplicate experiments.
Replication of SIVagm in cyclosporine A-treated Jurkat cells is Vif independent.
CsA is an immunosuppressive agent that binds to and inhibits the activity of CypA (47, 58). To further investigate the relationship between SIVagm Vif and CypA, we analyzed the effects of CsA on the replication of SIVagm or HIV-1 in Jurkat cells. Jurkat cells were infected with WT or vif-defective SIVagm or HIV-1 and cultured in the presence of 2.5 μM CsA (Fig. 5B). Virus replication was monitored by measuring the virus-associated reverse transcriptase activity. As noted before, Vif had no effect on the replication of HIV-1 in CsA-treated Jurkat cells (Fig. 5B, right panel). CsA treatment reduced the overall virus output about 2-fold compared to that of untreated cells (cf. with Fig. 1A, right panel), consistent with previous studies (57). Importantly, SIVagm replication in CsA-treated Jurkat cells was no longer Vif dependent (Fig. 5B, left panel), and virus output for both viruses was comparable to that of the WT virus in untreated cells (cf. Fig. 1A, left panel). These results further support our conclusion that CypA inhibits SIVagm replication.
To assess the effects of CsA treatment on viral infectivity, viruses from CsA-treated Jurkat cells (as shown in Fig. 5B) as well as from untreated control infections (not shown) were collected at peak virus production, normalized for equal reverse transcriptase activity, and used for the infection of untreated LuSIV indicator cells (Fig. 5C). Consistent with the results shown in Fig. 1B, the infectivity of SIVagm produced in the absence of CsA was reduced in the absence of Vif (Fig. 5C, left panel, CsA−), while the infectivity of HIV-1 was independent of the presence or absence of Vif (Fig. 5C, right panel CsA−). Importantly, vif-defective SIVagm produced in the presence of CsA was as infectious as WT SIVagm (Fig. 5C, left panel, CsA+) and was as infectious as the WT virus from untreated cells (Fig. 5C, left panel). These results support the conclusion that CypA in virus-producing cells but not the CypA present in the LuSIV target cells inhibits the infectivity of vif-defective SIVagm. In contrast, the infectivity of HIV-1 from CsA−-treated donor cells was reduced both for the WT and for the vif-defective virus relative to that for virus from untreated cells (Fig. 5C, right panel). Thus, donor cell CypA has a positive effect on HIV-1 but inhibits the replication of vif-defective SIVagm.
Target cell CypA does not inhibit infection by SIVagm.
Recent studies showed that target cell CypA is important for HIV-1 virion infectivity (25, 29, 36, 50, 57). To analyze the function of target cell CypA for SIVagm and HIV-1 infection, WT or vif-defective viruses produced from normal Jurkat cells in the absence of CsA were used to infect either normal LuSIV indicator cells or indicator cells pretreated for 24 h with CsA (2.5 μM). Infection efficiency was determined 24 h later by either real-time PCR (Fig. 6A) or by a standard luciferase assay (Fig. 6B). Results from real-time PCR and luciferase assays were comparable and demonstrated that SIVagm depended on Vif for efficient infection of untreated LuSIV cells (Fig. 6A and B, left panels, CsA−). Importantly, treatment of target cells with CsA did not ablate the Vif dependence for SIVagm infection (Fig. 6A and B, left panels, CsA+). Similar results were obtained when normal Jurkat cells were compared to CypA-null cells as target cells: both cell types exhibited a Vif-dependent inhibition of SIVagm (data not shown). In contrast, CsA treatment of target cells reduced HIV-1 infection but in a Vif-independent manner (Fig. 6A and B, right panels), consistent with previous reports on the role of target cell CypA for HIV-1 replication (25, 29, 36, 50, 57).
FIG. 6.
Target cell CypA does not affect infection by SIVagm. WT (Vif+) and Vif-deficient (Vif−) virus stocks were produced in untreated Jurkat cells and used to infect untreated LuSIV cells (CsA−) or LuSIV cells pretreated for 24 h with 2.5 μM CsA (CsA+). (A) Total DNA was isolated from a portion of the cells 24 h postinfection. Viral cDNA synthesis was quantified by real-time PCR, as described in Materials and Methods. Results from cells infected with the WT virus in the absence of CsA (Vif+, CsA−) were defined as 100%. (B) The remaining portion of infected LuSIV cells was used to determine the virus-induced luciferase activity as described in the legend to Fig. 1B. Luciferase activity induced by the WT virus in untreated target cells was defined as 100%. Error bars reflect the standard deviations calculated from triplicate experiments.
Target cell TRIM5α does not affect infection by SIVagm.
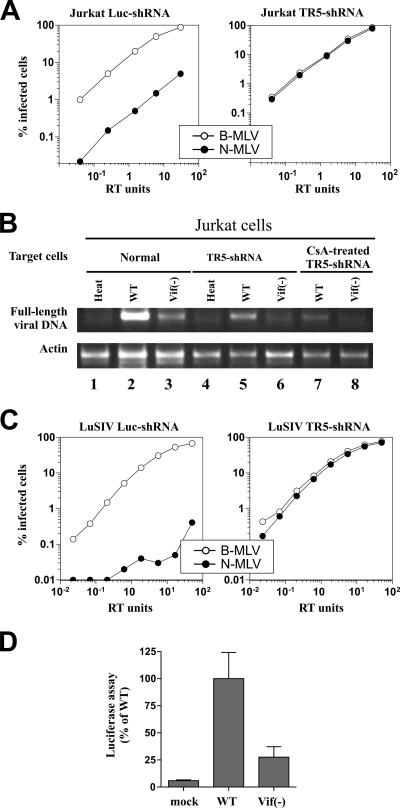
TRIM5α has emerged as a postentry restriction factor inhibiting virus replication at an early postentry step (52, 53). All of our previous results point to CypA as the host factor restricting replication of vif-defective SIV in human cells. To rule out a possible interference by human TRIM5α, we made use of a Jurkat cell line in which the expression of TRIM5α was silenced by shRNA (Fig. 7A). The successful inhibition of TRIM5α was validated by comparing the abilities of B-MLV and N-MLV to infect these cells. As shown in Fig. 7A, B-MLV efficiently infected normal and TRIM5α knockdown cells. In contrast, infection by N-MLV was inhibited in normal Jurkat cells but was similar to that of B-MLV in knockdown cells.
FIG. 7.
Target cell TRIM5α does not affect infection by SIVagm. (A) Jurkat cells were transduced with an HIV-1-based vector that confers puromycin resistance and delivers an shRNA expression construct specific either for human TRIM5α (TR5-shRNA) or for luciferase (Luc-shRNA). VSV-G pseudotyped N- or B-tropic MLVGFP virions were normalized for titer with nonrestrictive Mus dunni cells and then used to infect Jurkat Luc-shRNA cells or Jurkat TR5-shRNA cells. The percentage of GFP-positive (infected) cells was determined by flow cytometry. Shown are representative results of a single experiment. Identical results were obtained on three separate occasions using independently produced viral stocks. (B) WT and vif-deficient (Vif−) SIVagm stocks were produced with untreated Jurkat cells and used to infect untreated Jurkat cells (normal), Jurkat TR5-shRNA cells (TRIM5α-KD), or Jurkat TR5-shRNA cells pretreated for 24 h with 2.5 μM CsA (CsA-treated TRIM5α-KD). Total DNA was harvested 24 h postinfection. Accumulation of full-length viral cDNA was determined by DNA PCR amplification. A primer set for the amplification of actin DNA was included in each reaction as an internal control (Actin). Heat, heat-inactivated WT SIVagm. (C) LuSIV cells were transduced with TR5-shRNA or Luc-shRNA vectors as described in the legend to panel A. TRIM5α silencing was measured by determining the relative sensitivity of the cells to infection by VSV-G pseudotyped B-tropic or N-tropic MLVGFP virions. (D) LuSIV TR5-shRNA cells were infected with equal amounts of the WT or the vif-defective [Vif(−)] SIVagm derived from infected Jurkat cells. Mock-infected cells were analyzed in parallel (mock). Infected cells were harvested 24 h after infection, and virus-induced luciferase activity was measured as described in Materials and Methods. Error bars reflect standard deviations calculated from three independent experiments.
To assess the effect of TRIM5α on infection by SIVagm, normal Jurkat cells or TRIM5α knockdown cells were infected with either the WT virus (Fig. 7B, lanes 2, 5, and 7) or vif-defective SIVagm (Fig. 7B, lanes 3, 6, and 8). As a control, cells were also infected with heat-inactivated WT virus (Fig. 7B, lanes 1 and 4). Normal Jurkat cells (Fig. 7B, lanes 1 to 3), TRIM5α knockdown cells (Fig. 7B, lanes 4 to 6), and CsA-treated TRIM5α knockdown cells (Fig. 7B, lanes 7 to 8) were used as targets. Total DNA was harvested 24 h after infection, and the accumulation of full-length viral DNA was determined by semiquantitative DNA PCR (Fig. 7B, upper panel). A primer set for coamplification of actin DNA was included in each PCR (Fig. 7B, lower panel). As can be seen, infection by WT SIVagm was in all samples more efficient than infection by vif-deficient virus irrespective of TRIM5α expression or CsA treatment. These results suggest that TRIM5α does not contribute to the Vif-sensitive inhibition of SIVagm in human cells.
As an additional control for possible interference by TRIM5α, we employed a luciferase indicator cell line in which expression of TRIM5α was silenced by shRNA. The LuSIV TR5-shRNA cell line was derived from the parental LuSIV indicator cell line used for all other single-round infectivity experiments in this study. As for the Jurkat TR5-shRNA knock-down cells, successful inhibition of TRIM5α in LuSIV TR5-shRNA cells was validated by comparing the abilities of B-tropic and N-tropic MLVGFP to infect these cells (Fig. 7C). LuSIV TR5-shRNA cells were then infected with equal amounts of the WT or vif-defective SIVagm stocks produced from infected Jurkat cells (Fig. 7D). Consistent with the results shown in Fig. 7B, silencing of TRIM5α did not alleviate the CypA-induced, Vif-sensitive inhibition of SIVagm infection.
DISCUSSION
There is significant evidence that the ongoing, worldwide AIDS epidemic was caused by cross-species transmission of simian immunodeficiency viruses into the human population. How and when this transmission occurred are still debated, but it is now generally accepted that HIV-2 originated from SIV of sooty mangabeys (13, 28), while HIV-1 appears to have originated from SIV of chimpanzees (20). Zoonotic transmission of simian immunodeficiency viruses, however, is not common and is controlled by host factors that generally prohibit SIV replication in human hosts and many human-derived cell lines.
We have previously studied the replication of SIVagm in human cell lines and found that the virus was able to replicate in the human A3.01 T-cell line despite the presence of high levels of human APOBEC3G (54). Replication of SIVagm in A3.01 cells was Vif dependent, and the absence of Vif was associated with extensive hypermutation of the viral genome caused by A3G-induced cytidine deamination. This finding was surprising given the known species-specific function of Vif (48) and the inability of SIVagm Vif to target human A3G for proteasome-dependent degradation (6, 38, 45, 54). Thus, we concluded that A3G limited SIV replication in human cells but did not pose an absolute barrier for cross-species transmission of SIVagm (54).
One of the surprising findings of the current study is that efficient SIVagm replication was also dependent on a functional vif gene even in human cells lacking detectable cytidine deaminase activity. The inhibition of vif-deficient SIVagm was not due to the expression of low levels of A3G or the presence of other cytidine deaminases since the absence of Vif did not result in hypermutation of the viral genomes (Fig. 1C). Importantly, the Vif-dependent inhibition in Jurkat cells was specific for SIV and did not affect HIV-1 replication further, supporting the conclusion that the inhibition of SIV in these cells was not due to cytidine deamination.
Although the precise mechanism of inhibition remains to be investigated, we were able to demonstrate that inhibition of SIVagm occurred at the level of reverse transcription. Indeed, both early and late reverse transcription products were affected (Fig. 6A and Fig. 7B and data not shown). This is reminiscent of the activity of restriction factors such as TRIM5α. However, the fact that the inhibition of SIVagm in Jurkat cells is Vif sensitive and can be overcome by treatment with virus producer CsA but not with target cell CsA argues against an involvement of TRIM5α in our studies and instead points to CypA. This conclusion is further supported by the finding that the presence or absence of CypA in virus preparations was directly correlated to a Vif-dependent change in viral infectivity. Finally, knockdown of TRIM5α in Jurkat and LuSIV cells, which rendered the cells susceptible to N-MLV replication, did not relieve the Vif-dependent restriction of SIVagm (Fig. 7).
The ability of Vif to inhibit the packaging of CypA into SIVagm virions is reminiscent of its ability to prevent the encapsidation of A3G. Exactly how Vif prevents the packaging of A3G is still under investigation. There is strong evidence that Vif can induce proteasome degradation of A3G (for a review, see reference 17) and that intracellular depletion of A3G contributes to its absence from WT virions. Yet, there is also evidence that Vif can prevent the packaging of A3G into HIV virions through a degradation-independent mechanism (30, 31, 39). This latter activity of Vif, which remains to be explored in its molecular details, may be relevant to the Vif-dependent exclusion of CypA from SIVagm virions. This conclusion is justified by the observation that Vif had no effect on the intracellular steady-state level of CypA in any of the cell types analyzed in this study (Fig. 3A and data not shown) and did not reveal any evidence of increased protein degradation, using a pulse-chase experiment performed with HeLa cells (Fig. 4).
Previous reports showed that CypA is incorporated into HIV-1 virions through the interaction with viral capsid (1, 8, 9, 11, 18, 41, 55). In particular, two amino acids in HIV-1 Gag, Gly221 and Pro222, were found to be important for the binding of CypA (7). Although the Gag region of SIVagm contains a similar proline-rich region (Gly221 and Pro222 are in fact conserved in our SIVagm isolate), the same region is not conserved in SIVmac239, which also was found to encapsidate CypA in the absence of Vif (not shown). It seems therefore likely that CypA is packaged into vif-defective SIV through a mechanism that is distinct from HIV-1. Nevertheless, packaging of CypA into HIV-1 and SIVagm particles is, in both cases, sensitive to detergent treatment, suggesting that CypA is not associated with the viral core.
Our observation that CsA treatment of virus-producing cells but not target cells abolished the inhibitory effect of CypA on the infectivity of SIVagm clearly demonstrates that unlike HIV-1, the effect of CypA on SIVagm is restricted to virus-producing cells. The fact that CsA treatment elevates the infectivity of vif-defective viruses to that of WT controls (Fig. 5C) also demonstrates that the reduced infectivity of vif-defective virus is not due to the absence of Vif from virions but is a result of the presence of CypA. The fact that Vif specifically inhibits the packaging of CypA suggests that the inhibitory effect is exerted by the presence of CypA in SIVagm virions. It is unclear how virus-associated CypA inhibits SIVagm infectivity. However, we previously observed that packaging of excessive amounts of Vif can severely affect viral infectivity. In the case of Vif, we noted that the protein specifically interacted with the Gag precursor at or near the capsid/nucleocapsid cleavage site, thereby inhibiting proteolytic processing at the primary cleavage site (3). Due to the limiting amounts of virus-associated Vif, only a minor fraction of Gag precursor molecules was affected; however, this was sufficient to induce an assembly defect and cause a complete loss of viral infectivity (3). It is therefore conceivable that the interaction of virus-associated CypA with SIVagm Gag induces a similar assembly defect that reduces the infectivity of the viruses.
Acknowledgments
We thank Vanessa Hirsch for the SIVagm9063 clone and for SIV CA antibodies and Ronald Desrosiers for the SIVmac239 clone. We thank Michael Malim for the Vif monoclonal antibody (number 319) and Jason Roos and Janice Clements for the LuSIV indicator cell line, obtained through the NIH Research and Reference Reagent Program.
Part of this work was supported by a grant from the NIH Intramural AIDS Targeted Antiviral Program to K.S., by the Intramural Research Program of the NIH, NIAID to K.S., by NIH grant RO1AI36199 to J.L., and by Swiss National Science Foundation grant 3100A0-113558 to J.L.
Footnotes
Published ahead of print on 23 May 2007.
REFERENCES
- 1.Ackerson, B., O. Rey, J. Canon, and P. Krogstad. 1998. Cells with high cyclophilin A content support replication of human immunodeficiency virus type 1 Gag mutants with decreased ability to incorporate cyclophilin A. J. Virol. 72:303-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akari, H., M. Fujita, S. Kao, M. A. Khan, M. Shehu-Xhilaga, A. Adachi, and K. Strebel. 2004. High level expression of human immunodeficiency virus type-1 Vif inhibits viral infectivity by modulating proteolytic processing of the Gag precursor at the p2/nucleocapsid processing site. J. Biol. Chem. 279:12355-12362. [DOI] [PubMed] [Google Scholar]
- 4.Berthoux, L., S. Sebastian, E. Sokolskaja, and J. Luban. 2005. Cyclophilin A is required for TRIM5{alpha}-mediated resistance to HIV-1 in Old World monkey cells. Proc. Natl. Acad. Sci. USA 102:14849-14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. USA 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogerd, H. P., B. P. Doehle, H. L. Wiegand, and B. R. Cullen. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. USA 101:3770-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braaten, D., C. Aberham, E. K. Franke, L. Yin, W. Phares, and J. Luban. 1996. Cyclosporine A-resistant human immunodeficiency virus type 1 mutants demonstrate that Gag encodes the functional target of cyclophilin A. J. Virol. 70:5170-51706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braaten, D., E. K. Franke, and J. Luban. 1996. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J. Virol. 70:3551-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braaten, D., and J. Luban. 2001. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J. 20:1300-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briggs, J. A., M. N. Simon, I. Gross, H. G. Krausslich, S. D. Fuller, V. M. Vogt, and M. C. Johnson. 2004. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 11:672-675. [DOI] [PubMed] [Google Scholar]
- 11.Bukovsky, A. A., A. Weimann, M. A. Accola, and H. G. Gottlinger. 1997. Transfer of the HIV-1 cyclophilin-binding site to simian immunodeficiency virus from Macaca mulatta can confer both cyclosporin sensitivity and cyclosporin dependence. Proc. Natl. Acad. Sci. USA 94:10943-10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell, B. J., and V. M. Hirsch. 1997. Vpr of simian immunodeficiency virus of African green monkeys is required for replication in macaque macrophages and lymphocytes. J. Virol. 71:5593-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, Z., P. Telfier, A. Gettie, P. Reed, L. Zhang, D. D. Ho, and P. A. Marx. 1996. Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J. Virol. 70:3617-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowan, S., T. Hatziioannou, T. Cunningham, M. A. Muesing, H. G. Gottlinger, and P. D. Bieniasz. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA 99:11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cullen, B. R. 2006. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 80:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorfman, T., and H. G. Gottlinger. 1996. The human immunodeficiency virus type 1 capsid p2 domain confers sensitivity to the cyclophilin-binding drug SDZ NIM 811. J. Virol. 70:5751-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrlich, E. S., and X. F. Yu. 2006. Lentiviral Vif: viral hijacker of the ubiquitin-proteasome system. Int. J. Hematol. 83:208-212. [DOI] [PubMed] [Google Scholar]
- 18.Franke, E. K., H. E. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359-362. [DOI] [PubMed] [Google Scholar]
- 19.Fujita, M., H. Akari, A. Sakurai, A. Yoshida, T. Chiba, K. Tanaka, K. Strebel, and A. Adachi. 2004. Expression of HIV-1 accessory protein Vif is controlled uniquely to be low and optimal by proteasome degradation. Microbes Infect. 6:791-798. [DOI] [PubMed] [Google Scholar]
- 20.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 21.Gautsch, J. W., J. H. Elder, J. Schindler, F. C. Jensen, and R. A. Lerner. 1978. Structural markers on core protein p30 of murine leukemia virus: functional correlation with Fv-1 tropism. Proc. Natl. Acad. Sci. USA 75:4170-4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goff, S. P. 2004. Genetic control of retrovirus susceptibility in mammalian cells. Annu. Rev. Genet. 38:61-85. [DOI] [PubMed] [Google Scholar]
- 23.Harris, R. S., and M. T. Liddament. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4:868-877. [DOI] [PubMed] [Google Scholar]
- 24.Hatziioannou, T., S. Cowan, S. P. Goff, P. D. Bieniasz, and G. J. Towers. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatziioannou, T., D. Perez-Caballero, S. Cowan, and P. D. Bieniasz. 2005. Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells. J. Virol. 79:176-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc. Natl. Acad. Sci. USA 101:10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirsch, V. M., G. Dapolito, P. R. Johnson, W. R. Elkins, W. T. London, R. J. Montali, S. Goldstein, and C. Brown. 1995. Induction of AIDS by simian immunodeficiency virus from an African green monkey: species-specific variation in pathogenicity correlates with the extent of in vivo replication. J. Virol. 69:955-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirsch, V. M., R. A. Olmsted, M. Murphey-Corb, R. H. Purcell, and P. R. Johnson. 1989. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339:389-392. [DOI] [PubMed] [Google Scholar]
- 29.Ikeda, Y., L. M. Ylinen, M. Kahar-Bador, and G. J. Towers. 2004. Influence of gag on human immunodeficiency virus type 1 species-specific tropism. J. Virol. 78:11816-11822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kao, S., M. A. Khan, E. Miyagi, R. Plishka, A. Buckler-White, and K. Strebel. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 77:11398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kao, S., E. Miyagi, M. A. Khan, H. Takeuchi, S. Opi, R. Goila-Gaur, and K. Strebel. 2004. Production of infectious human immunodeficiency virus type 1 does not require depletion of APOBEC3G from virus-producing cells. Retrovirology 1:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karczewski, M. K., and K. Strebel. 1996. Cytoskeleton association and virion incorporation of the human immunodeficiency virus type 1 Vif protein. J. Virol. 70:494-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 101:10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan, M. A., C. Aberham, S. Kao, H. Akari, R. Gorelick, S. Bour, and K. Strebel. 2001. Human immunodeficiency virus type 1 Vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA. J. Virol. 75:7252-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khan, M. A., S. Kao, E. Miyagi, H. Takeuchi, R. Goila-Gaur, S. Opi, C. L. Gipson, T. G. Parslow, H. Ly, and K. Strebel. 2005. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J. Virol. 79:5870-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kootstra, N. A., C. Munk, N. Tonnu, N. R. Landau, and I. M. Verma. 2003. Abrogation of postentry restriction of HIV-1-based lentiviral vector transduction in simian cells. Proc. Natl. Acad. Sci. USA 100:1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozak, C. A., and A. Chakraborti. 1996. Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology 225:300-305. [DOI] [PubMed] [Google Scholar]
- 38.Mangeat, B., P. Turelli, S. Liao, and D. Trono. 2004. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 279:14481-14483. [DOI] [PubMed] [Google Scholar]
- 39.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 40.Nisole, S., C. Lynch, J. P. Stoye, and M. W. Yap. 2004. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. USA 101:13324-13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ott, D. E., L. V. Coren, D. G. Johnson, R. C. Sowder II, L. O. Arthur, and L. E. Henderson. 1995. Analysis and localization of cyclophilin A found in the virions of human immunodeficiency virus type 1 MN strain. AIDS Res. Hum. Retrovir. 11:1003-1006. [DOI] [PubMed] [Google Scholar]
- 42.Owens, C. M., B. Song, M. J. Perron, P. C. Yang, M. Stremlau, and J. Sodroski. 2004. Binding and susceptibility to postentry restriction factors in monkey cells are specified by distinct regions of the human immunodeficiency virus type 1 capsid. J. Virol. 78:5423-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Owens, C. M., P. C. Yang, H. Gottlinger, and J. Sodroski. 2003. Human and simian immunodeficiency virus capsid proteins are major viral determinants of early, postentry replication blocks in simian cells. J. Virol. 77:726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sayah, D. M., E. Sokolskaja, L. Berthoux, and J. Luban. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569-573.15243629 [Google Scholar]
- 45.Schrofelbauer, B., D. Chen, and N. R. Landau. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. USA 101:3927-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 47.Sigal, N. H., F. Dumont, P. Durette, J. J. Siekierka, L. Peterson, D. H. Rich, B. E. Dunlap, M. J. Staruch, M. R. Melino, S. L. Koprak, et al. 1991. Is cyclophilin involved in the immunosuppressive and nephrotoxic mechanism of action of cyclosporin A? J. Exp. Med. 173:619-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon, J. H., D. L. Miller, R. A. Fouchier, M. A. Soares, K. W. Peden, and M. H. Malim. 1998. The regulation of primate immunodeficiency virus infectivity by Vif is cell species restricted: a role for Vif in determining virus host range and cross-species transmission. EMBO J. 17:1259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sokolskaja, E., L. Berthoux, and J. Luban. 2006. Cyclophilin A and TRIM5α independently regulate human immunodeficiency virus type 1 infectivity in human cells. J. Virol. 80:2855-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sokolskaja, E., D. M. Sayah, and J. Luban. 2004. Target cell cyclophilin A modulates human immunodeficiency virus type 1 infectivity. J. Virol. 78:12800-12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strebel, K., D. Daugherty, K. Clouse, D. Cohen, T. Folks, and M. A. Martin. 1987. The HIV “A” (sor) gene product is essential for virus infectivity. Nature 328:728-730. [DOI] [PubMed] [Google Scholar]
- 52.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 53.Stremlau, M., M. Perron, M. Lee, Y. Li, B. Song, H. Javanbakht, F. Diaz-Griffero, D. J. Anderson, W. I. Sundquist, and J. Sodroski. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5{alpha} restriction factor. Proc. Natl. Acad. Sci. USA 103:5514-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takeuchi, H., S. Kao, E. Miyagi, M. A. Khan, A. Buckler-White, R. Plishka, and K. Strebel. 2005. Production of infectious SIVagm from human cells requires functional inactivation but not viral exclusion of human APOBEC3G. J. Biol. Chem. 280:375-382. [DOI] [PubMed] [Google Scholar]
- 55.Thali, M., A. Bukovsky, E. Kondo, B. Rosenwirth, C. T. Walsh, J. Sodroski, and H. G. Gottlinger. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372:363-365. [DOI] [PubMed] [Google Scholar]
- 56.Towers, G., M. Bock, S. Martin, Y. Takeuchi, J. P. Stoye, and O. Danos. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA 97:12295-12299.11027299 [Google Scholar]
- 57.Towers, G. J., T. Hatziioannou, S. Cowan, S. P. Goff, J. Luban, and P. D. Bieniasz. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138-1143. [DOI] [PubMed] [Google Scholar]
- 58.Tropschug, M., I. B. Barthelmess, and W. Neupert. 1989. Sensitivity to cyclosporin A is mediated by cyclophilin in Neurospora crassa and Saccharomyces cerevisiae. Nature 342:953-955. [DOI] [PubMed] [Google Scholar]
- 59.Xu, H., E. S. Svarovskaia, R. Barr, Y. Zhang, M. A. Khan, K. Strebel, and V. K. Pathak. 2004. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl. Acad. Sci. USA 101:5652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]