Abstract
Paramyxoviruses utilize both an attachment protein and a fusion (F) protein to drive virus-cell and cell-cell fusion. F exists functionally as a trimer of two disulfide-linked subunits: F1 and F2. Alignment and analysis of a set of paramyxovirus F protein sequences identified three conserved blocks (CB): one in the fusion peptide/heptad repeat A domain, known to play important roles in fusion promotion, one in the region between the heptad repeats of F1 (CBF1) (A. E. Gardner, K. L. Martin, and R. E. Dutch, Biochemistry 46:5094-5105, 2007), and one in the F2 subunit (CBF2). To analyze the functions of CBF2, alanine substitutions at conserved positions were created in both the simian virus 5 (SV5) and Hendra virus F proteins. A number of the CBF2 mutations resulted in folding and expression defects. However, the CBF2 mutants that were properly expressed and trafficked had altered fusion promotion activity. The Hendra virus CBF2 Y79A and P89A mutants showed significantly decreased levels of fusion, whereas the SV5 CBF2 I49A mutant exhibited greatly increased cell-cell fusion relative to that for wild-type F. Additional substitutions at SV5 F I49 suggest that both side chain volume and hydrophobicity at this position are important in the folding of the metastable, prefusion state and the subsequent triggering of membrane fusion. The recently published prefusogenic structure of parainfluenza virus 5/SV5 F (H. S. Yin et al., Nature 439:38-44, 2006) places CBF2 in direct contact with heptad repeat A. Our data therefore indicate that this conserved region plays a critical role in stabilizing the prefusion state, likely through interactions with heptad repeat A, and in triggering membrane fusion.
Paramyxoviruses are a diverse viral family including well-known human pathogens such as measles virus and respiratory syncytial virus (RSV), as well as the avian pathogen Newcastle disease virus (NDV) and the canine virus parainfluenza virus 5/simian virus 5 (PIV5/SV5). Hendra virus (HeV) and Nipah virus, two newly emergent zoonotic paramyxoviruses, are members of a new genus known as Henipavirus within the subfamily Paramyxovirinae, due in part to their larger genomes and antibody cross-reactivity (18, 54). HeV and Nipah virus also share approximately 83% amino acid sequence identity to each other and less than 30% to the rest of the family (18, 54). HeV was first identified in 1994 during an outbreak of severe respiratory illness near Brisbane, Australia, which resulted in the deaths of 14 horses and two humans, one of whom succumbed to respiratory illness and the other to viral meningoencephalitis (30, 35). Nipah virus was responsible for an outbreak of viral encephalitis in Malaysia in 1998, which resulted in the deaths of 105 people and the preventative destruction of more than 1 million swine (8, 9). In recent outbreaks of Nipah virus, resulting in fatality rates as high as 70%, human-to-human transmission was suspected (23). The distinction of HeV and Nipah virus as NIAID priority pathogens drives current research efforts toward the identification and optimization of antiviral therapies for these fatal viruses.
Most paramyxoviruses studied to date enter target host cells at a neutral pH via fusion of the viral envelope with the lipid bilayer of a target cell plasma membrane. However, recent data published by Schowalter et al. indicate that the fusion protein (F) of the paramyxovirus human metapneumovirus displays enhanced cell-cell fusion promotion at an acidic pH (46). Paramyxovirus membrane fusion is directed by the concerted efforts of two surface glycoproteins: F protein, which promotes membrane fusion, and an attachment protein, designated H (hemagglutinin), HN (hemagglutinin-neuraminidase), or G (glycoprotein), which allows for initial binding between the virus and the target cell. Although the trigger for F-mediated membrane fusion is still unknown, it is postulated that interactions between the attachment protein and F protein stimulate conformational changes in F that drive the merger of viral and host cell membranes (reviewed in references 26 and 29). For many paramyxoviruses, including HeV, the homotypic attachment protein is required for F-mediated membrane fusion (5, 24, 51). It has been shown, however, that a single point mutation within the NDV F protein allows for F-promoted, HN-independent fusion in the absence of the normally required attachment protein (48). Interestingly, some strains of SV5/PIV5 contain F proteins that can promote cell-cell membrane fusion in the absence of the HN protein, though the presence of HN stimulates fusion (12, 22, 40, 52, 55). Furthermore, the requirement for HN by some strains of SV5 can be eliminated when the F protein is destabilized by mutation or an elevated temperature (42).
Paramyxovirus F proteins initially exist as an inactive F0 precursor form, which must undergo N-linked glycosylation, homotrimerization, and subsequent proteolytic processing into the disulfide-linked heterodimer F1+F2 to be fusogenically active. The F1 subunit contains certain regions of known function, including an N-terminal fusion peptide domain, which inserts into the target cell membrane during fusion (1, 20, 32); a C-terminal transmembrane (TM) anchor; and two heptad repeats, heptad repeat A (HRA) and HRB, which are known to form a highly stable six-helix bundle that is critical for membrane fusion (2, 49). However, the ∼100-amino-acid F2 subunit currently has no defined function. In comparing paramyxovirus F proteins to other class I viral fusion proteins such as human immunodeficiency virus Env and influenza virus HA, F2 would be the equivalent of the non-TM-containing human immunodeficiency virus gp120 subunit and of influenza virus HA1, which are both known to be involved in attachment to host target cells prior to fusion initiation (3, 50). The function of the F2 subunit in paramyxoviruses is unknown, since the attachment proteins (H, HN, or G) are utilized for initial binding to target cells.
To identify amino acid conservation within paramyxovirus F proteins, and to better characterize regions of unknown function in F1 and F2, sequences from the F proteins of six paramyxovirus family members representing four different genera were aligned and analyzed for blocks of sequence conservation. Using two separate algorithms for sequence alignment, the Block Maker program (19) identified three conserved regions: one in the fusion peptide/HRA region, known to be involved in promotion of F-mediated membrane fusion; a second in the F2 subunit, designated the conserved block in F2 (CBF2); and a third within a large stretch of amino acids between the heptad repeats of F1, designated CBF1. Previous research in multiple laboratories has indicated a role for HRA in folding and fusion (47, 49, 56). We have recently demonstrated that there is a role for CBF1 in F protein folding and homotrimerization (15). Two previous studies of paramyxovirus F proteins have included mutations of a small number of residues that fall within CBF2. First, NDV F protein temperature-sensitive mutants containing mutations at the first residue of CBF2 and the residue preceding it (S60T and I61L) could overcome processing defects introduced by mutations in the HRA region, suggesting that these two regions may interact (53). Additionally, residues within the putative HRC region within F2, two of which fall within the C-terminal portion of CBF2, are important for measles virus F protein folding or fusion promotion (43). The finding of F protein sequence conservation among diverse members of the paramyxovirus family and the critical roles of HRA and CBF1 led us to hypothesize that CBF2 also has a conserved function, either in the promotion of membrane fusion or in the folding and processing of the F protein.
Since SV5 and HeV are two disparate members within the paramyxovirus family, we employed site-directed mutagenesis to introduce point mutations of the completely conserved residues within CBF2 of these two viral fusion proteins. The amino acids that define CBF2 include residues 49 to 88 in SV5 F and 57 to 95 in HeV F (see Fig. 1). In general, mutation of residues near the interior of CBF2 resulted in proteins that were defective in trimerization, trafficking, and cleavage, whereas mutations made to residues at the edge of CBF2 resulted in altered fusion promotion activity. However, the fusion phenotype of these mutants (hypo- versus hyperfusogenic) differed between SV5 and HeV F. The HeV CBF2 Y79A and P89A mutants exhibited a reduced level of F-promoted cell-cell fusion, as seen in syncytium assays and reporter gene assays. Interestingly, two of the SV5 CBF2 mutations (I49A and P82A) resulted in increased cell-cell fusion, with the SV5 F I49A mutant promoting membrane fusion at levels approximately three times greater than that for wild-type (wt) F. In contrast, a dramatic decrease in the level of fusion was observed with the SV5 F V50A mutant. Studies of additional substitutions at SV5 F residue I49 suggest that both side chain volume and hydrophobicity at this position greatly influence both initial protein folding and the triggering of membrane fusion. The recently published prefusogenic structure of PIV5/SV5 F (58) demonstrates that CBF2 interacts directly with HRA. Our data suggest that CBF2 may play a critical role in stabilizing the F protein and HRA in the prefusion state, likely through interactions with HRA, and in the triggering of membrane fusion.
FIG. 1.
Schematic of a paramyxovirus F protein and identification of CBF2. FP, fusion peptide. Backgrounds: solid, completely conserved amino acids; dark shading, residues conserved across most members; light shading, similar amino acids. Underlining indicates mutations made to alanine within SV5 F and HeV F.
MATERIALS AND METHODS
Cells and viruses.
CV-1, Vero, HeLa T4, BHK 21F, and BSR cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. BSR cells (provided by Karl-Klaus Conzelmann, Max Pettenkofer Institut) were derived from BHK cells to constitutively express the T7 polymerase (6). G-418 sulfate (Gibco Invitrogen, Carlsbad, CA) was added to the BSR medium to select for the T7-expressing cells. The recombinant vaccinia virus vTF7-3, expressing T7 polymerase (13), was propagated in CV-1 cells.
Plasmids.
Plasmids containing the HeV F and G genes were kindly provided by Lin-Fa Wang (Australian Animal Health Laboratory), and genes were ligated into the pCAGGS mammalian expression vector as described previously (38). HeV F was also ligated into pGEM4Z as described previously (10). Plasmids pGEM2X-SV5 F, pCAGGS-SV5 F, and pCAGGS-SV5 HN were provided by Robert Lamb (Howard Hughes Medical Institute, Northwestern University, Evanston, IL). Site-directed mutagenesis was performed on both the pGEM2X-SV5 F and pGEM4Z-HeV F plasmids using the QuikChange site-directed mutagenesis system (Stratagene, La Jolla, CA). The F genes of all mutants were sequenced to ensure that no secondary mutations were present. Specific SV5 F and HeV F mutants were subcloned into pCAGGS as previously described (10, 42).
Antibodies.
Robert Lamb kindly provided SV5 F antipeptide antibodies to amino acids 82 to 96 within the SV5 F2 subunit. Antipeptide antibodies (Genemed Custom Peptide Antibody Service, San Francisco, CA) were generated to amino acids 516 to 529 or 526 to 539 within the cytoplasmic tail of the SV5 F or HeV F protein, respectively. The SV5 F cytoplasmic tail antibody was utilized to examine proteolytic processing via pCAGGS expression (see Fig. 2C), since lack of antibody recognition was a concern for the SV5 F P82A mutant. Monoclonal antibody F1a, provided by Richard Randall (University of St Andrews, Fife, United Kingdom) (44), and a fluorescein isothiocyanate (FITC)-conjugated secondary antibody, goat anti-mouse immunoglobulin G (Jackson Immunoresearch Laboratories, West Grove, PA), were used for flow cytometry.
FIG. 2.
Homotrimerization and proteolytic processing of the CBF2 mutants. (A and B) wt SV5 F (A) and HeV F (B) and the CBF2 mutants were expressed in HeLa T4 cells by use of the recombinant vaccinia virus-T7 polymerase system. Three hours posttransfection, cells were labeled with Tran35S for 30 min and chased for 1 h. Samples were immunoprecipitated, separated on a 3.5% SDS-polyacrylamide gel in the absence of a reducing agent, and analyzed by use of the Storm or Typhoon phosphorimaging system. (C and D) wt SV5 F (C) and HeV F (D) and the CBF2 mutants were expressed in Vero cells using the pCAGGS expression plasmid. After overnight transfection, cells were starved, labeled with Tran35S for 1 h, and chased for 2 h, followed by immunoprecipitation and analysis by 15% SDS-polyacrylamide gel electrophoresis.
Expression of wt F and F protein mutants, metabolic labeling, and immunoprecipitation.
For expression of SV5 and HeV wt and mutant F proteins in the T7 promoter-driven pGEM plasmid vector system, subconfluent monolayers of HeLa T4 cells were first infected with recombinant vaccinia virus vTF7-3 (1 × 107 PFU/cell in DMEM plus 1% bovine serum albumin) for 60 min to allow for expression of the T7 polymerase (13). Cells were washed with phosphate-buffered saline (PBS) and transfected with 2 μg of pGEM2X-SV5 F or pGEM4Z-HeV F wt or mutant constructs by using Lipofectamine Plus reagents (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer's protocol.
For cross-linking analysis, following a 3-h incubation at 37°C, cells were washed with PBS, starved in cysteine- and methionine-deficient (Cys− Met−) DMEM for 30 min, and metabolically labeled for 30 min with Tran35S (100 μCi/ml; MP Biomedicals, Inc., Irvine, CA) in Cys− Met− DMEM. Cells were then washed with PBS and chased in cold (nonradioactive) DMEM for 1 h at 37°C. Cells were removed from culture dishes using PBS deficient in calcium and magnesium chloride (PBS−) plus 50 mM EDTA. Chemical cross-linking using the reagent 3,3′-dithiobi(sulfosuccinimidyl propionate) (DTSSP; 1 mM; Pierce Biotechnology Inc., Rockford, IL) was performed in the presence of 0.2% NP-40 as described previously (45). Cells were then lysed in radioimmunoprecipitation assay (RIPA) buffer supplemented with protease inhibitors (41) and 25 mM iodoacetamide. Immunoprecipitations were performed as described above (25) using SV5 F or HeV F antipeptide antibodies and protein A-conjugated Sepharose beads (Amersham/GE Healthcare Bio-Sciences, Piscataway, NJ) to pull down antibody-bound proteins. Samples were separated on a 3.5% polyacrylamide gel in the absence of the reducing agent dithiothreitol (Bio-Rad, Hercules, CA) to allow visualization of the precursor F0 form and were analyzed by storage phosphor autoradiography using a Storm or Typhoon imaging system (Amersham/GE Healthcare Bio-Sciences, Piscataway, NJ).
For expression of SV5 and HeV wt and mutant F genes in the pCAGGS vector, which utilizes a chicken actin promoter for high-level expression (31), subconfluent monolayers of Vero cells were transfected with 2 μg wt pCAGGS-SV5 F, pCAGGS-HeV F, or constructs containing the F protein mutant genes by use of Lipofectamine Plus reagents according to the manufacturer's specifications (Invitrogen Life Technologies, Carlsbad, CA). Following overnight incubation, cells were starved in Cys− Met− DMEM for 30 min and subsequently metabolically labeled with Tran35S (100 μCi/ml; MP Biomedicals, Inc., Irvine, CA) for 30 to 60 min. Cells were then washed twice with PBS and either lysed in RIPA buffer supplemented with protease inhibitors and 25 mM iodoacetamide or incubated in cold DMEM for 2 to 3 h at 37°C prior to lysis. Immunoprecipitations were performed as described above using SV5 F or HeV F antipeptide antibodies and protein A-conjugated Sepharose beads (Amersham, Piscataway, NJ) to pull down antibody-bound proteins. Samples were separated on a 15% polyacrylamide gel in the presence of a reducing agent to allow visualization of both the F1 and F2 subunits and were analyzed by storage phosphor autoradiography using a Storm or Typhoon imaging system (Amersham).
Flow cytometry.
SV5 F wt or mutant proteins were expressed in HeLa T4 cells using the pCAGGS expression vector with Lipofectamine Plus reagents. Cells were prepared for flow cytometry as described previously (21) with the primary monoclonal antibody F1a and the FITC-conjugated secondary antibody, goat anti-mouse immunoglobulin G (Jackson Immunoresearch Laboratories, West Grove, PA). The fluorescence intensity of 10,000 cells was measured by flow cytometry (FACSCalibur; Becton Dickinson, Mountain View, CA).
Biotinylation of cell surface proteins.
Following transient transfection of Vero cells with wt or mutant HeV F proteins as described above for the pCAGGS system, cells were starved for 30 min and metabolically labeled for 3 h. Cells were washed three times with ice-cold PBS− at pH 8. Cells were then incubated in 1 ml of the cell-impermeant analog of biotin EZ-Link sulfo-N-hydroxysuccinimide-biotin (sulfo-NHS-biotin; 1 mg/ml; Pierce, Rockford, IL) in PBS− (pH 8) by first rocking gently for 10 min at 4°C and then resting at 4°C for 25 min. Cells were then washed three times with ice-cold PBS− (pH 8), lysed in RIPA buffer, and immunoprecipitated as described for metabolic labeling experiments. One hundred microliters of 10% sodium dodecyl sulfate (SDS) was added to the protein A-Sepharose beads, and the samples were boiled for 10 min. The supernatant was removed and saved. Fifty microliters of 10% SDS was added to the protein A-Sepharose beads, and the samples were boiled for another 10 min. The supernatant was removed and added to the previous supernatant. Thirty microliters (one-fifth) of the combined supernatant was saved for analysis of the total F protein in the lysed cells. To the remaining 120 μl, 400 μl of biotinylation dilution buffer (20 mM Tris [pH 8], 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.2% bovine serum albumin) and 30 μl of immobilized streptavidin beads (Pierce) were added, and the samples were rocked at 4°C for 1 h. Samples were washed with RIPA buffer as described above. Both total and surface portions were analyzed on 15% polyacrylamide gels under reducing conditions and visualized by using the Storm or Typhoon imaging system (Amersham).
Syncytium assay for fusion.
wt or mutant SV5 or HeV F, along with either the SV5 attachment protein HN or the HeV attachment protein G, was expressed using the pCAGGS system in BHK 21F or Vero cells as described above. Syncytia were examined 15 to 29 h posttransfection using a Nikon TS100 inverted phase-contrast microscope (Nikon Inc., Garden City, NY), and pictures were taken at ×100 magnification using a Nikon Coolpix995 digital camera.
Luciferase reporter gene assay for fusion.
Subconfluent monolayers of Vero cells in six-well plates were transiently cotransfected with wt or mutant SV5 (0.8 μg) or HeV F (0.55 μg) along with either SV5 HN (0.8 μg) or HeV G (1.1 μg) and the T7 control plasmid (0.8 μg) containing luciferase cDNA under the control of the T7 promoter by using Lipofectamine Plus according to the manufacturer's protocol. At 24 h posttransfection, BSR cells, which constitutively express T7 polymerase (6), were overlaid on the transfected Vero cells at a ratio of ∼1:1 for 3 h. Luciferase activity was analyzed using a luciferase assay system (Promega) according to the manufacturer's protocol, with readings within the linear range for this assay. Light emission was read using an Lmax luminometer (Molecular Devices, Sunnyvale, CA).
RESULTS
Identification and mutagenesis of CBF2.
Sequences of the F proteins from the following paramyxovirus family members were aligned and analyzed for blocks of conservation: mumps virus (33), SV5/PIV5 (39), NDV (27), Sendai virus (14), Nipah virus (18), and HeV (17). Sequences were aligned using either the MOTIF program or the GIBBs sampler, and conserved blocks were identified by Block Maker (19). Both alignment programs, which utilize different sequence alignment algorithms, recognized a conserved region in the fusion peptide/HRA region, known to play critical roles in F-mediated membrane fusion. Both programs also identified blocks of conservation in the F2 subunit, designated CBF2 (Fig. 1), and in the large intervening region between the heptad repeats of F1 (CBF1). Our laboratory has demonstrated a function for CBF1 in F protein folding and homotrimerization (15). We hypothesized that CBF2 also has a conserved function, either in promotion of F-mediated membrane fusion or in the folding or processing of paramyxovirus F proteins.
A protein BLAST search was performed to identify sequences similar to CBF2 in other proteins in order to define its function, but this search resulted in no matches outside of paramyxovirus fusion proteins. Site-directed mutagenesis was therefore employed to introduce mutations into the seven completely conserved residues within CBF2 of SV5 F (I49A, V50A, K52A, P55A, Y72A, L80A, and P82A) and HeV F (I57A, V58A, K60A, P63A, Y79A, L87A, and P89A) (Fig. 1) in order to examine the effects on either protein folding or promotion of membrane fusion.
Folding and proteolytic processing of F proteins.
Paramyxovirus F proteins must undergo oligomerization into a homotrimer within the endoplasmic reticulum and subsequent proteolytic cleavage to be fusogenically active. Furin, a member of the proprotein convertase family, is responsible for cleaving SV5 F (16), as well as other paramyxovirus F proteins (4, 34, 59), within the trans-Golgi network (4). Cathepsin L, an endosomal/lysosomal protease, was recently identified as the protease responsible for cleavage of both HeV and Nipah virus F proteins (36, 37). HeV and Nipah virus F are hypothesized to be transported from the trans-Golgi network to the cell surface and subsequently internalized, cleaved, and then recycled to the cell surface (11, 28). All of these folding and processing events, along with a final targeting to the cell surface, must occur in order for the F protein to be fusogenically active.
To examine the effects of mutations in the F2 conserved block on expression and proper folding, the seven SV5 F and HeV F CBF2 mutants were analyzed against the wt to determine if homotrimerization had occurred, since formation of a trimer in the endoplasmic reticulum occurs prior to trafficking and subsequent proteolytic processing. Transient expression was performed using the recombinant vaccinia virus-T7 polymerase system (13). Cells were metabolically labeled for 30 min and chased for 1 h, followed by cross-linking of proteins with DTSSP, immunoprecipitation, and visualization via SDS-polyacrylamide gel electrophoresis and storage phosphor autoradiography. In the absence of a cross-linker, wt SV5 F forms a predominantly monomeric species, with some dimer and trimer forms visible. Upon the addition of a cross-linker, the dominant species is that of a discrete trimeric form (Fig. 2A). Among the SV5 F CBF2 mutants, only the I49A and V50A mutants form discrete trimers upon the addition of a cross-linker, while the P82A mutant displays an overall lower level of F protein expression and a detected trimeric species. The remainder of the SV5 CBF2 mutants formed a smear of higher-molecular-weight aggregates on the gels after the addition of a cross-linker, suggesting a malfolded protein (Fig. 2A). wt HeV F as well exhibited a predominantly monomeric species prior to cross-linker addition and an increase in the level of a discrete trimer after a cross-linker was added (Fig. 2B). It is difficult to detect a discrete trimeric band for any of the HeV CBF2 mutants, since a smear of higher-molecular-weight aggregates appears visible for all the mutants. The HeV F Y79A and P89A mutants display faint bands at the correct size for an F protein trimer, suggesting properly folded F proteins (Fig. 2B). Although some of the mutated residues that result in misfolded F proteins do not align between SV5 F and HeV F, they are, in general, located near the interior of CBF2 (SV5 F K52A, P55A, Y72A, and L80A; HeV F I57A, V58A, K60A, P63A, and L87A). In contrast, F protein mutants that form discrete trimers (SV5 F I49A, V50A, and P82A; HeV F Y79A and P89A) reside near the outer edges of CBF2 (Fig. 1 and 2A and B).
The CBF2 mutants in SV5 F or HeV F were also assayed for proteolytic processing. Only the SV5 F I49A, V50A, and P82A mutants and the HeV F Y79A and P89A mutants produced properly processed F proteins in the pGEM expression system (data not shown). Those mutants that were proteolytically cleaved (and the corresponding mutant in either SV5 F or HeV F) were subcloned into the pCAGGS mammalian expression vector. This plasmid allows for high-level expression from a chicken actin promoter in many cell types (31) and does not require prior viral infection of cells, thus making it ideal for assessing fusion promotion both qualitatively and quantitatively. Protein expression and processing after pCAGGS expression were examined by transient transfection of Vero cells, followed by a 1-h metabolic label and a 2-h chase. The SV5 F I49A, V50A, and P82A mutants were expressed and proteolytically processed (Fig. 2C), although the processing of both the I49A and P82A mutants was reduced: the percentage of cleaved F [F1/(F1 + F0)·100] was 21% for the I49A mutant and 14% for the P82A mutant, compared to 45% for the wt F protein. The V50A mutant, however, exhibited near-wt SV5 F levels of proteolytic processing, with 37% cleaved (Fig. 2C). The HeV F Y79A and P89A mutants were expressed at ∼ 30% of the level of wt HeV F, and the percentages of proteolytic cleavage measured 10% and 40%, respectively, compared to 45% cleaved wt HeV F (Fig. 2D).
Surface expression of F proteins.
Surface expression after proteolytic processing of paramyxovirus F proteins is required for cell-cell fusion promotion activity. Therefore, surface levels of the SV5 CBF2 mutants were measured by flow cytometry using the SV5 monoclonal antibody F1a and an FITC-conjugated secondary antibody. Mutants that were properly processed (the I49A, V50A, and P82A mutants [Fig. 2C]) were also present on the cell surface, although the mean fluorescence intensity was somewhat reduced, except for V50A, whose mean fluorescence intensity was slightly higher than that of wt SV5 F. Surface expression of the P82A mutant was reduced to less than 40% that of wt SV5 F (Table 1), which is consistent with the significant reduction in the level of proteolytic cleavage observed for this mutant. The I49A mutant, while cleaved at less than 50% of the level of wt SV5 F, was expressed on the cell surface at approximately 75% of the level of wt F. Flow cytometry was not performed on HeV F proteins due to the lack of an appropriate antibody.
TABLE 1.
Surface expression of the SV5 CBF2 mutantsa
| Mutation | % Positiveb | MFIc |
|---|---|---|
| Control (secondary antibody only) | 0.411 | 13.8 |
| Mock | 0.012 | 9.31 |
| wt SV5 F | 100 | 100 |
| I49A | 53.5 | 74.4 |
| V50A | 47.5 | 119 |
| Y72A | 0.026 | 14.2 |
| P82A | 36.9 | 34.8 |
Results are representative of three separate experiments and are normalized to the percentage for wt SV5 F, set at 100%.
Percentage of cells positive for F protein expression, determined by flow cytometry as described in Materials and Methods.
MFI, mean fluorescence intensity, determined by flow cytometry.
To quantitate the levels of surface expression of the HeV CBF2 mutants, cells were transfected and metabolically labeled as previously described. Surface F proteins were biotinylated with a cell-impermeant biotin analog, followed by immunoprecipitation and pull-down with streptavidin beads to visualize the surface-expressed proteins (Fig. 3). Only the HeV F Y79A and P89A mutants, the only mutations that produced cleaved F proteins, were expressed on the cell surface in the cleaved, fusogenically active form. The percentages of cleaved surface protein [F1/(F1 + F0)·100] were 28% (Y79A) and 49% (P89A), compared to 48% cleaved wt HeV F, yet the overall percentages of cleaved F (F1) expressed on the cell surface were only 17% (Y79A) and 23% (P89A) of the level of cleaved wt F (Fig. 3). These data suggest a defect in recycling to the cell surface after cleavage by cathepsin L in the endosomal pathway.
FIG. 3.
Surface expression of the HeV CBF2 mutants. wt HeV F and the CBF2 mutants were expressed in Vero cells by use of the pCAGGS system. After overnight transfection, cells were starved and labeled with Tran35S for 3 h. Samples were biotinylated and immunoprecipitated as total and surface populations as described in Materials and Methods. Samples were separated on a 15% SDS-polyacrylamide gel under reducing conditions and analyzed using the Storm or Typhoon phosphorimaging system.
Fusion promotion by the CBF2 mutants.
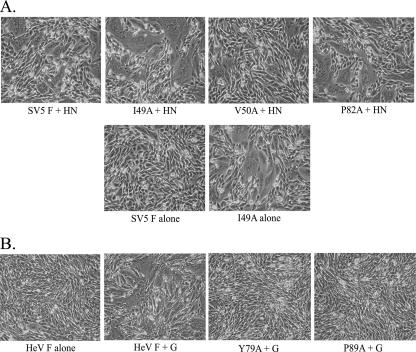
CBF2 mutants that were properly processed and trafficked to the cell surface were assayed for their ability to promote F-mediated cell-cell fusion. We first employed a syncytium assay to measure an overall level of cell-cell fusion. wt SV5 F, HeV F, and the CBF2 mutants were expressed in BHK cells with or without their attachment proteins by use of the pCAGGS vector expression system. Fifteen hours (SV5 F) or 29 h (HeV F) posttransfection, cells were examined for the formation of multinucleated giant cells, or syncytia.
The SV5 F V50A mutant, while processed similarly to wt SV5 F and expressed on the cell surface at levels slightly higher than that of wt F, was severely debilitated for syncytium formation (Fig. 4A). In contrast, the P82A mutant, for which cleavage and surface expression were significantly reduced, produced slightly larger syncytia than wt SV5 F. The SV5 F I49A mutant showed moderately reduced cleavage and slightly reduced surface expression yet displayed much larger syncytia than wt SV5 F when both were coexpressed with the attachment protein HN. SV5 F (strain W3A) is capable of inducing syncytium formation in the absence of the homotypic attachment protein HN, though HN does significantly stimulate fusion (12, 22, 40, 42, 52, 55). While expression of wt SV5 F in the absence of SV5 HN does not lead to syncytium formation by 15 h posttransfection, syncytia can be observed at ∼24 h (data not shown). The I49A mutant, however, promoted high levels of fusion in the absence of HN, exhibiting very large syncytia at 15 h posttranfection (also seen as early as 12 h posttransfection [data not shown]). Coexpression of HN appeared to enhance syncytium formation only slightly for this mutant.
FIG. 4.
Syncytium assay for cell-cell fusion. wt SV5 F (A) and HeV F (B) and the CBF2 mutants were expressed in BHK cells with or without their attachment proteins by use of the pCAGGS vector expression system. Fifteen hours (SV5 F) or 29 h (HeV F) posttransfection, pictures were taken of cells at ×100 magnification and examined for the formation of multinucleated giant cells termed syncytia.
For HeV F (Fig. 4B), the mutants that were properly processed (the HeV F Y79A and P89A mutants) displayed very little syncytium formation compared to wt HeV F. The reduction in the surface expression of cleaved F protein for these mutants may contribute to the decrease in F protein fusion promotion. However, a similar reduction in surface expression did not hinder the ability of the HeV F N464A mutant (7) to promote fusion at an enhanced level. It should be noted, in addition, that small syncytia were observed in Vero cells for the HeV F Y79A and P89A mutants (data not shown). These data demonstrate that mutations made to residues at the outer edges of CBF2 can modulate membrane fusion promotion by SV5 and HeV F proteins.
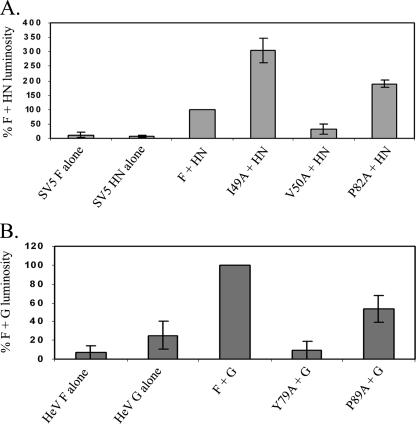
Membrane fusion activity was also examined using a reporter gene assay. wt SV5 F, HeV F, or the CBF2 mutants were expressed in Vero cells along with their homologous attachment proteins (required to facilitate cell-cell binding in this assay) by use of the pCAGGS vector expression system, and cells were also transfected with a plasmid containing the luciferase cDNA under the control of a T7 promoter. Twenty-four hours posttransfection, glycoprotein-expressing cells were overlaid for 3 h with BSR cells, which stably express T7 polymerase (6). Cell-cell fusion events allow for the accession of the T7 promoter in the Vero effector cell population by the T7 polymerase from the BSR target cells and subsequent production of luciferase. Upon the addition of a luciferase substrate and the subsequent reaction, the resultant luminosity reflects the level of cell-cell fusion. We observed a drastically decreased level of fusion for the SV5 F V50A mutant and an increased level of fusion for the P82A mutant (Fig. 5A), as seen in the syncytium data (Fig. 4A). For the I49A mutant, which displayed much larger syncytia than wt SV5 F (Fig. 4A), the level of fusion as measured by luminosity was approximately 300% that of wt F (Fig. 5A). This hyperfusogenic phenotype was even more apparent at 30°C (data not shown), a finding similar to results published for other hyperfusogenic SV5 F mutants (42, 56); at this temperature, the level of fusion promoted by wt SV5 F was much more drastically reduced than that promoted by the I49A mutant. These data indicate that surface expression alone does not dictate the fusogenic potential of F proteins and that SV5 F I49 may be a key residue in fusion promotion. Both of the HeV CBF2 mutants, which displayed negligible syncytium formation relative to that for wt F (Fig. 4B), also exhibited decreased levels of cell-cell fusion in the reporter gene assay, with Y79A mutant fusion near control levels and the P89A mutant at only ∼50% of the wt HeV F level of fusion promotion (Fig. 5B). Both mutants were cleaved and expressed on the cell surface at less than 30% of the wt HeV F level, suggesting that surface density may be a partial determinant of fusogenicity. The difference in cell-cell fusion promotion by the HeV F P89A mutant in syncytium versus reporter gene assays may be due to the different cell types utilized for these assays; the luciferase assay utilizes Vero cells, in which small syncytia were observed for both the Y79A and P89A mutants (data not shown).
FIG. 5.
Luciferase assay for cell-cell fusion. wt SV5 F (A) and HeV F (B) and the CBF2 mutants were expressed in Vero cells with their attachment proteins by use of the pCAGGS vector expression system, along with luciferase cDNA under the control of a T7 promoter. Twenty-four hours posttransfection, Vero cells were overlaid for 3 h with BSR cells, which stably express T7 polymerase. Luciferase transcription and reaction with the substrate luciferin produced light, which was used as a measurement for quantitating the level of cell-cell fusion. Numbers are normalized as percentages of the luminosity of wt F plus HN or of wt F plus G, set at 100%.
Further mutations made to SV5 F residue I49.
During the course of this research, the crystal structure of the prefusogenic, metastable form of PIV5/SV5 F was determined (58). Closer analysis of this structure (Fig. 6) revealed that CBF2 was adjacent to HRA, with certain residues making direct contact. Interestingly, in the prefusogenic, metastable form of SV5 F, HRA is compact and possesses only a partial helical structure. A portion of HRA constitutes two of three strands of a β-sheet, with a portion of CBF2 contributing the third β-strand (Fig. 6). SV5 F residues I49 and V50 fall within this third β-strand and are located in close proximity to L161, a residue displaying a hyperfusogenic phenotype when mutated to methionine (56). We hypothesized that the SV5 F V50A mutant may act to stabilize HRA, and hence the F protein, in the prefusion conformation, while the I49A mutant (and the L161M mutant) promotes triggering to the postfusion conformation. The mutation of isoleucine to alanine (I49A) is relatively conservative, with an exchange for a smaller, nonpolar side chain. We hypothesized that side chain packing was critical in stabilizing this β-sheet region and thus maintaining HRA in the prefusion conformation. Since the SV5 F I49A mutant exhibited such a pronounced hyperfusogenic phenotype, we employed site-directed mutagenesis to assess the effects of different residues at this position on F protein folding and fusion promotion. The following mutations were introduced at this position: I49L, I49V, I49K, I49G, I49F, I49S, and I49T. I49L, I49V, and I49F were chosen to examine the effects of other nonpolar side chains on packing in this region. I49K and I49G were introduced to examine the effects of charge and increased flexibility at this position, respectively. The I49T mutant would be similar in size to I49A but polar, whereas the I49S mutant is smaller and more polar. These mutants were examined for proper proteolytic cleavage (data not shown) and for cell surface expression by flow cytometry (Fig. 7A). The I49L, I49V, and I49T mutants displayed near-wt levels of cleaved protein (data not shown) and surface F protein (Fig. 7A), suggesting that hydrophobic side chains of similar volume do not disrupt initial protein folding into the prefusion metastable form. Not surprisingly, mutation from isoleucine to a charged lysine residue (I49K) resulted in an improperly processed and trafficked F protein, as did I49G, likely due to increased flexibility in CBF2 in a fold that is hypothesized to be rigid otherwise. The I49F and I49S mutants both displayed minimal proteolytic processing and surface expression, presumably due to a significant increase in side chain volume or a decrease in volume and an increase in polarity, respectively (data not shown and Fig. 7A).
FIG. 6.
CBF2 modeled within the prefusogenic form of PIV5/SV5 F. Purple, CBF2; light blue, HRA; gray, putative h4 helix of HRA; beige, fusion peptide; orange, L161; yellow, disulfide bond; green, completely conserved residues in CBF2. (Adapted from reference 58 using PyMOL 0.99 and color coded for clarification.).
FIG. 7.
Surface expression and fusion promotion by the SV5 F I49 mutants. (A) wt SV5 F and mutants with mutations of residue I49 were expressed in HeLa cells by use of the pCAGGS vector expression system and analyzed for surface expression by flow cytometry as described in Materials and Methods. Numbers are normalized as percentages of wt SV5 F surface expression (taken as 100%). (B) wt SV5 F and mutants with mutations of residue I49 were expressed in Vero cells with their attachment proteins by use of the pCAGGS vector expression system, along with luciferase cDNA under the control of a T7 promoter. Twenty-four hours posttransfection, Vero cells were overlaid for 3 h with BSR cells expressing T7 polymerase; they were subsequently analyzed for fusion promotion as described in the text. Numbers are normalized as percentages of the luminosity of wt F plus HN, set at 100%.
To assess the abilities of these mutants to induce F-mediated cell-cell fusion, we employed the luciferase reporter gene assay. wt SV5 F and the I49 mutants were expressed and examined for fusion promotion as described previously. The majority of the mutants that were not efficiently proteolytically processed or present on the cell surface at wt levels (the I49K, I49G, and I49S mutants) did not promote fusion above background levels. The I49A mutant again displayed a level of cell-cell fusion approximately 300% that of wt F. While the I49L and I49T mutants exhibited near-wt levels of fusion, both the I49V and I49F mutants promoted fusion above wt levels (Fig. 7B). This result was particularly unexpected for the I49F mutant, because the surface expression of this mutant was extremely low (Fig. 7A). Since increased surface density of SV5 F is known to increase levels of cell-cell fusion (12), this result suggests that higher surface levels of the I49F mutant would result in extremely high levels of cell-cell fusion.
A syncytium assay was utilized to confirm the reporter gene assay data. wt SV5 F and the I49 mutants were expressed in BHK cells along with the homotypic attachment protein HN as described previously, and at 24 h posttransfection, syncytia were examined (Fig. 8). The syncytium data are in general consistent with the luciferase data: extreme hyperfusogenic phenotypes were observed for the I49A and I49F mutants, and an increase in fusion over the wt level was observed for the I49V mutant. The I49L mutant displayed slightly increased fusion over the wt level in the syncytium assay, while fusion levels for this mutant were similar in the reporter gene assay. Similar small differences between the assays have been noted for a number of other mutants (28, 56) and may be due to the large difference in the incubation time for fusion between the two assays. The hyperfusogenic phenotypes were also observed for the I49A, I49F, I49L, and I49V mutants in the absence of HN (data not shown). These data indicate that mutations made to CBF2, particularly to SV5 F residue I49, can have dramatic effects on fusion promotion, resulting primarily from disruption of side chain packing, and thus stability, around this prefusion β-sheet.
FIG. 8.
Syncytium formation promoted by the SV5 F I49 mutants. wt SV5 F and mutants with mutations of residue I49 were expressed in BHK cells with the HN attachment protein by use of the pCAGGS vector expression system. Twenty-four hours posttransfection, pictures of cells were taken at ×100 magnification and examined for the formation of syncytia.
DISCUSSION
Sequence alignment and analysis of disparate paramyxovirus fusion proteins allowed us to identify three blocks of conserved amino acid sequence within the paramyxovirus F protein. The first conserved block spans the fusion peptide and HRA regions, two domains that play critical roles in F-mediated membrane fusion (1, 2, 20, 32, 49). The second conserved block identified resides within a stretch of approximately 250 amino acids between the two heptad repeats. We have demonstrated that this conserved block is required for proper folding of the F protein into its fusogenically active, trimeric form (15). Based on structural data of the prefusogenic form of PIV5/SV5 F (58), CBF1 is shown to pack tightly against the fusion peptide domain and is likely critical for promotion of proper folding of this extremely hydrophobic region.
Our mutational analysis of CBF2 revealed that this conserved region plays an important role both in the proper folding of paramyxovirus F proteins and in fusion regulation. Mutation of conserved residues within CBF2 resulted in either protein folding defects, as seen for mutants with changes near the interior of the region, or altered fusion promotion activities (summarized in Table 2). We observed reduced levels of cell-cell fusion for the HeV CBF2 mutants that were proteolytically processed and expressed on the cell surface (the Y79A and P89A mutants). This hypofusogenic phenotype could result from the reduced level of surface expression observed for these mutants, since surface densities have been shown to play an important role in F-mediated membrane fusion (12), or decreased fusion could be due to a defect in the triggering of membrane fusion. The HeV F mutant N464A (7) exhibited reduced surface expression, at a level similar to those observed for Y79A and P89A, yet promoted cell-cell fusion at levels higher than those for wt F, suggesting that some HeV F mutants that are expressed at reduced levels on the cell surface can promote fusion efficiently. These data demonstrate that mutations made to residues at the outer edges of CBF2 can modulate membrane fusion promotion by HeV F proteins.
TABLE 2.
Summary of the CBF2 alanine substitutionsa
CBF2 residues subjected to mutation are underlined in the sequences.
bBoldfaced mutations highlight differences between SV5 F and HeV F.
c−, negative result; +, significantly below wt; ++, reduced from wt; +++, similar to wt F; ++++, higher than wt F; +++++, significantly higher than wt F.
dFusion summary based on syncytium and luciferase data.
In contrast to the HeV F data, mutations made to SV5 CBF2 resulted in either a reduced level of fusion (V50A), an increased level of fusion (P82A), or an extremely hyperfusogenic phenotype (I49A). The hyperfusogenic phenotype observed for the SV5 F P82A mutant contrasts with the results for the corresponding mutation made to HeV F (P89A) and with those for the measles virus F P86A mutant, which has been shown to be fusion incompetent (43), suggesting a difference in structural stability within and around CBF2, contributed by nonconserved residues, for different F proteins. Further mutations made to SV5 CBF2 residue I49 demonstrated that this residue is a key amino acid involved in maintaining wt levels of fusion. Mutation of this isoleucine residue to alanine (I49A) resulted in slightly diminished surface expression compared to that for wt SV5 F but greatly increased cell-cell fusion. Mutation to phenylalanine (I49F) resulted in almost negligible surface expression, although fusion levels increased significantly. These data suggest that CBF2 may be a key region in stabilizing the prefusion conformation of the F protein. We hypothesize that mutations which destabilize the F protein may lead to increased fusion promotion (SV5 F I49A, I49L, I49V, I49F, and P82A), although too much destabilization would lead to malfolding of the F protein (SV5 F I49K and I49G and mutations near the interior residues of CBF2). Those mutations that stabilize the prefusion form of F (SV5 F V50A) would allow for proper folding yet inhibit the triggering of fusion.
The prefusogenic structure of PIV5/SV5 F provides significant insight into possible functions for CBF2. The tertiary structure/overall fold of CBF2 flanks that of HRA (Fig. 6). CBF2 appears to buttress HRA, with conserved residues making contacts with HRA. This fold allows for the formation of the intermolecular disulfide bond connecting the F1 and F2 subunits. SV5 F residue I49 is located in close proximity to L161, a residue promoting a hyperfusogenic phenotype when mutated to methionine (56). These two residues are likely involved in stabilizing HRA within each monomer and preventing its premature triggering into the fusion-active form. In the prefusogenic, metastable form of SV5 F, HRA possesses only a partial helical structure and is quite compact. A portion of HRA forms two of three strands of a β-sheet, with a portion of CBF2 contributing the third β-strand (Fig. 6). We hypothesize that the stability of this β-sheet is critical for promoting the proper folding of both HRA and the entire F protein (and is disrupted for trimerization- and processing-defective mutants) as well as for the regulation of membrane fusion levels, which are disrupted for both hypo- and hyperfusogenic mutants. The charge and side chain volume/packing of amino acids in and around this critical β-sheet affect its stability and thus the stability of the overall fold in CBF2 and in the entire F protein. It is only after the triggering of the F protein that F refolds into the putative prehairpin intermediate structure, in which the fusion peptide swings outward to insert into the target cell membrane and HRA forms an extended α-helix as part of a trimeric coiled-coil structure (58). This N-terminal coiled-coil forms the internal part of the postfusion six-helix bundle structure with C-terminal HRB (2, 57, 58). It is postulated that mutation of SV5 F I49 allows HRA to become destabilized such that less energy is required to promote the necessary conformational changes that result in fusion. We hypothesize that mutations which resulted in increased fusion promotion (I49A, I49L, I49V, and I49F) destabilized this critical β-sheet region just enough to promote the triggering of F to the postfusion state. However, greatly increased destabilization (I49K and I49G) would lead to malfolding of the F protein. The phenotype observed for the I49F mutant supports this hypothesis, because this result suggests that there is an extremely narrow transition between slight destabilization of this region and upregulated fusion, on the one hand, and significant instability and decreased folding and trafficking, on the other. The close proximity of residue I49 to another residue that has been shown to induce a hyperfusogenic phenotype after mutation (L161M) suggests that this is a triggering domain for SV5 F-promoted fusion.
It is clear that both folding and fusion effects are seen after mutation of CBF2 for both SV5 F and HeV F, but the effects of individual mutations are not the same. It is likely that this could be due, at least in part, to the differences in the triggering of fusion by these two F proteins. HeV, like many paramyxoviruses, requires binding of the attachment protein G to cell surfaces in order to initiate membrane fusion, and only the closely related Nipah virus G can substitute in order to trigger fusion. Low-pH pulses during HeV F expression, shown to enhance human metapneumovirus F-mediated fusion (46), do not permit cell-cell fusion in the absence of HeV G (data not shown). The SV5 F and HN proteins used in these studies were taken from SV5 strain W3A, and the wt F protein has been shown to induce syncytium formation in the absence of HN (12, 22, 40, 52, 55; also data not shown). These observations suggest that the SV5 F protein is more readily triggered for fusion than the HeV F protein, potentially due to differences in the stability of the prefusion, metastable state. It is possible that the structure of HeV F around HRA/CBF2 is more stable than that of SV5 F and that interactions with HeV G are needed to drive F conformational changes and initiate membrane fusion. The structure around SV5 HRA/CBF2, in contrast, may be in a sufficiently metastable state so that small perturbations in this region significantly reduce the requirement for SV5 HN in triggering F conformational changes and fusion promotion. No crystal structure for the entire HeV F protein ectodomain currently exists. Therefore, the amino acid sequence of HeV F was threaded onto the prefusion form of PIV5/SV5 F by using the Deep View Swiss-Pdb Viewer to predict the HeV F prefusion structure. HeV F aligns well overall with the prefusion form of PIV5/SV5 F and more specifically within CBF2 (A. Chang, M. A. Wurth, and R. E. Dutch, personal communication). Differing nonconserved residues within CBF2 between SV5 F and HeV F may therefore contribute to the phenotypic differences observed for corresponding residues in SV5 F and HeV F.
The data presented here indicate that the identified block in the F2 subunit (CBF2), conserved across disparate paramyxovirus family members, serves two important functions: (i) aiding in proper folding of the HRA region and (ii) stabilizing HRA (and the F protein) in its metastable, prefusion conformation. Thus, the F2 subunit has important roles in promoting the proper folding of the prefusion form of paramyxovirus F proteins and in regulating the necessary conformational changes within F to allow membrane fusion and the transition into the postfusion form.
Acknowledgments
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases (AI-51517) to R.E.D. A.E.G. was supported in part by a predoctoral fellowship from the American Heart Association, Ohio Valley Affiliate (0415223B).
We are grateful to Robert Lamb (HHMI, Northwestern University) for the pGEM2X-SV5 F, pCAGGS-SV5 F, and pCAGGS-SV5 HN plasmids and for SV5 F-specific antibodies. Lin-Fa Wang (Australian Animal Health Laboratory) kindly provided the HeV F and G plasmids. Richard Randall (University of St. Andrews, St. Andrews, Scotland) kindly provided the F1a monoclonal antibody. We thank David Rodgers (University of Kentucky) for assistance with figure construction. We also thank Trevor Creamer (University of Kentucky) and members of the Dutch lab for critical reviews of the manuscript.
Footnotes
Published ahead of print on 16 May 2007.
REFERENCES
- 1.Asano, K., and A. Asano. 1985. Why is a specific amino acid sequence of F glycoprotein required for the membrane fusion reaction between envelope of HVJ (Sendai virus) and target cell membranes? Biochem. Int. 10:115-122. [PubMed] [Google Scholar]
- 2.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 3.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 4.Bolt, G., and I. R. Pedersen. 1998. The role of subtilisin-like proprotein convertases for cleavage of the measles virus fusion glycoprotein in different cell types. Virology 252:387-398. [DOI] [PubMed] [Google Scholar]
- 5.Bossart, K. N., L.-F. Wang, B. Eaton, and C. C. Broder. 2001. Functional expression and membrane fusion tropism of the envelope glycoproteins of Hendra virus. Virology 290:121-135. [DOI] [PubMed] [Google Scholar]
- 6.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter, J. R., C. T. Pager, S. D. Fowler, and R. E. Dutch. 2005. Role of N-linked glycosylation of the Hendra virus fusion protein. J. Virol. 79:7922-7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chua, K. B., W. J. Bellini, P. A. Rota, B. H. Harcourt, A. Tamin, S. K. Lam, T. G. Ksiazek, P. E. Rollin, S. R. Zaki, W. Shieh, C. S. Goldsmith, D. J. Gulber, J. T. Roehrig, B. Eaton, A. R. Gould, J. Olson, H. Field, R. Daniels, A. E. Ling, C. J. Peters, L. J. Anderson, and B. W. Mahy. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432-1435. [DOI] [PubMed] [Google Scholar]
- 9.Chua, K. B., K. J. Goh, K. T. Wong, A. Kamarulzaman, P. S. Tan, T. G. Ksiazek, S. R. Zaki, G. Paul, S. K. Lam, and C. T. Tan. 1999. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet 354:1257-1259. [DOI] [PubMed] [Google Scholar]
- 10.Craft, W. W., Jr., and R. E. Dutch. 2005. Sequence motif upstream of the Hendra virus fusion protein cleavage site is not sufficient to promote efficient proteolytic processing. Virology 341:130-140. [DOI] [PubMed] [Google Scholar]
- 11.Diederich, S., M. Moll, H. D. Klenk, and A. Maisner. 2005. The Nipah virus fusion protein is cleaved within the endosomal compartment. J. Biol. Chem. 280:29899-29903. [DOI] [PubMed] [Google Scholar]
- 12.Dutch, R. E., S. B. Joshi, and R. A. Lamb. 1998. Membrane fusion promoted by increasing surface densities of the paramyxovirus F and HN proteins: comparison of fusion reactions mediated by simian virus 5 F, human parainfluenza virus type 3 F, and influenza virus HA. J. Virol. 72:7745-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujii, Y., K. Kiyotani, T. Yoshida, and T. Sakaguchi. 2001. Conserved and non-conserved regions in the Sendai virus genome: evolution of a gene possessing overlapping reading frames. Virus Genes 22:47-52. [DOI] [PubMed] [Google Scholar]
- 15.Gardner, A. E., K. L. Martin, and R. E. Dutch. 2007. A conserved region between the heptad repeats of paramyxovirus fusion proteins is critical for proper F protein folding. Biochemistry 46:5094-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garten, W., S. Hallenberger, D. Ortmann, W. Schafer, M. Vey, H. Angliker, E. Shaw, and H. D. Klenk. 1994. Processing of viral glycoproteins by the subtilisin-like endoprotease furin and its inhibition by specific peptidylchloroalkylketones. Biochimie 76:217-225. [DOI] [PubMed] [Google Scholar]
- 17.Gould, A. R. 1996. Comparison of the deduced matrix and fusion protein sequences of equine morbillivirus with cognate genes of the Paramyxoviridae. Virus Res. 43:17-31. [DOI] [PubMed] [Google Scholar]
- 18.Harcourt, B. H., A. Tamin, T. G. Ksiazek, P. E. Rollin, L. J. Anderson, W. J. Bellini, and P. A. Rota. 2000. Molecular characterization of Nipah virus, a newly emergent paramxyovirus. Virology 271:334-349. [DOI] [PubMed] [Google Scholar]
- 19.Henikoff, S., J. G. Henikoff, W. J. Alford, and S. Pietrokovski. 1995. Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene 163:GC17-GC26. [DOI] [PubMed] [Google Scholar]
- 20.Hernandez, L. D., R. J. Peters, S. E. Delos, J. A. T. Young, D. A. Agard, and J. M. White. 1997. Activation of a retroviral membrane fusion protein: soluble receptor-induced liposome binding of the ALSV envelope glycoprotein. J. Cell Biol. 139:1455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horvath, C. M., and R. A. Lamb. 1992. Studies on the fusion peptide of a paramyxovirus fusion glycoprotein: roles of conserved residues in cell fusion. J. Virol. 66:2443-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horvath, C. M., R. G. Paterson, M. A. Shaughnessy, R. Wood, and R. A. Lamb. 1992. Biological activity of paramyxovirus fusion proteins: factors influencing formation of syncytia. J. Virol. 66:4564-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu, V. P., M. J. Hossain, U. D. Parashar, M. M. Ali, T. G. Ksiazek, I. Kuzmin, M. Niezgoda, C. Rupprecht, J. Bresee, and R. F. Breiman. 2004. Nipah virus encephalitis reemergence, Bangladesh. Emerg. Infect. Dis. 10:2082-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu, X., R. Ray, and R. W. Compans. 1992. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J. Virol. 66:1528-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamb, R. A., P. R. Etkind, and P. W. Choppin. 1978. Evidence for a ninth influenza viral polypeptide. Virology 91:60-78. [DOI] [PubMed] [Google Scholar]
- 26.Lamb, R. A., R. G. Paterson, and T. S. Jardetzky. 2006. Paramyxovirus membrane fusion: lessons from the F and HN atomic structures. Virology 344:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGinnes, L. W., and T. G. Morrison. 1986. Nucleotide sequence of the gene encoding the Newcastle disease virus fusion protein and comparisons of paramyxovirus fusion protein sequences. Virus Res. 5:343-356. [DOI] [PubMed] [Google Scholar]
- 28.Meulendyke, K. A., M. A. Wurth, R. O. McCann, and R. E. Dutch. 2005. Endocytosis plays a critical role in proteolytic processing of the Hendra virus fusion protein. J. Virol. 79:12643-12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrison, T. G. 2003. Structure and function of a paramyxovirus fusion protein. Biochim. Biophys. Acta 1614:73-84. [DOI] [PubMed] [Google Scholar]
- 30.Murray, K., P. Selleck, P. Hooper, A. Hyatt, A. Gould, L. Gleeson, H. Westbury, L. Hiley, L. Selvey, B. Rodwell, et al. 1995. A morbillivirus that caused fatal disease in horses and humans. Science 268:94-97. [DOI] [PubMed] [Google Scholar]
- 31.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants by a novel eukaryotic vector. Gene 108:193-200. [DOI] [PubMed] [Google Scholar]
- 32.Novick, S. L., and D. Hoekstra. 1988. Membrane penetration of Sendai virus glycoproteins during the early stage of fusion with liposomes as determined by hydrophobic affinity labeling. Proc. Natl. Acad. Sci. USA 85:7433-7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okazaki, K., K. Tanabayashi, K. Takeuchi, M. Hishiyama, K. Okazaki, and A. Yamada. 1992. Molecular cloning and sequence analysis of the mumps virus gene encoding the L protein and the trailer sequence. Virology 188:926-930. [DOI] [PubMed] [Google Scholar]
- 34.Ortmann, D., M. Ohuchi, H. Angliker, E. Shaw, W. Garten, and H.-D. Klenk. 1994. Proteolytic cleavage of wild type and mutants of the F protein of human parainfluenza virus type 3 by two subtilisin-like endoproteases, furin and Kex2. J. Virol. 68:2772-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Sullivan, J. D., A. M. Allworth, D. L. Paterson, T. M. Snow, R. Boots, L. J. Gleeson, A. R. Gould, A. D. Hyatt, and J. Bradfield. 1997. Fatal encephalitis due to novel paramyxovirus transmitted from horses. Lancet 349:93-95. [DOI] [PubMed] [Google Scholar]
- 36.Pager, C. T., and R. E. Dutch. 2005. Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J. Virol. 79:12714-12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pager, C. T., W. W. Craft, Jr., J. Patch, and R. E. Dutch. 2006. A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology 346:251-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pager, C. T., M. A. Wurth, and R. E. Dutch. 2004. Subcellular localization and calcium and pH requirements for proteolytic processing of the Hendra virus fusion protein. J. Virol. 78:9154-9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paterson, R. G., T. J. R. Harris, and R. A. Lamb. 1984. Fusion protein of the paramyxovirus simian virus 5: nucleotide sequence of mRNA predicts a highly hydrophobic glycoprotein. Proc. Natl. Acad. Sci. USA 81:6706-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paterson, R. G., S. W. Hiebert, and R. A. Lamb. 1985. Expression at the cell surface of biologically active fusion and hemagglutinin-neuraminidase proteins of the paramyxovirus simian virus 5 from cloned cDNA. Proc. Natl. Acad. Sci. USA 82:7520-7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paterson, R. G., and R. A. Lamb. 1987. Ability of the hydrophobic fusion-related external domain of a paramyxovirus F protein to act as a membrane anchor. Cell 48:441-452. [DOI] [PubMed] [Google Scholar]
- 42.Paterson, R. G., C. J. Russell, and R. A. Lamb. 2000. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology 270:17-30. [DOI] [PubMed] [Google Scholar]
- 43.Plemper, R. K., and R. W. Compans. 2003. Mutations in the putative HR-C region of the measles virus F2 glycoprotein modulate syncytium formation. J. Virol. 77:4181-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Randall, R. E., D. F. Young, K. K. A. Goswami, and W. C. Russell. 1987. Isolation and characterization of monoclonal antibodies to simian virus 5 and their use in revealing antigenic differences between human, canine and simian isolates. J. Gen. Virol. 68:2769-2780. [DOI] [PubMed] [Google Scholar]
- 45.Russell, R., R. G. Paterson, and R. A. Lamb. 1994. Studies with cross-linking reagents on the oligomeric form of the paramyxovirus fusion protein. Virology 199:160-168. [DOI] [PubMed] [Google Scholar]
- 46.Schowalter, R. M., S. E. Smith, and R. E. Dutch. 2006. Characterization of human metapneumovirus F protein-promoted membrane fusion: critical roles for proteolytic processing and low pH. J. Virol. 80:10931-10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sergel, T. A., L. W. McGinnes, and T. G. Morrison. 2001. Mutations in the fusion peptide and adjacent heptad repeat inhibit folding or activity of the Newcastle disease virus fusion protein. J. Virol. 75:7934-7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sergel, T. A., L. W. McGinnes, and T. G. Morrison. 2000. A single amino acid change in the Newcastle disease virus fusion protein alters the requirement for HN protein in fusion. J. Virol. 74:5101-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sergel-Germano, T., C. McQuain, and T. Morrison. 1994. Mutations in the fusion peptide and heptad repeat regions of the Newcastle disease virus fusion protein block fusion. J. Virol. 68:7654-7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 51.Tanabayashi, K., and R. W. Compans. 1996. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J. Virol. 70:6112-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsurudome, M., M. Ito, M. Nishio, M. Kawano, H. Komada, and Y. Ito. 2001. Hemagglutinin-neuraminidase-independent fusion activity of simian virus 5 fusion (F) protein: difference in conformation between fusogenic and nonfusogenic F proteins on the cell surface. J. Virol. 75:8999-9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, C., G. Raghu, T. Morrison, and M. E. Peeples. 1992. Intracellular processing of the paramyxovirus F protein: critical role of the predicted amphipathic α-helix adjacent to the fusion domain. J. Virol. 66:4161-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, L. F., M. Yu, E. Hansson, L. I. Pritchard, B. Shiell, W. P. Michalski, and B. T. Eaton. 2000. The exceptionally large genome of Hendra virus: support for creation of a new genus within the family Paramyxoviridae. J. Virol. 74:9972-9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ward, C. D., R. G. Paterson, and R. A. Lamb. 1995. Mutants of the paramyxovirus SV5 fusion protein: regulated and extensive syncytium formation. Virology 209:242-249. [DOI] [PubMed] [Google Scholar]
- 56.West, D. S., M. S. Sheehan, P. K. Segeleon, and R. E. Dutch. 2005. Role of the simian virus 5 fusion protein N-terminal coiled-coil domain in folding and promotion of membrane fusion. J. Virol. 79:1543-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yin, H. S., R. G. Paterson, X. Wen, R. A. Lamb, and T. S. Jardetzky. 2005. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. USA 102:9288-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yin, H. S., X. Wen, R. G. Paterson, R. A. Lamb, and T. S. Jardetzky. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439:38-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zimmer, G., L. Budz, and G. Herrler. 2001. Proteolytic activation of respiratory syncytial virus fusion protein. Cleavage at two furin consensus sequences. J. Biol. Chem. 276:31642-31650. [DOI] [PubMed] [Google Scholar]