Abstract
The NS1A proteins of human influenza A viruses bind CPSF30, a cellular factor required for the processing of cellular pre-mRNAs, thereby inhibiting the production of all cellular mRNAs, including beta interferon mRNA. Here we show that the NS1A protein of the pathogenic H5N1 influenza A/Hong Kong/483/97 (HK97) virus isolated from humans has an intrinsic defect in CPSF30 binding. It does not bind CPSF30 in vitro and causes high beta interferon mRNA production and reduced virus replication in MDCK cells when expressed in a recombinant virus in which the other viral proteins are encoded by influenza A/Udorn/72. We traced this defect to the identities of amino acids 103 and 106 in the HK97 NS1A protein, which differ from the consensus amino acids, F and M, respectively, found in the NS1A proteins of almost all human influenza A virus strains. X-ray crystallography has shown that F103 and M106, which are not part of the CPSF30 binding pocket of the NS1A protein, stabilize the NS1A-CPSF30 complex. In contrast to the HK97 NS1A protein, the NS1A proteins of H5N1 viruses isolated from humans after 1998 contain F103 and M106 and hence bind CPSF30 in vitro and do not attenuate virus replication. The HK97 NS1A protein is less attenuating when expressed in a virus that also encodes the other internal HK97 proteins and under these conditions binds to CPSF30 to a substantial extent in vivo. Consequently, these internal HK97 proteins largely compensate for the absence of F103 and M106, presumably by stabilizing the NS1A-CPSF30 complex.
H5N1 influenza A viruses, which are highly pathogenic in humans, are prime candidates for the next pandemic influenza virus (8, 21). The first documented transmission of H5N1 viruses from chickens to humans occurred in 1997 in Hong Kong, resulting in the death of 6 out of 18 infected people (4, 27, 28). After all the chickens in the poultry markets in Hong Kong were killed, H5N1 viruses continued to circulate in poultry markets in China, but transmissions to humans were not documented for several years. This situation dramatically changed in 2003, when increasing numbers of fatal transmissions to humans resumed (23, 29). Since that time, more than 150 people have died from H5N1 virus infections, and the virus has now spread from Asia to Europe and Africa (32).
A major issue concerning H5N1 viruses is whether they are adapting to mammalian cells. Two such adaptations, in the PB2 subunit of the viral RNA polymerase (7) and in the receptor-binding hemagglutinin (HA) protein (34), have already been reported. H5N1 viruses that are virulent in mice encode lysine at position 627 in the PB2 protein, whereas H5N1 viruses that are not mice virulent, as well as other avian influenza A viruses, encode glutamic acid at this position (7). The presence of lysine at this position apparently enhances virus replication in mammalian cells (3, 26), but the mechanism of this enhancement has not been established. Amino acid changes at two positions in the HA enable some H5N1 viruses recently isolated from humans to bind to both avian and human sialic acid receptors (34). These HA changes may contribute to the acquisition of efficient human-to-human transmissibility, although changes in other viral proteins are also apparently needed (15).
Here we show that the H5N1 NS genes selected after 1998 enhance virus replication in mammalian cells. The NS1A protein encoded by the NS gene of human influenza A viruses is a small, multifunctional protein that participates in both protein-RNA and protein-protein interactions. Its N-terminal RNA-binding domain binds double-stranded RNA (dsRNA), albeit with low affinity (2, 6, 13, 31). The function of this dsRNA-binding activity during virus infection has been established by identifying the defect in virus replication exhibited by a recombinant influenza A/Udorn/72 (Ud) virus that encodes an NS1A protein that lacks dsRNA-binding activity (17). This analysis showed that the primary role of NS1A dsRNA-binding activity is the inhibition of the alpha/beta interferon (IFN-α/β)-induced oligo(A) synthetase/RNase L pathway and that NS1A dsRNA-binding activity has no detectable role in inhibiting the production of IFN-β mRNA or inhibiting the activation of protein kinase R (PKR) (11, 17). The rest of the NS1A protein, which is designated as the effector domain, has binding sites for several cellular proteins, including (i) CPSF30, a cellular factor required for the 3′-end processing of cellular pre-mRNAs, thereby inhibiting the production of all cellular mRNAs, including IFN-β mRNA (9, 12, 19, 22, 24); (ii) p85β, resulting in the activation of phosphatidylinositol-3-kinase signaling (5, 25); and (iii) PKR, resulting in the inhibition of PKR activation (18).
The CPSF30 binding site is crucial for virus replication, as shown by the fact that a recombinant human influenza virus expressing an NS1A protein lacking a binding site for CPSF30 is highly attenuated (22, 30). Further evidence for the importance of the CPSF30 binding site was obtained using a 61-amino-acid domain of CPSF30, comprising two of its zinc fingers (F2F3), which binds efficiently to the NS1A protein (30). The expression of the F2F3 domain in cells leads to the inhibition of influenza A virus replication, coupled with increased production of IFN-β mRNA, indicating that F2F3 inhibits virus replication by blocking the binding of NS1A to endogenous CPSF30 (30). These results indicate that the CPSF30 binding site of the NS1A protein is a potential target for the development of small-molecule antiviral drugs directed against influenza A virus.
In the present study, we show that the NS1A protein encoded by a pathogenic H5N1 virus that was transmitted to humans in 1997, H5N1 influenza A/Hong Kong/483/97 (HK97) virus, has an intrinsic defect in binding CPSF30. It does not bind the F2F3 fragment of CPSF30 in vitro and causes high IFN-β mRNA production and reduced virus replication in MDCK cells when expressed in a recombinant virus in which the other viral proteins are encoded by Ud. This intrinsic defect was traced to the identities of the amino acids at positions 103 and 106 in the NS1A protein, which differed from the consensus amino acids, F and M, respectively, that are found at these positions in the NS1A proteins of almost all influenza A virus strains isolated from humans. In contrast, the NS genes of H5N1 viruses isolated from humans after 1998 encode F103 and M106, thereby rectifying the intrinsic defect in NS1A protein-mediated binding of CPSF30. X-ray crystallography has shown that F103 and M106, which are not part of the CPSF30 binding pocket, stabilize the interaction of the NS1A protein with CPSF30 (K. Das, L.-C. Ma, R. Xiao, B. Radvansky, J. Aramini, L. Zhao, J. Marklund, R.-L. Kuo, K. Y. Twu, E. Arnold, R. M. Krug, and G. T. Montelione, submitted for publication). The HK97 NS1A protein is less attenuating when expressed in a virus that also encodes the other internal HK97 proteins and under these conditions binds to CPSF30 to a substantial extent, showing that these internal cognate (HK97) proteins largely compensate for the absence of F103 and M106, presumably by stabilizing the NS1A-CPSF30 complex.
(Our results demonstrating that F103 and M106 of the NS1A protein play important roles in the binding of CPSF30 by H5N1 NS1A proteins were presented at the Negative Strand RNA Virus meeting in Salamanca, Spain, 17 to 21 June 2006.)
MATERIALS AND METHODS
Generation of recombinant influenza A viruses.
The pHH21 plasmids expressing the PB1, PB2, PA, NP, M, and NS genes of HK97 and of influenza A/VN/1203/04 (VN04) were provided by Yoshihiro Kawaoka (7) and Ruben Donis, respectively, and a pHH21 plasmid encoding the A/Brevig Mission/1/18 virus (1918 virus) was provided by Christopher F. Basler (1). To generate recombinant influenza A viruses, a pHH21 plasmid encoding the NS gene indicated in the text was cotransfected into 293T cells with the two pHH21 plasmids encoding the Ud HA and neuraminidase (NA) genes plus five pHH21 plasmids encoding the PB1, PB2, PA, NP, and M genes indicated in the text. The four pcDNA3 plasmids encoding the Ud PB1, PB2, PA, and NP proteins were also cotransfected. At 12 h posttransfection, the medium was changed to Opti-MEM supplemented with 3 μg/ml of N-acetylated trypsin (NAT). After an additional 24 h, the 293T cells were overlaid onto MDCK cells for virus amplification. The recombinant viruses were amplified and their titers were determined as described previously (30). All eight genome RNA segments of the recombinant viruses were sequenced. All experiments using viruses containing any HK97, VN04, or 1918 gene were carried out in a biosafety level 3 laboratory using biosafety level 3-enhanced procedures.
Virus infections.
For multiple-cycle growth, MDCK cells were infected at a multiplicity of infection (MOI) of 0.001 PFU/cell of the indicated virus and incubated in serum-free Dulbecco's modified Eagle's medium supplemented with 2.5 μg/ml of NAT at 37°C. An aliquot of the medium was collected at the indicated times, and virus production was measured by plaque assay in MDCK cells. For the plaque reduction assays, monolayers of control and F2F3-expressing MDCK cells were infected with approximately 100 PFU of the indicated virus, and after 1 h of incubation at 37°C, the inoculum was removed and the cells were overlaid with 1% agarose containing Dulbecco's modified Eagle's medium plus 2.5 μg/ml NAT (30). For single-cycle infections, MDCK cells were infected with 5 PFU/cell of the indicated virus.
Glutathione-Sepharose affinity selection.
Glutathione transferase (GST)-F2F3 or GST was mixed with the indicated 35S-labeled NS1A protein and subjected to glutathione-Sepharose affinity selection as described previously (20). To prepare the labeled NS1A protein, the DNA encoding the appropriate NS1A protein was subcloned into pcDNA3 and translated using a Promega TNT coupled transcription/translation kit in the presence of [35S]methionine.
Assay for 3′-end cleavage of pre-mRNAs in vivo.
293 or quail embryo fibroblasts (kindly provided by Henry Bose, Jr.) were cotransfected with a pBC12 plasmid carrying a human β-globin gene and a pcDNA3 plasmid encoding the indicated NS1A protein using the FuGENE 6 transfection reagent for 293 cells and a CaCl2-containing reagent (20) for the quail cells. The transfected cells were collected at 40 h posttransfection, and RNA was extracted using TRIzol reagent (Invitrogen). An aliquot of the total RNA was analyzed by RNase protection using a uniformly labeled RNA probe, which was prepared as previously described (12). After this labeled RNA probe was annealed to the cellular RNA sample, followed by digestion with RNase A and phenol extraction, the protected RNA fragments were resolved by electrophoresis on a urea-polyacrylamide (5%) gel.
Measurement of IFN-β mRNA by real-time quantitative reverse transcription-PCR.
RNA was isolated from infected cells by using the TRIzol reagent at the indicated times after infection of MDCK cells. For each sample, 1-μg portions of total RNA, which correspond to equal cell equivalents, were reverse transcribed using an oligo(dT) primer to generate the DNA complementary to all mRNAs. The amount of IFN-β mRNA was determined using a TaqMan gene expression assay (Applied Biosystems) with 5′ and 3′ primers specific for canine IFN-β mRNA and a 6-carboxyfluorescein dye-labeled TaqMan MGB (minor groove binder) internal probe (30). Real-time PCR analysis was carried out using a Perkin-Elmer/Applied Biosystems 7900HT sequence detector.
Assay for the binding of CPSF30 to the NS1A protein in infected cells.
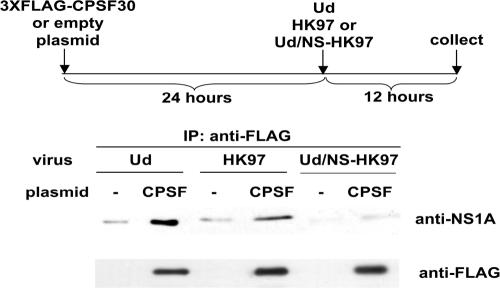
293T cells were transfected with a pcDNA3 plasmid encoding 3× FLAG-tagged CPSF30 (3×FLAG-CPSF30) or, as a control, an empty pcDNA3 plasmid. Twenty-four hours later, the cells were infected at a MOI of 2 PFU/cell with either Ud, HK97, or Ud/NS-HK97 virus (see Results for the description of the latter two viruses). Cells were collected at 12 h postinfection and were extracted using a solution containing 100 mM Tris-HCl, pH 8.0, 250 mM NaCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM phenylmethylsulfonyl fluoride. Cell extracts were immunoprecipitated with anti-FLAG M2 monoclonal antibody, and the immunoprecipitates were analyzed by immunoblotting using antibody directed against the Ud NS1A protein. Because the Ud NS1A antibody detected the HK97 NS1A protein fivefold less efficiently than the Ud NS1A in immunoblots, fivefold-larger aliquots of the immunoprecipitates from the HK97 and Ud/NS-HK97 virus-infected cells were analyzed in the immunoblots using this anti-NS1A antibody. To monitor the efficiency of immunoprecipitation, equal aliquots of each immunoprecipitate were analyzed by immunoblotting using anti-FLAG antibody.
RESULTS
The NS1A protein of a 2004 H5N1 virus, but not that of a 1997 H5N1 virus, binds the F2F3 fragment of CPSF30 and inhibits 3′-end processing of pre-mRNAs.
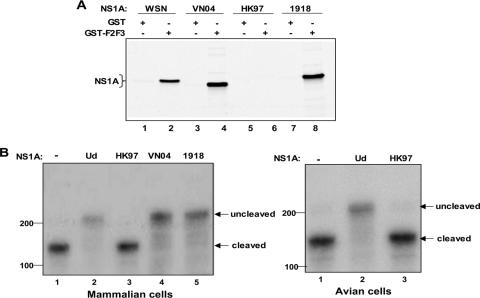
We determined whether H5N1 NS1A proteins bind CPSF30 in vitro, utilizing the 61-amino-acid sequence of two of its zinc fingers (F2F3), which is sufficient for binding to the NS1A protein of human influenza A viruses (30). Like the NS1A protein of Ud (30), the NS1A proteins of the mouse-adapted A/WSN/33 virus, the pathogenic H5N1 VN04 virus, and the 1918 virus bound to F2F3 in vitro, as determined in a GST pulldown assay (Fig. 1A). In contrast, no detectable binding to F2F3 was observed with the NS1A protein of the pathogenic HK97 virus (Fig. 1A, lanes 5 and 6).
FIG. 1.
The HK97 NS1A protein does not bind the F2F3 fragment of CPSF30 and does not inhibit 3′-end processing of cellular pre-mRNAs. (A) GST pulldown assay. GST-F2F3 or GST was mixed with 35S-labeled NS1A protein of the indicated virus. (B) 3′-end processing assay. 293 cells (left panel) or quail embryo fibroblasts (right panel) were cotransfected with a pBC12 plasmid carrying a human β-globin gene and either an empty pcDNA3 plasmid (lane 1) or a pcDNA3 plasmid encoding the indicated NS1A protein. RNA was analyzed by RNase protection using a 270-nucleotide-long RNA probe spanning the 3′ cleavage site of the β-globin pre-mRNA. The protected RNA fragment corresponding to the uncleaved pre-mRNA is 210 nucleotides long, and the RNA fragment corresponding to the 3′-end-cleaved pre-mRNA is 160 nucleotides long. No residual probe containing 270 nucleotides was detected.
As one approach to determine whether these two H5N1 NS1A proteins bind to CPSF30 in vivo, we used transient transfection assays that measure the inhibition of the 3′-end processing of β-globin pre-mRNA (12, 30) (Fig. 1B). Plasmid-driven expression of the Ud, VN04, or 1918 NS1A protein effectively inhibited the 3′-end processing of β-globin pre-mRNA in human 293 cells (Fig. 1B, left panel, lanes 2, 4, and 5), whereas the HK97 NS1A protein lacked this inhibitory activity (Fig. 1B, left panel, lane 3). Immunoblots showed that equivalent amounts of the four NS1A proteins were expressed (data not shown). In addition, the HK97 NS1A protein did not inhibit 3′-end processing in avian (quail) cells (Fig. 1B, right panel, lane 3), indicating that this NS1A protein does not bind either avian or mammalian CPSF30. The A/Teal/Hong Kong/W312/97 (Teal) NS1A protein, a presumed precursor to the HK97 NS1A protein (33), also did not bind to F2F3 or inhibit 3′-end processing (data not shown). In contrast, the Ud NS1A protein effectively inhibited 3′-end processing in avian as well as mammalian cells (Fig. 1B, right panel, lane 2), indicating that it is capable of binding avian as well as mammalian CPSF30.
An Ud reassortant virus containing the gene encoding the HK97 NS1A protein is attenuated and induces high levels of IFN-β mRNA.
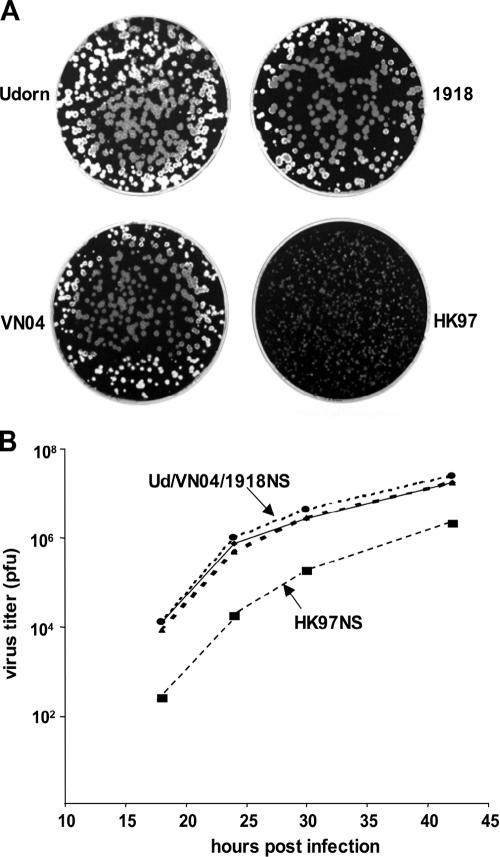
To determine how the HK97, VN04, and 1918 NS1A proteins affect virus replication, we generated recombinant Ud viruses in which the Ud NS gene was replaced by either the HK97, the VN04, or the 1918 NS gene. The recombinant viruses containing the VN04 or 1918 NS gene were indistinguishable from the Ud parent virus, as measured by plaque size in MDCK cells (Fig. 2A) and by multiple-cycle growth in MDCK cells (Fig. 2B). In contrast, the recombinant virus containing the HK97 NS gene formed pinpoint plaques, and the rate of replication and virus yield during multiple-cycle growth were approximately 100-fold lower than with the parent Ud virus. These results demonstrate that the HK97 NS gene attenuates virus replication in mammalian (MDCK) cells.
FIG. 2.
The Ud virus whose NS gene is replaced by the HK97 NS gene is attenuated. (A) Size of the plaques on MDCK cells of the Ud viruses containing the indicated NS gene. (B) Growth curves of the Ud viruses containing the indicated NS gene after infection of MDCK cells at a MOI of 0.001 PFU/cell.
We have shown that the expression of F2F3 in cells leads to the inhibition of influenza A virus replication, coupled with increased production of IFN-β mRNA (30), indicating that F2F3 blocks the binding of the NS1A protein to endogenous CPSF30. The most dramatic evidence for this inhibition is provided by plaque reduction assays (30). As is the case for the parental Ud virus (30), the number of plaques produced on F2F3-expressing cells by the recombinant Ud viruses containing either the 1918 or the VN04 NS gene was only 10% of that produced on control cells (Fig. 3A), verifying that the NS1A proteins encoded by the 1918 and VN04 NS genes bind to CPSF30 during virus infection. In contrast, no reduction in plaque number was observed for the F2F3-expressing cells with the Ud recombinant containing the HK97 NS gene, demonstrating that the encoded NS1A protein does not bind CPSF30 in infected cells.
FIG. 3.
The Ud virus whose NS gene is replaced by the HK97 NS gene does not bind CPSF30 and induces increased levels of IFN-β mRNA during infection. (A) Plaque reduction assays on control and F2F3-expressing MDCK cells using Ud viruses containing the indicated NS gene. The plaque assay for the Ud recombinant containing the HK97 NS gene was incubated for a day longer than the plaque assay shown in Fig. 2A to increase the size of the plaques. (B) The relative amounts of IFN-β mRNA produced during single-cycle growth in MDCK cells of the Ud viruses containing the indicated NS gene.
This lack of CPSF30 binding to the NS1A protein resulted in a substantial increase in the production of IFN-β mRNA during single-cycle virus growth, as measured by quantitative reverse transcription-PCR (Fig. 3B). We used these high-MOI conditions to ensure that we were measuring the amount of IFN-β mRNA produced per infected cell. The amount of IFN-β mRNA produced in cells infected by the Ud recombinant containing the HK97 NS gene was approximately fourfold greater than that produced in cells infected by either the Ud parental virus or the Ud recombinant containing the VN04 NS gene. These results indicate that the attenuation caused by the HK97 NS gene likely resulted from increased production of IFN-β mRNA due to the lack of CPSF30 binding by the encoded NS1A protein. Interestingly, the amount of IFN-β mRNA produced in cells infected by the Ud recombinant containing the 1918 NS gene was reproducibly fourfold lower than that produced in cells infected by either the Ud parental virus or the Ud recombinant containing the VN04 NS gene.
Two amino acid changes in the HK97 NS1A protein generate CPSF30 binding in vitro and in vivo.
Based on the above results, we predicted that we should be able to generate CPSF30 binding by the HK97 NS1A protein by changing one or more of its amino acids to the corresponding amino acid(s) in the VN04 NS1A protein. Alignment of NS1A amino acid sequences showed that the Teal and HK97 NS1A proteins shared 13 amino acids that differed from the corresponding amino acids that were shared by the VN04 and 1918 NS1A proteins, assuming that the deletion of amino acids 80 to 84 in the VN04 sequence is not relevant (Fig. 4). Of these 13 differences, we targeted 9, which were divided into the designated four groups. The specific amino acid changes introduced into the HK97 NS1A protein in each of these four groups are designated in the legend of Fig. 4.
FIG. 4.
Alignment of the protein sequences of Teal, HK97, VN04, and 1918 viruses (14). The positions of the groups (G1, G2, G3, and G4) of mutations introduced into the HK97 NS1A protein are indicated. G1 consists of R55E, E60A, and H63Q; G2 consists of L103F and I106M; G3 consists of N139D, A143T, and D152E; and G4 consists of T202A.
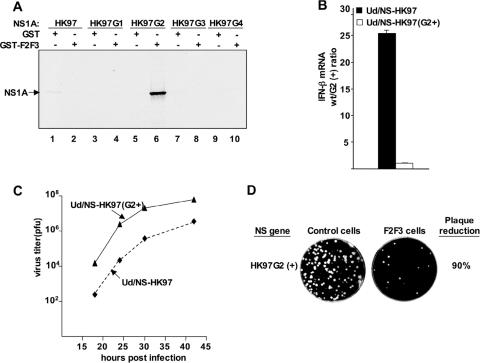
Changing the amino acids of the HK97 NS1A protein in group 2 (G2) to their corresponding amino acids in the VN04 and 1918 NS1A proteins (L103-to-F and I106-to-M) yielded a mutant HK97 NS1A protein that bound to F2F3 in vitro in a GST pulldown assay (Fig. 5A, lane 6). In contrast, the amino acid changes in the other three groups did not result in such a gain of function. It should be emphasized that the bases that were mutated to produce the G2 amino acid changes are in the intron that is removed by splicing to produce NS2 mRNA and hence do not change the amino acid sequence of the NS2 protein encoded by the HK97 NS gene (10).
FIG. 5.
The G2 (+) (I106M and L103F) mutations in the HK97 NS1A protein are required for the acquisition of CPSF30 binding that effectively suppresses IFN-β mRNA synthesis in infected cells and enhances virus replication. (A) GST pulldown assay. GST-F2F3 or GST was mixed with the wt or indicated mutant HK97 35S-labeled NS1A protein. (B) The relative amounts of IFN-β mRNA produced during single-cycle growth in MDCK cells of the Ud recombinants containing either the wt or G2 (+) mutant HK97 NS gene. (C) Growth curves of the Ud recombinant viruses containing the wt or G2 (+) mutant HK97 NS gene after infection of MDCK cells at low MOI. (D) Plaque reduction assay on control and F2F3-expressing MDCK cells using the Ud recombinant containing the G2 (+) mutant HK97 NS gene.
To determine the effects of these NS1A protein mutations during virus infection, we generated a recombinant Ud virus in which the Ud NS gene was replaced by an HK97 NS which contained the G2 (+) mutations L103F and I106M [Ud/NS-HK97(G2+) virus] and compared this recombinant virus to the recombinant Ud virus containing the wild-type (wt) HK97 NS gene [Ud/NS-HK97 virus]. During single-cycle growth, the amount of IFN-β mRNA produced by infection with the Ud/NS-HK97 virus was substantially (∼25-fold) greater than that produced by infection with the Ud/NS-HK97(G2+) virus (Fig. 5B), demonstrating that the mutational restoration of F2F3 binding to the NS1A protein resulted in reduced IFN-β mRNA production during infection. This reduction in IFN-β mRNA production resulted in an increased rate of virus replication during multiple-cycle growth: the rate of replication and virus yield of the Ud/NS-HK97(G2+) virus were 100-fold greater than those for the Ud/NS-HK97 virus (Fig. 5C). In addition, the number of plaques produced on F2F3-expressing cells by the Ud/NS-HK97(G2+) virus was only 10% of that produced on control cells (Fig. 5D), verifying that the NS1A protein encoded by this NS gene binds to CPSF30 during virus infection. Changing only one of these two amino acids, either L103 to F or I106 to M, in the HK97 NS1A protein had intermediate effects on both reduction of IFN-β mRNA production and increase in the rate of virus replication (data not shown).
When the HK97 NS1A protein is expressed by a recombinant virus encoding the other HK97 internal proteins, it is less attenuating and binds CPSF30 to a substantial extent.
The above results suggested that the HK97 NS gene would also attenuate virus replication in the context of the HK97 virus itself. This possibility is counterintuitive, because the HK97 virus is highly pathogenic in mice, chickens, ferrets, and humans (7, 35). To test this possibility, we generated recombinant viruses containing all HK97 genes except those encoding HA and NA, which were derived from the Ud virus. We did not include the HK97 HA and NA genes because of biosafety concerns and because we are evaluating virus replication in tissue culture cells rather than pathogenicity in animal models. One recombinant virus contained the wt HK97 NS gene, and the other recombinant contained an HK97 NS gene with the G2 (+) mutations (L103F and I106M). During multiple-cycle growth, the rate of virus replication of the HK97 recombinant virus containing the wt NS gene was approximately 20-fold lower than that of the recombinant virus containing the G2 (+) mutations (Fig. 6A). This reduced rate of replication was coupled with increased production of IFN-β mRNA production (∼9-fold), as assayed during single-cycle growth (Fig. 6B).
FIG. 6.
The HK97 NS gene also attenuates virus replication in the context of the other internal HK97 genes due to the identity of the amino acids at 103 and 106 in the NS1A protein. (A) Growth curves of recombinant viruses containing either the wt or G2 (+) mutant HK97 NS gene and all HK97 genes except those encoding HA and NA. (B) The amount of IFN-β mRNA produced during single-cycle growth of the recombinant containing the wt HK97 NS gene relative to that produced by the recombinant containing the G2 (+) mutant HK97 NS gene.
One explanation for these results is that the wt HK97 NS1A containing the G2 (−) amino acids at positions 103 and 106 (L and I, respectively) does not bind CPSF30 when it is expressed in a recombinant virus that encodes the other HK97 internal proteins (HK97 virus), as is the case when the HK97 NS1A protein is expressed in a recombinant virus that encodes internal Ud proteins (Fig. 2 to 5). However, this is not the case: the wt HK97 NS1A protein does indeed bind CPSF30 to a substantial extent when it is expressed in the HK97 virus. Several lines of evidence established this conclusion.
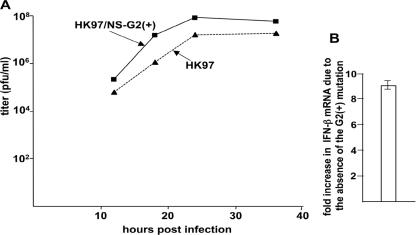
First, the replication of the HK97 virus is approximately 100-fold faster than that of the Ud/NS-HK97 virus (Fig. 7A), and the increased rate of replication of the HK97 virus is accompanied by a ninefold reduction in the production of IFN-β mRNA compared to that of the Ud/NS-HK97 virus (Fig. 7B). Second, the HK97 virus has a property that is not shared by the Ud/NS-HK97 virus, specifically, inhibition of replication in F2F3-expressing cells (Fig. 7C). The number of plaques on F2F3-expressing cells formed by the HK97 virus was reduced 70%, whereas no reduction in plaque number was observed for F2F3-expressing cells with the Ud/HK97-NS virus (Fig. 3A and 7C). The inhibition observed for F2F3-expressing cells is less than that observed when the HK97 NS1A protein contains F103 and M106, in the presence of either Ud or HK97 internal genes: plaque reduction in the F2F3-expressing cells was 90% (Fig. 3A, 5D, and 7C). These F2F3 inhibition results show that CPSF30 gains substantial access to the consensus CPSF30 binding site on the HK97 NS1A protein only when it is expressed in the presence of its cognate (HK97) internal proteins, despite the absence of F103 and M106 in the HK97 NS1A protein. Such access of CPSF30 to a binding site on the HK97 NS1A protein explains the decreased production of IFN-β mRNA by the HK97 virus compared to that of the Ud/NS-HK97 virus (Fig. 7A).
FIG. 7.
The HK97 virus is substantially less attenuated than the Ud/NS-HK97 virus and, unlike the Ud/NS-HK97 virus, is inhibited in F2F3-expressing cells. (A) Growth curves of the HK97 and Ud/NS-HK97 viruses after infection of MDCK cells at low MOI. (B) The amount of IFN-β mRNA produced during single-cycle growth of the HK97 virus relative to that produced by the Ud/NS-HK97 virus. (C) Plaque reduction assays on control and F2F3-expressing MDCK cells of the recombinant viruses expressing the indicated NS and internal (PA, PB1, PB2, NP, M) genes. The plaque assay for the Ud/NS-HK97 virus was incubated for a day longer than the other plaque assays to increase the size of the plaques.
To verify that the HK97 NS1A protein expressed by the HK97 virus binds CPSF30, we transfected 293T cells with a plasmid expressing 3×FLAG-CPSF30 or as a control with an empty plasmid. Twenty-four hours later, the cells were infected with either the Ud, the HK97, or the Ud/NS-HK97 virus for 12 h. Cell extracts were immunoprecipitated with anti-FLAG antibody, followed by immunoblotting using anti-NS1A antibody. As shown in Fig. 8, the Ud NS1A protein and the HK97 NS1A protein expressed by the HK97 virus coimmunoprecipitated with 3×FLAG-CPSF30. In contrast, little or none of the HK97 NS1A protein expressed by the Ud/NS-HK97 virus coimmunoprecipitated with 3×FLAG-CPSF30. We conclude that the presence of coexpressed cognate (HK97) internal proteins enables the HK97 NS1A protein that lacks F103 and M106 to bind CPSF30 in virus-infected cells. Nonetheless, CPSF30 binding is more effective when the HK97 NS1A protein expressed by the HK97 virus contains F103 and M106, because the presence of these two consensus amino acids results in a 20-fold increase in the rate of HK97 virus replication coupled with a decrease in IFN-β mRNA production (Fig. 6A and B) and also results in greater inhibition of virus replication in F2F3-expressing cells, i.e., 90% rather than 70% inhibition of plaque number (Fig. 3A, 5D, and 7C). Thus, the internal cognate (HK97) proteins substantially, but not completely, compensate for the absence of F103 and M106 in the HK97 NS1A protein (see Discussion).
FIG. 8.
The HK97 NS1A protein binds to CPSF30 in cells infected by the HK97 virus but not in cells infected by the Ud/NS-HK97 virus. 293T cells were transfected with the indicated plasmids and infected with the indicated recombinant influenza A viruses. Extracts from cells collected at 12 h after virus infection were immunoprecipitated with anti-FLAG M2 monoclonal antibody, and the immunoprecipitates were analyzed by immunoblotting with either anti-Ud NS1A antibody or anti-FLAG antibody.
DISCUSSION
In this study we show that the NS1A protein of the pathogenic HK97 virus that was transmitted to humans in 1997 has an intrinsic defect in binding CPSF30. It does not bind CPSF30 in vitro and causes high IFN-β mRNA production and reduced virus replication in MDCK cells when expressed in a recombinant virus in which the other viral proteins are encoded by Ud. We traced the defect in CPSF30 binding to the identity of two NS1A amino acids, L at position 103 and I at position 106, which were present in all the H5N1 viruses isolated from humans in 1997 and 1998 (Table 1). In contrast, the NS1A protein of the 2004 pathogenic VN04 virus has F and M at positions 103 and 106, respectively, and as a result binds CPSF30 in vitro and does not attenuate virus replication in MDCK cells. All the NS1A proteins of H5N1 viruses isolated from humans after 2003 have F and M at these positions (Table 1). These two amino acid changes in the NS1A protein result from two single-base mutations: an A-to-C mutation that converts a UUA codon (L) to a UUC codon (F), and a U-to-G mutation that converts an AUU codon (I) to an AUG codon (M) (underlining indicates base mutation) (14). It is unlikely that the functional significance of these two single-base changes could be deduced from the sequence analysis of the HK97 and VN04 NS genes. Essentially all (>98%) of the influenza A viruses (H3N2, H2N2, and H1N1) that have been isolated from humans, including the H3N2 Ud and 1918 H1N1 viruses, contain F and M at positions 103 and 106, respectively, in their NS1A proteins (14) (see Table 3). The high conservation of these two amino acids strongly suggests that replication in humans provides selective pressure for efficient binding of CPSF30 by the NS1A protein to suppress the production of IFN-β mRNA.
TABLE 1.
Identities of the amino acids at 106 and 103 in H5N1 NS1A proteins isolated from humans since 1997 (14)
| Yr of isolation | No. of virus isolates with indicated amino acid at NS1A positiona:
|
|||
|---|---|---|---|---|
| 106
|
103
|
|||
| I | M | L | F | |
| 1997 | 16 | 0 | 16 | 0 |
| 1998 | 1 | 0 | 1 | 0 |
| 1999 | 0 | 0 | 0 | 0 |
| 2000 | 0 | 0 | 0 | 0 |
| 2001 | 0 | 0 | 0 | 0 |
| 2002 | 0 | 0 | 0 | 0 |
| 2003 | 0 | 5 | 0 | 5 |
| 2004 | 0 | 16 | 0 | 16 |
| 2005 | 0 | 15 | 0 | 15 |
| 2006 | 0 | 44 | 0 | 44 |
The positions corresponding to amino acids 103 and 106 are based on the sequence of the HK97 NS1A protein.
TABLE 3.
The amino acids at 106 and 103 in the NS1A proteins of human and avian (H9N2 and H6N1) influenza A viruses (14)
| Virus strain | Amino acid (% of total) at positiona:
|
||||||
|---|---|---|---|---|---|---|---|
| 106
|
103
|
||||||
| M | I | Other | F | Y | L | Other | |
| Human H3N2 | 99.9 | 0.1 | 0 | 99.5 | 0 | 0.4 | 0.1 |
| Human H2N2 | 100 | 0 | 0 | 100 | 0 | 0 | 0 |
| Human H1N1 | 99.5 | 0.5 | 0 | 98.5 | 0 | 1 | 0.5 |
| Avian H9N2 | 14.2 | 85.2 | 0.6 | 23.9 | 1.1 | 73.9 | 1.1 |
| Avian H6N1 | 78.3 | 21.7 | 0 | 19.5 | 2.2 | 78.3 | 0 |
The positions corresponding to amino acids 103 and 106 are based on the sequence of the HK97 and Ud virus NS1A proteins. The number of virus strains included in this analysis are as follows: for human H3N2, 945 (1968 to 2005); for human H2N2, 49 (1957 to 1974); for human H1N1, 200 (1918 to 2004); for avian H9N2, 176 (1997 to 2006); and for avian H6N1, 46 (1997 to 2006). Of the avian H9N2 and H6N1 NS1A proteins, 11% and 17%, respectively, contain both M at 106 and F at 103. The two mouse-adapted H1N1 strains, A/WSN/33 and A/PR8/34, were not included in the human strains.
Surprisingly, the NS1A protein of several H5N1 viruses isolated from avian species in 1997, unlike the NS1A protein of H5N1 viruses isolated from humans at that time, had F and M at 103 and 106, respectively (14) (Table 2). In addition, from 1999 to 2002, a period during which no H5N1 viruses were isolated from humans, the vast majority of the H5N1 viruses isolated from avian species encoded NS1A proteins with F and M at these positions. We presume that these avian H5N1 viruses were the source of the H5N1 viruses that were transmitted to humans in 2003. The predominance of these avian H5N1 viruses after 1998 suggests two possibilities: (i) that replication of H5N1 viruses in avian species, as well as in humans, provides selective pressure for efficient binding of CPSF30 by the NS1A protein to suppress the production of IFN-β mRNA; and/or (ii) that during the 1999-to-2002 period there were a significant number of undocumented H5N1 virus infections of mammalian species, where the selective pressure for efficient suppression of IFN-β production occurred. Consistent with the latter possibility, the single H5N1 virus isolate from mammals in 2001 (A/swine/Fujian/F1/2001) encodes an NS1A protein with the consensus mammalian sequence at positions 106 (M) and 103 (F) (14). In contrast to the avian H5N1 NS1A proteins, there is considerable variability in the identities of the amino acids at positions 103 and 106 in the NS1A proteins of the avian H9N2 and H6N1 viruses that were cocirculating with avian H5N1 viruses from 1997 to 2006 (14) (Table 3). Although the majority (78%) of the H6N1 NS1A proteins contain M at 106, only 17% contain both M106 and F103. Only 11% of the H9N2 NS1A proteins contain both M106 and F103. Apparently, replication of H6N1 and H9N2 viruses in avian species tolerates different efficiencies of CPSF30 binding and different levels of IFN-β mRNA production.
TABLE 2.
Identities of the amino acids at 106 and 103 in H5N1 NS1A proteins isolated from avian species since 1997 (14)
| Yr of isolation | No. of virus isolates with indicated amino acid at NS1A positiona:
|
|||
|---|---|---|---|---|
| 106
|
103
|
|||
| I | M | L | F | |
| 1997 | 11 | 5 | 11 | 4 |
| 1998 | 0 | 0 | 0 | 0 |
| 1999 | 0 | 5 | 0 | 1 |
| 2000 | 2 | 13 | 1 | 4 |
| 2001 | 1 | 49 | 2 | 47 |
| 2002 | 4 | 37 | 3 | 37 |
| 2003 | 1 | 50 | 2 | 47 |
| 2004 | 3 | 171 | 1 | 172 |
| 2005 | 3 | 208 | 3 | 207 |
| 2006 | 0 | 109 | 0 | 109 |
The positions corresponding to amino acids 103 and 106 are based on the sequence of the HK97 NS1A protein.
The recently obtained X-ray crystal structure of the complex of the Ud NS1A effector domain with F2F3 has identified the CPSF30 binding pocket on the NS1A protein, which is comprised of amino acids that are highly conserved among human influenza A viruses, including the 1918 virus and all H5N1 viruses (e.g., HK97 and VN04) (Das et al., submitted). The crystal structure also shows that F103 and M106 are not part of the CPSF30 binding pocket but rather participate in critical intermolecular interactions that stabilize the NS1A:F2F3 complex, and their replacement by other amino acids, L at 103 and I at 106, disrupts the complex. These structural results explain why the HK97 NS1A protein, which contains L103 and I106, does not bind F2F3 in vitro and does not bind CPSF30 in vivo when it is expressed in a virus that encodes the internal proteins of a different influenza A virus, i.e., the Ud virus.
However, CPSF30 does bind to a substantial extent to the consensus binding pocket on the HK97 NS1A protein when it is expressed in a virus that also encodes the other internal HK97 internal proteins. Consequently, these cognate (HK97) internal proteins, unlike their Ud counterparts, are capable of substantially, though not completely, compensating for the absence of F103 and M106 and thus enabling CPSF30 to bind to the HK97 NS1A protein. This result indicates that one or more of these cognate (HK97) internal proteins most likely interact with, and hence substantially stabilize, the complex containing the HK97 NS1A protein and CPSF30. Preliminary results show that such binding of CPSF30 to the HK97 NS1A protein requires the HK97 polymerase complex (PB1, PB2, PA, and NP) but not the HK97 M protein. These results strongly suggest a specific NS1A-polymerase interaction, i.e., that the HK97 polymerase complex, unlike its Ud counterpart, interacts with the HK97 NS1A protein to stabilize the structure of the HK97NS1A:CPSF30 complex in vivo in the absence of M at 106 and F at 103. As a consequence, the F2F3 region of CPSF30 would be able to access the consensus binding pocket on the HK97 NS1A protein, albeit not as effectively as when these two NS1A amino acids are present. It is reasonable to propose that a similar NS1A-polymerase interaction also occurs with other influenza A viruses, but such an interaction would not be evident when the NS1A protein contains the consensus F103 and M106 amino acids that by themselves are sufficient to stabilize the NS1A:CPSF30 complex. We are currently carrying out experiments to elucidate the molecular mechanisms underlying this apparent NS1A-polymerase interaction.
Despite encoding an NS1A protein with an intrinsic CPSF30 binding defect, the 1997 HK97 virus is still highly pathogenic in birds, humans, mice, and ferrets (7, 35). In addition, the NS1A protein of the mouse-adapted influenza A/PR8/34 (PR8) virus, which is pathogenic in mice (33), lacks the human F103 and M106 consensus sequence (14) and does not bind F2F3 in vitro (R.-L. Kuo, K. Y. Twu, J. Marklund, and R. M. Krug, unpublished experiments). These findings indicate that pathogenicity does not require a fully functional NS1A protein and that other viral proteins are sufficient to confer a pathogenic phenotype, consistent with other results indicating that pathogenicity/virulence is polygenic (8, 21, 33). In other words, virulence cannot be ascribed to a single specific viral gene but rather requires a combination of several viral genes. Consequently, more than one virus-induced effect on the host may be required for virulence and/or virulence may require that two or more of the viral proteins undergo specific interactions with each other (21). The latter situation is likely the case for the HK97 virus, because the intrinsic CPSF30 binding defect of the HK97 NS1A protein is apparently ameliorated by an interaction with its cognate (HK97) polymerase complex. Without this amelioration of CPSF30 binding, it is not likely that the HK97 virus would be pathogenic in mammals. We are currently determining whether the PR8 NS1A protein and the small number (<2%) of the human virus NS1A proteins that lack F103 and M106 bind to CPSF30 when they are expressed by a virus that also encodes their cognate internal proteins.
In addition, our results may explain why the VN04 virus is more pathogenic than the HK97 virus (16). The VN04 NS1A protein, unlike the HK97 NS1A protein, is not intrinsically defective in binding CPSF30, and hence the VN04 NS1A protein is more effective than the HK97 NS1A in blocking the production of IFN-β mRNA and is not attenuating for virus replication. The superior activity of the VN04 NS1A protein may be responsible for rendering the VN04 virus more pathogenic than the HK97 virus. We are currently testing this hypothesis in animal models.
Acknowledgments
This investigation was supported by National Institutes of Health grant AI-11772 to R.M.K.
We thank Chen Zhao and Gaetano Montelione for valuable discussions.
Footnotes
Published ahead of print on 23 May 2007.
REFERENCES
- 1.Basler, C. F., A. H. Reid, J. K. Dybing, T. A. Janczewski, T. G. Fanning, H. Zheng, M. Salvatore, M. L. Perdue, D. E. Swayne, A. Garcia-Sastre, P. Palese, and J. K. Taubenberger. 2001. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc. Natl. Acad. Sci. USA 98:2746-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chien, C. Y., Y. Xu, R. Xiao, J. M. Aramini, P. V. Sahasrabudhe, R. M. Krug, and G. T. Montelione. 2004. Biophysical characterization of the complex between double-stranded RNA and the N-terminal domain of the NS1 protein from influenza A virus: evidence for a novel RNA-binding mode. Biochemistry 43:1950-1962. [DOI] [PubMed] [Google Scholar]
- 3.Crescenzo-Chaigne, B., S. van der Werf, and N. Naffakh. 2002. Differential effect of nucleotide substitutions in the 3′ arm of the influenza A virus vRNA promoter on transcription/replication by avian and human polymerase complexes is related to the nature of PB2 amino acid 627. Virology 303:240-252. [DOI] [PubMed] [Google Scholar]
- 4.de Jong, J. C., E. C. Claas, A. D. Osterhaus, R. G. Webster, and W. L. Lim. 1997. A pandemic warning? Nature 389:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hale, B. G., D. Jackson, Y. H. Chen, R. A. Lamb, and R. E. Randall. 2006. Influenza A virus NS1 protein binds p85beta and activates phosphatidylinositol-3-kinase signaling. Proc. Natl. Acad. Sci. USA 103:14194-14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatada, E., and R. Fukuda. 1992. Binding of influenza A virus NS1 protein to dsRNA in vitro. J. Gen. Virol. 73:3325-3329. [DOI] [PubMed] [Google Scholar]
- 7.Hatta, M., P. Gao, P. Halfmann, and Y. Kawaoka. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840-1842. [DOI] [PubMed] [Google Scholar]
- 8.Horimoto, T., and Y. Kawaoka. 2005. Influenza: lessons from past pandemics, warnings from current incidents. Nat. Rev. Microbiol. 3:591-600. [DOI] [PubMed] [Google Scholar]
- 9.Kim, M. J., A. G. Latham, and R. M. Krug. 2002. Human influenza viruses activate an interferon-independent transcription of cellular antiviral genes: outcome with influenza A virus is unique. Proc. Natl. Acad. Sci. USA 99:10096-10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamb, R. A., and R. M. Krug. 2001. Orthomyxoviridae: the viruses and their replication, p. 1487-1532. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 11.Li, S., J. Y. Min, R. M. Krug, and G. C. Sen. 2006. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 349:13-21. [DOI] [PubMed] [Google Scholar]
- 12.Li, Y., Z. Y. Chen, W. Wang, C. C. Baker, and R. M. Krug. 2001. The 3′-end-processing factor CPSF is required for the splicing of single-intron pre-mRNAs in vivo. RNA 7:920-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu, Y., M. Wambach, M. G. Katze, and R. M. Krug. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology 214:222-228. [DOI] [PubMed] [Google Scholar]
- 14.Macken, C., H. Lu, J. Goodman, and L. Boykin. 2001. The value of a database in surveillance and vaccine selection, p. 103-106. In A. Osterhaus, N. Cox, and A. W. Hampson (ed.), Options for the control of influenza IV. Elsevier Science, Amsterdam, The Netherlands.
- 15.Maines, T. R., L.-M. Chen, Y. Matsuoka, H. Chen, T. Rowe, J. Ortin, A. Falcon, N. T. Hien, L. Q. Mai, E. R. Sedyaningsih, S. Harun, T. M. Tumpey, R. O. Donis, N. J. Cox, K. Subbarao, and J. M. Katz. 2006. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc. Natl. Acad. Sci. USA 103:12121-12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maines, T. R., X. H. Lu, S. M. Erb, L. Edwards, J. Guarner, P. W. Greer, D. C. Nguyen, K. J. Szretter, L. M. Chen, P. Thawatsupha, M. Chittaganpitch, S. Waicharoen, D. T. Nguyen, T. Nguyen, H. H. Nguyen, J. H. Kim, L. T. Hoang, C. Kang, L. S. Phuong, W. Lim, S. Zaki, R. O. Donis, N. J. Cox, J. M. Katz, and T. M. Tumpey. 2005. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J. Virol. 79:11788-11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Min, J.-Y., and R. M. Krug. 2006. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2′-5′ OAS/RNase L pathway. Proc. Natl. Acad. Sci. USA 103:7100-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min, J.-Y., S. Li, G. C. Sen, and R. M. Krug. 2007. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology 363:236-243. [DOI] [PubMed] [Google Scholar]
- 19.Nemeroff, M. E., S. M. Barabino, Y. Li, W. Keller, and R. M. Krug. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell 1:991-1000. [DOI] [PubMed] [Google Scholar]
- 20.Nemeroff, M. E., X. Qian, and R. M. Krug. 1995. The influenza virus NS1 protein forms multimers in vitro and in vivo. Virology 212:422-428. [DOI] [PubMed] [Google Scholar]
- 21.Noah, D. L., and R. M. Krug. 2005. Influenza virus virulence and its molecular determinants. Adv. Virus Res. 65:121-145. [DOI] [PubMed] [Google Scholar]
- 22.Noah, D. L., K. Y. Twu, and R. M. Krug. 2003. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAs. Virology 307:386-395. [DOI] [PubMed] [Google Scholar]
- 23.Peiris, J. S., W. C. Yu, C. W. Leung, C. Y. Cheung, W. F. Ng, J. M. Nicholls, T. K. Ng, K. H. Chan, S. T. Lai, W. L. Lim, K. Y. Yuen, and Y. Guan. 2004. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet 363:617-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimizu, K., A. Iguchi, R. Gomyou, and Y. Ono. 1999. Influenza virus inhibits cleavage of the HSP70 pre-mRNAs at the polyadenylation site. Virology 254:213-219. [DOI] [PubMed] [Google Scholar]
- 25.Shin, Y. K., Q. Liu, S. K. Tikoo, L. A. Babiuk, and Y. Zhou. 2007. Influenza A virus NS1 protein activates the phosphatidylinositol 3-kinase (PI3K)/Akt pathway by direct interaction with the p85 subunit of PI3K. J. Gen. Virol. 88:13-18. [DOI] [PubMed] [Google Scholar]
- 26.Shinya, K., S. Hamm, M. Hatta, H. Ito, T. Ito, and Y. Kawaoka. 2004. PB2 amino acid at position 627 affects replicative efficiency, but not cell tropism, of Hong Kong H5N1 influenza A viruses in mice. Virology 320:258-266. [DOI] [PubMed] [Google Scholar]
- 27.Suarez, D. L., M. L. Perdue, N. Cox, T. Rowe, C. Bender, J. Huang, and D. E. Swayne. 1998. Comparisons of highly virulent H5N1 influenza A viruses isolated from humans and chickens from Hong Kong. J. Virol. 72:6678-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subbarao, K., A. Klimov, J. Katz, H. Regnery, W. Lim, H. Hall, M. Perdue, D. Swayne, C. Bender, J. Huang, M. Hemphill, T. Rowe, M. Shaw, X. Xu, K. Fukuda, and N. Cox. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279:393-396. [DOI] [PubMed] [Google Scholar]
- 29.Tran, T. H., T. L. Nguyen, T. D. Nguyen, T. S. Luong, P. M. Pham, V. C. Nguyen, T. S. Pham, C. D. Vo, T. Q. Le, T. T. Ngo, B. K. Dao, P. P. Le, T. T. Nguyen, T. L. Hoang, V. T. Cao, T. G. Le, D. T. Nguyen, H. N. Le, K. T. Nguyen, H. S. Le, V. T. Le, D. Christiane, T. T. Tran, J. de Menno, C. Schultsz, P. Cheng, W. Lim, P. Horby, and J. Farrar. 2004. Avian influenza A (H5N1) in 10 patients in Vietnam. N. Engl. J. Med. 350:1179-1188. [DOI] [PubMed] [Google Scholar]
- 30.Twu, K. Y., D. L. Noah, P. Rao, R.-L. Kuo, and R. M. Krug. 2006. The CPSF30 binding site on the NS1A protein of influenza A virus is a potential antiviral target. J. Virol. 80:3957-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, W., K. Riedel, K. Lynch, C. Chien, G. T. Montelione, and R. M. Krug. 1999. RNA-binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA 5:195-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.WHO. 2007. Pandemic influenza. http://www.who.int/csr/disease/avian_influenza/en/index.html.
- 33.Wright, P. F., and R. G. Webster. 2001. Orthomyxoviruses, p. 1533-1579. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 34.Yamada, S., Y. Suzuki, T. Suzuki, M. Q. Le, C. A. Nidom, Y. Sakai-Tagawa, Y. Muramoto, M. Ito, M. Kiso, T. Horimoto, K. Shinya, T. Sawada, M. Kiso, T. Usui, T. Murata, Y. Lin, A. Hay, L. F. Haire, D. J. Stevens, R. J. Russell, S. J. Gamblin, J. J. Skehel, and Y. Kawaoka. 2006. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature 444:378-382. [DOI] [PubMed] [Google Scholar]
- 35.Zitzow, L. A., T. Rowe, T. Morken, W. J. Shieh, S. Zaki, and J. M. Katz. 2002. Pathogenesis of avian influenza A (H5N1) viruses in ferrets. J. Virol. 76:4420-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]