Abstract
The NS3 protein of hepatitis C virus (HCV) possesses protease activity responsible for the proteolytic cleavage of the viral polyprotein at the junctions of nonstructural proteins downstream of NS3. The NS3 protein was also found to be internally cleaved. In this study, we demonstrated that internal cleavages occurred on the NS3 protein of genotype 1b in the presence of NS4A, both in culture cells and with a mouse model system. No internal cleavage products were detected with the NS3 and NS4A proteins of genotype 2a. Three potential cleavage sites were detected in the NS3 protein (genotype 1b), with IPT402|S being the major one. The internal cleavage requires the polyprotein processing activity of NS3 protease, but when supplemented in trans, the internal cleavage efficiency is reduced. In addition, several mutations in NS4A disrupted the internal cleavage of NS3 but did not affect polyprotein processing, indicating that NS4A contributes differently to these two proteolytic activities. Furthermore, Ile-25, Val-26, and Ile-29 of the NS4A protein, important for the NS4A-dependent internal cleavages, were also shown to be critical for the transforming activity of NS3, but mutations at these critical residues resulted only in a slight increase of HCV replicating efficiency. The internal cleavage-associated enhancement of the transforming activity of NS3 was reduced when a T402A substitution at the major internal cleavage site was introduced. The multiple roles of NS4A in viral multiplication and pathogenesis make NS4A an ideal molecular target for HCV therapy.
Hepatitis C virus (HCV) is a member of the family Flaviviridae with a single-stranded, positive-sense RNA of 9.6 kb that encodes a polyprotein of approximately 3,000 amino acid residues (8, 18, 35). The polyprotein precursor is further processed into structural and nonstructural proteins by cellular signal peptidase and viral proteases. Cleavage at the NS2-NS3 junction occurs autoproteolytically with the NS2-3 protease (15), whereas cleavage of the downstream nonstructural proteins is mediated by the viral NS3 serine protease (3, 10, 14).
The viral NS3 protein has multiple functions. The N-terminal-one-third region possesses protease activity, and the C-terminal-two-thirds region contains NTPase and RNA helicase activities. In addition, previous studies indicate that the N-terminal domain of the NS3 protein has the potential for cell transformation (30, 41, 43). Subcutaneous inoculation of nude mice with cells previously transfected with a plasmid that expresses a C-terminal deletion NS3 protein induced tumor formation (16). Mechanisms of NS3 involved in cell transformation are not fully understood. Nevertheless, the transforming ability of NS3 was abolished when cells were treated with the protease inhibitors phenylmethylsulfonyl fluoride and tosylsulfonyl phenylalanyl chloromethyl ketone (43). This indicates that protease activity is required for the transforming activity of the HCV NS3 protein. Furthermore, internal cleavages of the NS3 protein have been reported. One study demonstrated that the cleavage occurs at the QRR462|G site in the NS3 helicase domain and is probably mediated by a cellular protease (33). However, another study demonstrated that the internal NS3 cleavage is NS3-4A protease dependent and occurs at FCH369|S and IPT402|S sites (41). In addition, the naturally truncated form of NS3 spanning amino acid residues 1 to 402 has a greater oncogenic potential than full-length NS3 (41).
HCV NS4A acts as a cofactor of the NS3 serine protease and interacts with the N terminus of NS3 through its central hydrophobic domain (2, 4, 11, 31, 36, 39). The interaction increases the stability of both the NS3 and NS4A proteins. Several hydrophobic residues in the central region of NS4A involved in the interactions were demonstrated to be important for its cofactor function (6, 20, 23, 32).
In this study, we demonstrated that internal cleavage of the HCV NS3 protein has genotype specificity and is mediated by an NS4A-dependent NS3 protease activity. In addition, amino acid residues of the NS4A protein involved in the internal cleavage of NS3 are different from those required for the cofactor activity of the NS3 serine protease. Single amino acid substitutions at Ile-25, Val-26, and Ile-29 of the NS4A protein affected the interaction between NS3 and NS4A and abolished the internal cleavage of NS3 but retained the polyprotein processing activity. The key residues for the internal cleavage of NS3 were also shown to be critical for the transforming activity of NS3. We propose that through binding to the NS3 protein, NS4A mediates the internal cleavage of NS3 and enhances the transforming activity of NS3.
MATERIALS AND METHODS
Construction of expression plasmids. (i) Plasmids pcDNA-HCV-SG and pcDNA-HCV-SG-154.
Plasmids pcDNA-HCV-SG (21) and pcDNA-HCV-SG-154 represent HCV subgenomic replicons that consist of the HCV internal ribosome entry site, the neomycin resistance gene, and the encephalomyocarditis virus internal ribosome entry site fused to the HCV sequences of genomic type 1b (Taiwanese strain) from NS3 to NS5B, including the 3′ noncoding region, as shown in Fig. 1. Both plasmids encode a viral polyprotein from NS3 to NS5B with identical sequences except for three amino acid residues in the NS3. Plasmid pcDNA-HCV-SG has Leu-94, Leu-104, and Ile-153 in the NS3 protease domain, whereas pcDNA-HCV-SG-154 was derived from a variant clone that has Met-94, Pro-104, and Asn-153.
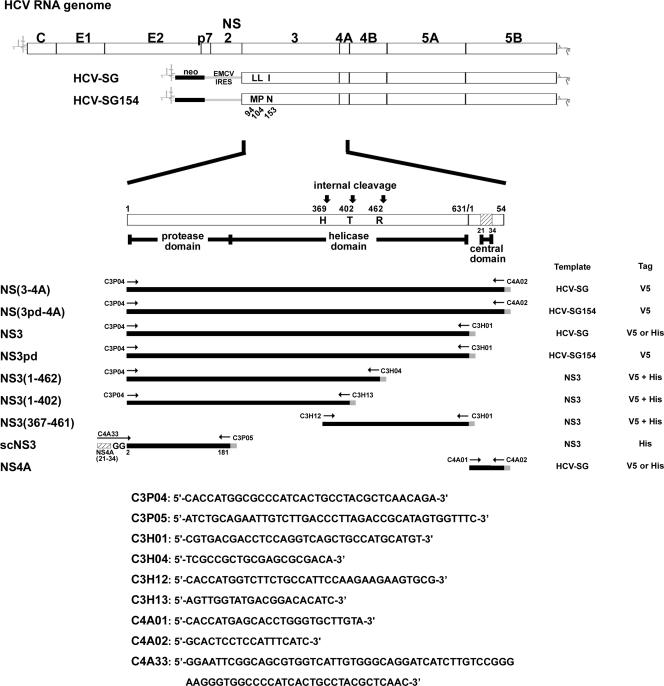
FIG. 1.
Schematic representation of the HCV RNA genome and NS(3-4A) constructs. Open bars represent the coding regions of the HCV genomic and subgenomic RNAs. Variable residues within the NS3 protease domain of the HCV subgenomic replicons HCV-SG and HCV-SG154 are indicated. Numbers mark the positions of the amino acid residues. The NS(3-4A) region is enlarged to represent the domains of the NS(3-4A) constructs used in this study. Templates and primers used to generate the NS(3-4A) constructs are indicated. cDNA fragments are shown as black lines, and tags at the C termini are shown as gray lines. Constructs with V5, His, or both V5 and His tag are indicated. Internal cleavage sites in the helicase domain of NS3 are marked.
(ii) Plasmid pcDNA-NS(4B-5An)-V5HisTopo.
For the construction of plasmid pcDNA-NS(4B-5An)-V5HisTopo, plasmid pcDNA-HCV-SG was used as a template to perform PCR to generate an NS(4B-5An) DNA fragment encompassing the HCV cDNA sequences of the full-length NS4B and the N-terminal 155 amino acid residues of NS5A. The NS(4B-5An) DNA fragment was cloned into pcDNA3.1D/V5-His-TOPO (Invitrogen) to generate plasmid pcDNA-NS(4B-5An)-V5HisTopo encoding a V5His-tagged NS(4B-5An) protein.
(iii) Plasmids pcDNA-X-V5HisTopo, pcDNA-X-V5, pcDNA-X-HisTopo, pcDNA-X(J)-V5HisTopo, and pcDNA-X(J)-V5.
X represents the HCV NS(3-4A) protease of HCV genotype 1b and a series of its derivatives, whereas X(J) represents those of genotype 2a. Generation of plasmids pcDNA-NS3-V5HisTopo and pcDNA-NS4A-V5HisTopo has been described previously (21). Similar approaches were taken to generate other pcDNA-X-V5HisTopo plasmids. In brief, DNA fragments were amplified from templates as shown in Fig. 1 with various primers and cloned into pcDNA3.1D/V5-His-TOPO. For construction of plasmid pcDNA-X-V5, plasmid pcDNA-X-V5HisTopo was digested with AgeI restriction endonuclease and treated with the Klenow fragment of DNA polymerase I prior to a further digestion with HindIII restriction endonuclease. The resultant X-V5 fragments were cloned into pcDNA3.1(+), from which the polylinker sequences between XbaI and HindIII had been deleted and the XbaI end blunted. For generation of the series of plasmids pcDNA-X(J)-V5HisTopo and pcDNA-X(J)-V5, DNA fragments were amplified from replicon JFH-1 (19) with genotype 2a-specific primers. To construct plasmid pcDNA-X-HisTopo, pcDNA-X-V5HisTopo was first digested with XhoI and AgeI restriction endonucleases and then treated with the Klenow fragment of DNA polymerase I prior to self-ligation. Plasmid pcDNA-scNS3-HisTopo encodes His-tagged single-chain NS3, NS4A(21-34)-GG-NS3(2-181)-His, as described elsewhere (9, 37).
(iv) Substitution mutants of pcDNA-NS3-V5, pcDNA-NS4A-V5, and pcDNA-HCV-SG.
Amino acid substitutions were generated by the QuikChange Multi site-directed mutagenesis kit (Stratagene) with primers that introduce specific mutations into the wild-type plasmids. The T402A mutation was introduced into the wild-type plasmid pcDNA-NS3-V5 to generate pcDNA-NS3(T402A)-V5, while the mutations G8AV9A, Y16AC17A, G21A, S22A, V23A, V24A, I25A, V26A, R28A, I29A, L31A, S32A, and K34A were independently introduced into the plasmid pcDNA-NS4A-V5 to generate a series of NS4A mutant plasmids. To generate pcDNA-HCV-SG-NS4A(I25AV26A), the I25AV26A double mutation was introduced into the plasmid pcDNA-HCV-SG.
(v) Plasmid pAdTrack-NS3-V5His.
Plasmid pAdTrack-NS3-V5His contains a dual cytomegalovirus (CMV) promoter positioned in opposite orientations to drive the expression of green fluorescent protein (GFP) and a V5His-tagged NS3 protein. For generation of plasmid pAdTrack-NS3-V5His, pcDNA-NS3-V5HisTopo (21) was digested with HindIII and PmeI restriction endonucleases. The resultant NS3-V5His DNA fragment was cloned into the HindIII-EcoRV sites of pAdTrack-CMV.
Cell lines and expression of recombinant proteins in culture cells.
Huh7 cells (a human hepatoma cell line) and NIH 3T3 cells (a mouse fibroblast cell line) were maintained at 37°C in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum plus 100 units of penicillin and 100 μg of streptomycin/ml. Expression of recombinant proteins in Huh7 cells was performed by infecting cells with recombinant vaccinia virus (vTF7-3) harboring the T7 RNA polymerase gene, followed by DNA transfection with the Lipofectin reagent as described previously (17). Expression of recombinant proteins in NIH 3T3 cells was performed by DNA transfection with the Lipofectamine 2000 reagent (Invitrogen).
In vitro transcription and RNA transfection.
In vitro transcription was carried out with T7 RNA polymerase and DNA templates, pcDNA-HCV-SG and pcDNA-HCV-SG-NS4A(I25AV26A), previously linearized with XbaI. Following RNase-free DNase (Promega) treatment and phenol-chloroform extraction, the RNA transcripts were used to perform RNA transfection in Huh7 cells with Lipofectin reagent (Invitrogen). Plasmid pCMV-Luc (25) linearized with BamHI was cotransfected as an internal control.
Reverse transcription, PCR, and real-time PCR.
Forty-eight hours after RNA transfection, total RNA was isolated from the transfected cells. Reverse transcription was performed as described previously (21) with the primer 5′-GCTCTAGAATTCGCCAGCCCCCTGATGG-3′, which can specifically anneal to the antigenomic strand of HCV subgenomic RNA. PCR was carried out with the primer set 5′-AGTACCACAAGGCCTTTCGC-3′ and 5′-CTGTCTTCACGCAGAAAGCG-3′ to amplify the antigenomic strand of HCV subgenomic RNA. In addition, the primer set 5′-GAGATCTCACGCAGGCAGTT-3′ and 5′-TTTTGTGCCAGAGTCCTTCGA-3′ was used in parallel to detect luciferase mRNA as a control of transfection efficiency. Real-time PCR was carried out using the same sets of primers and Power SYBR Green PCR Master Mix (Applied Biosystems) and analyzed with the ABI Prism 7900HT sequence detection system (Applied Biosystems).
Expression of transgenes in mice by hydrodynamics injection.
Hydrodynamics injection was performed as previously described (24) with modifications. In brief, 20 μg each of the plasmids pAdTrack-NS3-V5His and pcDNA-NS4A-V5 was injected into the tail vein of 4- to 5-week-old BALB/c mice in a volume of saline equivalent to 10% of the body mass of the mouse (2 ml for a mouse of 20-g body mass). The total volume was delivered within 5 to 8 s. Eight to twenty-four hours after injection, mice were sacrificed. Expression of the viral proteins in the mouse livers and generation of internal cleavage products of the NS3 protein were analyzed by Western blot analysis.
Coimmunoprecipitation and Western blot analysis.
Coimmunoprecipitation and Western blot analysis were performed as described previously (21).
Polyprotein processing activity of NS3 protease.
The polyprotein processing activity of the NS3 protease was examined by cotransfecting Huh7 cells with plasmids encoding V5-tagged NS3 protease and NS4A cofactor and a plasmid representing the substrate protein NS(4B-5An)-V5His, unless indicated otherwise. Two days posttransfection, the transfected cells were harvested and subjected to Western blot analysis. Antibody specific to the V5 tag was used for the detection of V5-tagged NS3, NS4A, NS(4B-5An)-V5His, and NS5A(1-155)-V5His.
Soft agar assay.
To analyze colony formation in soft agar, 2 ml bottom agar mixture containing 0.7% agar in F12 medium with 10% fetal calf serum was layered over each well of a six-well plate. NIH 3T3 cells (5 × 104) harvested 24 h posttransfection were resuspended in Dulbecco's modified Eagle's medium containing 0.35% agar and overlaid onto the bottom agar. Colonies were counted after incubation at 37°C for 4 weeks in a CO2 incubator.
Nucleotide sequence accession number.
The cDNA sequence encoding the NS3 to NS5B proteins of HCV genotype 1b (Taiwanese strain) has been deposited in EMBL under accession number AM494937.
RESULTS
NS4A is required for internal cleavage of NS3 both in culture cells and with the mouse model.
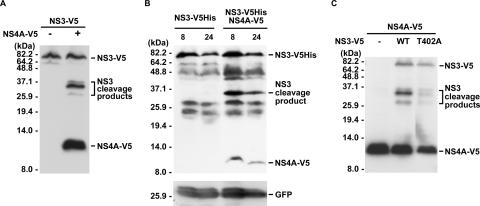
Internal cleavages of the HCV NS3 protein have been reported previously. One study reported that an internal cleavage occurs at the QRR462|G site and is mediated by a cellular protease (33), while the other demonstrated two other sites of internal NS3 cleavage, at IPT402|S and FCH369|S, that are dependent on NS3-4A protease activity (41). In this study, we found that internal cleavages of the NS3 protein occur only in the presence of the viral NS4A protein (Fig. 2). In addition, one major and two minor cleavage products of the NS3 protein with a C-terminal V5 tag were detected by Western blot analysis when Huh7 cells were cotransfected with NS3-V5- and NS4A-V5-encoding plasmids (Fig. 2A). The internal cleavage of the NS3 protein was also observed when hydrodynamics injection was performed to coexpress NS3 and NS4A proteins in mouse liver. Nevertheless, only one major cleavage product was detected (Fig. 2B). Based on the molecular size and its comigration (data not shown) with the major cleavage product detected in Fig. 2A, we propose that the major cleavage product represents the V5His-tagged C-terminal domain of the NS3-V5His protein generated from cleavage at the IPT402|S site.
FIG. 2.
Internal cleavage of the NS3 protein in the presence of NS4A. (A) Internal cleavage of NS3 in Huh7 cells. Plasmids encoding V5-tagged HCV NS3 and NS4A proteins were cotransfected into Huh7 cells. Cell lysates were harvested 2 days posttransfection for Western blot analysis with anti-V5 antibody. (B) Internal cleavage of NS3 in mouse liver tissue. Plasmids pAdTrack-NS3-V5His and pcDNA-NS4A-V5 were introduced into mouse liver tissue by hydrodynamics injection. Western blot analysis was performed with tissue homogenates prepared at 8 or 24 h after injection as indicated with antibody against the V5 epitope. GFP coexpressed with NS3 from pAdTrack-NS3-V5His was analyzed in parallel as an internal control. (C) Effect of T402A mutation on the internal cleavage activity of NS3. Plasmids encoding V5-tagged HCV proteins as indicated were transfected into Huh7 cells. Cell lysates were harvested 2 days posttransfection for Western blot analysis with anti-V5 antibody.
Sequence analysis of the HCV polyprotein revealed the substrate specificity of the NS3 protease to be D/EXXXXC/T↓S/A (14). When analyzing the putative sites of internal cleavage of the genotype 1b NS3 protein used in this study, both the P1 (Thr-402) and P1′ (Ser-403) positions in the VSVIPT402|S cleavage site matched the conserved sequences. However, only the residue at the P1′ position (Ser-370) of HLIFCH369|S coincided with the conserved NS3 protease cleavage sequence, while no sequence conservation was observed with SRSQRR462|G. Thus, amino acid mutation at Thr-402 was introduced into the NS3-expressing plasmid. Huh7 cells were cotransfected with the NS3(T402A) mutant plasmid and the NS4A-V5 expression plasmid, followed by Western blot analysis. As shown in Fig. 2C, mutation at Thr-402 nearly abolished the generation of the major cleavage product of NS3. The results indicate that the major cleavage product is generated from the cleavage at IPT402|S.
Polyprotein processing activity of the NS3 protease is required for NS4A-dependent internal NS3 cleavage.
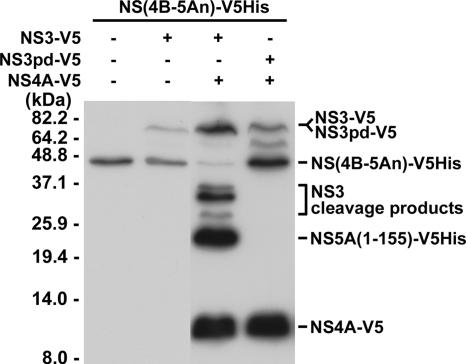
To examine whether the polyprotein processing activity of the NS3 protease is required for the internal cleavage of NS3, an NS3 polyprotein processing system was established. Huh7 cells were cotransfected with plasmids encoding the NS3 protease and the NS4A cofactor and a plasmid representing the V5His-tagged HCV protein encompassing the full-length NS4B and the N-terminal 155 amino acid residues of the NS5A protein, designated NS(4B-5An)-V5His. The NS(4B-5An)-V5His fusion protein served as a substrate that underwent proteolytic cleavage by the NS3 protease to produce NS4B and NS5A(1-155)-V5His in the presence of NS4A (Fig. 3). Furthermore, NS3pd-V5, an NS3 variant that does not process the polyprotein substrate, was used in this study. As shown in Fig. 3, neither the proteolytic processing product NS5A(1-155)-V5His nor the internal cleavage products were detected with the NS3pd protein even in the presence of NS4A. These results indicate that in addition to NS4A, polyprotein processing activity of the NS3 protease is required for the internal cleavage of NS3.
FIG. 3.
Requirement of NS3 polyprotein processing activity for internal cleavage of NS3. Plasmids encoding V5-tagged NS3, NS3pd, NS4A, and the substrate NS(4B-5An) proteins as indicated were cotransfected into Huh7 cells. Two days posttransfection, cells were harvested to perform Western blot analysis with antibody specific to the V5 epitope.
Internal NS3 cleavage requires polyprotein processing activity that can be supplemented in trans and can occur in the context of NS(3-4A).
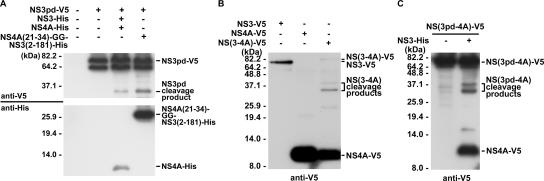
Although no internal cleavage products were detected with the NS3pd protein, we found that the function-deficient NS3pd protein can be internally cleaved by a functional NS3 protease supplemented in trans. In the presence of NS4A and a functional NS3 protease, a major cleavage product was detected by antibodies specific to the V5 tag of NS3pd (Fig. 4A). Nevertheless, the efficiency of internal cleavage was significantly reduced. Moreover, the internal cleavage of NS3pd also occurred by the supplementing of a single-chain NS3 protease that contains the NS4A sequences from amino acid residues 21 to 34 fused to the N terminus of NS3 [NS4A(21-34)-GG-NS3(2-181)-His] (Fig. 4A). A single-chain NS3 protease was previously shown to be enzymatically active and have a higher protease activity than the noncovalent complex of NS3 and NS4A, presumably due to the short linker that connects NS4A and NS3 protease peptides covalently (9, 37).
FIG. 4.
NS3 protease activity required for internal cleavage can be supplemented in trans. Plasmids encoding proteins as indicated in panels A to C were cotransfected into Huh7 cells. Two days posttransfection, cells were harvested for Western blot analysis with antibodies specific to the V5 epitope and the His tag of the recombinant proteins as indicated. (A) Internal cleavage of NS3pd protein in the presence of functional NS3 supplemented in trans. (B) Autoprocessing of NS(3-4A)-V5 protein. (C) Internal cleavage of NS(3pd-4A) protein by functional NS3 supplemented in trans.
To further examine whether NS4A can facilitate the internal cleavage of NS3 in cis, plasmids encoding NS(3-4A) and NS(3pd-4A) fusion proteins were used to transfect Huh7 cells. The results demonstrated that the junction between NS3 and NS4A in the NS(3-4A) protein can be spontaneously cleaved. In addition, proteins representing the internal cleavage products of NS3 fused to NS4A-V5 were detected (Fig. 4B). When the NS(3pd-4A) protein was applied, no internal cleavage products were generated. Nevertheless, similar to the case of NS3pd, internal cleavage of NS(3pd-4A) became detectable when a functional NS3 was coexpressed in trans (Fig. 4C). Taken together, these results indicate that the internal cleavage of NS3 requires NS4A and functional NS3 protease. It can also occur in the context of NS(3-4A), but when NS3 is supplemented in trans, the internal cleavage efficiency is reduced. In addition, the result from the study of single-chain NS3 indicates that only the middle domain of the NS4A protein, from amino acid residues 21 to 34, is required to facilitate the internal cleavage of NS3.
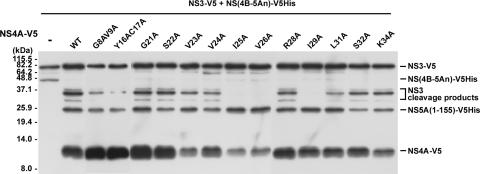
Critical residues of NS4A involved in the internal cleavage of NS3 are separable from its cofactor activity for polyprotein processing.
NS4A interacts with NS3 through its middle domain and regulates the polyprotein processing activity of the NS3 protease (23). In this study, we have shown that internal cleavage of NS3 requires the polyprotein processing activity of NS3 in the presence of NS4A (Fig. 3). However, it is not clear whether the requirement of NS4A for the internal cleavage is fully dependent on its cofactor activity for the NS3 protease. To identify critical residues of NS4A involved in the NS4A-dependent NS3 internal cleavage and to examine the correlation between the occurrence of internal cleavage and the NS4A-dependent polyprotein processing activity of NS3, a series of substitution mutations in the middle domain of NS4A, spanning amino acid residues 21 to 34, were introduced into the wild-type NS4A-expressing plasmid (pcDNA-NS4A-V5). Plasmids representing the V5-tagged NS3 protease, the NS4A cofactor, and a V5His-tagged polyprotein processing substrate, NS(4B-5An), were cotransfected into Huh7 cells. Two days posttransfection, cells were harvested and Western blot analysis was performed with anti-V5 antibody to detect the internal cleavage products of NS3 and the substrate processing product NS5A(1-155)-V5His. Surprisingly, NS4A-dependent cleavage at the junction of NS4B and NS5A was processed efficiently under all conditions examined, whereas NS4A(I25A), NS4A(V26A), and NS4A(I29A) completely blocked the generation of the internal cleavage products (Fig. 5). These results clearly demonstrate that residues Ile-25, Val-26, and Ile-29 of NS4A are important for NS4A-dependent NS3 internal cleavage activity. The observation that amino acid residues of NS4A involved in the internal cleavage of NS3 are separable from those required for the cofactor activity of the NS3 serine protease indicates that different mechanisms of NS4A are involved in internal NS3 cleavage and polyprotein processing activities.
FIG. 5.
Effects of NS4A mutation on internal NS3 cleavage and polyprotein processing at the NS4B/NS5A boundary. Huh7 cells were cotransfected with plasmids encoding the NS(4B-5An)-V5His substrate, NS3-V5, and the wild type (WT) or a mutant form of the NS4A-V5 protein as indicated. Western blot analysis was performed with anti-V5 antibody 2 days posttransfection to detect the substrate NS(4B-5An)-V5His, the processing product NS5A(1-155)-V5His, and the V5-tagged NS4A, NS3, and NS3 internal cleavage products.
Interactions between NS3 and NS4A mutant proteins in culture cells.
NS4A is known to complex with and stabilize NS3 (6, 23). Whether the interaction between NS3 and NS4A directly involves the internal cleavage of NS3 or facilitates the cleavage mediated by an unidentified mechanism is not clear. To examine possible effects of the NS4A mutations on the interactions between NS4A and NS3, coimmunoprecipitation-Western blot analysis was performed following a cotransfection of Huh7 cells with plasmids representing the His-tagged NS3 protein and a V5-tagged mutant form of NS4A. As shown in Fig. 6, NS4A(I25A), NS4A(V26A), and NS4A(I29A) mutants failed to coprecipitate with the NS3 protein, indicating that mutations at residues Ile-25, Val-26, and Ile-29 of the NS4A protein affected the interaction between NS3 and NS4A in culture cells. However, as shown in Fig. 5, these three NS4A mutants retained cofactor activity to facilitate NS3-mediated polyprotein processing at the NS4B/NS5A boundary. These results indicate that even though NS4A is required for both polyprotein processing and internal cleavage of NS3, their requirements for the strength of NS3-NS4A interactions are quite different. It is possible that the internal cleavage requires a stronger or a more stable association between NS3 and NS4A. With the Ile-25, Val-26, or Ile-29 mutation, the interaction between NS4A and NS3 becomes either unstable or too weak to be detected in the assay system, but this weak interaction could still support proteolytic cleavage at the NS4B/NS5A boundary.
FIG. 6.
Interactions between NS3 and NS4A mutant proteins in culture cells. Huh7 cells were cotransfected with plasmids encoding the NS3-His protein and NS4A-V5 or its mutant protein as indicated. Cell lysates were harvested 2 days posttransfection to perform coimmunoprecipitation (IP) with anti-His antibody, followed by Western blot (WB) analysis with anti-V5 antibody. Inputs represent 5% of the cell lysates used in the coimmunoprecipitation assay.
Effects of NS4A mutations on transforming activity of NS3 internal cleavage products.
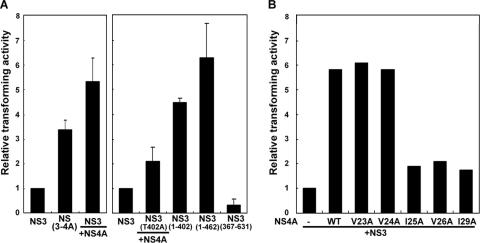
In Fig. 2C, we demonstrate that the T402A mutation diminishes the production of the major internal cleavage product of NS3. The NS3(1-402) protein, which represents the N-terminal domain of NS3 from amino acid residue 1 to 402, was previously demonstrated to have an oncogenic potential greater than that of wild-type NS3 (41). We proposed that through binding to the NS3 protein, NS4A mediates internal cleavage of NS3, which in turn enhances the transforming activity of NS3. Transforming activity of NS3 with or without coexpression of NS4A and their mutant forms was examined by soft agar assay. As shown in Fig. 7A, NIH 3T3 cells expressing the NS(3-4A) protein had a 3.4-fold-greater transforming activity than the cells expressing the NS3 protein alone. Coexpression of NS4A with wild-type NS3 enhanced the transforming activity up to 5.3-fold. Nevertheless, when NS4A was coexpressed with the NS3(T402A) mutant, the relative transforming activity was reduced (compare lane 5 to lane 3). In addition, NS3(1-402) showed a transforming activity of 4.5-fold. Previous studies also suggested a cellular protease-mediated internal cleavage of NS3 at QRR462|G (33), but biological significances of the internal cleavage were not determined. In this study, we showed that the N-terminal NS3 protein, NS3(1-462), had a transforming activity up to 6.2-fold that of the full-length NS3, and only negligible transforming activity was detected with the C-terminal NS3 protein, NS3(367-631).
FIG. 7.
Transforming activity of NS3 in the presence of wild-type NS4A and its mutants. (A) NS4A enhances the transforming activity of the NS3 protein. Plasmids encoding the wild-type NS3 protein or its mutant forms were independently transfected into NIH 3T3 cells with or without coexpression of wild-type NS4A. Fifty-thousand transiently transfected cells were used in the soft agar assay. Relative transforming activities were calculated by normalization of the colony numbers to that with the NS3 protein. (B) Effects of NS4A mutations on transforming activity of NS3. NIH 3T3 cells were cotransfected with plasmids encoding NS3 and NS4A or its mutants as indicated for the soft agar assay. Relative transforming activities were calculated by normalization of the colony numbers with coexpression of NS3 and NS4A to that with the NS3 protein alone. Results shown represent averages for three (A) or two (B) independent experiments.
To further understand the effects of NS4A mutation on the transforming activity of NS3, plasmids encoding NS4A mutants with a single amino acid substitution were independently cotransfected into NIH 3T3 cells with an NS3-encoding plasmid, followed by soft agar assay. As shown in Fig. 7B, neither the V23A mutation nor the V24A mutation had an effect on the ability of NS4A in enhancing the transforming activity of NS3. However, the transforming activity was significantly reduced when NS4A(I25A), NS4A(V26A), or NS4A(I29A) was coexpressed with NS3. The transforming activity of NS3 correlated very well with the presence of the NS3 internal cleavage products shown in Fig. 2 and 5, indicating an association of the internal cleavage products of NS3 with the transforming activity.
Internal cleavage and transforming activity of the NS3/4A protein of HCV genotype 2a.
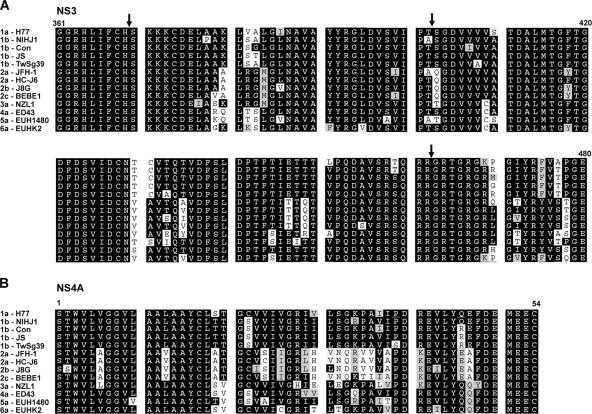
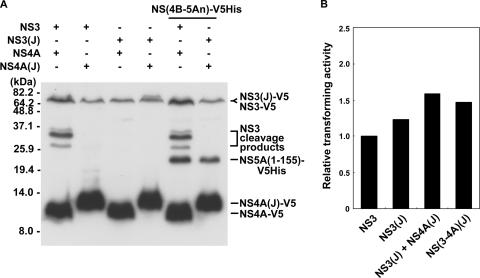
Amino acid sequence comparison among NS(3-4A) proteins of different HCV genotypes revealed a conservation at QRR462|G, but the conservation at the IPT402|S internal cleavage site was not observed among genotypes 2, 3, and 5 (Fig. 8A). In addition, amino acid residues Val-26 and Ile-29 of the NS4A protein, identified as critical for the internal cleavage activity of the NS3 protein of genotype 1b (Fig. 5), were not conserved among genotypes 2, 4, and 6, either (Fig. 8B). It is not clear whether the sequence difference correlates to the internal cleavage activity and the variation of clinical severity of liver diseases in patients infected with different HCV genotypes. The HCV JFH-1 clone that belongs to genotype 2a has IPA402|Q in the NS3 protein and Ile-26 and Leu-29 in the NS4A protein. JFH-1 has previously been demonstrated to replicate efficiently in Huh7 cells (19) and to produce secretable viral particles that are infectious for both Huh7 cells and chimpanzees (40). Whether internal cleavage of the NS3 protein could occur in the genotype 2a JFH-1 clone was examined by coexpressing NS3 and NS4A proteins of either genotype 2a or genotype 1b in Huh7 cells. The results showed that internal cleavage of NS3 occurred only when NS3 and NS4A were both from genotype 1b (Fig. 9A). However, with NS3 and NS4A proteins both from genotype 2a, the NS(4B-5An) polyprotein substrate of genotype 1b was cleaved at an efficiency comparable to that of the activity of the genotype 1b protease. Altogether, these results suggest that both IPT402|S of the NS3 protein and Val-26 and Ile-29 of the NS4A protein are required for the internal cleavage of NS3. Transforming activity of the NS3/4A protein of the genotype 2a JFH-1 clone was further examined. As shown in Fig. 9B, NS3 of genotype 2a had transforming activity slightly greater than that of genotype 1b, but with the coexistence of the genotype 2a NS4A protein, the transforming activity increased only slightly. The results clearly demonstrate a correlation between genotype-specific internal cleavage activity and transforming activity of NS3. They also suggest a possible contribution of the internal NS3 cleavage to the significant differences in clinical severity for patients infected with different HCV genotypes (26, 34, 42). The minor increase of the transforming activity of NS3(JFH-1) in the presence of NS4A may result from the function of NS4A in stabilizing the NS3 protein.
FIG. 8.
Conservation of the critical amino acid residues involved in internal NS3 cleavage. Alignment of a partial sequence of the NS3 protein (residues 361 to 480) (A) or full-length NS4A (B) from various HCV genotypes, including 1a-H77 (NC_004102), 1b-NIHJ1 (D89815), 1b-Con (AJ238799), 1b-JS (D85516), 1b-TwSg39 (AM494937), 2a-JFH-1 (AB047639), 2a-HC-J6 (D00944), 2b-J8G (D10988), 2c-BEBE1 (D50409), 3a-NZL1 (D17763), 4a-ED43 (Y11604), 5a-EUH1480 (Y13184), and 6a-EUHK2 (Y12083). Arrows in panel A indicate the sites of internal NS3 cleavage.
FIG. 9.
Internal cleavage and transforming activity of NS3 protein of genotype 2a JFH-1 clone. (A) Internal cleavage activity of NS3 protein of genotype 2a JFH-1 clone. Plasmids encoding NS3 and NS4A from genotype 1b and genotype 2a were transfected into Huh7 cells with or without coexpression of NS(4B-5An)-V5His. Western blot analysis was performed 2 days posttransfection with antibody specific to the V5 epitope. “(J)” indicates proteins of genotype 2a JFH-1 clone. (B) Effect of NS4A protein on transforming activity of NS3 (JFH-1). NIH 3T3 cells were transfected with plasmids as indicated. Twenty-four hours posttransfection, a soft agar assay was carried out as described in the legend to Fig. 7. Results represent averages for two independent experiments.
Effect of internal NS3 cleavage on replication of HCV subgenomic RNA.
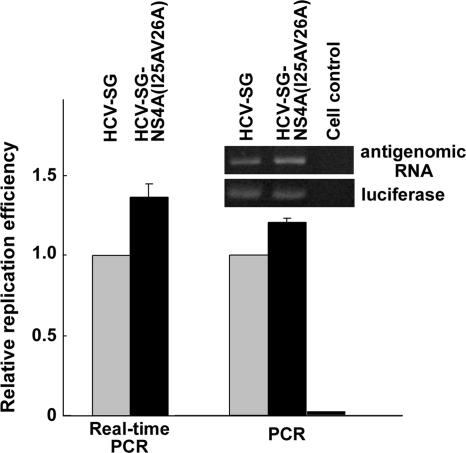
Internal cleavage of NS3 disrupts the structure of the helicase domain known to be important for HCV replication. To analyze whether internal cleavage of the NS3 protein has any effect on HCV RNA replication, a double mutation, I25AV26A, was introduced into the NS4A sequences of plasmid pcDNA-HCV-SG, which represents the subgenomic replicon of HCV genotype 1b. Two days posttransfection of the wild-type and mutant replicon RNA into Huh7 cells, reverse transcription was performed, followed by PCR/real-time PCR to analyze the production of the antigenomic RNA as an indicator of RNA replication. As shown in Fig. 10, the level of antigenomic RNA in cells transfected with the NS4A(I25A V26A) mutant replicon was slightly higher than that of the wild-type-transfected cells. This suggests that internal cleavage of the NS3 protein would downregulate the replication efficiency of HCV RNA, possibly due to a disruption of the RNA helicase activity of NS3. Nevertheless, the effect was limited. Since the internal cleavage of NS3 is always incomplete even in the presence of a higher dose of NS4A (data not shown), it is likely that the uncleaved NS3 provides the helicase activity sufficient for the replication of HCV genomic RNA.
FIG. 10.
Effect of internal NS3 cleavage on the replication of HCV genomic RNA. HCV subgenomic replicon (HCV-SG) and mutant HCV-SG-NS4A(I25AV26A) RNA were independently cotransfected into Huh7 cells with control luciferase RNA. Two days posttransfection, total RNA was isolated to perform reverse transcription and PCR/real-time PCR as described in Materials and Methods. PCR products representing the antigenomic strand of HCV RNA were normalized against control luciferase RNA. A representative image of reverse transcription-PCR products and relative replication efficiency compared to those of the wild-type replicon are shown.
DISCUSSION
In the present study, we demonstrate a genotype-specific internal cleavage of the HCV NS3 protein (Fig. 9A). The HCV NS3 protein of genotype 1b could be internally cleaved in the presence of NS4A (Fig. 2). Using an NS3 variant (NS3pd) that lacks proteolytic activity on polyprotein processing, we found that the internal cleavage of NS3 requires a functional NS3 protease (Fig. 3). In fact, the catalytic triad (His-57, Asp-81, and Ser-139) remained unchanged in the NS3pd protein. Whether the functional deficiency of NS3pd resulted from an incorrect conformation of the NS3 protein is not clear. Nevertheless, NS3pd could still act as a substrate and be internally cleaved when a functional NS3 protein was supplemented in trans (Fig. 4). In addition, we found that single amino acid substitutions at Ile-25, Val-26, and Ile-29 of NS4A affect the interaction between NS3 and NS4A (Fig. 6) and abolish the internal cleavage of NS3 in transfected cells but retain the ability to facilitate the proteolytic cleavage at the NS4B/NS5A boundary (Fig. 5). These results strongly indicate that NS4A has differential roles involved in the internal NS3 cleavage and the proteolytic processing of HCV polyprotein. We propose that internal cleavage requires a more stable interaction between NS3 and NS4A than that of polyprotein processing. Details of mechanisms remain to be elucidated.
Internal cleavage of viral NS3 protease/helicase has been detected in cells infected with dengue virus 2 (1). Internal cleavage requires both NS3 and its cofactor NS2B (1) and occurs within the conserved QRR458|GRIGR helicase motif (38). Internal cleavage was also observed in tick-borne encephalitis virus (29), West Nile virus (5), and plant potyvirus (28). In a difference from the internal cleavage of the HCV NS3 protein, which can occur in trans, Bera et al. demonstrated that the NS2B-NS3 protease of West Nile virus could occur only in cis (5). Effects of NS3 internal cleavage on the replication of dengue virus and West Nile virus are not clear. In the case of potyvirus, internal cleavages separate the N-terminal VPg domain from the C-terminal proteinase domain of the NIa proteinase (7) and release a 24-amino-acid peptide from the C terminus (28). The proteolytic activity of the C-terminally truncated proteinase was less efficient than that of the full-length proteinase. In this study, we found that in the case of HCV, the NS4A-dependent internal cleavage of NS3 slightly downregulated the replication of the viral subgenome (Fig. 10) and produced the N-terminal product NS3(1-402), which has a greater oncogenic potential than full-length NS3 (Fig. 7).
Although the influence of viral genotypes on clinical severity for hepatitis C patients remains controversial, most studies correlate HCV genotype 1b to severe hepatitis and advanced liver disease (26, 34, 42). In HCV-related cirrhosis and hepatocellular carcinoma, genotype 1b is predominant. In the present study, we found that NS4A-dependent internal cleavage of NS3 occurred in genotype 1b but not genotype 2a (Fig. 9A). The soft agar assay with NS3 and NS4A mutants (Fig. 7) linked the NS4A-dependent internal cleavage to the oncogenic potential of the NS3 protein. Interestingly, the NS4A protein of genotype 2a had a limited effect on the transforming activity of NS3 that lacks internal cleavage activity (Fig. 9B). Thus, the hepatocarcinogenic effect of HCV genotype 1b may associate with the oncogenic potential of NS3 with internal cleavage activity.
NS3/4A is responsible for proteolytic processing of the viral polyprotein for generation of nonstructural proteins important for viral multiplication. The question is whether further cleavage of NS3 within the helicase domain disrupts the helicase activity and thus affects replication of the viral genome. In this study, we found that NS4A plays different roles in supporting HCV polyprotein processing and internal NS3 cleavage (Fig. 5). In addition, the internal cleavage never reached completion even in the presence of higher levels of the NS4A protein (data not shown). It is possible that the differential requirements of NS4A for these two proteolytic activities ensure a maintaince of helicase activity from the uncleaved NS3 protein to support the replication of the viral genome. Critical residues of NS4A involved in the internal cleavage and transforming activity of NS3 were functionally correlated (Fig. 7). However, mutations at the critical residues of internal cleavage increased only slightly the efficiency of viral replication (Fig. 10), indicating that uncleaved NS3 could provide sufficient helicase activity for viral replication.
On the other hand, NS3/4A was demonstrated to cleave cellular proteins, Toll-interleukin 1 receptor domain-containing adaptor inducing beta interferon (TRIF) and CARD adaptor inducing beta interferon (Cardif), to facilitate immune evasion of HCV (12, 22, 27). In addition, the intrahepatic subsets of immune cells were altered and protection of hepatocytes from Fas-mediated apoptosis was observed with an NS3/4A expression transgenic mouse model (13). Whether the NS3 cleavage products are involved in immune modulation needs to be further studied. Taken together, the multiple functions of NS3/4A may contribute to the low titer, chronic infection, and hepatocarcinogenesis of HCV. This makes NS4A an ideal target for HCV therapy.
Acknowledgments
We thank Shun-Chi Wu and Tzu-Ling Tseng (National Taiwan University) for technical assistance and helpful comments on this article. We are grateful to Takaji Wakita (Tokyo Metropolitan Institute for Neuroscience) for providing the HCV JFH-1 replicon and to Jen-Yang Chen (National Health Research Institutes and National Taiwan University) for providing plasmid pCMV-Luc.
This work was supported in part by research grants NSC 94-2752-B-002-009-PAE, NSC 95-2752-B-002-009-PAE, and NSC 95-2320-B-002-094 from the National Science Council of the Republic of China.
Footnotes
Published ahead of print on 23 May 2007.
REFERENCES
- 1.Arias, C. F., F. Preugschat, and J. H. Strauss. 1993. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology 193:888-899. [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1994. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J. Virol. 68:5045-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1993. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J. Virol. 67:3835-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., V. Lohmann, T. Wilkinson, and J. O. Koch. 1995. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J. Virol. 69:7519-7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bera, A. K., R. J. Kuhn, and J. L. Smith. 2007. Functional characterization of cis and trans activity of the flavivirus NS2B-NS3 protease. J. Biol. Chem. 282:12883-12892. [DOI] [PubMed] [Google Scholar]
- 6.Butkiewicz, N. J., M. Wendel, R. Zhang, R. Jubin, J. Pichardo, E. B. Smith, A. M. Hart, R. Ingram, J. Durkin, P. W. Mui, M. G. Murray, L. Ramanathan, and B. Dasmahapatra. 1996. Enhancement of hepatitis C virus NS3 proteinase activity by association with NS4A-specific synthetic peptides: identification of sequence and critical residues of NS4A for the cofactor activity. Virology 225:328-338. [DOI] [PubMed] [Google Scholar]
- 7.Carrington, J. C., R. Haldeman, V. V. Dolja, and M. A. Restrepo-Hartwig. 1993. Internal cleavage and trans-proteolytic activities of the VPg-proteinase (NIa) of tobacco etch potyvirus in vivo. J. Virol. 67:6995-7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, A. Medina-Selby, P. J. Barr, A. J. Weiner, D. W. Bradley, G. Kuo, and M. Houghton. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimasi, N., A. Pasquo, F. Martin, S. Di Marco, C. Steinkuhler, R. Cortese, and M. Sollazzo. 1998. Engineering, characterization and phage display of hepatitis C virus NS3 protease and NS4A cofactor peptide as a single-chain protein. Protein Eng. 11:1257-1265. [DOI] [PubMed] [Google Scholar]
- 10.Eckart, M. R., M. Selby, F. Masiarz, C. Lee, K. Berger, K. Crawford, C. Kuo, G. Kuo, M. Houghton, and Q. L. Choo. 1993. The hepatitis C virus encodes a serine protease involved in processing of the putative nonstructural proteins from the viral polyprotein precursor. Biochem. Biophys. Res. Commun. 192:399-406. [DOI] [PubMed] [Google Scholar]
- 11.Failla, C., L. Tomei, and R. De Francesco. 1995. An amino-terminal domain of the hepatitis C virus NS3 protease is essential for interaction with NS4A. J. Virol. 69:1769-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 13.Frelin, L., E. D. Brenndorfer, G. Ahlen, M. Weiland, C. Hultgren, M. Alheim, H. Glaumann, B. Rozell, D. R. Milich, J. G. Bode, and M. Sallberg. 2006. The hepatitis C virus and immune evasion: non-structural 3/4A transgenic mice are resistant to lethal tumour necrosis factor-α-mediated liver disease. Gut 55:1475-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67:2832-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. A second hepatitis C virus-encoded proteinase. Proc. Natl. Acad. Sci. USA 90:10583-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He, Q. Q., R. X. Cheng, Y. Sun, D. Y. Feng, and H. Zheng. 2003. Hepatocyte transformation and tumor development induced by hepatitis C virus NS3 N-terminal protein. Zhonghua Bing Li Xue Za Zhi 32:255-259. (In Chinese.) [PubMed]
- 17.Hsieh, P. K., S. C. Chang, C. C. Huang, T. T. Lee, C. W. Hsiao, Y. H. Kou, I. Y. Chen, C. K. Chang, T. H. Huang, and M. F. Chang. 2005. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 79:13848-13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato, N., M. Hijikata, Y. Ootsuyama, M. Nakagawa, S. Ohkoshi, T. Sugimura, and K. Shimotohno. 1990. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl. Acad. Sci. USA 87:9524-9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 20.Kim, J. L., K. A. Morgenstern, C. Lin, T. Fox, M. D. Dwyer, J. A. Landro, S. P. Chambers, W. Markland, C. A. Lepre, E. T. O'Malley, S. L. Harbeson, C. M. Rice, M. A. Murcko, P. R. Caron, and J. A. Thomson. 1996. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87:343-355. [DOI] [PubMed] [Google Scholar]
- 21.Kou, Y. H., S. M. Chou, Y. M. Wang, Y. T. Chang, S. Y. Huang, M. Y. Jung, Y. H. Huang, M. R. Chen, M. F. Chang, and S. C. Chang. 2006. Hepatitis C virus NS4A inhibits cap-dependent and the viral IRES-mediated translation through interacting with eukaryotic elongation factor 1A. J. Biomed. Sci. 13:861-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, K., E. Foy, J. C. Ferreon, M. Nakamura, A. C. Ferreon, M. Ikeda, S. C. Ray, M. Gale, Jr., and S. M. Lemon. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. USA 102:2992-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, C., J. A. Thomson, and C. M. Rice. 1995. A central region in the hepatitis C virus NS4A protein allows formation of an active NS3-NS4A serine proteinase complex in vivo and in vitro. J. Virol. 69:4373-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, F., Y. Song, and D. Liu. 1999. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene. Ther. 6:1258-1266. [DOI] [PubMed] [Google Scholar]
- 25.Liu, M. T., Y. R. Chen, S. C. Chen, C. Y. Hu, C. S. Lin, Y. T. Chang, W. B. Wang, and J. Y. Chen. 2004. Epstein-Barr virus latent membrane protein 1 induces micronucleus formation, represses DNA repair and enhances sensitivity to DNA-damaging agents in human epithelial cells. Oncogene 23:2531-2539. [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Labrador, F. X., S. Ampurdanes, X. Forns, A. Castells, J. C. Saiz, J. Costa, J. Bruix, J. M. Sanchez Tapias, M. T. Jimenez de Anta, and J. Rodes. 1997. Hepatitis C virus (HCV) genotypes in Spanish patients with HCV infection: relationship between HCV genotype 1b, cirrhosis and hepatocellular carcinoma. J. Hepatol. 27:959-965. [DOI] [PubMed] [Google Scholar]
- 27.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 28.Parks, T. D., E. D. Howard, T. J. Wolpert, D. J. Arp, and W. G. Dougherty. 1995. Expression and purification of a recombinant tobacco etch virus NIa proteinase: biochemical analyses of the full-length and a naturally occurring truncated proteinase form. Virology 210:194-201. [DOI] [PubMed] [Google Scholar]
- 29.Pugachev, K. V., N. Nomokonova, O. V. Morozova, and A. G. Pletnev. 1992. A short form of the tick-borne encephalitis virus NS3 protein. FEBS Lett. 297:67-69. [DOI] [PubMed] [Google Scholar]
- 30.Sakamuro, D., T. Furukawa, and T. Takegami. 1995. Hepatitis C virus nonstructural protein NS3 transforms NIH 3T3 cells. J. Virol. 69:3893-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Satoh, S., Y. Tanji, M. Hijikata, K. Kimura, and K. Shimotohno. 1995. The N-terminal region of hepatitis C virus nonstructural protein 3 (NS3) is essential for stable complex formation with NS4A. J. Virol. 69:4255-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimizu, Y., K. Yamaji, Y. Masuho, T. Yokota, H. Inoue, K. Sudo, S. Satoh, and K. Shimotohno. 1996. Identification of the sequence on NS4A required for enhanced cleavage of the NS5A/5B site by hepatitis C virus NS3 protease. J. Virol. 70:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shoji, I., T. Suzuki, M. Sato, H. Aizaki, T. Chiba, Y. Matsuura, and T. Miyamura. 1999. Internal processing of hepatitis C virus NS3 protein. Virology 254:315-323. [DOI] [PubMed] [Google Scholar]
- 34.Stankovic-Djordjevic, D., N. Djordjevic, G. Tasic, M. Dinic, A. Karanikolic, and M. Pesic. 2007. Hepatitis C virus genotypes and the development of hepatocellular carcinoma. Chin. J. Dig. Dis. 8:42-47. [DOI] [PubMed] [Google Scholar]
- 35.Takamizawa, A., C. Mori, I. Fuke, S. Manabe, S. Murakami, J. Fujita, E. Onishi, T. Andoh, I. Yoshida, and H. Okayama. 1991. Structure and organization of the hepatitis C virus genome isolated from human carriers. J. Virol. 65:1105-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanji, Y., M. Hijikata, S. Satoh, T. Kaneko, and K. Shimotohno. 1995. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J. Virol. 69:1575-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taremi, S. S., B. Beyer, M. Maher, N. Yao, W. Prosise, P. C. Weber, and B. A. Malcolm. 1998. Construction, expression, and characterization of a novel fully activated recombinant single-chain hepatitis C virus protease. Protein Sci. 7:2143-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teo, K. F., and P. J. Wright. 1997. Internal proteolysis of the NS3 protein specified by dengue virus 2. J. Gen. Virol. 78:337-341. [DOI] [PubMed] [Google Scholar]
- 39.Tomei, L., C. Failla, R. L. Vitale, E. Bianchi, and R. De Francesco. 1996. A central hydrophobic domain of the hepatitis C virus NS4A protein is necessary and sufficient for the activation of the NS3 protease. J. Gen. Virol. 77:1065-1070. [DOI] [PubMed] [Google Scholar]
- 40.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang, S. H., C. G. Lee, M. K. Song, and Y. C. Sung. 2000. Internal cleavage of hepatitis C virus NS3 protein is dependent on the activity of NS34A protease. Virology 268:132-140. [DOI] [PubMed] [Google Scholar]
- 42.Zein, N. N., and D. H. Persing. 1996. Hepatitis C genotypes: current trends and future implications. Mayo Clin. Proc. 71:458-462. [DOI] [PubMed] [Google Scholar]
- 43.Zemel, R., S. Gerechet, H. Greif, L. Bachmatove, Y. Birk, A. Golan- Goldhirsh, M. Kunin, Y. Berdichevsky, I. Benhar, and R. Tur-Kaspa. 2001. Cell transformation induced by hepatitis C virus NS3 serine protease. J. Viral Hepat. 8:96-102. [DOI] [PubMed] [Google Scholar]