Abstract
All plus-strand RNA viruses encode an RNA-dependent RNA polymerase (RdRp) that functions as the catalytic subunit of the viral replication/transcription complex, directing viral RNA synthesis in concert with other viral proteins and, sometimes, host proteins. RNA synthesis essentially can be initiated by two different mechanisms, de novo initiation and primer-dependent initiation. Most viral RdRps have been identified solely on the basis of comparative sequence analysis, and for many viruses the mechanism of initiation is unknown. In this study, using the family prototype equine arteritis virus (EAV), we address the mechanism of initiation of RNA synthesis in arteriviruses. The RdRp domains of the members of the arterivirus family, which are part of replicase subunit nsp9, were compared to coronavirus RdRps that belong to the same order of Nidovirales, as well as to other RdRps with known initiation mechanisms and three-dimensional structures. We report here the first successful expression and purification of an arterivirus RdRp that is catalytically active in the absence of other viral or cellular proteins. The EAV nsp9/RdRp initiates RNA synthesis by a de novo mechanism on homopolymeric templates in a template-specific manner. In addition, the requirements for initiation of RNA synthesis from the 3′ end of the viral genome were studied in vivo using a reverse genetics approach. These studies suggest that the 3′-terminal nucleotides of the EAV genome play a critical role in viral RNA synthesis.
Central in the life cycle of plus-strand RNA viruses is the process of RNA-templated RNA synthesis, which is required to replicate and transcribe the viral genome and which occurs in association with cytoplasmic membranes in the infected host cell. However, the enzymes required for this activity are not present in the eukaryotic host cell. Consequently, all plus-strand RNA viruses encode an RNA-dependent RNA polymerase (RdRp) that functions as the catalytic subunit for viral RNA synthesis, and it operates in concert with other viral and/or host proteins. Correct initiation of RNA synthesis is essential for the integrity of the viral genome. Although diverse RNA viruses use an amazing variety of replication scenarios, there are only two principally different mechanisms by which RNA synthesis can be initiated: initiation can occur either de novo or in a primer-dependent fashion (reviewed in references 27 and 61). Upon de novo initiation, the starting nucleotide provides the 3′ hydroxyl group for the addition of the next nucleotide. Primer-dependent initiation requires the use of either an oligonucleotide or a protein primer to provide the hydroxyl nucleophile. For primer-dependent initiation in RNA viruses, the most commonly used primer is either (i) an oligonucleotide that is covalently linked to a protein, as used, for example, by picornaviruses like poliovirus (41), or (ii) a capped primer that is cleaved from the 5′ end of a host mRNA by a so-called cap-snatching mechanism, which is, e.g., used for transcription by some of the segmented minus-strand RNA viruses such as influenza virus (21).
Nidoviruses are enveloped, plus-strand RNA viruses that have been grouped together on the basis of similarities in genome organization, the use of similar strategies for nonstructural and structural protein expression, and the presumed common ancestry of key replicative enzymes, including the RdRp (for recent reviews, see references 18 and 54). The order Nidovirales currently includes the families Arteriviridae, Coronaviridae, and Roniviridae. Nidovirus genomes carry a cap structure at their 5′ end (33, 46) and are 3′ polyadenylated (58). RNA synthesis in infected cells entails both amplification of the viral genome and the production of an extensive set of subgenomic mRNAs (sg mRNAs), presumably each from their specific minus-strand intermediates (for recent reviews, see references 40, 48, and 49). Thus, the 3′ terminus of the viral genome must be used to initiate the synthesis of both full-length and subgenome-length minus-strand RNA. RNA synthesis is driven by a complex set of nidovirus enzymes generated by autoproteolysis of two large polyproteins, pp1a and pp1ab, which are produced by the translation of open reading frames (ORFs) 1a and 1ab, respectively. Both polyproteins are expressed from the genomic RNA, with pp1ab synthesis requiring a ribosomal frameshift near the 3′ end of ORF1a. Although the enzyme complex formed after pp1a and pp1ab processing contains both replicase and transcriptase activity, it is usually referred to as replicase.
The replicase polyproteins of equine arteritis virus (EAV), the arterivirus prototype, are autocatalytically cleaved at 11 sites by three internal proteinases (57, 71). The resulting cleavage products, nsp1 to nsp12, associate into a membrane-bound replication/transcription complex (42, 60). The catalytic core of this complex is thought to be formed by the subunits containing (predicted) RdRp and helicase activities (11), nsp9 and nsp10, respectively. The helicase function of EAV nsp10 was previously verified using an in vitro assay (51, 52). However, the identification of nsp9 (residues 1678 to 2370 of pp1ab) as the arterivirus RdRp has been based solely on the presence of characteristic sequence motifs in its C-terminal part (63, 64). Only the SDD signature in RdRp motif C (see below), which discriminates all nidovirus RdRps from those of other virus groups containing a GDD motif in this position (11), has been targeted using an EAV reverse genetics system, and it was found to be critical for virus replication (63). The exact borders of the RdRp domain and the function of the large upstream domain that forms the N-terminal part of the RdRp-containing nsp9 cleavage product have not been defined for any nidovirus. It was previously reported that the nidovirus RdRp domain is evolutionarily clustered with the RdRps of plus-strand RNA viruses that use a genome-linked viral protein (VPg) to prime RNA synthesis (30).
The structures of the RdRps solved to date resemble a cupped right hand, with finger, palm, and thumb domains, providing the correct geometrical arrangement of substrate molecules and metal ions at the active site for catalysis (14). In contrast to other polymerases adopting this fold, RdRps are rather compact molecules, with finger and thumb domains making contact and enclosing the active site. High-resolution structures of RdRps of positive- and double-strand RNA viruses (14, 16, 35, 68) revealed clear similarities despite sharing little sequence identity, suggesting an evolutionary link between the RdRps of these RNA viruses (6, 30). A comparison of RdRp structures shows that the enzymes of viruses using a VPg-primer-dependent mechanism of initiation, such as picornaviruses, have a more accessible active site than RdRps of viruses with a de novo initiation mechanism, such as flaviviruses, reoviruses, and bacteriophage Φ6 (14, 61). De novo-initiating RdRps use additional structural elements (priming loops) in the form of extensions protruding from the thumb domain (flavivirus RdRps and the RdRp of Φ6) or a loop within the palm domain (reovirus RdRp) to partially occlude the active site in order to create an initiation platform (14). The structure of the de novo initiation complex of the bacteriophage Φ6 RdRp demonstrated that the priming nucleotide is positioned by stacking to a tyrosine residue in the priming loop (6). It has been shown that alteration of this tyrosine decreases the de novo initiation ability of the RdRp (34). Interestingly, the paradigm that RdRps are either primer dependent or de novo initiating seems to be changing. It was recently demonstrated that the norovirus RdRp (of the family Caliciviridae) uses both initiation mechanisms on specific RNA templates in vitro and also may do so in vivo (45). Accordingly, the structures of two calicivirus RdRps revealed that the C terminus folds back on the active center, thus providing a putative temporary initiation platform (16, 37).
Like all other RdRps, nidovirus RdRps contain the conserved catalytic motifs A, B, and C in the palm subdomain, as well as motif F in the finger subdomain, which contacts the incoming nucleotides. They also contain motif E, a structural (not sequence) motif found in both primer-dependent and de novo-initiating RdRps, forming the connection between the palm and thumb domains. Motif G, also present in nidovirus RdRps, is a sequence signature characteristic of primer-dependent RdRps (19). Evidence for a primer-dependent in vitro activity of the coronavirus (CoV) RdRp (nsp12) came from a study of the severe acute respiratory syndrome CoV (SARS-CoV) enzyme, which was found to be active on poly(A) templates by using oligo(U) to prime initiation (8). In a recent report, a second SARS-CoV RdRp (nsp8) was identified that was able to initiate RNA synthesis de novo with low processivity and low fidelity (25). This second RdRp was proposed to act as a primase, possibly generating the primers for the canonical primer-dependent nsp12/RdRp.
There are no data on the structure, function, or in vitro activity of the RdRps of arteriviruses, which are distantly related to those of CoVs (for recent reviews, see references 18 and 54). In this study, we report the expression in Escherichia coli and purification of the active EAV nsp9/RdRp and address the mechanism of initiation of arterivirus RNA synthesis. The arterivirus RdRp sequences were compared to those of their CoV homologues and to other RdRps with known initiation mechanisms and three-dimensional structures. In an in vitro assay, the EAV RdRp was able to initiate RNA synthesis on homopolymeric templates using a de novo mechanism. Initiation of RNA synthesis was also studied in vivo by using a reverse genetics approach, which demonstrated the importance of the 3′-terminal nucleotides of the EAV genome. This work provides a basis for further characterization of the arterivirus RdRp and dissection of the molecular basis of RNA polymerization. Such detailed molecular knowledge will be essential for developing therapeutics that selectively inhibit viral RdRps in vivo.
MATERIALS AND METHODS
Amino acid sequence alignments.
The RdRps of Arteriviridae and Coronaviridae were first aligned using Multialin (10). The alignment was then adjusted manually using Seaview (17). A pairwise structural alignment of all RdRps with known structures was generated with the Swiss-Prot Protein Data Bank (PDB) viewer (http://www.expasy.org/spdbv/), and conserved residues in motifs A, B, and C were aligned using the hepatitis C virus RdRp domain (PDB code 1C2P) as a reference. A secondary-structure prediction for the EAV RdRp was generated by PredictProtein. The final complete alignment was adjusted manually using Seaview (17), and a graphic visualization was generated by Espript (20). The complete alignment that was the basis for Fig. 1 is available from the authors upon request.
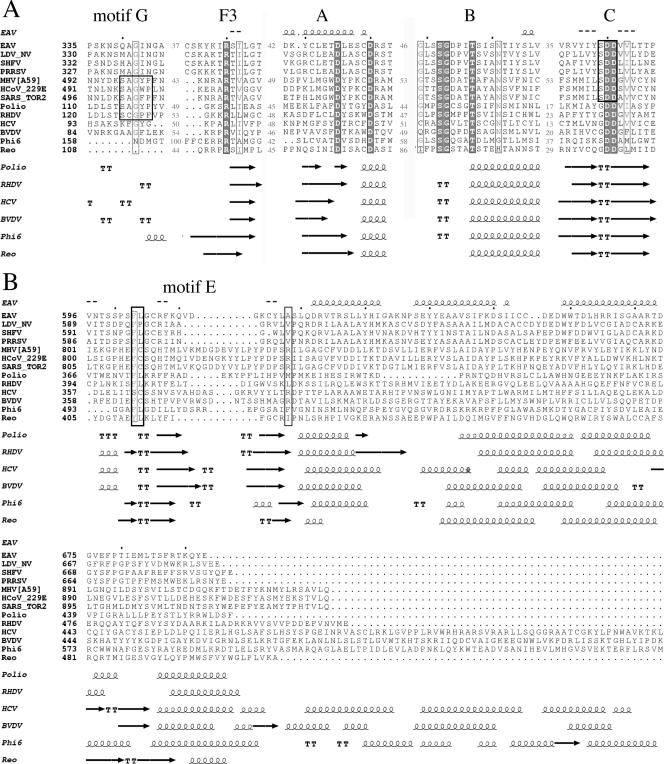
FIG. 1.
Structure-based sequence alignment of RdRp domains of four arteriviruses, three CoVs, poliovirus (Picornaviridae), rabbit hemorrhagic disease virus (RHDV; Caliciviridae), hepatitis C virus (HCV; Flaviviridae), bovine viral diarrhea virus (BVDV; Flaviviridae), bacteriophage Φ6 (Cystoviridae), and mammalian orthoreovirus (Reoviridae). The picornavirus and calicivirus enzymes are VPg-primer-dependent RdRps, whereas the flavivirus, cystovirus, and reovirus RdRps initiate RNA synthesis de novo. Details on the alignment are given in Material and Methods. The complete alignment is available from the authors upon request. (A) Alignment of motifs G and F3 (finger domain) as well as A, B, and C (palm domain). The predicted secondary structure for the EAV RdRp is depicted above the sequence. Secondary-structure elements of proteins with known structures are given below the alignment. Residues conserved in all RdRps are in white on a dark gray background. Residues conserved in more than 75% of the sequences are boxed and have a light gray background. Black boxes highlight motif G (T/SX1-2GX0-1P), which is conserved in Coronaviridae, Picornaviridae, and Caliciviridae, and the SDD signature sequence of motif C that is typical of nidovirus RdRps. (B) Alignment of RdRp thumb domains, starting with motif E. Residues near the N-terminal and C-terminal ends of the conserved β-hairpin are boxed in black. The numbering of BVDV and reovirus RdRp sequences starts with the first residue considered to be part of the RdRp core domain; the sequence of the reovirus RdRp ends with the last residue of the RdRp domain. The RdRp domains were aligned, and the alignments were adjusted according to the predicted secondary structure, as depicted. The aligned sequences and NCBI accession numbers are the following: EAV (NP_127506), lactate dehydrogenase-elevating virus neurovirulent type C (LDV_NV; NP_065670), simian hemorrhagic fever virus (SHFV; NP_742092), porcine reproductive and respiratory syndrome virus (PRRSV; NP_066135), mouse hepatitis virus, strain A59 (MHV-A59; NP_068668), human CoV 229E (HCoV-229E; NP_068668), and SARS-CoV, strain Toronto 2 (SARS_Tor2; NP_828849). The RdRps with known three-dimensional structures are poliovirus (Polio; PDB code 1RA6), RHDV (PDB code 1KHW), HCV (PDB code 1C2P), BVDV (PDB code 1S48), Φ6 (PDB code 1HHS), and mammalian orthoreovirus (PDB code 1MUK).
Expression and purification of the EAV nsp9/RdRp.
The central part of the cDNA sequence encoding the EAV nsp9/RdRp was taken from expression vector pL1ab100 (63), in which the ORF1a/ORF1b ribosomal frameshift site had been removed by mutating nucleotide A-5404 to C and inserting a C between G-5399 and T-5400 (numbers refer to the EAV-Bucyrus reference sequence; NCBI Entrez Genome Refseq no. NC_002532). Using small PCR products, the termini of the nsp9-coding sequence were tailored to supply a starting methionine codon and a C-terminal hexahistidine tag. The gene was first engineered between the NcoI and AvaI restriction sites of Gateway entry vector pENTR11 (Invitrogen), sequenced, and subsequently placed downstream of the T7 promoter in Gateway expression vector pDEST14 (Invitrogen), giving rise to construct pDEST14-nsp9CHis. The nsp9 protein expressed from this vector consisted of an N-terminal methionine, amino acids Gly-1678 to Glu-2370 of EAV replicase pp1ab, and six C-terminal histidines.
The protein was expressed in E. coli C41pROS cells after overnight induction with 1 mM isopropyl-β-d-thiogalactopyranoside at 25°C. This optimized expression condition was obtained after screening different bacterial strains, culture media, and induction temperatures according to the procedure described by Abergel et al. (1). The bacterial pellet was resuspended in lysis buffer (50 mM Tris [pH 8.0], 10 mM imidazole [pH 8.0], 150 mM NaCl, 20 mM MgSO4, 0.25 mg/ml lysozyme, and 0.1 mg/ml DNase I), incubated for 30 min at 4°C, and finally sonicated. The protein was purified from the soluble fraction of the bacterial lysate by two sequential chromatography steps. The lysate was first loaded onto a nickel-nitrilotriacetic acid (Ni-NTA) column (Amersham-Biosciences). The protein was eluted with 250 mM imidazole and then loaded onto a Superdex 200 gel filtration column (Amersham Biosciences). The protein was eluted from the second column in buffer GF1 (20 mM HEPES [pH 8.0], 100 mM NaCl, 1 mM dithiothreitol [DTT], 5% glycerol), and peak fractions containing EAV nsp9/RdRp-His were pooled. The yield was approximately 10 mg/liter of E. coli culture.
Polymerase activity assays.
Homopolymeric templates were obtained from Amersham Biosciences. For the generation of viral templates, the 3′-proximal region of the EAV genome was PCR amplified with the upstream primer containing a 5′-flanking T7 promoter sequence. The PCR products were used directly as the templates for in vitro transcription using T7 RNA polymerase. For generation of the templates encompassing the 3′-terminal 303 nucleotides of the EAV genome, sense primer E801 (genome positions 12401 to 12420) was used together with either antisense primer E804 [representing the complement of 3′-terminal nucleotides 12703 to 12704 and the first 20 residues of the poly(A) tail] or E154 (complementary to positions 12680 to 12704). For the template encompassing the 3′-terminal 58 nucleotides, sense primer E803 (positions 12646 to 12665) was used. All DNA and RNA oligonucleotides were obtained from Eurogentec.
To analyze RdRp activity by filter-binding assays, reactions were performed in 50 μl of RdRp buffer (50 mM HEPES [pH 8.0], 10 mM KCl, 5 mM DTT, 100 μM nucleoside triphosphate [NTP], 2 mM MnCl2, and 4 mM MgCl2) containing 400 nM template, 2 μM EAV nsp9/RdRp, and 0.5 μCi [3H]NTP (either ATP, UTP, or GTP, depending on the template used). To test primer-dependent initiation of RNA synthesis, DNA or RNA oligonucleotides (4 μM) were annealed to the template by heating to 65°C for 10 min, followed by slow cooling to room temperature. Samples were taken from the assays at different time points, the reaction was stopped by addition of an equal volume of 50 mM EDTA, and the mixture was spotted onto DE-81 filter paper (Whatman). The filters were dried and washed three times with 0.3 M ammonium formate (pH 8.0) and once with ethanol, and then they were dried. Liquid scintillation fluid was added, and incorporation was measured in counts per minute (cpm) using a Wallac MicroBeta liquid scintillation counter.
To analyze the RNA products generated in these assays by polyacrylamide gel electrophoresis, reactions were performed in the presence of 100 μM NTP and 1 μCi [α-32P]NTP. The reactions were stopped by addition of an equal volume of formamide-EDTA-containing electrophoresis loading buffer, and products were separated in 6% or 14% acrylamide-7 M urea sequencing gels (see the legend to Fig. 5). Radiolabeled products were visualized using a Bio-Rad Molecular Imager FX.
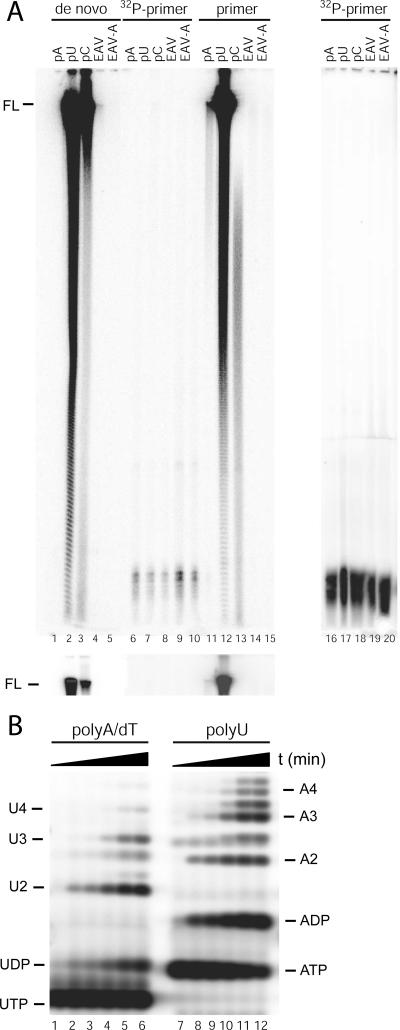
FIG. 5.
Visualization of RNA products derived from EAV nsp9/RdRp activity. (A) De novo-initiated synthesis and primer-dependent RNA synthesis were tested on the homopolymeric templates poly(A) (pA), poly(C) (pC), and poly(U) (pU) and on viral RNA templates representing the 3′-proximal domain of the EAV genome either with (EAV-A) or without (EAV) a poly(A)20 tail. Reaction products were analyzed on a 6% polyacrylamide-7 M urea gel. For the reactions analyzed in lanes 1 to 5 and 11 to 15, α-32P-labeled nucleotides were used. For lanes 6 to 10 and 16 to 20, complementary γ-32P-labeled 18-nucleotide DNA oligonucleotides were annealed onto the templates poly(A)/oligo(dT), poly(U)/oligo(dA), poly(C)/oligo(dG), EAV-A/oligo(dT), and EAV/oligo(dT) [although oligo(dT) is not complementary to the EAV template]. Subsequently, these RdRp assays were performed in the presence of unlabeled nucleotides only. The full-length product is marked FL, and for better visualization of this product a shorter exposure of the top part of the gel is shown below. (B) Demonstration of de novo initiation using a poly(U) template (lanes 7 to 12) and a poly(A)/oligo(dT) primer-template combination (lanes 1 to 6). The reactions were stopped at different time points (0, 5, 10, 30, 60, and 120 min) and analyzed on a 14% polyacrylamide-7 M urea gel. Several short reaction products were detected, corresponding to dinucleotides (U2 and A2), trinucleotides (U3 and A3), and tetranucleotides (U4 and A4). In addition, some diphosphate products were detected, migrating slower than their corresponding triphosphate products (compare ATP and UTP to ADP and UDP).
Site-directed mutagenesis, RNA transfections, and analysis of virus replication.
Mutations in the 3′ terminus of the EAV genome were engineered in a small shuttle plasmid by standard site-directed PCR mutagenesis, verified by sequence analysis, and transferred to the EAV full-length cDNA clone pEAV211 (59, 62). Following in vitro transcription by T7 RNA polymerase, the concentration of full-length RNA was measured by UV spectroscopy, and the integrity of the RNA was verified by agarose gel electrophoresis. BHK-21 cells were transfected with equal amounts (6 μg) of full-length EAV RNA using a Nucleofector electroporation device (Amaxa Biosystems) according to the supplier's protocol (kit T, program T-20). Immunofluorescence dual-labeling assays with EAV-specific antisera for nsp3 (rabbit) and nucleocapsid protein (mouse monoclonal antibody) were performed as described previously (60) at different time points after transfection.
Virus titration using plaque assays was done as described previously (36). BHK-21 cells were grown to subconfluence and infected with a virus dilution series. Infection was performed for 1 h at 39.5°C, after which an agarose overlay was applied. Plaques were detected after incubation at 39.5°C for 72 h. For RNA isolation, cells were lysed at 14 h posttransfection, and intracellular RNA was isolated using the acidic phenol method, as described previously (39). Viral RNA was analyzed using denaturing formaldehyde-agarose gels and hybridization with radioactively labeled oligonucleotide probe E868 (antisense, positions 12270 to 12289), which recognizes both the genome and all sg mRNAs. For the analysis of the revertant of mutant cg, the 3′-terminal region of the EAV genome was amplified by reverse transcription-PCR (RT-PCR) from the total intracellular RNA from infected cells. For the RT reaction, an oligo(dT) primer complementary to the poly(A) tail was used. In the subsequent PCR, the reverse primer oligo(dT) and the forward primer E817 (positions 12288 to 12308) were used. The RT-PCR product was used for direct population sequencing.
RESULTS
Sequence analysis of the EAV nsp9/RdRp.
To obtain an initial theoretical assessment of the structural and functional features of the EAV nsp9/RdRp, a structure-based sequence alignment was generated that included the four known arteriviruses, three CoVs, two VPg-primer-dependent RdRps (from Picornaviridae and Caliciviridae) with known three-dimensional structures, and four structurally characterized de novo-initiating RdRps of Flaviviridae, Cystoviridae, and Reoviridae origin (see the Fig. 1 legend for details). A secondary-structure prediction of the 693-amino-acid-long EAV nsp9/RdRp was generated and used to align regions where no sequence conservation was observed with RdRps of other virus families (the complete alignment is available from the authors upon request). The alignment suggested that the RdRp domain in EAV nsp9 starts around residue 250 and consists of approximately 440 residues, a size corresponding to that of small VPg-primer-dependent RdRps.
The alignment for catalytic motifs A, B, and C and for motif F3 (5) is shown in Fig. 1A. Note that the arterivirus RdRp contains an SDD signature in motif C, a property shared with RdRps of CoVs and other nidoviruses, in contrast to all other groups of plus-strand RNA viruses. The role of this serine residue in nidovirus RdRps has not been studied, but it was shown that the corresponding glycine residue is essential for poliovirus RdRp and other RdRps carrying a GDD signature (38). Residues in motif G of the finger domain (Fig. 1A), which previously was proposed to be present in nidovirus RdRps (19), contact the template in an RdRp-RNA primer-template complex (13). A conserved proline in motif G of the poliovirus RdRp was found to be essential for activity on poly(A)/ oligo(dT) and was proposed to be involved in the correct positioning of the primer-template complex to allow formation of a stable elongation complex (55). Motif G can be defined as (T/S)-X1-2-G-X0-1-P and is present in CoV RdRps, but it is incomplete in the arterivirus RdRp.
The alignment of motif E and the entire thumb domain is shown in Fig. 1B. In a picornavirus, residues in motif E were found to contact the primer in an RNA primer-template complex (13) as well as in a complex with the VPg primer (12). Motif E residues have been proposed to be part of the initiation platform in de novo-initiating RdRps (4, 15), and therefore motif E seems to be involved in initiation in general. Nevertheless, there is no recognizable sequence conservation in motif E that could correspond to the initiation mechanism used by an RdRp. Structural conservation of the β-hairpin can be observed for both strictly VPg-primer-dependent and de novo-initiating RdRps.
Finally, Fig. 1B clearly illustrates that arterivirus RdRps contain a rather short thumb region, as also observed for the VPg-primer-dependent RdRps. This region does not contain extra elements that could function as an initiation platform, a feature described for de novo-initiating RdRps. Furthermore, no differences in spacing between the RdRp motifs in the finger or palm domain were observed (Fig. 1), suggesting that no additional elements are present in this region, as were reported for the de novo-initiating reovirus RdRp. In conclusion, our examination of the limited number of functionally and structurally characterized viral RdRps that are available indicated that EAV nsp9 contains a relatively small RdRp domain resembling that of primer-dependent RdRps.
Expression and purification of EAV nsp9/RdRp-His.
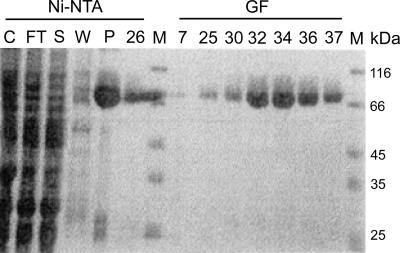
A gene encoding the full-length EAV nsp9/RdRp, tagged with six histidines at its C terminus, was cloned in the Gateway pDEST14 expression vector for expression in E. coli. Optimal conditions for soluble protein expression were determined according to the procedure described by Abergel et al. (1), which resulted in production of a predominant protein of the expected size (76 kDa) (Fig. 2). The nsp9/RdRp-His protein was purified to near homogeneity using a Ni-NTA chromatography column and subsequently an S200 gel filtration column. EAV nsp9/RdRp-His eluted at its expected molecular size, demonstrating its monomeric state. The five peak fractions (Fig. 2, GF fractions 32 to 36) were pooled, yielding approximately 10 mg of protein per liter of E. coli culture. During the later stages of the project, several N-terminally truncated mutants of nsp9 were designed in an attempt to improve its in vitro activity. Although these deletions were based on alignments and structures of various other RdRps and also took into account data on the activity of truncated forms of some of these enzymes, none of these variants could be stably expressed.
FIG. 2.
Expression and purification of EAV nsp9/RdRp-His. Coomassie brilliant blue staining of a sodium dodecyl sulfate-polyacrylamide gel showing the two-step purification of the enzyme by Ni-NTA affinity chromatography (Ni-NTA) and S200 gel filtation (GF). C, total protein in the cell lysate; FT, flowthrough; S, soluble fraction; W, waste fraction; P, peak fraction; 26, fraction 26 of the Ni-NTA eluate; M, molecular size marker. On the right side of the gel, different fractions of the run on the S200 gel filtration column are shown. Fractions 32 to 36 were pooled to obtain the nsp9/RdRp-His preparation used in this study.
Initial characterization of EAV nsp9/RdRp activity.
The activity of the purified EAV nsp9/RdRp was first examined using filter-binding assays to monitor incorporation of [3H]UTP by using a poly(A) RNA template and an oligo(dT) DNA primer. At first, a relatively low level of polymerase activity was observed after incubation at 30°C, and incorporation of the substrate increased over time (Fig. 3A). The RdRp activity was not inhibited by either rifampin (20 μg/ml) (Fig. 3A) or actinomycin D (50 μg/ml) (data not shown), indicating that it was not based on contaminating bacterial RNA polymerase or DNA polymerase.
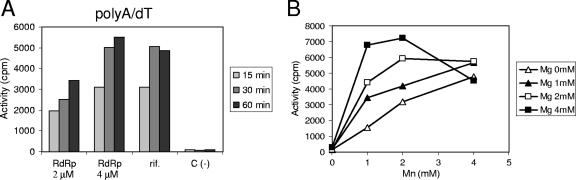
FIG. 3.
Activity of the purified EAV nsp9/RdRp on a poly(A)/oligo(dT) primer-template complex. (A) Time course of [3H]UTP incorporation (in cpm) on poly(A)/oligo(dT) using two concentrations of EAV RdRp (2 and 4 μM). As a specificity control, the reaction with 4 μM of enzyme was also carried out in the presence of rifampin (rif.), an inhibitor of E. coli DNA-dependent RNA polymerase. RNA synthesis was monitored in a filter-binding assay, followed by liquid scintillation counting. The data set on the right (C-) shows the results of a control reaction without the enzyme. (B) Optimization of the concentration of the catalytic ions Mg2+ and Mn2+ for RNA synthesis on poly(A)/oligo(dT). [3H]UTP incorporation (in cpm) after 30 min in the presence of increasing concentrations of Mg2+ and Mn2+ was measured by a filter-binding assay and liquid scintillation counting.
The initial RdRp reaction buffer contained 50 mM HEPES (pH 8.0), 10 mM KCl, 10 mM DTT, 10 μM UTP, 2 mM MnCl2, and 5 mM MgCl2. Various other conditions were tested to optimize RdRp activity (data not shown). The influence of temperature on polymerase activity was tested by comparing reactions performed at 23, 30, and 37°C. Activity at 30°C was twofold higher than that at both 23°C and 37°C, with the latter probably being due to nsp9/RdRp instability at this elevated temperature. Different KCl and NaCl concentrations in the reaction buffer (0, 10, and 50 mM) were compared, but differences between the effects of KCl and NaCl were not detected. Both salts were found to inhibit RdRp activity at a 50 mM concentration, and similar activities were observed in the absence of KCl or NaCl or in the presence of 10 mM of either salt.
The effects of bovine serum albumin (20 μg/ml) and 1% or 5% glycerol as stabilizing agents during the reaction were tested, as were the effects of 0.1% Triton X-100 and a lower DTT concentration (5 mM), but none of these reagents affected polymerase activity. Subsequently, the optimal UTP concentration was determined (1, 10, 100, and 200 μM of UTP), and 100 μM appeared optimal, increasing polymerase activity by approximately fivefold compared to that found in the initial assays, in which 10 μM UTP was used. Finally, the optimal concentrations of the catalytic Mg2+ and Mn2+ ions were determined (Fig. 3B). Polymerase activity was detected only when Mn2+ ions were present in the reaction mixture. In the presence of Mn2+, increasing Mg2+ concentrations were found to stimulate polymerase activity, but above 4 mM Mg2+, inhibition was observed (data not shown). The optimal combination of both ions was determined to be 2 mM of Mn2+ and 4 mM of Mg2+. Therefore, the subsequent experiments on homopolymeric templates were performed at 30°C in an RdRp buffer containing 50 mM HEPES (pH 8.0), 10 mM KCl, 5 mM DTT, 100 μM NTP, 2 mM MnCl2, and 4 mM MgCl2.
Activity of the EAV nsp9/RdRp on homopolymeric templates.
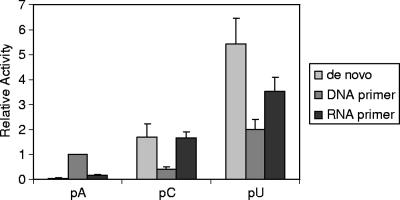
The purified EAV nsp9/RdRp was analyzed for its ability to initiate RNA synthesis on different homopolymeric RNA templates, either by de novo initiation or by a primer-dependent mechanism using 18-nucleotide DNA and RNA primers (Fig. 4). Poly(G) was not tested as a template, since these RNAs are known to contain stable higher order structures. In the absence of a primer, a high level of polymerase activity was found using poly(U) and poly(C) templates, whereas no significant activity was detected on the poly(A) template. This indicated that the EAV RdRp can initiate RNA synthesis by using a de novo mechanism in a template-specific manner, with poly(U) being the most efficient template. As observed above (Fig. 3), a low level of polymerase activity was obtained on a poly(A) template using an oligo(dT) DNA primer, and a very low level of activity was observed using an oligo(U) RNA primer. Annealing of cDNA or RNA primers reduced the level of polymerase activity on both poly(U) and poly(C) templates compared to that following de novo initiation in the reaction without primer. Possibly, the annealed primers interfere with ongoing RNA synthesis resulting from de novo initiation, as they need to be displaced from the template.
FIG. 4.
De novo and primer-dependent activity of the purified EAV nsp9/RdRp on homopolymeric RNA templates. [3H]UTP incorporation was measured by a filter-binding assay and liquid scintillation counting after a 30-min reaction on the homopolymeric templates poly(A) (pA), poly(C) (pC), and poly(U) (pU). Besides de novo-initiated RNA synthesis (the left bar of each set), primer-dependent activity was tested using 18-nucleotide DNA or RNA oligonucleotides complementary to the template. Activity was measured in three independent experiments, and incorporation on poly(A)/oligo(dT) was arbitrarily set at 1.
To characterize the reaction products in further detail, RdRp assays were performed using 32P-labeled NTPs and were analyzed on a denaturing acrylamide gel (Fig. 5A). Consistent with the results from the filter-binding assays, no products were generated on the poly(A) template in the absence of a primer (Fig. 5A, lane 1). Using the poly(C) and poly(U) templates, full-length products of approximately 300 nucleotides were generated (Fig. 5A, lanes 2 and 3). In addition, a ladder of shorter products was observed that likely resulted from either premature termination of RNA synthesis or runoff polymerization from the heterogeneously sized templates that were used. This ladder was most obvious using the poly(U) template. The poly(G) products synthesized from the poly(C) template may form higher order RNA structures that interfered with separation of the bands. Upon annealing of oligo(dT) to poly(A), a small amount of full-length product was generated (Fig. 5A, lane 11), whereas annealing of a cDNA primer reduced the amount RNA synthesis on poly(C) and poly(U) templates compared to the amount of RNA synthesis of the de novo-initiated reaction (Fig. 5A, lanes 12 and 13). In a control experiment, the poly(A) template was incubated with [32P]GTP and [32P]ATP, and the poly(U) template was incubated with [32P]GTP and [32P]UTP (data not shown). None of these reactions resulted in labeled products, suggesting that the observed incorporation was not caused by terminal transferase activity associated with the EAV nsp9/RdRp.
Primer-dependent versus de novo RNA synthesis by the EAV nsp9/RdRp.
To test if primers, and especially oligo(dT), were incorporated into the generated RNA products, we performed reactivity assays similar to the experiments described above using unlabeled nucleotides and 32P-end-labeled primers that were annealed onto homopolymeric templates. As shown in Fig. 5A (lanes 6 to 8), no labeled extension products were generated on these templates. In a second experiment, even prolonged exposure failed to reveal significant amounts of RNA products (Fig. 5A, lanes 16 to 18). Together, these results suggested that the EAV RdRp is unable to use DNA primers to initiate RNA synthesis. Although oligo(dT) was found to stimulate polymerase activity on a poly(A) template, the primer is apparently not incorporated into the generated RNA product. Also, the DNA primers oligo(dA) and oligo(dG) and the RNA primers oligo(A), oligo(G), and oligo(U) were tested for their ability to activate RNA synthesis on a poly(A) template (data not shown). Of these, only oligo(dA) was found to stimulate RNA synthesis, although it did so at less than 50% of the level of activation induced by oligo(dT).
To visualize the early products of de novo initiation on the poly(U) template, the reaction products were analyzed on a denaturing high-percentage polyacrylamide gel. Besides the full-length RNA product running in the top of the gel (data not shown), the lower part of this gel (Fig. 5B, lanes 7 to 12) clearly revealed generation of di-, tri-, and tetranucleotide products, which are the first products synthesized during de novo initiation. The initiation products increased with time, concomitant with the appearance of longer products in the top of the gel (data not shown). Increasing with longer reaction times, we detected some diphosphate products migrating slower than their corresponding triphosphate products (compare ATP to ADP in Fig. 5B, lanes 7 to 12). Interestingly, di-, tri-, and tetranucleotide products were also observed with the poly(A)/oligo(dT) primer-template combination and must, thus, result from de novo initiation on the poly(A) template. These results further confirmed that, under the conditions of this assay, the EAV nsp9/RdRp is able to initiate RNA synthesis de novo.
Activity of the EAV nsp9/RdRp on viral templates.
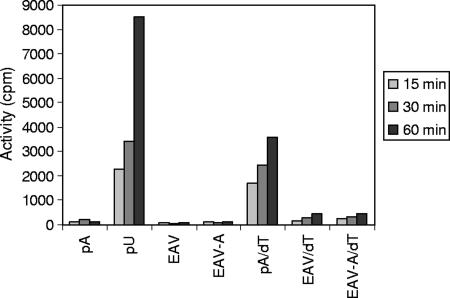
Subsequently, we analyzed RNA synthesis by the purified EAV nsp9/RdRp on viral templates, in particular using the 3′ end of the viral genome, which is the natural initiation site for minus-strand RNA synthesis. De novo initiation and primer-dependent initiation were studied using in vitro-transcribed RNA templates encompassing the 3′-terminal 303 nucleotides of the genome, either with (EAV-A) or without (EAV) a poly(A) tail of 20 nucleotides (Fig. 6). No de novo initiation on viral templates was observed, although (as described above) a high level of RdRp activity was found using a poly(U) control template. Upon the annealing of an oligo(dT) DNA primer, a low level of incorporation was observed using the viral template both with and without a poly(A) tail. However, the level of incorporation measured was lower than that obtained with the poly(A)/oligo(dT) template-primer combination. Again, the reaction products were analyzed on a denaturing acrylamide gel (Fig. 5A). No labeled reaction products could be detected upon de novo initiation (lanes 4 and 5) or in the presence of oligo(dT) (lanes 14 and 15), possibly because the level of polymerase activity was very low. The same was also true when 32P-end-labeled primers were added (Fig. 5A, lanes 9 and 10 as well as 19 and 20).
FIG. 6.
Time course analysis of de novo and primer-dependent initiation of RNA synthesis by the purified EAV nsp9/RdRp. [3H]UTP incorporation was measured using the homopolymeric templates poly(A) (pA) and poly(U) (pU) and viral RNA templates encompassing the EAV 3′ genome terminus with (EAV-A) or without (EAV) a poly(A)20 tail. De novo-initiated activity and activity stimulated by the presence of an oligo(dT) DNA primer were monitored using filter-binding assays and liquid scintillation counting.
During our attempts to achieve de novo initiation by the EAV RdRp on viral templates, multiple conditions and templates were tested (data not shown). Since de novo initiation is known to depend on a high concentration of the initiating nucleotide, we tested increased concentrations of all four nucleotides (0.2, 0.5, 1, and 5 mM NTP) or a higher concentration of the 32P-labeled nucleotide only. We also reevaluated the effect of different concentrations of the catalytic ions Mn2+ and Mg2+ (between 1 and 5 mM). However, none of the conditions tested resulted in de novo initiation on viral templates. A shorter viral template, representing the 3′-terminal 58 nucleotides of the EAV genome, was also tested. Finally, since for several plus-strand RNA viruses a long-distance interaction between the genomic 5′ and 3′ termini has been found to be essential for RNA synthesis, we generated a viral template RNA in which the 5′-terminal 306 nucleotides were fused to the 3′-terminal 303 nucleotides of the EAV genome. However, also using these templates, de novo initiation of RNA synthesis was not detected. Thus, although template-specific de novo initiation of RNA synthesis was observed on homopolymeric templates, the requirements for initiation from templates containing viral sequences remain to be elucidated.
Initiation of EAV RNA synthesis in vivo.
We used a reverse genetics approach to study the initiation of RNA synthesis in the context of the viral life cycle. The plus-strand RNA genome of EAV is polyadenylated at its 3′ end, but in vitro the purified EAV RdRp was not capable of de novo initiation on homopolymeric poly(A) templates. Therefore, in vivo RNA synthesis may initiate immediately upstream of the poly(A) tail, in particular, on the two C residues (nucleotides 12703 to 12704) that form the 3′ terminus of the genome. To test this hypothesis, mutations were introduced in an EAV full-length cDNA clone (62). Both C residues were replaced with either G or U (Fig. 7A). In addition, a mutant lacking the poly(A) tail was generated.
FIG. 7.
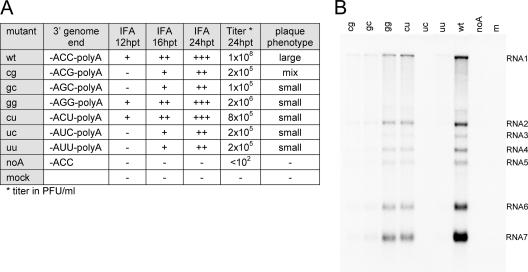
Analysis of EAV mutants carrying mutations in the 3′ terminus of the genome. (A) BHK-21 cells were transfected with a series of mutants in which the two terminal C residues were replaced with either G (cg, gc, and gg) or U (cu, uc, uu). Also, a mutant lacking the poly(A) tail (noA) was included. The production of viral proteins was monitored by immunofluorescence microscopy using antisera recognizing replicase subunit nsp3 and nucleocapsid protein (N). In addition, supernatants harvested from the transfected cell cultures at 24 h posttransfection were tested for the presence of progeny virus using plaque assays. (B) Viral RNA synthesis of EAV mutants in the 3′ genome terminus. Infectious RNA was transfected into BHK-21 cells, and intracellular RNA was isolated at 14 h posttransfection. The RNA was separated in a denaturing agarose gel and analyzed by hybridization to an oligonucleotide detecting all plus-strand viral RNAs. The positions of the genome (RNA1) and sg mRNAs (RNA2 to RNA7) are indicated. wt, wild type; hpt, hours posttransfection.
These mutations were tested in vivo for their effects on viral RNA synthesis by transfection of BHK-21 cells with full-length RNA transcribed in vitro from wild-type and mutant EAV cDNA clones. After approximately one cycle of virus replication (at 14 h posttransfection), viral protein synthesis was monitored by immunofluorescence assays using antisera recognizing nsp3 and the nucleocapsid (N) protein, which served as indicators for genome replication and sg mRNA synthesis, respectively (Fig. 7A). For all mutants, immunofluorescence assays revealed clearly reduced levels of both viral proteins, suggesting impaired virus viability, whereas no signal was observed for the poly(A) deletion mutant. In addition, supernatants harvested from the transfected cell cultures at 24 h posttransfection were tested for the presence of progeny virus using plaque assays. No plaques were detected for the poly(A) deletion mutant. All other 3′-end mutants showed reduced virus replication, with a small-plaque phenotype and a 100- to 1,000-fold reduction in virus titer (Fig. 7A). For mutant cg, a mixture of small and large plaques was observed, suggesting that a faster replicating revertant was emerging. To analyze the genomic 3′ end of this revertant, total cellular RNA was isolated from infected cells, and this region was amplified by RT-PCR. Subsequently, population-based sequencing of the PCR product was performed that revealed reversion to the 3′-terminal CC doublet present in the wild-type EAV genome.
To analyze the synthesis of viral RNA (genomic and subgenomic) directly, intracellular RNA was isolated from transfected cells at 14 h posttransfection. The isolated RNA was separated in denaturing formaldehyde-agarose gels and hybridized to an oligonucleotide probe detecting all virus-specific plus-strand RNAs. As shown in Fig. 7B, for the control transfection with the wild-type viral RNA, abundant synthesis of both genome RNA (RNA1) and sg mRNAs 2 to 7 was detected. For all mutants, the production of both genomic and sg RNA was severely reduced. No signal was detected for the poly(A) deletion mutant, although we cannot exclude the possibility that this transcript suffered from reduced stability due to the lack of a 3′ poly(A) tail. These results suggest that, in vivo, the EAV 3′-terminal sequence plays a critical role in viral RNA synthesis, most likely during initiation of minus-strand RNA synthesis.
DISCUSSION
We report here the first successful expression and purification of an arterivirus RdRp. Like all plus-strand RNA viruses, arteriviruses carry an RdRp enzyme that functions as the catalytic subunit of the viral replication/transcription complex, directing viral RNA synthesis in concert with other viral/cellular proteins. Although common viral RdRp motifs were identified long ago (30), the mechanisms governing initiation of RNA synthesis have remained ill defined. RNA synthesis can be initiated by two different mechanisms, either de novo or depending on a primer (27, 61). The bacteriophage Φ6 RdRp represents one of the best-studied model systems for de novo initiation (6, 47, 61). Among animal plus-strand RNA viruses, catalytic RdRp activity has been demonstrated and characterized for members of the Picornaviridae, Flaviviridae, and, more recently, Caliciviridae families. For the RdRps of many other families, though, experimental data are lacking or characterization of the enzyme has only just started, as in the case of SARS-CoV (8).
The EAV nsp9/RdRp was expressed in E. coli and purified to near homogeneity using a two-step purification procedure. Initial activity was demonstrated using a poly(A)/oligo(dT) primer-template combination (Fig. 3A), after which the assay conditions were optimized for temperature (30°C optimum, as found for many other RdRps [50, 66, 70]) and salt concentration. Different KCl and NaCl concentrations were compared, and the effect of several potentially stabilizing agents was tested, but none of them significantly improved RdRp activity. A nucleotide concentration of 100 μM appeared optimal for activity. Finally, the importance of the catalytic Mg2+ and Mn2+ ions was tested (Fig. 3B), with activity being detected only in the presence of Mn2+ ions but also being dependent on the concentration of Mg2+. The optimal combination of both ions was determined to be 2 mM of Mn2+ and 4 mM of Mg2+.
The EAV nsp9/RdRp was found to be catalytically active in the absence of other viral or cellular proteins and initiated RNA synthesis de novo on homopolymeric templates in a template-specific manner. This de novo activity was found on both poly(U) and poly(C) templates but not on a poly(A) template. This is consistent with the observation that de novo RNA synthesis generally initiates with a purine as the starting nucleotide. Analysis of the reaction products (Fig. 5A) demonstrated that a full-length product was generated, in addition to a ladder of bands that likely results from either premature termination of RNA synthesis or runoff polymerization from the heterogeneously sized templates. This observation indicates that in this assay the EAV RdRp is a rather distributive RNA polymerase, although obviously in the context of the viral life cycle its processivity could be enhanced by, e.g., other viral or cellular proteins in the replication/transcription complex. The generation of dinucleotide and other short oligonucleotide products resulting from de novo initiation on poly(U) templates was demonstrated (Fig. 5B). Some polymerase activity also was found on poly(A) templates upon addition of an oligo(dT) primer, generating a full-length product (Fig. 5A). Surprisingly, this DNA primer was not incorporated into the synthesized RNA product. However, in the presence of the oligo(dT) primer, we observed de novo initiation by the EAV nsp9/RdRp on the poly(A) template, as was again demonstrated by detection of the dinucleotide product and other short oligonucleotide products (Fig. 5B). Therefore, the oligo(dT) primer must activate RNA synthesis by an alternative mechanism, possibly by acting as a cofactor stimulating de novo initiation. Of the other DNA and RNA primers tested, only oligo(dA) moderately stimulated polymerase activity. We currently have no mechanistic explanation for the observed stimulatory effect of these DNA oligonucleotides.
Our study suggests that the initiation mechanism of arteriviruses may differ from that of CoVs, which are thought to rely on primer-dependent initiation of RNA synthesis (8, 25). Evidence for primer-dependent activity of a CoV RdRp was presented in a study on the SARS-CoV nsp12/RdRp, which was found to be active in vitro on poly(A) templates with oligo(U) as the primer (8). However, this study did not confirm the incorporation of the primer into the RNA product, and de novo initiation was not tested (8). Another indirect argument for a primer-dependent mechanism of RNA synthesis comes from a report on a putative SARS-CoV primase (nsp8) (25) that may generate the primers for the canonical primer-dependent nsp12/RdRp. The CoV and arterivirus RdRps cluster with RdRps of plus-strand RNA viruses using a genome-linked viral protein for priming RNA synthesis (30). However, note that the 5′ ends of their genomes do not bear such a VPg type of protein but bear a cap structure, implying that the nature of a putative primer would have to be different. No such RdRp has been functionally or structurally characterized in any detail. Our sequence analysis (Fig. 1) indicated that the RdRp domain in EAV nsp9, identified on the basis of our current knowledge of RdRp structures, is rather small, as in the case of RdRps that preferentially use VPg-linked primers. On the other hand, EAV nsp9 does not contain a complete motif G, which has been implicated in primer-dependent initiation and is shared by the RdRps of picornaviruses, caliciviruses, and CoVs. Our data show that the EAV nsp9/RdRp can use a de novo initiation mechanism for RNA synthesis. Like other RdRps that have this capability, the arterivirus RdRp might require additional structural elements for initiation platform formation. In this context, one might speculate that the N-terminal domain of nsp9 could contribute to the formation of an initiation platform.
Despite considerable efforts and testing of different reaction conditions and templates, no de novo polymerase activity could be measured on viral templates. This suggests that the initiation of RNA synthesis requires additional factors that are absent in this in vitro assay using the purified EAV nsp9/RdRp. Though the enzyme was able to initiate RNA synthesis de novo from homopolymeric templates, the use of viral templates may depend on an unknown protein or oligonucleotide primer. Alternatively, a long-distance RNA-RNA interaction, e.g., mediating circularization of the genomic template, may be required for initiation of RNA synthesis, as was reported for flaviviruses (2, 22, 29, 69). No RNA synthesis was found using an EAV template containing both the 5′- and 3′-terminal 300 nucleotides. However, sequences elsewhere in the genome or protein factors may be required to mediate circularization, as was described for several plus-strand RNA viruses (24, 26, 43). Finally, additional viral or cellular proteins may act in concert with the RdRp to initiate RNA synthesis on viral templates. Viral RNA templates are highly structured, and one obvious factor absent in this in vitro assay is the viral helicase (nsp10), which may be required to unwind the RNA secondary structure and was reported to be essential for viral RNA synthesis in vivo (65).
A viral genome terminus consisting of a stem-loop structure and a single-stranded sequence is a common feature in initiation of viral RNA synthesis. The 3′-proximal domain of the EAV genome was recently reported to contain such an RNA hairpin structure, which is required for viral RNA synthesis (3). This stem-loop structure is followed by three single-stranded nucleotides (-ACC) and the poly(A) tail at the genomic 3′ end. De novo initiation requires the stacking of the two initiating nucleotides and their correct spatial orientation in the specialized priming platform of the RdRp enzyme. This highly specific process is guided by the affinity of the polymerase for both template and initiation nucleotides. Therefore, replacement of the ultimate nucleotide(s) of the template RNA usually results in a severe reduction of viral RNA synthesis (7, 23, 28, 31, 53, 56). Since no significant de novo initiation of RNA synthesis was observed on a poly(A) homopolymeric RNA template, we postulated that EAV minus-strand RNA synthesis could initiate upstream of the poly(A) tail at the two C residues (nucleotides 12703 and 12704) that form the genomic 3′ terminus. To test this hypothesis, these C residues were targeted by site-directed mutagenesis and replaced by either G or U residues (Fig. 7). According to an RNA secondary structure prediction made using the Mfold program (72, 73), these mutations did not affect the folding of the upstream stem-loop structure (data not shown). Replacement of the 3′-terminal C nucleotides with either G or U residues severely impaired virus replication, presumably by inhibiting the initiation of minus-strand RNA synthesis. The importance of the terminal C residue was further supported by reversion of the cg mutant, which rapidly regained the wild-type terminal CC doublet. Most likely, the EAV nsp9/RdRp initiation site is loaded preferentially with GTP, and 3′-terminal mutations in the template can revert within one round of replication following incorporation of a mismatched GTP as the starting nucleotide for minus-strand RNA synthesis. Strikingly, the other known arteriviruses all have genomes with two 3′-terminal U residues. In addition, the purified EAV RdRp was found to be active on poly(U) templates. The fact that replacement of the terminal CC by UU dramatically affected EAV viability and RNA synthesis suggests that, in vivo, the initiation of EAV RNA synthesis is a highly specific process, in which the two single-stranded C residues likely play a critical role.
Our data also suggested that the 3′ poly(A) tail is required for EAV viability. Upon transfection of a mutant genome lacking a poly(A) tail, the production of viral RNA and protein could not be detected. However, the stage in the viral life cycle for which the poly(A) tail is critical remains unknown, since it almost certainly also plays a role in RNA stability and efficient genome translation. If RNA synthesis was initiated internally, e.g., on the 3′-terminal C doublet, this would require the subsequent addition of a poly(A) tail by a nontemplated mechanism. Viral or cellular terminal transferase activity could be responsible, although we did not detect such an activity associated with the EAV nsp9/RdRp in this study. Alternatively, the genome may be polyadenylated by the cellular cytoplasmic polyadenylation machinery (reviewed in reference 44), despite the fact that the 3′-terminal sequence preceding the EAV poly(A) tail does not contain the polyadenylation consensus signal (AAUAAA). Template-independent polyadenylation was also reported for the genomes of Sindbis virus (23), bamboo mosaic potexvirus (9), hepatitis A virus (32), and hepatitis C virus (67).
The purified EAV nsp9/RdRp was able to initiate RNA synthesis de novo in a template-specific manner, but no activity was found on poly(A) templates and more structured heteropolymeric viral RNA templates. This template specificity may be a mechanism to prevent initiation of RNA synthesis on the polyadenylated structured mRNAs that are abundantly present in the host cell's cytoplasm. Specific initiation of RNA synthesis may be regulated by critical RNA-RNA or protein-RNA interactions involving the viral RNA template, and ongoing studies using the purified EAV nsp9/RdRp aim to identify such determinants of initiation and template specificity. For example, the recently described hairpin near the 3′ terminus of the EAV genome is a likely target for such interactions (3). The ability to purify an enzymatically active EAV RdRp will facilitate its further functional and structural characterization and will provide insight into the molecular basis of RNA polymerization. Such detailed molecular knowledge of viral RdRps also may contribute to the identification of selective inhibitors of this important class of viral enzymes.
Acknowledgments
We thank Danny Nedialkova for generation of the nsp9/RdRp expression construct.
This study was supported, in part, by the integrated project VIZIER (LSHG-CT-2004-511960) of the European Union Sixth PCRDT. Nancy Beerens was supported by VENI grant 700.53.405 from the Council for Chemical Sciences of The Netherlands Organization for Scientific Research (NWO-CW).
Footnotes
Published ahead of print on 30 May 2007.
REFERENCES
- 1.Abergel, C., B. Coutard, D. Byrne, S. Chenivesse, J. B. Claude, C. Deregnaucourt, T. Fricaux, C. J. S. Gianesini-Boutreux, R. Lebrun, C. Maza, C. Notredame, O. Poirot, K. Suhre, M. Varagnol, and J. M. Claverie. 2006. Structural genomics of highly conserved microbial genes of unknown function in search of new antibacterial target. J. Struct. Funct. Genomics 4:141-157. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez, D. E., M. F. Lodeiro, S. J. Luduena, L. I. Pietrasanta, and A. V. Gamarnik. 2005. Long-range RNA-RNA interactions circularize the Dengue virus genome. J. Virol. 79:6631-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beerens, N., and E. J. Snijder. 2006. RNA signals in the 3′ terminus of the genome of equine arteritis virus are required for viral RNA synthesis. J. Gen. Virol. 87:1977-1983. [DOI] [PubMed] [Google Scholar]
- 4.Bressanelli, S., L. Tomei, F. A. Rey, and R. De Francesco. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruenn, J. A. 2003. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 31:1821-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. L. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. [DOI] [PubMed] [Google Scholar]
- 7.Chapman, M. R., and C. C. Kao. 1999. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J. Mol. Biol. 286:709-720. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, A., W. Zhang, Y. Xie, W. Jiang, E. Arnold, S. G. Sarafianos, and J. Ding. 2005. Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology 335:165-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, J. H., C. W. Peng, Y. H. Hsu, and C. H. Tsai. 2002. The synthesis of minus-strand RNA of bamboo mosaic potexvirus initiates from multiple sites within the poly(A) tail. J. Virol. 76:6114-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corpet, F. 1988. Multiple sequence alignment with hierarchical-clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.den Boon, J. A., E. J. Snijder, E. D. Chirnside, A. A. de Vries, M. C. Horzinek, and W. J. M. Spaan. 1991. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J. Virol. 65:2910-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrer-Orta, C., A. Arias, R. Agudo, R. Perez-Luque, C. Escarmis, E. Domingo, and N. Verdaguer. 2006. The structure of a protein primer-polymerase complex in the initiation of genome replication. EMBO J. 25:880-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrer-Orta, C., A. Arias, R. Perez-Luque, C. Escarmis, E. Domingo, and N. Verdaguer. 2004. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J. Biol. Chem. 279:47212-47221. [DOI] [PubMed] [Google Scholar]
- 14.Ferrer-Orta, C., A. Arias, C. Escarmis, and N. Verdaguer. 2006. A comparison of viral RNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 16:27-34. [DOI] [PubMed] [Google Scholar]
- 15.Ferron, F., C. Bussetta, H. Dutartre, and B. Canard. 2005. The modeled structure of the RNA dependent RNA polymerase of GBV-C virus suggests a role for motif E in Flaviviridae RNA polymerases. BMC Bioinformatics 6:255-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fullerton, S. W. B., M. Blaschke, B. Coutard, J. Gebhardt, A. Gorbalenya, B. Canard, P. A. Tucker, and J. Rohayem. 2007. Structural and functional characterization of sapovirus RNA-dependent RNA polymerase. J. Virol. 81:1858-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galtier, N., M. Gouy, and C. Gautier. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comp. Appl. Biosci. 12:543-548. [DOI] [PubMed] [Google Scholar]
- 18.Gorbalenya, A. E., L. Enjuanes, J. Ziebuhr, and E. J. Snijder. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorbalenya, A. E., F. M. Pringle, J. L. Zeddam, B. T. Luke, C. E. Cameron, J. Kalmakoff, T. N. Hanzlik, K. H. J. Gordon, and V. K. Ward. 2002. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J. Mol. Biol. 324:47-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gouet, P., X. Robert, and E. Courcelle. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31:3320-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagen, M., L. Tiley, T. D. Y. Chung, and M. Krystal. 1995. The role of template-primer interactions in cleavage and initiation by the influenza-virus polymerase. J. Gen. Virol. 76:603-611. [DOI] [PubMed] [Google Scholar]
- 22.Hahn, C. S., Y. S. Hahn, C. M. Rice, E. Lee, L. Dalgarno, E. G. Strauss, and J. H. Strauss. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 198:33-41. [DOI] [PubMed] [Google Scholar]
- 23.Hardy, R. W. 2006. The role of the 3′ terminus of the Sindbis virus genome in minus-strand initiation site selection. Virology 345:520-531. [DOI] [PubMed] [Google Scholar]
- 24.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imbert, I., J. C. Guillemot, J. M. Bourhis, C. Bussetta, B. Coutard, M. P. Egloff, F. Ferron, A. E. Gorbalenya, and B. Canard. 2006. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 25:4933-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isken, O., C. W. Grassmann, R. T. Sarisky, M. Kann, S. Zhang, F. Grosse, P. N. Kao, and S. E. Behrens. 2003. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J. 22:5655-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kao, C. C., P. Singh, and D. J. Ecker. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:251-260. [DOI] [PubMed] [Google Scholar]
- 28.Khromykh, A. A., N. Kondratieva, J. Y. Sgro, A. Palmenberg, and E. G. Westaway. 2003. Significance in replication of the terminal nucleotides of the flavivirus genome. J. Virol. 77:10623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75:6719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72:2197-2206. [DOI] [PubMed] [Google Scholar]
- 31.Kuo, L., R. Fearns, and P. L. Collins. 1997. Analysis of the gene start and gene end signals of human respiratory syncytial virus: quasi-templated initiation at position 1 of the encoded mRNA. J. Virol. 71:4944-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kusov, Y. Y., R. Gosert, and V. Gauss-Muller. 2005. Replication and in vivo repair of the hepatitis A virus genome lacking the poly(A) tail. J. Gen. Virol. 86:1363-1368. [DOI] [PubMed] [Google Scholar]
- 33.Lai, M. M., C. D. Patton, and S. A. Stohlman. 1982. Further characterization of mRNAs of mouse hepatitis virus: presence of common 5′-end nucleotides. J. Virol. 41:557-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laurila, M. R. L., E. V. Makeyev, and D. H. Bamford. 2002. Bacteriophage phi 6 RNA-dependent RNA polymerase—molecular details of initiating nucleic acid synthesis without primer. J. Biol. Chem. 277:17117-17124. [DOI] [PubMed] [Google Scholar]
- 35.Malet, H., M. P. Egloff, B. Selisko, R. E. Butcher, P. J. Wright, M. Roberts, A. Gruez, G. Sulzenbacher, C. Vonrhein, G. Bricogne, J. M. Mackenzie, A. A. Khromykh, A. D. Davidson, and B. Canard. 2007. Crystal structure of the RNA polymerase domain of the west nile virus non-structural protein 5. J. Biol. Chem. 282:10678-10689. [DOI] [PubMed] [Google Scholar]
- 36.Molenkamp, R., S. Greve, W. J. M. Spaan, and E. J. Snijder. 2000. Efficient homologous RNA recombination and requirement for an open reading frame during replication of equine arteritis virus defective interfering RNAs. J. Virol. 74:9062-9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng, K. K. S., N. Pendas-Franco, J. Rojo, J. A. Boga, A. Machin, J. M. M. Alonso, and F. Parra. 2004. Crystal structure of Norwalk virus polymerase reveals the carboxyl terminus in the active site cleft. J. Biol. Chem. 279:16638-16645. [DOI] [PubMed] [Google Scholar]
- 38.O'Reilly, E. K., and C. C. Kao. 1998. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology 252:287-303. [DOI] [PubMed] [Google Scholar]
- 39.Pasternak, A. O., A. P. Gultyaev, W. J. M. Spaan, and E. J. Snijder. 2000. Genetic manipulation of arterivirus alternative mRNA leader-body junction sites reveals tight regulation of structural protein expression. J. Virol. 74:11642-11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pasternak, A. O., W. J. M. Spaan, and E. J. Snijder. 2006. Nidovirus transcription: how to make sense? J. Gen. Virol. 87:1403-1421. [DOI] [PubMed] [Google Scholar]
- 41.Paul, A. V., J. H. van Boom, D. Filippov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 42.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piron, M., P. Vende, J. Cohen, and D. Poncet. 1998. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 17:5811-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richter, J. D. 1999. Cytoplasmic polyadenylation in development and beyond. Microbiol. Mol. Biol. Rev. 63:446-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rohayem, J., I. Robel, K. Jager, U. Scheffler, and W. Rudolph. 2006. Protein-primed and de novo initiation of RNA synthesis by norovirus 3Dpol. J. Virol. 80:7060-7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sagripanti, J. L., R. O. Zandomeni, and R. Weinmann. 1986. The cap structure of simian hemorrhagic-fever virion RNA. Virology 151:146-150. [DOI] [PubMed] [Google Scholar]
- 47.Salgado, P. S., E. V. Makeyev, S. J. Butcher, D. H. Bamford, D. I. Stuart, and J. M. Grimes. 2004. The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase. Structure 12:307-316. [DOI] [PubMed] [Google Scholar]
- 48.Sawicki, S. G., and D. Sawicki. 2005. Coronavirus transcription, a perspective, p. 31-55. In L. Enjuanes (ed.), Coronavirus replication and reverse genetics. Springer, Berlin, Germany. [DOI] [PMC free article] [PubMed]
- 49.Sawicki, S. G., D. L. Sawicki, and S. G. Siddell. 2007. A contemporary view of coronavirus transcription. J. Virol. 81:20-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selisko, B., H. Dutartre, J. C. Guillemot, C. Debarnot, D. Benarroch, A. Khromykh, P. Despres, M. P. Egloff, and B. Canard. 2006. Comparative mechanistic studies of de novo RNA synthesis by flavivirus RNA-dependent RNA polymerases. Virology 351:145-158. [DOI] [PubMed] [Google Scholar]
- 51.Seybert, A., C. C. Posthuma, L. C. van Dinten, E. J. Snijder, A. E. Gorbalenya, and J. Ziebuhr. 2005. A complex zinc finger controls the enzymatic activities of nidovirus helicases. J. Virol. 79:696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seybert, A., L. C. van Dinten, E. J. Snijder, and J. Ziebuhr. 2000. Biochemical characterization of the equine arteritis virus helicase suggests a close functional relationship between arterivirus and coronavirus helicases. J. Virol. 74:9586-9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siegel, R. W., S. Adkins, and C. C. Kao. 1997. Sequence-specific recognition of a subgenomic RNA promoter by a viral RNA polymerase. Proc. Natl. Acad. Sci. USA 94:11238-11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Snijder, E. J., S. G. Siddell, and A. E. Gorbalenya. 2005. The order Nidovirales, p. 390-404. In B. W. Mahy and V. ter Meulen (ed.), Topley and Wilson's microbiology and microbial infections. Hodder Arnold, London, United Kingdom.
- 55.Thompson, A. A., and O. B. Peersen. 2004. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 23:3462-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tilgner, M., and P. Y. Shi. 2004. Structure and function of the 3′ terminal six nucleotides of the West Nile virus genome in viral replication. J. Virol. 78:8159-8171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Aken, D., J. Zevenhoven-Dobbe, A. E. Gorbalenya, and E. J. Snijder. 2006. Proteolytic maturation of replicase polyprotein pp1a by the nsp4 main proteinase is essential for equine arteritis virus replication and includes internal cleavage of nsp7. J. Gen. Virol. 87:3473-3482. [DOI] [PubMed] [Google Scholar]
- 58.van Berlo, M. F., M. C. Horzinek, and B. A. M. van der Zeijst. 1982. Equine arteritis virus-infected cells contain 6 polyadenylated virus-specific RNAs. Virology 118:345-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van den Born, E., A. P. Gultyaev, and E. J. Snijder. 2004. Secondary structure and function of the 5′-proximal region of the equine arteritis virus RNA genome. RNA 10:424-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van der Meer, Y., H. van Tol, J. Krijnse Locker, and E. J. Snijder. 1998. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J. Virol. 72:6689-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Dijk, A. A., E. V. Makeyev, and D. H. Bamford. 2004. Initiation of viral RNA-dependent RNA polymerization. J. Gen. Virol. 85:1077-1093. [DOI] [PubMed] [Google Scholar]
- 62.van Dinten, L. C., J. A. den Boon, A. L. M. Wassenaar, W. J. M. Spaan, and E. J. Snijder. 1997. An infectious arterivirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc. Natl. Acad. Sci. USA 94:991-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Dinten, L. C., S. Rensen, A. E. Gorbalenya, and E. J. Snijder. 1999. Proteolytic processing of the open reading frame 1b-encoded part of arterivirus replicase is mediated by nsp4 serine protease and is essential for virus replication. J. Virol. 73:2027-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Dinten, L. C., A. L. M. Wassenaar, A. E. Gorbalenya, W. J. M. Spaan, and E. J. Snijder. 1996. Processing of the equine arteritis virus replicase ORF1b protein: identification of cleavage products containing the putative viral polymerase and helicase domains. J. Virol. 70:6625-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Dinten, L. C., H. van Tol, A. E. Gorbalenya, and E. J. Snijder. 2000. The predicted metal-binding region of the arterivirus helicase protein is involved in subgenomic mRNA synthesis, genome replication, and virion biogenesis. J. Virol. 74:5213-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Dyke, T. A., and J. B. Flanegan. 1980. Identification of poliovirus polypeptide P63 as a soluble RNA-dependent RNA polymerase. J. Virol. 35:732-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Leeuwen, H. C., J. M. P. Liefhebber, and W. J. M. Spaan. 2006. Repair and polyadenylation of a naturally occurring hepatitis C virus 3′ nontranslated region-shorter variant in selectable replicon cell lines. J. Virol. 80:4336-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yap, T. L., T. Xu, Y. L. Chen, H. Malet, M. P. Egloff, B. Canard, S. G. Vasudevan, and J. Lescar. 2007. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J. Virol. 81:4753-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.You, S., B. Falgout, L. Markoff, and R. Padmanabhan. 2001. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 276:15581-15591. [DOI] [PubMed] [Google Scholar]
- 70.Zhong, W., A. S. Uss, E. Ferrari, J. Y. N. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]
- 72.Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244:48-52. [DOI] [PubMed] [Google Scholar]
- 73.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]