Abstract
Hepatitis C virus (HCV) infection is a global health concern affecting an estimated 3% of the world's population. Recently, cell culture systems have been established, allowing recapitulation of the complete virus life cycle for the first time. Since the HCV proteins p7 and NS2 are not predicted to be major components of the virion, nor are they required for RNA replication, we investigated whether they might have other roles in the viral life cycle. Here we utilize the recently described infectious J6/JFH chimera to establish that the p7 and NS2 proteins are essential for HCV infectivity. Furthermore, unprocessed forms of p7 and NS2 were not required for this activity. Mutation of two conserved basic residues, previously shown to be important for the ion channel activity of p7 in vitro, drastically impaired infectious virus production. The protease domain of NS2 was required for infectivity, whereas its catalytic active site was dispensable. We conclude that p7 and NS2 function at an early stage of virion morphogenesis, prior to the assembly of infectious virus.
Hepatitis C virus (HCV) is a major causative agent of severe liver disease, with an estimated 170 million people infected worldwide (36). HCV is the sole member of the genus Hepacivirus, which, together with the Flavivirus and Pestivirus genera, comprise the family Flaviviridae (reviewed in reference 21). The HCV genome is approximately 9,600 nucleotides in length and is translated from an internal ribosome entry site (IRES) to generate a polyprotein in the order NH2-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COOH. Co- and posttranslational processing of the polyprotein by viral and cellular proteases yields the individual viral proteins. Core (C) and envelope proteins (E1 and E2) are the major structural proteins, which, together with a host derived lipid bilayer and the viral RNA, comprise the virion. The nonstructural (NS) proteins, NS3 to NS5B, are essential components of the viral replicase. NS3 possesses helicase and NTPase activities and, along with its cofactor NS4A, comprises the major viral protease. NS4B and NS5A play essential, but as yet undefined roles in RNA replication. NS5B is the RNA-dependent RNA polymerase (RdRp).
p7 and NS2 are dispensable for RNA replication, since subgenomic replicons that lack the entire C to NS2 coding region replicate autonomously (2, 22). p7 is a small (63 amino acids) hydrophobic protein that is predicted to span the membrane twice. Both the N and C termini of p7 reside within the endoplasmic reticulum (ER) lumen, with a short, basic intervening loop exposed to the cytoplasm (4). The N and C termini of p7 are released from the polyprotein by host signal peptidase. Incomplete and delayed processing leads to the accumulation of E2-p7 and p7-NS2 precursors, respectively. (10, 19, 25). While sequence determinants within both p7 and NS2 have been shown to modulate E2-p7 and p7-NS2 cleavage efficiencies (3), the functional importance of these precursors is not known. In chimpanzees, p7 was found to be essential for HCV infectivity (31). Moreover, disruption of either uncleaved E2-p7 or p7-NS2 using an encephalomyocarditis virus (EMCV) IRES abolished infectivity in this system. In vitro, p7 oligomerizes to form ion-conducting channels (12, 28, 29), the activity of which can be disrupted by mutation of conserved basic residues within the cytoplasmic loop (13). Mutation of these residues in the context of the chimpanzee model system was also found to block infectivity (31).
NS2 is a membrane-associated cysteine protease (10, 23). The N terminus of NS2 consists of one or more transmembrane domains, the exact number of which remains controversial. The C-terminal domain of NS2, together with the N-terminal third of NS3, forms the NS2-3 protease, an enzyme that catalyzes a single cleavage between the two proteins (10). The crystal structure of the C-terminal domain of NS2 has recently been determined and reveals a dimeric protease containing two composite active sites (23). Although NS2 itself is not required for RNA replication, its cleavage from NS3 is essential (18, 35). The significance of sequences within NS2 to the infectivity of intergenotypic chimeras has suggested that NS2 may play a role in infectious virus production (30, 38).
Recently, systems allowing the study of the complete HCV life cycle in cell culture (HCVcc) have been developed (20, 34, 38-40). HCVcc relies on a unique genotype 2a patient isolate, JFH-1, which exhibits robust replication and infectious virus production in specially developed human hepatoma cells (Huh-7.5). Our laboratory developed a chimeric genome termed J6/JFH that expresses the J6 structural proteins (20) and exhibits enhanced infectious virus production relative to the JFH-1 parent (30). With the advent of the HCVcc system the determinants of infectious virus production can be studied for the first time. Here, we report an analysis of the importance of p7 and NS2 for HCVcc infectivity. Using bicistronic genomes, we demonstrate that unprocessed forms of p7 (E2-p7 and p7-NS2) are not essential for infectious virus production. Utilizing a novel monocistronic reporter virus, we demonstrate that p7 itself is essential for infectivity and that mutations introduced into the basic cytoplasmic loop of p7 are deleterious to this function. We report that NS2 is also required for HCVcc infectivity. Using a bicistronic genome to separate NS2 from NS3, we find that unprocessed NS2-3 is not required for infectious virus production and that the NS2 protease domain, but not its active site, is essential for infectious virus production. Finally, we provide evidence that both p7 and NS2 function prior to the formation of intracellular infectious virus.
MATERIALS AND METHODS
Cell culture.
Huh-7.5 cells were propagated in Dulbecco modified Eagle medium (Gibco) supplemented with 10 mM nonessential amino acids and 10% fetal bovine serum, referred to below as complete medium. Cells were grown at 37°C in 5% CO2.
Plasmid constructs.
Plasmids were constructed by standard methods. Constructs were verified by restriction enzyme digestion and sequencing. Descriptions of the cloning strategies are provided below, plasmid and primer sequences are available upon request. Unless otherwise noted, positions are based on nucleotide (nt) numbering of the J6/JFH chimeric genotype 2a genome (20). All constructs are based on this genome or its derivative, J6/JFH1.1, which includes unique silent restrictions sites at positions 2392 (BglII) and 2955 (NotI).
(i) E2-IRES-p7 and p7-IRES-NS2.
E2-IRES-p7 contains the EMCV IRES inserted after nt 2590 in J6/JFH1.1, with the addition of a stop codon and a unique PmeI site after E2 and a start codon before p7. p7-IRES-NS2 contains the EMCV IRES inserted after nt 2779 in J6/JFH1.1 with the addition of a stop codon and a unique PmeI site after p7 and a start codon before NS2. For all bicistronic constructs the EMCV IRES sequence was obtained from a previously described subgenomic JFH-1 replicon (16) by PCR amplification.
(ii) J6/JFH(p7-Rluc2A) and mutant derivatives.
To construct the reporter virus genome J6/JFH(p7-Rluc2A), a unique silent restriction site was introduced between the p7 and NS2 coding sequence at position 2784 (MluI) in J6/FH1.1 using overlapping PCR mutagenesis. This site was used to insert a 995-bp DNA cassette encoding the Renilla luciferase gene (Rluc) in tandem with the foot-and-mouth disease virus (FMDV) 2A peptide (Rluc2A), PCR amplified from FL-J6/JFH-C19′Rluc2AUbi (33) using primers containing MluI restriction sites. To construct Δp7-Rluc2A, an in-frame deletion of the entire p7 coding sequence (nt 2591 to 2779) was introduced into J6/JFH(p7-Rluc2A) by using PCR deletion mutagenesis. Mutations in p7 (N17A, N17/Y21F, Y21F, K33A/R35A, and K33Q/R35Q, p7 protein numbering) were introduced into J6/JFH(p7-Rluc2A) by overlapping PCR mutagenesis using primers containing the desired changes.
(iii) NS2-IRES-NS3 and mutant derivatives.
NS2-IRES-NS3 contains the EMCV IRES inserted after nt 3430 in J6/JFH with the addition of a stop codon and a unique PmeI site after NS2 and a start codon before NS3. ΔNS2-IRES-NS3 contains a deletion of the complete NS2 coding sequence (nt 2780 to 3430) in NS2-IRES-NS3 and includes a stop codon and a unique PmeI site after p7. NS2Δpro-IRES-NS3 contains a partial deletion encompassing the C-terminal protease domain of NS2 (nt 3077 to 3430) in NS2-IRES-NS3. A mutation of the NS2 protease active site catalytic cysteine residue (C184A, NS2 protein numbering) was introduced into J6/JFH and NS2-IRES-NS3 by overlapping PCR mutagenesis using primers encoding the desired change.
(iv) NS2-IRES-Gluc2AUbi and mutant derivatives.
The NS2 bicistronic reporter genome encodes the Gaussia luciferase (Gluc) (32) in the second cistron of NS2-IRES-NS3. The coding sequence for Gluc, excluding the first 16 residues, was PCR amplified from pCMV-GLuc (New England Biolabs, Ipswich MA). A Gluc2AUbi reporter gene cassette, encoding Gluc in tandem with FMDV 2A and ubiquitin (Ubi) sequences obtained from FL-J6/JFH-C19′Rluc2AUbi (33), was fused to the N terminus of NS3 in NS2-IRES-NS3 using overlapping PCR. ΔNS2-IRES-NS3 and NS2Δpro-IRES-NS3 derivatives of NS2-IRES-Gluc2AUbi were obtained by ligation of EcoRI/PmeI fragments containing NS2 deletions from their respective nonreporter bicistronic constructs, with the 10,294-bp EcoRI/PmeI fragment from NS2-IRES-Gluc2AUbi.
(v) ΔE1E2 and GNN containing genomes.
ΔE1E2 genomes were constructed essentially as described previously (34) and contain a large in-frame deletion within the E1E2 coding sequence (nt 989 to 2041). GNN genomes contain a double mutation within the RdRp motif of NS5B (GDD to GNN).
RNA transcription.
In vitro transcripts were generated as previously described (20). Briefly, plasmids were linearized by XbaI and purified by using a Minelute column (QIAGEN, Valencia, CA). RNA was transcribed from 1 μg of purified template by using the T7 Megascript kit (Ambion, Austin, TX). Reactions were incubated at 37°C for 3 h, followed by a 15-min digestion with 3 U of DNase I (Ambion). RNA was purified by using an RNeasy kit (QIAGEN) with an additional on-column DNase treatment. RNA was quantified by absorbance at 260 nm and diluted to 0.5 μg/μl. Prior to storage at −80°C, RNA integrity was determined by agarose gel electrophoresis and visualization by ethidium bromide staining.
RNA electroporation.
Huh-7.5 cells were electroporated with RNA as previously described (20). Briefly, Huh-7.5 cells were treated with trypsin, washed twice with ice-cold RNase-free AccuGene phosphate-buffered saline (PBS; BioWhittaker, Rockland ME), and resuspended at 1.75 × 107 cells/ml in PBS. Then, 5 μg of each RNA was combined with 0.4 ml of cell suspension and immediately pulsed using a BTX ElectroSquare Porator ECM 830 (820 V, 99 μsec, five pulses). Electroporated cells were incubated at room temperature for 10 min prior to resuspension in 15 ml of complete medium. Resuspended cells were plated into 24-well and P100 tissue culture dishes.
Assays for RNA replication.
At 8, 24, 48, and 72 h postelectroporation, cells in 24-well plates were washed with Dulbecco PBS (DPBS) and lysed by the addition of Renilla lysis buffer (Promega, Madison WI) or RLT buffer (QIAGEN) containing 0.14 M β-mercaptoethanol for assay of replication by luciferase activity or quantitative reverse transcription-PCR (qRT-PCR), respectively. For luciferase assay, lysates stored at −80°C were thawed completely prior to addition of Renilla substrate (Promega) according to the manufacturer's instructions. The luciferase activity was measured by using a Berthold Centro LB 960 96-well luminometer (Berthold, Bundoora, Australia). For qRT-PCR analysis, prior to storage at −80°C, lysates were homogenized by centrifugation through a QiaShredder column (QIAGEN) for 2 min at 14,000 × g. Total RNA was isolated by RNeasy kit (QIAGEN) and quantified by determining the absorbance at 260 nm. A total of 50 ng of total cellular RNA was used per reaction. qRT-PCRs were performed on a LightCycler 480 (Roche, Basel Switzerland) using the LightCycler amplification kit (Roche) with primers directed against the viral 5′ untranslated region. We assembled 20-μl reactions according to the manufacturer's instructions.
Assays for infectious virus production.
At 8, 24, 48, and 72 h postelectroporation, the media in P100 dishes were harvested and replaced with complete media. Harvested cell culture supernatants were clarified by using a 0.22-μm-pore-size filter and stored in aliquots at −80°C. For detection of infectious virus production by luciferase assay, naive cells were infected with clarified cell culture supernatants and incubated for 48 h prior to analysis. For cotransfection of RNA, after removal and replacement of the media at 8 h posttransfection, cell culture supernatants were harvested at 72 h posttransfection and assayed for infectious virus. For cell-free passage, naive cells were infected with cell culture supernatants harvested at 72 h posttransfection and incubated for an additional 72 h prior to assay for infectious virus production by luciferase assay. Determination of infectious virus production by limiting dilution assay was performed as described previously (20). Briefly, clarified cell culture supernatants were serially diluted and used to infect approximately 3 × 103 cells plated in 96-well dishes. At 4 days postinfection, cells were washed with DPBS, fixed with ice-cold methanol, and stained for the presence of NS5A expression as described previously (20). The 50% tissue culture infectious dose (TCID50) was calculated (20).
Core ELISA.
Ortho HCV antigen enzyme-linked immunosorbent assay to quantify core protein (ELISA; Ortho Clinical Diagnostics, Raritan, NJ) was performed according to the manufacturer's instructions. Briefly, cell culture supernatants harvested at 72 h postelectroporation were appropriately diluted and then applied to plates coated with a mixture of mouse anti-core monoclonal antibodies (MAbs). Antigen was detected by the addition of a second MAb cocktail conjugated to horseradish peroxidase. After a washing step, the wells were developed by the addition of substrate and subsequent stop solution; the absorbance at 490 nm was measured. Core was quantified by comparison to a standard curve using Softmax software.
Assay for intracellular infectivity.
At 72 h postelectroporation, cells were washed with DPBS twice, harvested by treatment with trypsin, and pelleted at 500 × g for 5 min. Pellets were resuspended in complete medium and lysed by four freeze-thaw cycles. Cell lysates were clarified twice by centrifugation at 1,500 × g for 5 min. The supernatant was collected and used for infection of naive cells in the presence of anti-CD81 MAb (α-CD81; JS81; Pharmingen, San Diego, CA) or an isotype control (α-IgG1; anti-mouse IgG1; Pharmingen) at a final concentration of 10 μg/ml. After 4 h of infection, the medium was removed and replaced with complete medium. At 48 h postinfection, the cells were assayed for luciferase activity.
RESULTS
p7 processing intermediates are not required for infectious virus production.
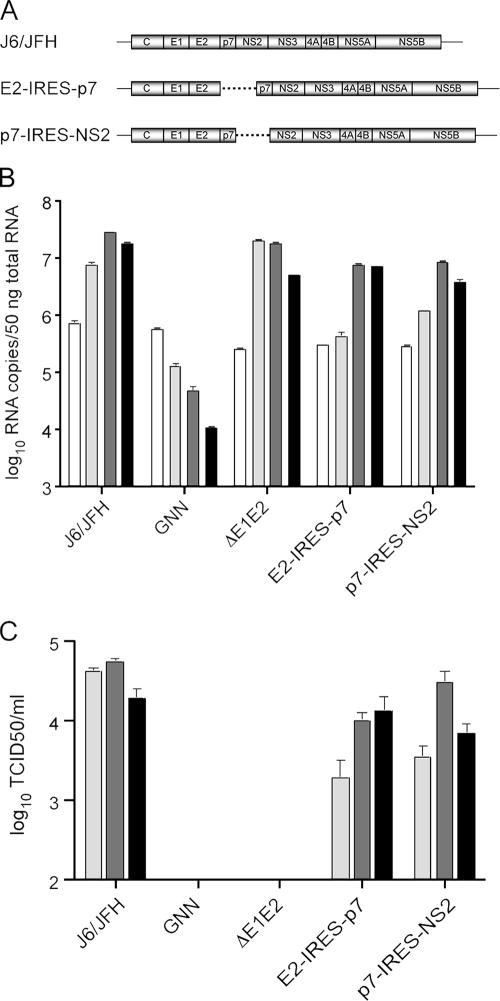
Incomplete or delayed processing has been reported to lead to the accumulation of E2-p7 and p7-NS2 precursors, respectively. (10, 19, 25). The functional significance of these uncleaved proteins, however, is not known. To assess the importance of these intermediates to the viral life cycle, an EMCV IRES was used to separate p7 from either E2 or NS2 (Fig. 1A). In the bicistronic genome E2-IRES-p7, the first cistron, C through E2, is translated from the HCV IRES, while in the second cistron the EMCV IRES initiates translation at the N terminus of p7. A similar bicistronic genome, p7-IRES-NS2, encodes C through p7 in the first cistron and begins translation of the second cistron at the N terminus of NS2. To assess the viability of the E2-IRES-p7 and p7-IRES-NS2 genomes, Huh-7.5 cells were electroporated with in vitro-generated transcripts, and RNA replication was examined by qRT-PCR at several time points posttransfection (Fig. 1B). Although both bicistronic genomes replicated similarly, they were moderately impaired relative to the monocistronic genome J6/JFH or J6/JFHΔE1E2, which contains an in-frame deletion of the envelope proteins. J6/JFH(GNN), which encodes a lethal mutation of the conserved NS5B RdRp motif (GDD to GNN), did not replicate and served as a negative control.
FIG. 1.
Unprocessed forms of p7 are dispensable for infectious virus production. (A) Schematic representation of full-length monocistronic genome J6/JFH (top) and bicistronic genomes E2-IRES-p7 (middle) and p7-IRES-NS2 (bottom). The dotted line indicates the location of the EMCV IRES in bicistronic genomes. (B) Replication of mono- and bicistronic genomes at 8, 24, 48, and 72 h posttransfection (white to black bars, respectively) as determined by qRT-PCR. HCV RNA copies normalized to 50 ng of total RNA. (C) Infectious virus production of mono- and bicistronic genomes determined by TCID50 assay at 24, 48, and 72 h posttransfection (light gray to black bars, respectively). The means and standard errors of the mean (SEM) of duplicate electroporations are shown. GNN, J6/JFH(GNN); ΔE1E2, J6/JFHΔE1E2.
Infectious virus production by cells harboring the transfected HCV genomes was quantified by a limiting dilution (TCID50) assay (Fig. 1C). Relative to J6/JFH, E2-IRES-p7 and p7-IRES-NS2 demonstrated a decrease of approximately 10- to 20-fold in infectious virus production at 24 h posttransfection. At later time points, however, titers approached J6/JFH, suggesting that neither unprocessed E2-p7 nor p7-NS2 play an essential role in infectious virus production. In addition, the viability of both genomes indicates that neither p7 nor NS2 require their native signal sequences, normally provided by the C terminus of E2 and p7, respectively, to function in the virus life cycle.
Characterization of a novel J6/JFH reporter virus encoding luciferase.
The ability to functionally separate p7 from its processing intermediates suggested the possibility of inserting a reporter in this region of the genome. A monocistronic reporter virus was created in which the Renilla luciferase (Rluc) gene was inserted between the p7 and NS2 coding sequences (Fig. 2A). The signal sequence at the C terminus of p7 was predicted to mediate translocation of the reporter into the ER lumen, allowing its subsequent release from the polyprotein by signal peptidase. To ensure correct processing of Rluc at its C terminus, the FMDV 2A peptide was included (7). 2A mediates a cotranslational, autocatalytic cleavage at its own C terminus, leaving a non-native proline residue at the N terminus of NS2. The liberated Rluc protein remains fused to the FMDV 2A peptide. This reporter genome was designated J6/JFH(p7-Rluc2A) (Fig. 2A).
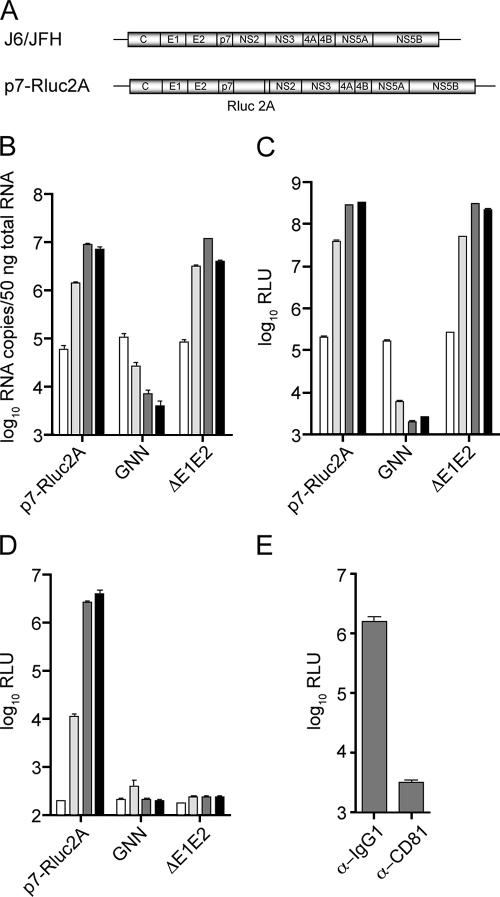
FIG. 2.
Characterization of a novel monocistronic reporter virus. (A) Schematic representation of full-length J6/JFH (top) and the reporter virus J6/JFH(p7-Rluc2A) (bottom). The J6/JFH(p7-Rluc2A) genome contains the coding sequence for Renilla luciferase followed by the FMDV (2A) peptide between p7 and NS2. RNA replication of J6/JFH(p7-Rluc2A) as determined by qRT-PCR (B) and luciferase assay (C) at 8, 24, 48, and 72 h posttransfection (white to black bars, respectively). For qRT-PCR analysis, HCV RNA copies were normalized to 50 ng of total RNA. (D) Infectious virus production of J6/JFH(p7-Rluc2A) at 8, 24, 48, and 72 h posttransfection (white to black bars, respectively). (E) Virus neutralization of J6/JFH(p7-Rluc2A). Virus-containing supernatants were treated with the isotype control antibody, α-IgG1, or the neutralizing antibody, α-CD81. The luciferase activity was determined at 48 h postinfection. The means and SEM of at least duplicate experiments are shown. GNN, J6/JFH(p7-Rluc2A)GNN; ΔE1E2, J6/JFH(p7-Rluc2A)ΔE1E2.
To assess the viability of J6/JFH(p7-Rluc2A), RNA replication was measured at various time points after electroporation of Huh-7.5 cells with in vitro-generated transcripts. Both qRT-PCR analysis of viral RNA accumulation and quantification of the luciferase activity in cell lysates indicated that J6/JFH(p7-Rluc2A) and J6/JFH(p7-Rluc2A)ΔE1E2 replicated, whereas J6/JFH(p7-Rluc2A)GNN did not (Fig. 2B and C). A correlation was observed between the accumulation of HCV-specific RNA and the level of reporter gene expression over time, indicating that luciferase activity could be used as a sensitive measure of HCV RNA replication.
To evaluate the ability of J6/JFH(p7-Rluc2A) to produce infectious virus, clarified cell culture supernatants, harvested at various time points posttransfection, were used to infect naive Huh-7.5 cells. Assay of infected cells for luciferase activity after 48 h indicated that J6/JFH(p7-Rluc2A) was capable of infectious virus production, whereas J6/JFH(p7-Rluc2A)ΔE1E2 and J6/JFH(p7-Rluc2A)GNN were not (Fig. 2D). To determine whether the observed transduction of luciferase activity was the result of authentic HCV infection, cell culture supernatants were treated with an MAb previously shown to block essential interactions of HCV with its receptor molecule CD81 (20). Treatment of supernatant with α-CD81, but not with an isotype control antibody (α-IgG1), resulted in a significant reduction in luciferase activity, indicating that J6/JFH(p7-Rluc2A) virions infected cells via native entry pathways (Fig. 2E). The titer of J6/JFH(p7-Rluc2A) determined by TCID50 assay reflected the transduced luciferase activity and indicated that the reporter virus produced approximately 5- to 10-fold less infectious virus than J6/JFH at 48 h posttransfection (data not shown). These results demonstrate that an Rluc insertion in the p7-NS2 coding region allows efficient infectious virus production and that reporter gene expression could be used to assess levels of RNA replication and virus infection.
p7 is essential for infectious virus production.
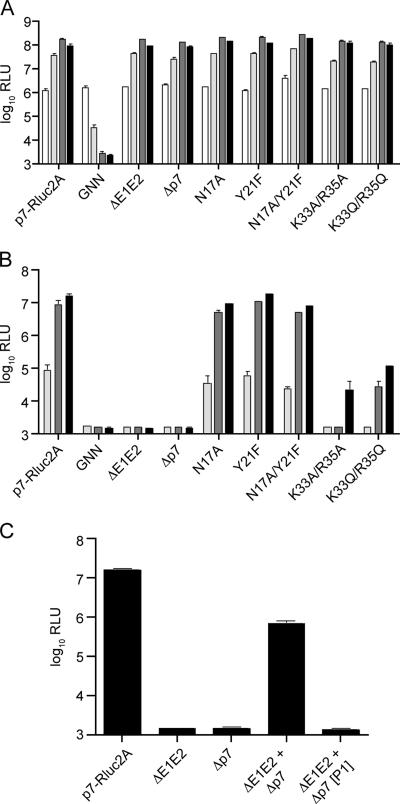
Using the J6/JFH(p7-Rluc2A) reporter virus, we investigated the importance of p7 in the viral life cycle. This genome had the advantages of the sensitive reporter activity, as well as an organization that allowed p7 to be deleted without creating a potentially deleterious fusion of E2 and NS2 sequences. An in-frame deletion of the entire p7 coding sequence was introduced into J6/JFH(p7-Rluc2A), resulting in Δp7-Rluc2A. Assay for intracellular luciferase activity at various time points posttransfection revealed no significant differences in RNA replication between J6/JFH(p7-Rluc2A) and Δp7-Rluc2A (Fig. 3A). These results indicated that neither the p7 coding sequence, nor the protein itself, was required for RNA replication. To assay for infectious virus production, naive cells were infected with clarified cell culture supernatants harvested at 24, 48, and 72 h posttransfection. In contrast to J6/JFH(p7-Rluc2A), Δp7-Rluc2A failed to produce detectable levels of infectious virus, suggesting that p7 was essential for infectivity (Fig. 3B).
FIG. 3.
p7 is essential for infectious virus production. (A) RNA replication of J6/JFH(p7-Rluc2A) genomes by luciferase assay at 8, 24, 48, and 72 h posttransfection (white to black bars, respectively). (B) Infectious virus production at 24, 48, and 72 h posttransfection (light gray to black bars, respectively). The deletions and mutations in the J6/JFH(p7-Rluc2A) genome are designated below. The positions of the point mutants are based on p7 numbering. (C) Infectious virus production at 72 h posttransfection of cells cotransfected with ΔE1E2 and Δp7 genomes (ΔE1E2+Δp7) and after one cell-free passage of the supernatant (ΔE1E2+Δp7[P1]). The means and SEM of duplicate experiments are shown. GNN, J6/JFH(p7-Rluc2A)GNN; ΔE1E2, J6/JFH(p7-Rluc2A)ΔE1E2; Δp7, Δp7-Rluc2A.
Since Rluc is C-terminal to E2 in this genome, it was possible that the introduced deletion of p7 disrupted structural protein function. To determine whether the Δp7-Rluc2A structural proteins were functional, Huh-7.5 cells were cotransfected with Δp7-Rluc2A and J6/JFH(p7-Rluc2A)ΔE1E2 genomes. Whereas transfection with either genome alone did not allow infectious virus production, cotransfection resulted in detectable levels of infectivity, suggesting that the structural proteins encoded by Δp7-Rluc2A were indeed functional (Fig. 3C). Importantly, infectious virus produced by cotransfection of Δp7-Rluc2A and J6/JFH(p7-Rluc2A)ΔE1E2 was able to initiate only a single round of infection, indicating that genetic recombination had not occurred (Fig. 3C). These data show that p7 was not required for RNA replication but was essential for infectious virus production.
Mutation of residues affecting the putative ion channel activity of p7.
p7 has been reported to form oligomers that can function as ion-conductive channels in vitro. Hexameric and heptameric models of the ion channel have been proposed in which the first transmembrane domains of adjacent p7 monomers form a hydrophilic pore through which ions pass (5, 27). In addition, basic residues within a short conserved cytoplasmic loop between the transmembrane domains may form an ion gate and are apparently critical for the ion channel activity of p7 in vitro (13). We investigated the importance of two residues predicted to line the hydrophilic pore, as well as two basic residues within the cytoplasmic loop of p7 in the context of the J6/JFH(p7-Rluc2A) reporter virus. Mutations affecting the putative hydrophilic pore were introduced at positions 17 (N17A) and 21 (Y21F) of p7 (p7 protein numbering), either separately or together. The two basic cytoplasmic loop residues were mutated together to either alanine (K33A/R35A) or glutamine (K33Q/R35Q).
No significant differences in RNA replication were observed between J6/JFH(p7-Rluc2A) and the mutant genomes (Fig. 3A). Moreover, whereas N17A and N17A/Y21F showed very slight decreases in infectious virus production, Y21F was not impaired (Fig. 3B). In contrast, mutation of the basic loop residues significantly impaired infectious virus production. K33A/R35A exhibited the most drastic phenotype, with only low levels of infectious virus detectable at the latest time point examined (72 h). K33Q/R35Q, although also significantly reduced in infectious virus production, exhibited a less severe phenotype. These results suggested that the basic cytoplasmic loop residues of p7 are functionally important, whereas the residues predicted to line the hydrophilic pore are not.
p7 acts at an early stage of virion morphogenesis.
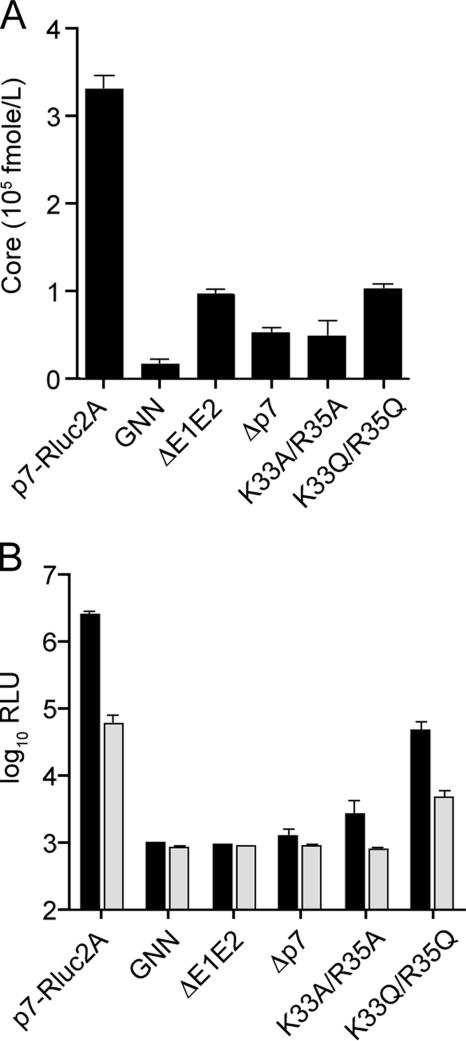
It was possible that the defects observed upon deletion of p7 or mutation of its basic cytoplasmic loop residues were the result of a drastic reduction in the infectivity of released virions. To investigate this, we utilized a sensitive ELISA to determine the amount of core protein released into cell culture supernatants. Δp7-Rluc2A, the highly impaired mutants K33A/R35A and K33Q/R35Q, and J6/JFH(p7-Rluc2A)ΔE1E2 showed similar levels of HCV core protein release at 72 h posttransfection and were all significantly reduced from J6/JFH(p7-Rluc2A) (Fig. 4A). These data suggested that the loss of infectious virus production observed upon deletion or mutation of p7 was not the result of decreased infectivity of secreted particles but rather caused by a general block in virion release.
FIG. 4.
p7 functions at an early stage of virion morphogenesis. HCV core release into cell culture supernatants (A) and intracellular infectious virus accumulation (B) at 72 h posttransfection. HCV core release was determined by using a sensitive core-based ELISA (see Materials and Methods for details) on clarified cell culture supernatants harvested at 72 h posttransfection. For panel B, an analysis of infectious intracellular virus accumulation at 72 h posttransfection was performed. Lysates generated by multiple rounds of freeze-thaw were combined with isotype control antibody, α-IgGI (▪), or the HCV neutralizing antibody, α-CD81 (□), during infection of naive Huh-7.5 cells. Luciferase activity was determined at 48 h postinfection. The means and SEM of duplicate experiments are shown. GNN, J6/JFH(p7-Rluc2A)GNN; ΔE1E2, J6/JFH(p7-Rluc2A)ΔE1E2; Δp7, Δp7-Rluc2A.
To further investigate the stage of virion production at which p7 might be involved, we determined the effects of its deletion or mutation on intracellular infectious virus accumulation. At 72 h posttransfection, cells were washed and lysed by multiple freeze-thaw cycles to release intracellular particles (8). Cells infected with the resultant lysates were assayed for luciferase activity at 48 h postinfection. Lysis of cells harboring J6/JFH(p7-Rluc2A) resulted in the release of intracellular infectious virus (Fig. 4B). Furthermore, this infectivity could be neutralized with an α-CD81 antibody but not with an isotype control, confirming that the luciferase transduction was the result of authentic viral infection. In contrast to J6/JFH(p7-Rluc2A) but similar to J6/JFH(p7-Rluc2A)ΔE1E2, lysates derived from Δp7-Rluc2A transfected cells transduced only background levels of luciferase activity, indicating that no intracellular infectivity was released. The p7 mutants K33A/R35A and K33Q/R33Q showed low levels of intracellular infectious virus (Fig. 4B), corresponding to the low levels extracellular infectious virus observed (Fig. 3B). Taken together, these data indicate that p7 functions at an early stage in virus morphogenesis, prior to the assembly and release of infectious intracellular virions.
NS2, but not uncleaved NS2-3, is required for infectious virus production.
The tolerance of the E2-p7 and p7-NS2 regions to insertion of foreign sequences led us to investigate whether other regions of the HCV genome were as flexible. In particular, we were interested in examining the importance of the junction between NS2 and NS3, since in the case of the related Pestivirus, bovine viral diarrhea virus (BVDV), uncleaved NS2-3 has been shown to be essential for infectious virus production (1). To determine whether unprocessed NS2-3 was required for HCV infectivity, this precursor was disrupted by insertion of an EMCV IRES. The resultant bicistronic genome was named NS2-IRES-NS3 (Fig. 5A). Replication of NS2-IRES-NS3 was monitored at various time points posttransfection by qRT-PCR analysis and was found to be moderately impaired relative to J6/JFH (Fig. 5B). Determination of infectious virus titers by limiting dilution assay indicated that NS2-IRES-NS3 was competent for infectious virus production (Fig. 5C). The reduction in titers relative to J6/JFH, particularly at early time points, was consistent with the decreased replication of NS2-IRES-NS3. These data indicated that unprocessed NS2-3 was not required for HCV infectivity.
FIG. 5.
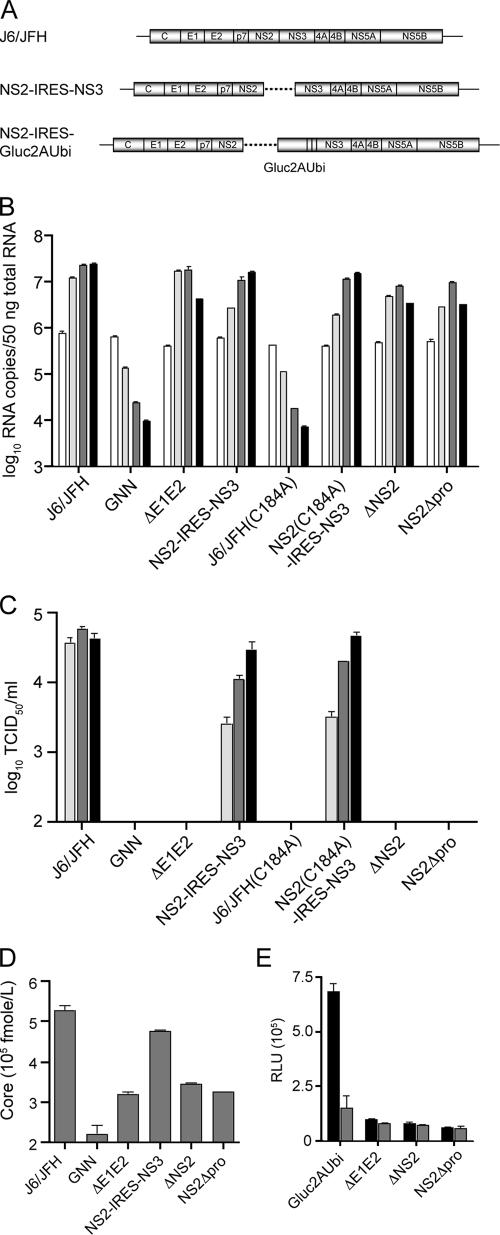
NS2, but not NS2-3, is required for infectious virus production. (A) Schematic representation of full-length monocistronic J6/JFH and bicistronic NS2-IRES-NS3 genomes. A dotted line indicates location of an EMCV IRES in NS2-IRES-NS3. (B) RNA replication of mono- and bicistronic genomes by qRT-PCR at 8, 24, 48, and 72 h posttransfection (white to black bars, respectively). HCV RNA copies normalized to 50 ng of total RNA. (C) Infectious virus production of mono- and bicistronic genomes as determined by TCID50 assay at 24, 48, and 72 h posttransfection (light gray to black bars, respectively). (D) HCV core release of mono- and bicistronic genomes at 72 h posttransfection. GNN, J6/JFH(GNN); ΔE1E2, J6/JFHΔE1E2; ΔNS2, ΔNS2-IRES-NS3; NS2Δpro, NS2Δpro-IRES-NS3. (E) Intracellular infectious virus accumulation of reporter bicistronic genomes at 72 h posttransfection. Lysates generated by multiple rounds of freeze-thaw were combined with isotype control antibody, α-IgGI (▪), or the HCV neutralizing antibody, α-CD81 (□), during infection of naive Huh-7.5 cells. The luciferase activity was determined at 48 h postinfection. Gluc2AUbi, NS2-IRES-Gluc2AUbi; ΔE1E2, NS2-IRES-Gluc2AUbiΔE1E2; ΔNS2, ΔNS2-IRES-Gluc2AUbi; NS2Δpro, NS2Δpro-IRES-Gluc2AUbi. The means and SEM of duplicate experiments are shown.
To determine the importance of NS2 itself in the viral life cycle, bicistronic genomes encoding either a complete deletion of the protein (ΔNS2-IRES-NS3) or a partial deletion encompassing the C-terminal protease domain of NS2 (NS2Δpro-IRES-NS3) were constructed. Replication of each construct was monitored at 8, 24, 48, and 72 h postelectroporation by qRT-PCR analysis. Similar to NS2-IRES-NS3, replication of each bicistronic genome was moderately decreased relative to the parental monocistronic J6/JFH (Fig. 5B). These results were consistent with previous findings that NS2 is not essential for RNA replication. Infectious virus production was determined by limiting dilution assay of clarified cell culture supernatants harvested at 24, 48, and 72 h postelectroporation. Neither ΔNS2-IRES-NS3 nor NS2Δpro-IRES-NS3 produced detectable levels of infectious virus, indicating that even when separated from NS3, at least the protease domain of NS2 was essential for HCV infectivity.
To examine the importance of the NS2-3 protease activity, the active-site cysteine of the NS2 protease was mutated to alanine (C184A) in the context of both mono- and bicistronic genomes. Mutation of the NS2 protease active site within the monocistronic, J6/JFH(C184A), but not the bicistronic genome, NS2(C184A)-IRES-NS3, drastically impaired RNA replication (Fig. 5B). These results are consistent with the essential role of NS2-3 cleavage for RNA replication (18, 35). Analysis of infectious virus production at various time points posttransfection indicated that NS2(C184A)-IRES-NS3 produced infectious titers similar to those of NS2-IRES-NS3. These data suggest that the catalytic activity of the postcleavage form of NS2 was not required for infectious virus production. Taken together, these results indicate that NS2, but not its uncleaved precursor, is essential for infectious virus production and that the NS2 protease domain, but not its catalytic activity, is important for this function.
NS2 acts at an early stage of virion morphogenesis.
To further define the role of NS2 in HCV infectivity, we investigated whether disruption of this protein resulted in noninfectious particle production. Similar amounts of core protein release into cell culture supernatants were seen for J6/JFHΔE1E2, ΔNS2-IRES-NS3, and NS2Δpro-IRES-NS3 at 72 h postelectroporation, suggesting that deletion of NS2 did not result in the production of noninfectious particles and that these defective genomes exhibited a general block in virion release (Fig. 5D).
We next investigated whether deletion of NS2 affected the accumulation of infectious intracellular virus. To facilitate this experiment, we constructed a reporter derivative of NS2-IRES-NS3, in which Gaussialuciferase (Gluc) was inserted at the beginning of the second cistron, immediately upstream of NS3. To ensure proper processing, FMDV 2A and a ubiquitin (Ubi) monomer were included downstream of Gluc. The resulting bicistronic reporter genome was named NS2-IRES-Gluc2AUbi (Fig. 5A). Comparison of NS2-IRES-NS3 and NS2-IRES-Gluc2AUbi by limiting dilution assay indicated that the two genomes produced similar levels of infectious virus at 48 h postelectroporation (data not shown). Similarly, replication and infectious virus production of the NS2 deletion mutants in the context of the reporter genome were consistent with results from the nonreporter background (data not shown).
To examine the accumulation of intracellular infectious virus, cells transfected with NS2-IRES-Gluc2AUbi genomes were washed and then lysed by multiple rounds of freeze-thawing. Cells inoculated with the resulting lysates were assayed for luciferase activity after 48 h. Infection of naive cells with lysates derived from NS2-IRES-Gluc2AUbi resulted in transduced luciferase activity that was neutralized in the presence α-CD81, but not α-IgG1 antibodies, thus confirming the release of authentic viral particles (Fig. 5E). In contrast to NS2-IRES-Gluc2AUbi but similar to NS2-IRES-Gluc2AUbiΔE1E2, neither ΔNS2-IRES-nsGluc2AUbi nor NS2Δpro-IRES-Gluc2AUbi derived lysates transduced significant luciferase activity, indicating an absence of intracellular infectivity (Fig. 5E). These results indicated that NS2 functions at an early stage of virus morphogenesis and that at least the protease domain is an essential determinant of this activity.
DISCUSSION
The recent development of a full infectious system has allowed the complete life cycle of HCV to be studied for the first time and the roles of proteins not required for RNA replication to be assigned. p7 and NS2 are two NS proteins that are dispensable for RNA replication and whose roles are not fully understood. Here we carried out a deletion and mutation analysis of p7 and NS2 and report that these proteins perform essential roles in the production of infectious virions in the HCVcc system.
The small, hydrophobic nature of p7 prompted its classification as a viroporin, a class of virally encoded proteins that includes influenza virus M2 and which are generally thought to be involved in various stages of virus assembly (see reference 9 for a review). A common characteristic of these proteins is their ability to self-organize in lipid membranes to form ion conducting channels. Several laboratories have demonstrated this property of p7, suggesting that, like M2, one of its functions may be to protect glycoproteins from undergoing rearrangements during virus egress by modulating the pH of acidic intracellular membrane compartments (12, 28, 29). Here we report that, consistent with this hypothesis, p7 does function in infectious virus production. In addition, mutation of two basic residues in the cytoplasmic loop of the protein, previously shown to be required for ion channel activity (13), severely impaired infectivity.Mutation of hydrophilic residues predicted to line the interior of the channel did not have as drastic impact on infectivity, although the effects of these mutations on ion channel activity in vitro are not known.
Although p7 was found to be essential for the production of infectious virus, our data are not entirely consistent with a role for p7 solely as an ion channel involved in egress. The production of extracellular infectious virus has recently been shown to be preceded by the accumulation of intracellular infectious virus, likely transiting the secretory pathway (8). If p7 is functioning solely as an ion channel to protect glycoproteins from inactivation late in egress, infectious intracellular virus that had not yet been inactivated would be predicted to be liberated by freeze-thawing. We did not detect such virus, suggesting that if p7 does play a role in egress, it likely also acts early in morphogenesis prior to infectious particle production. Unfortunately, details regarding infectious virus production for HCV are not well understood, and it is possible that the assembly of infectious virions and their subsequent egress to late-stage compartments may occur with sufficiently rapid kinetics that, in the absence of p7, intracellular infectious virions are promptly inactivated and therefore undetectable. Recent studies of HCV entry, however, provide further evidence that ion channel activity may not be the primary function of p7. By analogy with other members of the Flaviviridae, it had been presumed that virions undergo a maturation step during egress, prior to release, in which particles become acid sensitive and as a consequence primed for receptor-mediated endocytosis. Recent evidence, however, suggests that HCV virions are not primed but rather are acid resistant until triggered by unknown factors during entry (33). Although it is not known whether intracellular virions are also acid resistant, this finding might obviate the need for the protective ion channel activity of p7 during egress.
Another possibility is that the ion channel activity of p7 may be required during virus entry. In this model, virion incorporated p7 would facilitate virion disassembly by acidification of the particle during uptake, similar to the role of M2 protein in influenza virus. Although p7 does not appear to be a major component of the virion, it has been suggested that E2-p7 is capable of incorporation into virus-like particles in insect cells (15). However, if p7 does play an essential role as a component of the HCV virion, its incorporation is not dependent on E2-p7, since we found this unprocessed form of p7 was dispensable for infectious virus production. Moreover, in the absence of p7, virus structural protein release was not significantly different from an assembly defective control, suggesting that the observed loss of infectivity was not due to aberrant production of noninfectious virions. Thus, it is unlikely for p7 to a have a role solely in virus entry.
We therefore hypothesize that p7 acts early in infectious virus particle assembly rather than in late egress or entry. In addition, it is possible that mutations within the basic cytoplasmic loop may affect other aspects of p7 that are essential to its function including membrane topology, localization, and/or interactions with other viral proteins. Accumulating evidence suggests that p7 likely interacts with other proteins important for infectivity. We have reported that compensatory mutations in p7 overcome core protein mutants that are defective early in infectious virus assembly (C. L. Murray, C. T. Jones, J. Tassello, and C. M. Rice, submitted for publication). In addition, growing evidence suggests a genetic interaction between p7 and NS2 (30, 38).
Here we report that NS2 is also required for infectious virus production and, similar to p7, acts early in virion morphogenesis prior to the accumulation of infectious intracellular virus. NS2, together with NS3, forms the NS2-3 protease, which is responsible for a single cleavage of the HCV polyprotein at the C terminus of NS2 (10). The recently determined structure of the protease domain of NS2 suggests that, once cleaved, retention of the C terminus of NS2 within its active-site substrate-binding pocket might render the enzyme inactive (23). Indeed, we found that once removed from NS3, the catalytic activity of the postcleavage form of NS2 does not appear to play an essential role in either RNA replication or infectious virus production.
Recent reports of intergenotypic chimeras have suggested genetic interactions between the N-terminal transmembrane domain of NS2 and upstream sequences, while the protease domain required compatibility with other NS proteins (30). This indicates that the NS2 protease domain may form important interactions with other NS proteins during the process of virion assembly. In BVDV, it has been shown that uncleaved NS2-3 is essential for infectious virus production (1). In HCVcc we observed that preventing the production of uncleaved NS2-3 using the bicistronic genome NS2-IRES-NS3, resulted in only a moderate impairment of infectious virus production. It is still possible that once cleaved, NS2 and NS3 may associate to form a complex with a function analogous to that of unprocessed NS2-3 in BVDV. Indeed, a physical interaction between NS2 and NS3 has been reported (17).
Similar to uncleaved NS2-3, uncleaved forms of p7 were found to be dispensable for infectious virus production in HCVcc. This observation is consistent with previous work in which a BVDV bicistronic genome lacking unprocessed E2-p7 was fully infectious; p7-NS2 was not investigated in the Pestivirus system (14). These results suggest that, although the unusual processing phenotype of the E2-p7-NS2 region appears to be conserved between the Hepacivirus and Pestivirus genera, its regulation does not appear to play an essential role in infectious virus production in vitro. It should be noted, however, that similar genotype 1a bicistronic genomes were reported to be noninfectious in a chimpanzee study (31). This discrepancy may be explained by our observation that bicistronic genomes were moderately impaired in RNA replication and infectious virus production, which may result in an inability to reach a threshold of infectivity in vivo. Moreover, significant differences in RNA replication efficiencies between genotype 2a JFH-1-derived genomes, such as J6/JFH, and those of genotype 1 origin, likely affect the tolerance of these genomes to genetic manipulation. Alternatively, it is possible that the importance of unprocessed forms of p7 may be genotype specific. Indeed, differences in the processing efficiency of the E2-p7 region between genotype 1a and 1b sequences suggests that at least the regulation of processing exhibits genotype dependence (15, 19). Finally, it is possible that unprocessed forms of p7 may play functional roles in vivo that are dispensable in vitro.
Interestingly, the observation that unprocessed intermediates of p7 were not essential for infectivity indicated that both p7 and NS2 proteins did not require N-terminal signal sequences for their functions in this respect. Initial studies of p7 membrane topology have suggested a model in which the N and C termini are oriented toward the ER lumen (4). Two recent studies, however, propose that p7 may adopt a second membrane topology in which its C terminus is cytoplasmically oriented, suggesting that the topology of p7 may be somewhat dynamic (11, 15). More importantly, the localization of p7 to intracellular membranes is independent of an N-terminal signal sequence, suggesting the existence of internal targeting signals (11). Similarly, in vitro studies of NS2 found that its N-terminal signal sequence was not required for membrane association, suggesting that NS2 also contains internal signals that mediate its proper targeting and subsequent insertion into membranes (37). Moreover, it is possible that the N-terminal transmembrane domains of NS2 may adopt multiple membrane topologies, as suggested above for p7, which have distinct functions in the virus life cycle. Indeed, this idea has been suggested for other HCV proteins including E1, E2 and NS4B (6, 24, 26). It will be of great interest to determine not only the membrane topology of p7 and NS2 in HCVcc but also how the dynamics of these topologies may affect the function of these proteins in the viral life cycle.
In conclusion, we have demonstrated that the p7 and NS2 proteins are essential for infectious virus production in HCVcc. We suggest that these proteins likely act in concert at an early stage in virion morphogenesis. Their identification in this regard solidifies these proteins as bona fide targets for novel antiviral therapies.
Acknowledgments
We thank Cristian Cruz, Patricia Holst, Michelle Hunter, Erica Machlin, Michelle Moh, Maryline Panis, Merna Torres, Hana You, and Anesta Webson for laboratory support and technical assistance. We are grateful to Matthew Evans, Ivo Lorenz, and Margaret MacDonald for helpful discussions and for critical reading of the manuscript. We thank Brett Lindenbach for providing the plasmid J6/JFH(GNN).
This study was supported by The Greenberg Medical Research Institute and the Gates Foundation (FNIH/GCGH 6-37980-G03-07).
Footnotes
Published ahead of print on 30 May 2007.
REFERENCES
- 1.Agapov, E. V., C. L. Murray, I. Frolov, L. Qu, T. M. Myers, and C. M. Rice. 2004. Uncleaved NS2-3 is required for production of infectious bovine viral diarrhea virus. J. Virol. 78:2414-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for hepatitis C virus genomic and subgenomic RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrere-Kremer, S., C. Montpellier, L. Lorenzo, B. Brulin, L. Cocquerel, S. Belouzard, F. Penin, and J. Dubuisson. 2004. Regulation of hepatitis C virus polyprotein processing by signal peptidase involves structural determinants at the p7 sequence junctions. J. Biol. Chem. 279:41384-41392. [DOI] [PubMed] [Google Scholar]
- 4.Carrere-Kremer, S., C. Montpellier-Pala, L. Cocquerel, C. Wychowski, F. Penin, and J. Dubuisson. 2002. Subcellular localization and topology of the p7 polypeptide of hepatitis C virus. J. Virol. 76:3720-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke, D., S. Griffin, L. Beales, C. S. Gelais, S. Burgess, M. Harris, and D. Rowlands. 2006. Evidence for the formation of a heptameric ion channel complex by the hepatitis C virus p7 protein in vitro. J. Biol. Chem. 281:37057-37068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cocquerel, L., A. Op de Beeck, M. Lambot, J. Roussel, D. Delgrange, A. Pillez, C. Wychowski, F. Penin, and J. Dubuisson. 2002. Topological changes in the transmembrane domains of hepatitis C virus envelope glycoproteins. EMBO J. 21:2893-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Felipe, P., L. E. Hughes, M. D. Ryan, and J. D. Brown. 2003. Co-translational, intraribosomal cleavage of polypeptides by the foot-and-mouth disease virus 2A peptide. J. Biol. Chem. 278:11441-11448. [DOI] [PubMed] [Google Scholar]
- 8.Gastaminza, P., S. B. Kapadia, and F. V. Chisari. 2006. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J. Virol. 80:11074-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez, M. E., and L. Carrasco. 2003. Viroporins. FEBS Lett. 552:28-34. [DOI] [PubMed] [Google Scholar]
- 10.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. A second hepatitis C virus-encoded proteinase. Proc. Natl. Acad. Sci. USA 90:10583-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin, S., D. Clarke, C. McCormick, D. Rowlands, and M. Harris. 2005. Signal peptide cleavage and internal targeting signals direct the hepatitis C virus p7 protein to distinct intracellular membranes. J. Virol. 79:15525-15536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffin, S. D., L. P. Beales, D. S. Clarke, O. Worsfold, S. D. Evans, J. Jaeger, M. P. Harris, and D. J. Rowlands. 2003. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, amantadine. FEBS Lett. 535:34-38. [DOI] [PubMed] [Google Scholar]
- 13.Griffin, S. D., R. Harvey, D. S. Clarke, W. S. Barclay, M. Harris, and D. J. Rowlands. 2004. A conserved basic loop in hepatitis C virus p7 protein is required for amantadine-sensitive ion channel activity in mammalian cells but is dispensable for localization to mitochondria. J. Gen. Virol. 85:451-461. [DOI] [PubMed] [Google Scholar]
- 14.Harada, T., N. Tautz, and H. J. Thiel. 2000. E2-p7 region of the bovine viral diarrhea virus polyprotein: processing and functional studies. J. Virol. 74:9498-9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isherwood, B. J., and A. H. Patel. 2005. Analysis of the processing and transmembrane topology of the E2p7 protein of hepatitis C virus. J. Gen. Virol. 86:667-676. [DOI] [PubMed] [Google Scholar]
- 16.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 17.Kiiver, K., A. Merits, M. Ustav, and E. Zusinaite. 2006. Complex formation between hepatitis C virus NS2 and NS3 proteins. Virus Res. 117:264-272. [DOI] [PubMed] [Google Scholar]
- 18.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin, C., B. D. Lindenbach, B. M. Pragai, D. W. McCourt, and C. M. Rice. 1994. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J. Virol. 68:5063-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 21.Lindenbach, B. D., H. J. Thiel, and C. M. Rice. 2007. Flaviviridae: the viruses and their replication, p. 1101-1152. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 1. Lippincott-Raven Publishers, Philadelphia, PA. [Google Scholar]
- 22.Lohmann, V., F. Korner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 23.Lorenz, I. C., J. Marcotrigiano, T. G. Dentzer, and C. M. Rice. 2006. Structure of the catalytic domain of the hepatitis C virus NS2-3 protease. Nature 442:831-835. [DOI] [PubMed] [Google Scholar]
- 24.Lundin, M., M. Monne, A. Widell, G. Von Heijne, and M. A. Persson. 2003. Topology of the membrane-associated hepatitis C virus protein NS4B. J. Virol. 77:5428-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizushima, H., H. Hijikata, S.-I. Asabe, M. Hirota, K. Kimura, and K. Shimotohno. 1994. Two hepatitis C virus glycoprotein E2 products with different C termini. J. Virol. 68:6215-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakai, K., T. Okamoto, T. Kimura-Someya, K. Ishii, C. K. Lim, H. Tani, E. Matsuo, T. Abe, Y. Mori, T. Suzuki, T. Miyamura, J. H. Nunberg, K. Moriishi, and Y. Matsuura. 2006. Oligomerization of hepatitis C virus core protein is crucial for interaction with the cytoplasmic domain of E1 envelope protein. J. Virol. 80:11265-11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patargias, G., N. Zitzmann, R. Dwek, and W. B. Fischer. 2006. Protein-protein interactions: modeling the hepatitis C virus ion channel p7. J. Med. Chem. 49:648-655. [DOI] [PubMed] [Google Scholar]
- 28.Pavlovic, D., W. Fischer, M. Hussey, D. Durantel, S. Durantel, N. Branza-Nichita, S. Woodhouse, R. A. Dwek, and N. Zitzmann. 2005. Long alkyl chain iminosugars block the HCV p7 ion channel. Adv. Exp. Med. Biol. 564:3-4. [DOI] [PubMed] [Google Scholar]
- 29.Pavlovic, D., D. C. Neville, O. Argaud, B. Blumberg, R. A. Dwek, W. B. Fischer, and N. Zitzmann. 2003. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc. Natl. Acad. Sci. USA 100:6104-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pietschmann, T., A. Kaul, G. Koutsoudakis, A. Shavinskaya, S. Kallis, E. Steinmann, K. Abid, F. Negro, M. Dreux, F. L. Cosset, and R. Bartenschlager. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 103:7408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakai, A., M. S. Claire, K. Faulk, S. Govindarajan, S. U. Emerson, R. H. Purcell, and J. Bukh. 2003. The p7 polypeptide of hepatitis C virus is critical for infectivity and contains functionally important genotype-specific sequences. Proc. Natl. Acad. Sci. USA 100:11646-11651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tannous, B. A., D. E. Kim, J. L. Fernandez, R. Weissleder, and X. O. Breakefield. 2005. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol. Ther. 11:435-443. [DOI] [PubMed] [Google Scholar]
- 33.Tscherne, D. M., C. T. Jones, M. J. Evans, B. D. Lindenbach, J. A. McKeating, and C. M. Rice. 2006. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 80:1734-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welbourn, S., R. Green, I. Gamache, S. Dandache, V. Lohmann, R. Bartenschlager, K. Meerovitch, and A. Pause. 2005. Hepatitis C virus NS2/3 processing is required for NS3 stability and viral RNA replication. J. Biol. Chem. 280:29604-29611. [DOI] [PubMed] [Google Scholar]
- 36.WHO. 1997. Hepatitis C: global prevalence. Wkly. Epidemiol. Rec. 72:341-344. [PubMed] [Google Scholar]
- 37.Yamaga, A. K., and J. H. Ou. 2002. Membrane topology of the hepatitis C virus NS2 protein. J. Biol. Chem. 277:33228-33234. [DOI] [PubMed] [Google Scholar]
- 38.Yi, M., Y. Ma, J. Yates, and S. M. Lemon. 2007. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J. Virol. 81:629-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yi, M., R. A. Villanueva, D. L. Thomas, T. Wakita, and S. M. Lemon. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. USA 103:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]