Abstract
Certain major histocompatibility complex (MHC) class I alleles are associated with the control of human immunodeficiency virus and simian immunodeficiency virus (SIV) replication. We have designed sequence-specific primers for detection of the rhesus macaque MHC class I allele Mamu-B*08 by PCR and screened a cohort of SIV-infected macaques for this allele. Analysis of 196 SIVmac239-infected Indian rhesus macaques revealed that Mamu-B*08 was significantly overrepresented in elite controllers; 38% of elite controllers were Mamu-B*08 positive compared to 3% of progressors (P = 0.00001). Mamu-B*08 was also associated with a 7.34-fold decrease in chronic phase viremia (P = 0.002). Mamu-B*08-positive macaques may, therefore, provide a good model to understand the correlates of MHC class I allele-associated immune protection and viral containment in human elite controllers.
Certain HLA class I alleles strongly influence whether individuals become slow or rapid progressors after human immunodeficiency virus (HIV) infection (8, 18-20, 28, 30, 32, 39). Long-term nonprogressor/elite controller (EC) cohorts are enriched for HLA-B27 and HLA-B57. Numerous studies have implicated these molecules in the presentation of epitopes that elicit effective HIV-specific CD8+ T-lymphocyte responses (2, 3, 13, 17, 21, 22, 24, 31, 32). Unfortunately, there are many difficulties associated with understanding the basis for these HLA associations in HIV-infected people, including inoculum variability, allelic diversity, and access to acute-phase samples. Studying simian immunodeficiency virus (SIV)-infected rhesus macaques that control viral replication may shed some light on why certain HLA-B27- and HLA-B57-positive humans control HIV infection.
Although certain major histocompatibility complex (MHC) class I alleles, including Mamu-A*01 and Mamu-B*17, are associated with slow disease progression in SIV-infected macaques (33-35, 38, 41, 42), independently, the presence of these alleles is not predictive for disease outcome. In the case of Mamu-B*17, only one-fourth of Mamu-B*17-positive macaques become ECs, controlling SIV replication below 1,000 viral RNA (vRNA) copies/ml of plasma during the chronic phase of SIV infection (41). Even Mamu-B*17-positive macaques containing identical MHC class I haplotypes have widely divergent disease courses (40).
Recently, we defined novel Vif- and Nef-specific CD8+ T-cell responses in three EC macaques. We discovered that these epitopes were recognized by T cells restricted by the MHC class I molecule, Mamu-B*08 (J. T. Loffredo et al., unpublished data). Therefore, we investigated the impact of Mamu-B*08 on SIV disease progression in a cohort of 196 SIVmac239-infected Indian rhesus macaques.
Development of PCR-SSP for Mamu-B*08.
To identify Mamu-B*08-positive animals, rhesus macaques (Macaca mulatta) of Indian descent were genotyped for the MHC class I allele, Mamu-B*08, using PCR amplification with sequence-specific DNA priming (PCR-SSP) methodology as previously described (27, 36). The nucleotide sequences of the primers used for typing Mamu-B*08 were as follows: forward, 5′-CGT GAG GCG GAG CAG GTC-3′; and reverse, 5′-CCA CAG CTC CGA TGA ACA CAG-3′. Because Mamu-B*08 and Mamu-B*03 are strikingly similar in the regions encoding the alpha-1 to alpha-3 domains (6), we designed the reverse primer for the Mamu-B*08 PCR-SSP reaction to target multiple unique polymorphisms present in the exon encoding the transmembrane region of Mamu-B*08 (Fig. 1A and B). Primers were diluted to a working concentration of 1 μM.
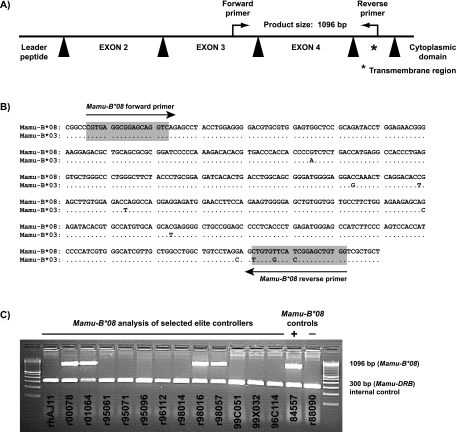
FIG. 1.
Design of PCR-sequence specific primers (PCR-SSP) to amplify the MHC class I allele Mamu-B*08 from genomic DNA. (A) Primers were designed to amplify a 1,096-bp product that contains part of exon 3 (77 bp), intron 3 (573 bp), exon 4 (276 bp), intron 4 (106), and 64 bp of the transmembrane region (exon 5) of Mamu-B*08. Black triangles represent Mamu-B*08 introns. (B) Sequence similarity between Mamu-B*08 and Mamu-B*03. To avoid amplification of Mamu-B*03, the Mamu-B*08 primers were designed such that the forward primer amplifies Mamu-B*08 or Mamu-B*03 and the reverse primer takes advantage of three unique polymorphisms in the transmembrane region (exon 5), allowing for specific detection of Mamu-B*08 only. (C) PCR-SSP genotyping of Mamu-B*08 from genomic DNA of selected elite controller macaques. Macaques 84557 and r88090 were used as Mamu-B*08 positive and negative controls, respectively. Previously, a cDNA library isolated from 84557 indicated that this macaque expresses Mamu-B*08. Mamu-B*08 was not found in a cDNA library from macaque r88090 (6). PCR-SSP primers that amplify a portion of Mamu-DRB (∼300 bp) were included as an internal control to minimize false negatives. A 100-bp DNA ladder was used to interpret amplicon sizes.
Thermal cycling conditions were identical to methods previously published (27), with the exception of the second annealing temperature being 67.9°C for 21 cycles. At the completion of 30 thermal cycles, the PCR underwent a final extension at 72°C for 8 min, followed by a terminal hold at 25°C. Subsequently, PCR products were electrophoresed on 2% agarose gels at a constant voltage in 0.5× sodium borate buffer (7). The corresponding Mamu-B*08 allele-specific amplicon was ∼1,096 bp (Fig. 1C). Each Mamu-B*08 typing reaction also included primers to target an ∼300-bp fragment of the class II Mamu-DRB as an internal control (23). MHC class I typing reactions were not considered valid without the presence of this internal control amplification product. Amplicon length was measured relative to a 100-bp DNA ladder (Invitrogen, Carlsbad, CA). Amplification specificity of the Mamu-B*08 typing primers was confirmed by using previously established sequencing methods (18a) in 44 Mamu-B*08-positive Indian rhesus macaques.
The remaining MHC class I alleles (Mamu-A*01, -A*02, -A*08, -A*11, -B*01, -B*03, -B*04, -B*17, and -B*29) were typed as previously described (18a, 27).
Colony frequencies of Mamu-B*08.
After completion of PCR-SSP development, we screened eight different Indian rhesus macaque colonies to determine the frequency of Mamu-B*08 (Table 1). After testing >2,900 macaques, we found that the overall frequency of Mamu-B*08 was ∼5.8%. At the Wisconsin National Primate Research Center (WNPRC), the frequency of Mamu-B*08 was 6.8% (1,271 macaques tested). Overall, the frequency ranged from 0.9 to 14.1% across the various macaque colonies.
TABLE 1.
Frequency of Mamu-B*08 in different Indian rhesus macaque colonies
| Colonya | No. of Mamu-B*08+ animals | Total no. of animals | Frequency (%) |
|---|---|---|---|
| Southern Research Institute (Birmingham, AL) | 1 | 113 | 0.9 |
| Yerkes NPRC | 3 | 141 | 2.1 |
| Various NIH/NCI-funded projects | 8 | 380 | 2.1 |
| Tulane NPRC | 3 | 122 | 2.5 |
| Caribbean Primate Research Center | 18 | 451 | 4.0 |
| California NPRC | 11 | 163 | 6.7 |
| Wisconsin NPRC | 86 | 1271 | 6.8 |
| Oregon NPRC | 41 | 290 | 14.1 |
| Total | 171 | 2,931 | 5.8 |
NPRC, National Primate Research Center; NIH/NCI, National Institutes of Health and National Cancer Institute.
Mamu-B*08 is significantly enriched in a cohort of elite controller macaques and is associated with lower viremia in the chronic phase of SIV infection.
We next investigated the frequency of Mamu-B*08 in a cohort of previously identified EC macaques (41). Four of the thirteen ECs expressed Mamu-B*08 (Fig. 1C). Interestingly, three of the four Mamu-B*17-negative ECs, r00078, r01064, and r98057, were Mamu-B*08 positive. After testing a cohort of 192 SIV-infected macaques for Mamu-B*08, we identified seven Mamu-B*08-positive macaques, four of which were from this initial EC cohort.
To investigate the role of Mamu-B*08 in control of pathogenic SIV replication, we recently infected four additional Mamu-B*08-positive Indian rhesus macaques with SIVmac239 (100 50% tissue culture infective doses, intravenously). Two of these Mamu-B*08-positive macaques, r00032 and r02019, had viral loads below 1,000 vRNA copies/ml at 22 weeks postinfection. We have included them in Table 2 as ECs due to their low plasma viremia early into the chronic phase of SIV infection. All four recently infected Mamu-B*08-positive macaques were included in the statistical analysis.
TABLE 2.
Mamu-B*08 is significantly overrepresented among elite controller macaquesa
| Animal | Presence (+) or absence (−) of MHC class I allele or value for parameter
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A*01 | A*02 | A*08 | A*11 | B*01 | B*03 | B*04 | B*08 | B*17 | B*29 | |
| rhAJ11 | - | + | - | + | - | - | - | - | + | + |
| r00078 | - | - | + | - | - | - | - | + | - | + |
| r01064 | - | + | - | - | - | - | - | + | - | - |
| r95061b | + | + | - | - | - | - | - | - | + | + |
| r95071 | - | + | - | - | - | - | - | - | + | + |
| r95096c | + | - | - | + | - | - | - | - | + | + |
| r96112d | - | - | + | - | - | - | - | - | + | + |
| r98014e | - | + | - | + | - | - | - | - | + | + |
| r98016 | - | + | - | - | - | - | - | + | + | + |
| r98057 | - | - | - | + | - | - | - | + | - | - |
| 99C051 | - | + | - | - | - | - | - | - | + | + |
| 99x032f | - | + | + | - | - | - | - | - | - | - |
| 96C114g | + | - | - | - | - | - | - | - | + | + |
| C59Zh | - | - | + | - | - | + | + | - | - | - |
| r00032 | - | + | - | - | - | - | - | + | - | - |
| r02019 | - | - | + | - | - | - | - | + | - | - |
| Frequency in elite controller cohort (%) (n = 16)i | 19 | 56 | 31 | 25 | 0 | 6 | 6 | 38 | 56 | 63 |
| Frequency in entire cohort (%) (n = 196) | 38 | 27 | 26 | 7 | 33 | 2 | 2 | 6 | 21 | 22 |
Sixteen SIVmac239-infected Indian rhesus macaques that maintained chronic phase (≥10 weeks) plasma virus concentrations of <1,000 vRNA copies/ml were identified as ECs. This 1,000 vRNA copies/ml EC threshold represents a >2-log reduction in chronic-phase plasma virus concentrations compared to the chronic-phase geometric mean of the entire cohort of 196 SIV-infected macaques (223,800 vRNA copies/ml). Viral control was maintained for >1 year during the chronic phase of SIVmac239 infection or from week 10 until the animal was assigned to another study. All ECs were infected for >39 weeks (9 months) with the exception of r96112 (38 weeks), along with r00032 and r02019 (22 weeks each in an ongoing study). Animals were vaccine naive except where indicated.
DNA/MVA Gag181-189CM9 epitope only. The vaccine results were previously published (1).
Lipopeptide Gag181-189CM9 epitope only (35).
CpG in incomplete Freund adjuvant (41).
Intramuscular injections of 107 irradiated PBMC from SIV-naive animal rh2055 (41).
Whole inactivated SIV virions. The vaccine results were previously published (25).
CpG in PBS (J. Lifson, unpublished results).
Significant enrichments of the elite controller cohort for expression of a particular MHC class I allele are indicated in bold. Enrichment of this elite controller group for MHC class I alleles was determined with PROC LOGISTIC in SAS 9.1 (SAS Institute, Cary, NC). Mamu-A*02, P = 0.01; Mamu-A*11, P = 0.01; Mamu-B*08, P = 0.00001; Mamu-B*17, P = 0.001; Mamu-B*29, P = 0.0003.
PROC LOGISTIC regression analysis was performed on 196 SIV-infected Indian rhesus macaques from two independent cohorts (143 animals from D. Watkins' studies and 53 animals from J. Lifson's studies) to determine whether the frequency of Mamu-B*08 was enriched in EC macaques. The analysis methods were performed as previously described (41), except the program used was SAS 9.1 (SAS Institute, Cary, NC). All macaques were infected with the molecular clone SIVmac239, and plasma virus concentrations were monitored as previously described (10, 26) at multiple time points for a minimum of 10 weeks postinfection. Although vaccinated animals are included in the 196 animal cohort, none of these macaques received a vaccine regimen that demonstrated effective and durable control of SIV replication.
Including the two recently infected Mamu-B*08-positive EC animals, 16 SIVmac239-infected macaques were identified as ECs, defined as maintaining a chronic phase (≥10 weeks) plasma viremia geometric mean below 1,000 vRNA copies/ml (Table 2). Mamu-B*08 was dramatically enriched in this EC group. Thirty-eight percent of this EC cohort expressed Mamu-B*08 compared to just three percent of the 180 SIV-infected macaques that did not control viral replication to the EC threshold (P = 0.00001). Overall, 6 of 11 (∼55%) macaques identified as Mamu-B*08-positive became ECs. Two of the eleven Mamu-B*08-positive macaques also displayed what we refer to as controller status, limiting viral replication to <22,000 viral RNA copies/ml (one log reduction of the cohort's chronic phase geometric mean, 223,800 vRNA copies/ml) for at least 10 weeks in the chronic stage of SIV infection. Hence, a majority of Mamu-B*08-positive macaques (8 of 11 [73%]) displayed better-than-typical viral control of pathogenic SIVmac239 replication.
Animal C59Z, a Mamu-B*03-positive macaque, was also recently identified as 1 of the 16 EC macaques (Table 2). Although Mamu-B*03 was previously associated with slow SIV disease progression (12), it has been difficult to conduct additional studies with Mamu-B*03 due to the low frequency of this allele (overall, <1%; 18a). Interestingly, a previous study demonstrated that Mamu-B*03 and HLA-B27 bind peptides with similar motifs (11). Mamu-B*03 and Mamu-B*08 are almost identical in amino acid sequence (6). There are only two amino acid differences between Mamu-B*03 and Mamu-B*08 in regions that influence peptide binding and antigen recognition (5, 16). Both differences reside in the alpha-1 domain (exon two); the alpha-2 domains (exon 3) of Mamu-B*03 and Mamu-B*08 are identical. Therefore, it is likely that Mamu-B*08 also shares this HLA-B27 binding profile.
We analyzed the association between MHC class I alleles in both acute- and chronic-phase viremia for each MHC class I allele in the 196 SIVmac239-infected macaque cohort. With the exception of the program used (SAS 9.1; SAS Institute, Cary, NC), the analysis methods were performed as previously described (41). PROC REG was used to estimate the relative log geometric mean acute phase peak (single highest value from weeks 1 to 3 for each macaque) plasma virus concentration. PROC MIXED was used to estimate the relative log geometric mean for chronic-phase (≥10 weeks) plasma viremia. All typed MHC class I alleles, EC status, and vaccine status were included as covariates in the models.
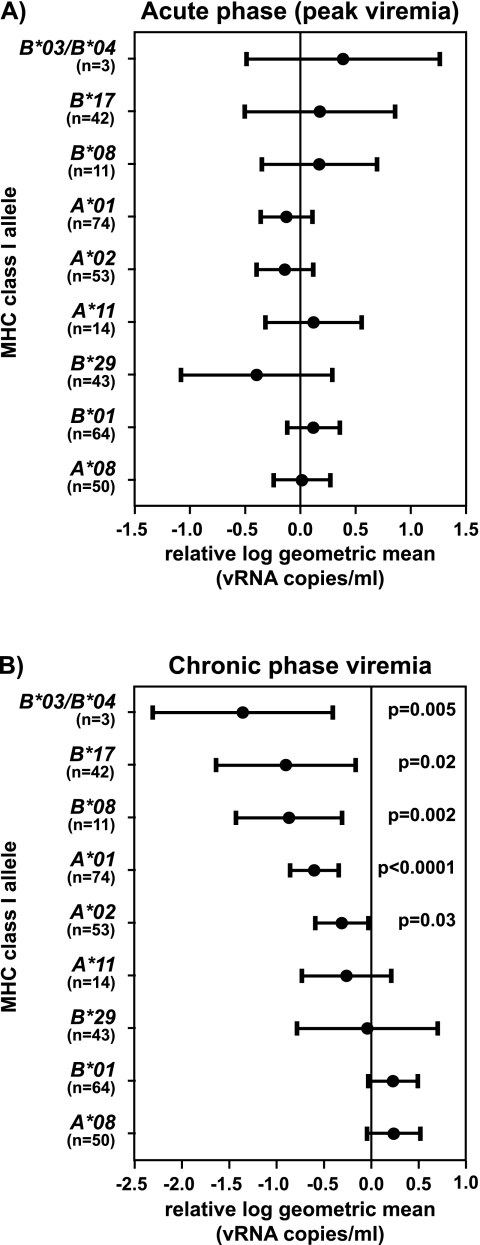
Although none of the MHC class I alleles correlated with changes in peak viremia during the acute phase of SIV infection (Fig. 2A), several MHC class I alleles affected chronic phase viremia (Fig. 2B). The 11 Mamu-B*08-positive macaques had a 7.34-fold decrease in geometric mean chronic phase plasma virus concentrations compared to the rest of the cohort (P = 0.002). The reduction in the relative log geometric mean of Mamu-B*08-positive macaques was similar to that of macaques expressing Mamu-B*17 (7.93-fold), an allele previously associated with slow disease progression (35, 41).
FIG. 2.
Mamu-B*08 influences chronic-phase plasma virus concentrations in SIVmac239-infected Indian rhesus macaques. (A) Acute-phase peak viremia (highest plasma virus concentration between weeks 1 and 3 postinfection) is not affected by the presence of the typed MHC class I alleles in 196 SIVmac239-infected Indian rhesus macaques. (B) Contribution of typed MHC class I alleles to variation in chronic-phase plasma viremia in the same cohort of 196 macaques. The chronic-phase geometric mean was calculated, and the plasma virus concentration time points ranged from ≥10 weeks to <7.16 years postchallenge. MHC class I effects on SIV plasma viremia are shown with the changes in geometric mean displayed as dots for each MHC class I allele (Mamu-A*01, -A*02, -A*08, -A*11, -B*01, -B*03/-B*04, -B*08, -B*17, and -B*29) relative to the whole cohort and 95% confidence intervals (bars). Probabilities (P values) for MHC class I alleles with statistically significant reductions from the relative log geometric mean are displayed. Mamu-B*03 and -B*04 were analyzed together because the three Mamu-B*03-positive macaques were also positive for Mamu-B*04.
We identified another Mamu-B*08-positive EC macaque (r99006) not included in the 196 SIV-infected animal cohort described in the present study. Animal r99006 was part of a separate study in which Mamu-A*01-positive and Mamu-B*17-positive macaques were infected with an engineered CD8+ T-cell escape variant virus (14, 15) to prevent the development of immunodominant CD8+ T-cell responses thought to be associated with viral control (35). Macaque r99006 controlled viremia to <1,000 vRNA copies/ml for almost 4 years after SIV infection before being assigned to another study. We have also identified three additional Mamu-B*08-positive macaques from other investigators (D. Evans and G. Franchini, personal communications). At 25 weeks postinfection, two of the three Mamu-B*08-positive macaques limited replication of SIVmac239 or SIVmac251 isolates at or below ∼1,000 vRNA copies/ml. In total, 11 of 15 (73%) Mamu-B*08-positive macaques controlled replication of pathogenic strains of SIV.
It has been previously reported that Mamu-B*08 segregates on the same chromosome as Mamu-B*06 in rhesus macaques (37). Interestingly, when investigating MHC class I cDNA libraries from several Mamu-B*08-positive Indian rhesus macaques at the WNPRC, we also noticed that all of these macaques possessed Mamu-B*06 transcripts (data not shown). None of the cDNA libraries from Mamu-B*08-negative macaques possessed Mamu-B*06 transcripts. However, control of SIV replication is unlikely due to Mamu-B*06. From our studies, Mamu-B*06 does not appear to be involved in the presentation of SIV epitopes (unpublished data). Mamu-B*08, but not Mamu-B*06, was detected by one-dimensional isoelectric focusing (6). This suggests that Mamu-B*06 may not encode an expressed MHC class I protein.
Mamu-B*08-positive Indian rhesus macaques might provide an insightful model for understanding the influence of particular MHC class I alleles on viral control. We have recently identified several Mamu-B*08-restricted CD8+ T-cell epitopes in the chronic phase of SIV infection (Loffredo et al., unpublished). We are currently determining whether CD8+ immune responses restricted by Mamu-B*08 exert strong patterns of immunodominance similar to those associated with HLA-B27 and HLA-B57 (2, 4, 22). Mamu-B*08-restricted SIV epitopes should be useful for future pathogenesis studies, including the generation of MHC class I tetramers, to study differences in the CD8+ T-cell responses between progressor and EC Mamu-B*08-positive macaques. Finally, given the independent influence of this allele on the outcome of SIV infection, it might be prudent to remove Mamu-B*08-positive macaques from vaccine studies.
Acknowledgments
We thank Debra L. Fisk for MHC class I sequencing assistance and Richard Rudersdorf and Lyle Wallace for the construction of MHC class I cDNA libraries. We are also grateful to Danilo R. Casimiro, David T. Evans, Genoveffa Franchini, Adrian McDermott, Eva Rakasz, Thomas Friedrich, Levi Yant, and Matthew Reynolds for providing plasma virus concentrations and SIV disease status of macaques used in their studies. Sean Spencer, Laura Valentine, and David O'Connor provided helpful discussions. We also thank the virology, genetics, and immunology core laboratories at the National Primate Research Center, University of Wisconsin-Madison (WNPRC) for technical assistance.
This research was supported by a National Institutes of Health (NIH) contract HHSN266200400088C and NIH grants R24 RR015371, R24 RR016038, R01 AI049120, and R01 AI052056 to D.I.W. in addition to P51 RR000167 to the WNPRC. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. This study was conducted in part at a facility constructed with support from Research Facilities Improvement grant numbers RR15459-01 and RR020141-01 (WNPRC).
The content of this publication does not necessarily reflect the views or policies of the U.S. Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print on 30 May 2007.
REFERENCES
- 1.Allen, T. M., P. Jing, B. Calore, H. Horton, D. H. O'Connor, T. Hanke, M. Piekarczyk, R. Ruddersdorf, B. R. Mothe, C. Emerson, N. Wilson, J. D. Lifson, I. M. Belyakov, J. A. Berzofsky, C. Wang, D. B. Allison, D. C. Montefiori, R. C. Desrosiers, S. Wolinsky, K. J. Kunstman, J. D. Altman, A. Sette, A. J. McMichael, and D. I. Watkins. 2002. Effects of cytotoxic T lymphocytes (CTL) directed against a single simian immunodeficiency virus (SIV) Gag CTL epitope on the course of SIVmac239 infection. J. Virol. 76:10507-10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altfeld, M., E. T. Kalife, Y. Qi, H. Streeck, M. Lichterfeld, M. N. Johnston, N. Burgett, M. E. Swartz, A. Yang, G. Alter, X. G. Yu, A. Meier, J. K. Rockstroh, T. M. Allen, H. Jessen, E. S. Rosenberg, M. Carrington, and B. D. Walker. 2006. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8+ T cell response against HIV-1. PLoS Med. 3:e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey, J. R., T. M. Williams, R. F. Siliciano, and J. N. Blankson. 2006. Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J. Exp. Med. 203:1357-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bihl, F., N. Frahm, L. Di Giammarino, J. Sidney, M. John, K. Yusim, T. Woodberry, K. Sango, H. S. Hewitt, L. Henry, C. H. Linde, J. V. R. Chisholm, T. M. Zaman, E. Pae, S. Mallal, B. D. Walker, A. Sette, B. T. Korber, D. Heckerman, and C. Brander. 2006. Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J. Immunol. 176:4094-4101. [DOI] [PubMed] [Google Scholar]
- 5.Bjorkman, P. J., M. A. Saper, B. Samraoui, W. S. Bennett, J. L. Strominger, and D. C. Wiley. 1987. The foreign antigen binding site and T-cell recognition regions of class I histocompatibility antigens. Nature 329:512-518. [DOI] [PubMed] [Google Scholar]
- 6.Boyson, J. E., C. Shufflebotham, L. F. Cadavid, J. A. Urvater, L. A. Knapp, A. L. Hughes, and D. I. Watkins. 1996. The MHC class I genes of the rhesus monkey: different evolutionary histories of MHC class I and II genes in primates. J. Immunol. 156:4656-4665. [PubMed] [Google Scholar]
- 7.Brody, J. R., and S. E. Kern. 2004. Sodium boric acid: a Tris-free, cooler conductive medium for DNA electrophoresis. BioTechniques 36:214-216. [DOI] [PubMed] [Google Scholar]
- 8.Carrington, M., G. W. Nelson, M. P. Martin, T. Kissner, D. Vlahov, J. J. Goedert, R. Kaslow, S. Buchbinder, K. Hoots, and S. J. O'Brien. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283:1748-1752. [DOI] [PubMed] [Google Scholar]
- 9.Casimiro, D. R., F. Wang, W. A. Schleif, X. Liang, Z. Q. Zhang, T. W. Tobery, M. E. Davies, A. B. McDermott, D. H. O'Connor, A. Fridman, A. Bagchi, L. G. Tussey, A. J. Bett, A. C. Finnefrock, T. M. Fu, A. Tang, K. A. Wilson, M. Chen, H. C. Perry, G. J. Heidecker, D. C. Freed, A. Carella, K. S. Punt, K. J. Sykes, L. Huang, V. I. Ausensi, M. Bachinsky, U. Sadasivan-Nair, D. I. Watkins, E. A. Emini, and J. W. Shiver. 2005. Attenuation of simian immunodeficiency virus SIVmac239 infection by prophylactic immunization with DNA and recombinant adenoviral vaccine vectors expressing Gag. J. Virol. 79:15547-15555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cline, A. N., J. W. Bess, M. Piatak, Jr., and J. D. Lifson. 2005. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J. Med. Primatol. 34:303-312. [DOI] [PubMed] [Google Scholar]
- 11.Dzuris, J. L., J. Sidney, E. Appella, R. W. Chesnut, D. I. Watkins, and A. Sette. 2000. Conserved MHC class I peptide binding motif between humans and rhesus macaques. J. Immunol. 164:283-291. [DOI] [PubMed] [Google Scholar]
- 12.Evans, D. T., D. H. O'Connor, P. Jing, J. L. Dzuris, J. Sidney, J. da Silva, T. M. Allen, H. Horton, J. E. Venham, R. A. Rudersdorf, T. Vogel, C. D. Pauza, R. E. Bontrop, R. DeMars, A. Sette, A. L. Hughes, and D. I. Watkins. 1999. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 5:1270-1276. [DOI] [PubMed] [Google Scholar]
- 13.Feeney, M. E., Y. Tang, K. A. Roosevelt, A. J. Leslie, K. McIntosh, N. Karthas, B. D. Walker, and P. J. Goulder. 2004. Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. J. Virol. 78:8927-8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedrich, T. C., E. J. Dodds, L. J. Yant, L. Vojnov, R. Rudersdorf, C. Cullen, D. T. Evans, R. C. Desrosiers, B. R. Mothe, J. Sidney, A. Sette, K. Kunstman, S. Wolinsky, M. Piatak, J. Lifson, A. L. Hughes, N. Wilson, D. H. O'Connor, and D. I. Watkins. 2004. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat. Med. 10:275-281. [DOI] [PubMed] [Google Scholar]
- 15.Friedrich, T. C., A. B. McDermott, M. R. Reynolds, S. Piaskowski, S. Fuenger, I. P. De Souza, R. Rudersdorf, C. Cullen, L. J. Yant, L. Vojnov, J. Stephany, S. Martin, D. H. O'Connor, N. Wilson, and D. I. Watkins. 2004. Consequences of cytotoxic T-lymphocyte escape: common escape mutations in simian immunodeficiency virus are poorly recognized in naive hosts. J. Virol. 78:10064-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrett, T. P., M. A. Saper, P. J. Bjorkman, J. L. Strominger, and D. C. Wiley. 1989. Specificity pockets for the side chains of peptide antigens in HLA-Aw68. Nature 342:692-696. [DOI] [PubMed] [Google Scholar]
- 17.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 18.Hendel, H., S. Caillat-Zucman, H. Lebuanec, M. Carrington, S. O'Brien, J. M. Andrieu, F. Schachter, D. Zagury, J. Rappaport, C. Winkler, G. W. Nelson, and J. F. Zagury. 1999. New class I and II HLA alleles strongly associated with opposite patterns of progression to AIDS. J. Immunol. 162:6942-6946. [PubMed] [Google Scholar]
- 18a.Kaizu, M., G. J. Borchardt, C. E. Glidden, D. L. Fisk, J. T. Loffredo, D. I. Watkins, and W. M. Rehrauer. Molecular typing of major histocompatibility complex class I alleles in the Indian rhesus macaque which restricts SIV CD8+ T cell epitopes. Immunogenetics, in press. [DOI] [PubMed]
- 19.Kaslow, R. A., M. Carrington, R. Apple, L. Park, A. Munoz, A. J. Saah, J. J. Goedert, C. Winkler, S. J. O'Brien, C. Rinaldo, R. Detels, W. Blattner, J. Phair, H. Erlich, and D. L. Mann. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405-411. [DOI] [PubMed] [Google Scholar]
- 20.Kaslow, R. A., C. Rivers, J. Tang, T. J. Bender, P. A. Goepfert, R. El Habib, K. Weinhold, and M. J. Mulligan. 2001. Polymorphisms in HLA class I genes associated with both favorable prognosis of human immunodeficiency virus (HIV) type 1 infection and positive cytotoxic T-lymphocyte responses to ALVAC-HIV recombinant canarypox vaccines. J. Virol. 75:8681-8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiepiela, P., A. J. Leslie, I. Honeyborne, D. Ramduth, C. Thobakgale, S. Chetty, P. Rathnavalu, C. Moore, K. J. Pfafferott, L. Hilton, P. Zimbwa, S. Moore, T. Allen, C. Brander, M. M. Addo, M. Altfeld, I. James, S. Mallal, M. Bunce, L. D. Barber, J. Szinger, C. Day, P. Klenerman, J. Mullins, B. Korber, H. M. Coovadia, B. D. Walker, and P. J. Goulder. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769-775. [DOI] [PubMed] [Google Scholar]
- 23.Knapp, L. A., E. Lehmann, M. S. Piekarczyk, J. A. Urvater, and D. I. Watkins. 1997. A high frequency of Mamu-A*01 in the rhesus macaque detected by polymerase chain reaction with sequence-specific primers and direct sequencing. Tissue Antigens 50:657-661. [DOI] [PubMed] [Google Scholar]
- 24.Leslie, A. J., K. J. Pfafferott, P. Chetty, R. Draenert, M. M. Addo, M. Feeney, Y. Tang, E. C. Holmes, T. Allen, J. G. Prado, M. Altfeld, C. Brander, C. Dixon, D. Ramduth, P. Jeena, S. A. Thomas, A. S. John, T. A. Roach, B. Kupfer, G. Luzzi, A. Edwards, G. Taylor, H. Lyall, G. Tudor-Williams, V. Novelli, J. Martinez-Picado, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282-289. [DOI] [PubMed] [Google Scholar]
- 25.Lifson, J. D., M. J. Piatak, J. L. Rossio, J. J. Bess, E. Chertova, D. Schneider, R. Kiser, V. Coalter, B. Poore, R. Imming, R. C. Desrosiers, L. E. Henderson, and L. O. Arthur. 2002. Whole inactivated SIV virion vaccines with functional envelope glycoproteins: safety, immunogenicity, and activity against intrarectal challenge. J. Med. Primatol. 31:205-216. [DOI] [PubMed] [Google Scholar]
- 26.Loffredo, J. T., B. J. Burwitz, E. G. Rakasz, S. P. Spencer, J. J. Stephany, J. P. Vela, S. R. Martin, J. Reed, S. M. Piaskowski, J. Furlott, K. L. Weisgrau, D. S. Rodrigues, T. Soma, G. Napoe, T. C. Friedrich, N. A. Wilson, E. G. Kallas, and D. I. Watkins. 2007. The antiviral efficacy of simian immunodeficiency virus-specific CD8+ T cells is unrelated to epitope specificity and is abrogated by viral escape. J. Virol. 81:2624-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loffredo, J. T., J. Sidney, S. Piaskowski, A. Szymanski, J. Furlott, R. Rudersdorf, J. Reed, B. Peters, H. D. Hickman-Miller, W. Bardet, W. M. Rehrauer, D. H. O'Connor, N. A. Wilson, W. H. Hildebrand, A. Sette, and D. I. Watkins. 2005. The high frequency Indian rhesus macaque MHC class I molecule, Mamu-B*01, does not appear to be involved in CD8+ T lymphocyte responses to SIVmac239. J. Immunol. 175:5986-5997. [DOI] [PubMed] [Google Scholar]
- 28.Mann, D. L., R. P. Garner, D. E. Dayhoff, K. Cao, M. A. Fernandez-Vina, C. Davis, N. Aronson, N. Ruiz, D. L. Birx, and N. L. Michael. 1998. Major histocompatibility complex genotype is associated with disease progression and virus load levels in a cohort of human immunodeficiency virus type 1-infected Caucasians and African Americans. J. Infect. Dis. 178:1799-1802. [DOI] [PubMed] [Google Scholar]
- 29.McDermott, A. B., D. H. O'Connor, S. Fuenger, S. Piaskowski, S. Martin, J. Loffredo, M. Reynolds, J. Reed, J. Furlott, T. Jacoby, C. Riek, E. Dodds, K. Krebs, M. E. Davies, W. A. Schleif, D. R. Casimiro, J. W. Shiver, and D. I. Watkins. 2005. Cytotoxic T-lymphocyte escape does not always explain the transient control of simian immunodeficiency virus SIVmac239 viremia in adenovirus-boosted and DNA-primed Mamu-A*01-positive rhesus macaques. J. Virol. 79:15556-15566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNeil, A. J., P. L. Yap, S. M. Gore, R. P. Brettle, M. McColl, R. Wyld, S. Davidson, R. Weightman, A. M. Richardson, and J. R. Robertson. 1996. Association of HLA types A1-B8-DR3 and B27 with rapid and slow progression of HIV disease. QJM 89:177-185. [DOI] [PubMed] [Google Scholar]
- 31.Migueles, S. A., A. C. Laborico, W. L. Shupert, M. S. Sabbaghian, R. Rabin, C. W. Hallahan, D. Van Baarle, S. Kostense, F. Miedema, M. McLaughlin, L. Ehler, J. Metcalf, S. Liu, and M. Connors. 2002. HIV-specific CD8+ T-cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 3:1061-1068. [DOI] [PubMed] [Google Scholar]
- 32.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long-term nonprogressors. Proc. Natl. Acad. Sci. USA 97:2709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mothe, B. R., J. Weinfurter, C. Wang, W. Rehrauer, N. Wilson, T. M. Allen, D. B. Allison, and D. I. Watkins. 2003. Expression of the major histocompatibility complex class I molecule Mamu-A*01 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 77:2736-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muhl, T., M. Krawczak, P. Ten Haaft, G. Hunsmann, and U. Sauermann. 2002. MHC class I alleles influence set-point viral load and survival time in simian immunodeficiency virus-infected rhesus monkeys. J. Immunol. 169:3438-3446. [DOI] [PubMed] [Google Scholar]
- 35.O'Connor, D. H., B. R. Mothe, J. T. Weinfurter, S. Fuenger, W. M. Rehrauer, P. Jing, R. R. Rudersdorf, M. E. Liebl, K. Krebs, J. Vasquez, E. Dodds, J. Loffredo, S. Martin, A. B. McDermott, T. M. Allen, C. Wang, G. G. Doxiadis, D. C. Montefiori, A. Hughes, D. R. Burton, D. B. Allison, S. M. Wolinsky, R. Bontrop, L. J. Picker, and D. I. Watkins. 2003. Major histocompatibility complex class I alleles associated with slow simian immunodeficiency virus disease progression bind epitopes recognized by dominant acute-phase cytotoxic-T-lymphocyte responses. J. Virol. 77:9029-9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olerup, O., and H. Zetterquist. 1991. HLA-DRB1*01 subtyping by allele-specific PCR amplification: a sensitive, specific and rapid technique. Tissue Antigens 37:197-204. [DOI] [PubMed] [Google Scholar]
- 37.Otting, N., C. M. Heijmans, R. C. Noort, N. G. de Groot, G. G. Doxiadis, J. J. van Rood, D. I. Watkins, and R. E. Bontrop. 2005. Unparalleled complexity of the MHC class I region in rhesus macaques. Proc. Natl. Acad. Sci. USA 102:1626-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pal, R., D. Venzon, N. L. Letvin, S. Santra, D. C. Montefiori, N. R. Miller, E. Tryniszewska, M. G. Lewis, T. C. VanCott, V. Hirsch, R. Woodward, A. Gibson, M. Grace, E. Dobratz, P. D. Markham, Z. Hel, J. Nacsa, M. Klein, J. Tartaglia, and G. Franchini. 2002. ALVAC-SIV-gag-pol-env-based vaccination and macaque major histocompatibility complex class I (A*01) delay simian immunodeficiency virus SIVmac-induced immunodeficiency. J. Virol. 76:292-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang, J., S. Tang, E. Lobashevsky, A. D. Myracle, U. Fideli, G. Aldrovandi, S. Allen, R. Musonda, and R. A. Kaslow. 2002. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J. Virol. 76:8276-8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wojcechowskyj, J. A., L. J. Yant, R. W. Wiseman, S. L. O'Connor, and D. H. O'Connor. 2007. Control of simian immunodeficiency virus SIVmac239 is not predicted by inheritance of Mamu-B*17-containing haplotypes. J. Virol. 81:406-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yant, L. J., T. C. Friedrich, R. C. Johnson, G. E. May, N. J. Maness, A. M. Enz, J. D. Lifson, D. H. O'Connor, M. Carrington, and D. I. Watkins. 2006. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 80:5074-5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, Z. Q., T. M. Fu, D. R. Casimiro, M. E. Davies, X. Liang, W. A. Schleif, L. Handt, L. Tussey, M. Chen, A. Tang, K. A. Wilson, W. L. Trigona, D. C. Freed, C. Y. Tan, M. Horton, E. A. Emini, and J. W. Shiver. 2002. Mamu-A*01 allele-mediated attenuation of disease progression in simian-human immunodeficiency virus infection. J. Virol. 76:12845-12854. [DOI] [PMC free article] [PubMed] [Google Scholar]