Abstract
An infectious hepatitis C virus (HCV) cDNA clone (JFH1) was generated recently. However, quantitative analysis of HCV infection and observation of infected cells have proved to be difficult because the yield of HCV in cell cultures is fairly low. We generated infectious HCV clones containing the convenient reporters green fluorescent protein (GFP) and Renilla luciferase in the NS5a-coding sequence. The new viruses responded to antiviral agents in a dose-dependent manner. Responses of individual cells containing HCV to alpha interferon (IFN-α) were monitored using GFP-tagged HCV and time-lapse confocal microscopy. Marked variations in the response to IFN-α were observed among HCV-containing cells.
It is estimated that 170 million individuals worldwide and about 1% of the population in developed countries are chronically infected with hepatitis C virus (HCV) (17). Most acute HCV infections become chronic, and some progress to liver cirrhosis or hepatocellular carcinoma (1, 7). A protective vaccine does not yet exist, and therapeutic options are limited. Alpha interferon (IFN-α) in combination with ribavirin is the only recommended therapy (5).
HCV contains a single-stranded, positive-sense RNA genome of approximately 9.6 kb, which encodes a polyprotein that is flanked by nontranslated regions at its 5′ and 3′ ends (2). The polyprotein precursor is co- and posttranslationally processed by cellular and viral proteases to yield the mature structural and nonstructural proteins, which are arranged in the sequence NH2-C-E1-E2-P7-NS2-NS3-NS4a-NS4b-NS5a-NS5b-COOH.
The availability of a cell culture system is a prerequisite to study the proliferation cycle of a virus and to devise strategies for prophylactic and therapeutic interventions (3). The most recent advance is the development of a virus production system that is based on transfection of the human hepatoma cell line Huh 7 with genomic HCV RNA (JFH1) isolated from a patient with fulminant hepatitis (11, 16, 18). This cell culture-based model allows the study of all stages of the HCV life cycle. Recently, two studies demonstrated the mechanism of HCV entry into tissue culture cells using this infection system (9, 15). To facilitate studies of HCV infection, those authors generated luciferase reporter viruses using heterologous controlling elements such as the internal ribosome entry site of encephalomyocarditis virus and the 2A protease of foot-and-mouse disease virus.
In the present study, we generated new infectious HCV clones containing reporters without addition of heterologous sequences to facilitate expression of viral gene products. We incorporated the green fluorescent protein (GFP) and Renilla luciferase (Rluc) reporters after amino acid 2394 (amino acid 418 of NS5a) of the JFH construct (Fig. 1A). Insertion of heterologous sequences at this insertion site has previously been shown to allow replication of subgenomic replicons (13).
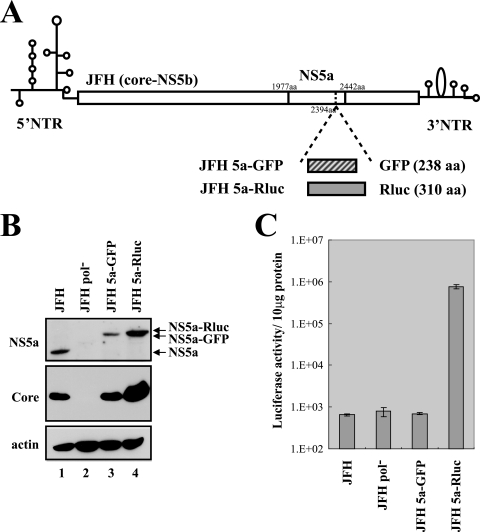
FIG. 1.
Generation of infectious HCV containing GFP or Rluc fused with NS5a. (A) Schematic diagrams of JFH and its derivatives containing the GFP or Rluc gene within the NS5a gene. NTR, nontranslated region; aa, amino acids. (B) Western blot analysis of Huh 7.5.1 cells transfected with JFH, JFH Pol−, JFH 5a-GFP, or JFH 5a-Rluc RNA. Protein levels were analyzed by Western blotting with anti-NS5a, anticore, or antiactin antibodies. (C) The expression of NS5a-Rluc was monitored by measuring luciferase activity in the cells transfected with JFH, JFH Pol−, JFH 5a-GFP, or JFH 5a-Rluc RNA. The luciferase activities in the cells were normalized by the protein concentrations determined by the Bradford assay. Experiments were performed three times. Means and standard deviations are shown.
First, we compared the parental JFH1 genome with its derivatives containing GFP or Rluc reporter genes (JFH 5a-GFP and JFH 5a-Rluc) in terms of viral protein expression in transfected cells (Fig. 1B). RNAs transcribed in vitro were introduced into the Huh 7.5.1 cell line (18) by electroporation as described by Wakita et al. (16). Three days after transfection, cell lysates were prepared and the levels of the NS5a protein and core protein were assessed by Western blot analysis using anti-NS5a and anticore antibodies (gifts from Ralf Bartenschlager, University of Heidelberg) (Fig. 1B). NS5a-GFP and NS5a-Rluc fusion proteins with the predicted molecular masses were well expressed, as shown in Fig. 1B (lanes 3 and 4, respectively). Similar levels of core protein were expressed in the cells transfected with JFH and JFH 5a-GFP RNAs (Fig. 1B, lanes 1 and 3). Interestingly, greater amounts of core protein were observed in cells transfected with JFH 5a-Rluc RNA than in cells transfected with JFH RNA (compare lane 4 with lane 1 in Fig. 1B). More cells transfected with JFH and JFH 5a-GFP RNAs died compared with cells transfected with JFH 5a-Rluc RNA, for unknown reasons (data not shown). This cytopathic effect of JFH and JFH 5a-GFP RNAs may be attributed to the difference in the amount of viral protein. However, neither NS5a nor core protein was detected in cells transfected with JFH Pol− RNA, which was used as a negative control because it contains a mutation at the catalytic site of the RNA polymerase NS5b (Fig. 1B, lane 2).
Replication of HCV was also monitored using the reporter Rluc integrated into NS5a in the JFH 5a-Rluc construct (Fig. 1C). At 3 days after transfection, luciferase activity in the cells transfected with JFH 5a-Rluc RNA increased by about 1,000-fold compared with the activity in the cells transfected with the JFH, JFH Pol−, or JFH 5a-GFP RNA (Fig. 1C).
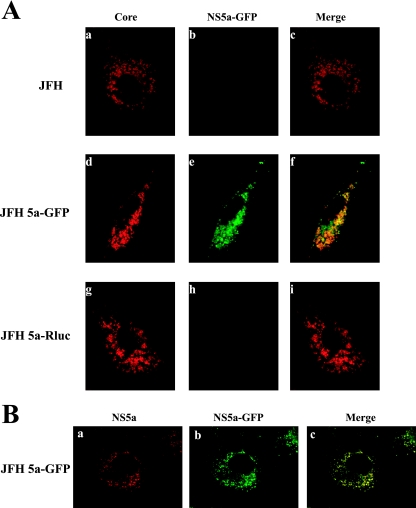
We then examined the subcellular localization of core and NS5a-GFP proteins in cells transfected with JFH, JFH 5a-GFP, or JFH 5a-Rluc RNAs (Fig. 2A). Three days after transfection, cells were fixed and fluorescence microscopy was performed as described previously (8). Core protein was visible in a ring-like pattern around lipid droplets in the JFH RNA-transfected cells (Fig. 2A, panel a; see Fig. S1A at http://www.postech.ac.kr/dept/life/mv1/figure_s1.pdf), as previously reported (14). The same patterns of localization of core protein were observed in the cells transfected with JFH 5a-GFP and JFH 5a-Rluc RNAs (compare panels d and g with panel a in Fig. 2A). No signal was detected by the antibody against core protein in cells transfected with JFH Pol− RNA (data not shown). Huh 7.5.1 cells transfected with JFH 5a-GFP RNA displayed bright punctate patterns in the cytoplasm (Fig. 2A, panel e). This 5a-GFP fluorescence was perfectly colocalized with the signal of NS5a displayed by an anti-NS5 monoclonal antibody (AUSTRAL Biologicals) (Fig. 2B). Similar cytoplasmic punctate patterns of NS5a protein were observed in the cells transfected with JFH and JFH 5a-Rluc RNAs (compare panels a and b in Fig. S1B at http://www.postech.ac.kr/dept/life/mv1/figure_s1.pdf with panel a in Fig. 2B). GFP fluorescence was not detected in cells transfected with other constructs (Fig. 2A, panels b and h). A partial colocalization of core and NS5a-GFP, shown as yellow signals, was observed when the core and NS5a-GFP images were merged (Fig. 2A, panel f). The pattern of colocalization of core and NS5a proteins was similar to that of core and NS3 proteins reported by Rouille et al. (14).
FIG. 2.
Subcellular localizations of core and 5a-GFP proteins. (A) Huh7.5.1 cells transfected with JFH, JFH 5a-GFP, and JFH 5a-Rluc RNAs were grown on coverslips for 3 days. Cells were fixed, permeabilized, and treated with anticore monoclonal antibody (Affinity Bioreagents) and secondary antibody (tetramethyl rhodamine isocyanate-conjugated donkey anti-mouse immunoglobulin G). The localization patterns of core and NS5a-GFP are shown in red and green, respectively. The merged images are also shown. (B) Following transfection of Huh 7.5.1 cells with JFH 5a-GFP RNA, immunocytochemistry was performed with an anti-NS5 antibody (AUSTRAL Biologicals) and a tetramethyl rhodamine isocyanate-conjugated donkey anti-mouse immunoglobulin G. The NS5a signal is shown in red, and the NS5a-GFP signal is shown in green. The merged image is also shown.
Next, we assessed the ability of the reporter constructs to release infectious virion particles and whether it was possible to quantify their infectivity using fluorescence microscopy and a luciferase assay. We transfected the JFH, JFH Pol−, JFH 5a-GFP, and JFH 5a-Rluc RNAs into Huh 7.5.1 cells and then harvested the culture supernatants at 8 days after transfection. Infection was performed as described previously by using medium (100 μl) of the transfected cell culture (16). At 3 days after infection, total cellular RNA was isolated from infected cells and the level of HCV RNA was measured by quantitative real-time PCR (Fig. 3A). Similar levels of HCV RNAs were detected in cells infected with JFH, JFH 5a-GFP, and JFH 5a-Rluc viruses (Fig. 3A, lanes 1, 3, and 4, respectively). By contrast, HCV RNA was not detectable in cells infected with culture supernatant obtained from cells transfected with JFH Pol− RNA (Fig. 3A, lane 2). Luciferase activity was detected in cells inoculated with culture supernatant containing JFH 5a-Rluc virus and increased for up to 3 days postinfection (Fig. 3B).
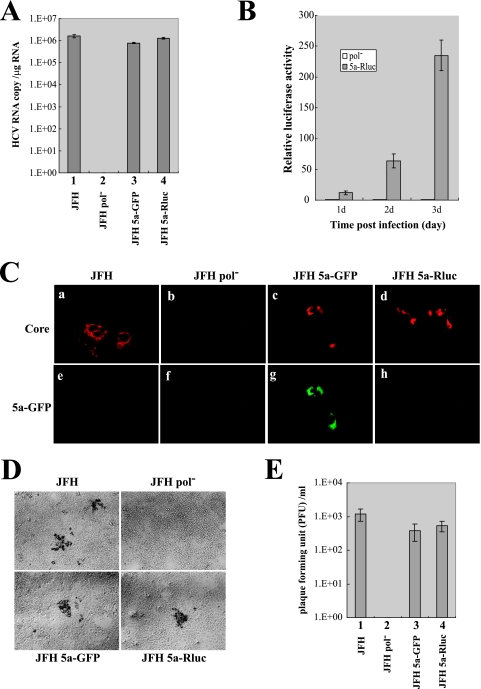
FIG. 3.
Infectivity of assayable viruses. (A) Cell-free culture fluids were collected 8 days after transfection. Supernatants were used to inoculate naïve Huh 7.5.1 cells. Total RNAs were isolated from infected cells, and the levels of HCV RNA were measured by real-time reverse transcription-PCR. The levels of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA were used as internal mRNA controls. The HCV RNA levels shown are the copy number per 1 μg of cellular RNA. Experiments were performed three times, and the values are depicted as described for Fig. 1. (B) Huh 7.5.1 cells were infected with JFH 5a-Rluc virus for 3 days. Each day, cells were harvested and luciferase activities were measured. Luciferase activities were normalized to those obtained from cells inoculated with culture supernatants of cells transfected with JFH Pol− RNA, which were set to 1. Experiments were performed three times, and the values are shown as described for Fig. 1. (C) Huh 7.5.1 cells were fixed at 3 days postinfection with JFH, JFH Pol−, JFH 5a-GFP, or JFH 5a-Rluc virus, and the core-expressing cells are shown in red as in Fig. 2A (panels a to d). The NS5a-GFP signal was directly visualized by fluorescence microscopy (green) of the same cells (panels e to h). (D) Plaques generated by infection of JFH, JFH 5a-GFP, and JFH 5a-Rluc viruses were observed by phase-contrast microscopy (magnification, ×100). (E) Plaques were counted, and the viral titers are depicted as PFU per milliliter of medium. Experiments were performed four times, and the values are shown as described in the legend to Fig. 1C.
Infectivity of HCV derivatives was also shown by fluorescence microscopy. Naïve Huh 7.5.1 cells were inoculated with culture supernatants, and productive infections were demonstrated by an immunocytochemical method using an antibody against HCV core (Fig. 3C). As shown in Fig. 3C, infection was readily detectable with JFH, JFH 5a-GFP, and JFH 5a-Rluc viruses (Fig. 3C, panels a, c, and d), whereas no core-expressing cells were found after inoculation with the polymerase mutant (JFH Pol−) (Fig. 3C, panel b). Moreover, in the same core-expressing cells, 5a-GFP fluorescence was observed after inoculation with JFH 5a-GFP virus (Fig. 3C, panel g).
To determine the infectivity of the JFH, JFH 5a-GFP, and JFH 5a-Rluc viruses, plaque assays were performed by the method used in determination of the infectivity of bovine viral diarrhea virus (4). After incubation of viral stocks at 37°C for 3 h, the Huh 7.5.1 cells were overlaid with semisolid medium containing gum tragacanth (Sigma). At 5 days after infection, virus-infected cells were visualized by immunostaining with NS5a-specific antibody (AUSTRAL Biologicals) as described previously (11) (Fig. 3D). Viral titers in the medium on Huh 7.5.1 cells transfected with RNAs of HCV variants are depicted in Fig. 3E. The titers of JFH 5a-GFP and JFH 5a-Rluc virus were lower than that of JFH by 30% and 50%, respectively. These data indicate that the insertions of GFP and Rluc into NS5a moderately impaired the viral infectivity of JFH. Taking together the immunocytochemical data and viral infectivity tests, we concluded that the HCV derivatives containing GFP or Rluc produce functional proteins and replicate properly. Therefore, 5a-GFP fluorescence and 5a-Rluc activity can be used to visualize HCV-infected cells (JFH 5a-GFP) and quantify the level of virus infection (JFH 5a-Rluc).
Taking advantage of the ability to quantify the JFH 5a-Rluc virus, we examined the antiviral activities of IFN-α, ribavirin, and BILN 2061 (10) (Fig. 4). Dose-response experiments showed that IFN-α, ribavirin, and BILN 2061 inhibited proliferation of JFH 5a-Rluc virus in the infected cells (Fig. 4A, B, and C, respectively). The median effective concentrations of IFN-α and BILN 2061 against JFH 5a-Rluc virus were similar to those against J6/JFH virus as previously reported by Lindenbach et al. (11). This indicates that the modified virus JFH 5a-Rluc, which contains a heterologous polypeptide, responds to antiviral agents in a similar manner as the JFH virus.
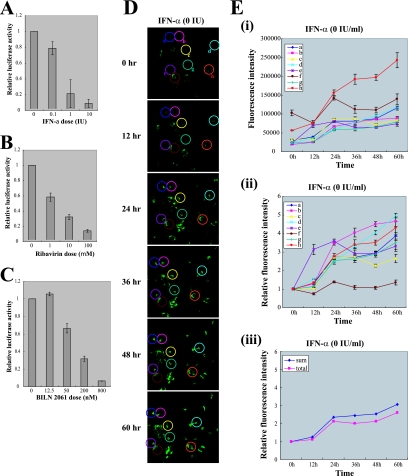
FIG. 4.
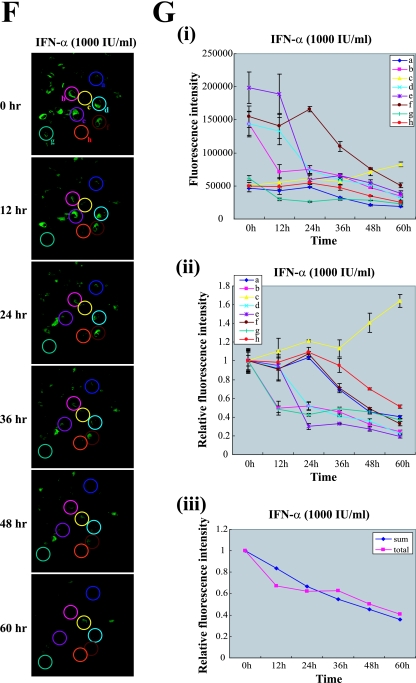
Monitoring the antiviral effects of antiviral agents with infectious HCV containing reporter genes. (A, B, and C) Huh 7.5.1 cells were incubated with the indicated concentration of IFN-α (A), ribavirin (B), or BILN 2061 (C) for 8 h before infection with JFH 5a-Rluc virus. After inoculation with culture supernatant, the same concentrations of drugs were maintained for 3 days. Three days after inoculation, cells were harvested and luciferase activities were measured. Luciferase activities were normalized to those obtained from mock-treated cells, which were set to 1. Cytotoxic effects of drugs were monitored with the protein concentration as determined by the Bradford assay. Experiments were performed three times, and the values are shown as described for Fig. 1. (D) Huh 7.5.1 cells that were transfected with JFH 5a-GFP RNA without treatment with IFN-α were monitored every 12 h up to 60 h by time-lapse confocal microscopy (Zeiss LSM 5 Live). Eight cells (circled and labeled a, b, c, d, e, f, g, and h) were selected for quantitative analyses of the 5a-GFP fluorescence. Five z-stack images were taken at the indicated times, and representative images are shown. (E) Fluorescence intensities of the eight cells circled in panel D are plotted as arbitrary fluorescence units versus time in panel i and as relative fluorescence intensities versus time in panel ii. The relative fluorescence intensities were obtained by dividing the intensity at each time point by that at the starting time point. Mean values of five z-stack images with standard deviations are plotted. The fluorescence intensities obtained from the whole area of images reflecting whole cells in the panels (total) and the average intensities of eight cells (sum) are plotted in panel iii. All image analyses were performed using MetaMorph software. (F) Huh 7.5.1 cells transfected with JFH 5a-GFP RNA were treated with IFN-α (1000 IU/ml) and monitored every 12 h up to 60 h by time-lapse confocal microscopy (Zeiss LSM 5 Live). Eight cells (circled and labeled a, b, c, d, e, f, g, and h) were selected for quantitative analyses of the 5a-GFP fluorescence. Five z-stack images were taken at the indicated times, and representative images are shown. (G) Fluorescence intensities of eight cells circled in panel F are plotted as arbitrary fluorescence units versus time in panel i and as relative fluorescence intensities versus time in panel ii. The relative fluorescence intensities were obtained by dividing the intensity at each time point by that at the starting time point. Mean values of five z-stack images with standard deviations are plotted. The fluorescence intensities obtained from whole area of images reflecting whole cells in the panels (total) and the average intensities of eight cells (sum) are plotted in panel iii. All image analyses were performed using MetaMorph software.
Even though IFN-α is used as therapy for HCV infections (6), many patients do not respond to IFN-α treatment (12). The mechanism of this IFN-α resistance is poorly understood. Moreover, the antiviral activity of IFN-α against HCV in individual HCV-infected cells has not yet been investigated due to technical limitations. We tried to monitor the anti-HCV effect of IFN-α in individual cells by using a derivative of HCV, JFH 5a-GFP, the replication of which can be microscopically monitored in individual cells in real time. Huh 7.5.1 cells transfected with JFH 5a-GFP RNA were treated with IFN-α or mock treated, and GFP fluorescence was monitored every 12 h up to 60 h by time-lapse confocal microscopy (Zeiss LSM 5 Live). For time-lapse imaging, coverslips were mounted onto the microscope stage, which was equipped with a temperature- and gas-controlled chamber (Chamlide IC; Live Cell Instrument, Korea). Quantitative analyses of the fluorescence images were performed using MetaMorph software. In cells that were not treated with IFN-α, the total intensity of 5a-GFP fluorescence increased with increasing cultivation time (Fig. 4E, panel iii). Analyses of the fluorescence intensities of eight cells (circled in Fig. 4D) showed that the fluorescence intensity of each cell increased by various amounts as the cultivation time increased (Fig. 4E, panels i and ii). The fluorescence intensities of eight cells were averaged (Fig. 4E, panel iii) and showed increases over time similar to that of the total intensity of whole images (Fig. 4E, panel iii). These results indicate that the selected eight cells represent the viral replication pattern of all cells on the coverslip. In cells treated with IFN-α, the total fluorescence intensity decreased in a time-dependent manner (Fig. 4G, panel iii). Analyses of fluorescence intensities of the eight cells circled in Fig. 4F showed that the antiviral effect of IFN-α varied in each cell (Fig. 4G, panels i and ii). 5a-GFP fluorescence intensities in seven cells (circles a, b, d, e, f, g, and h in Fig. 4F) gradually reduced even though the actual kinetics of the intensity reductions differed among individual cells (Fig. 4G, panels i and ii). However, the GFP signal in one cell (circle c in Fig. 4D) increased in the presence of IFN-α (Fig. 4G, panels i and ii). The averaged intensities of eight cells reduced in a manner similar to that for the total intensities of whole images (Fig. 4G, panel iii). These results indicate that the selected eight cells represent the IFN-α sensitivity of all cells on the coverslip. Taken together, the data indicate that the rates of replication of HCV RNA in HCV-infected cells and the IFN-α sensitivity of HCV-infected cells vary markedly and that HCV-infected cells showing IFN-α-resistance are present at the early stages of viral infection. The IFN-α resistance may be due to a putative variation in the host cell or a putative mutation in the viral genome. The molecular basis for these variations remains to be determined.
In this work, we generated novel reporter viruses that exhibited 5a-GFP fluorescence and 5a-Rluc activity in infected cells without the addition of a heterologous controlling element such as the internal ribosome entry site element of encephalomyocarditis virus or the foot-and-mouse disease virus 2A protease. Therefore, these systems reflect the HCV infection cycle and will be useful in investigating the viral life cycle and in development of new anti-HCV drugs.
Acknowledgments
We are grateful to Ralf Bartenschlager (University of Heidelberg) for the core and NS5a antibodies and to Francis Chisari (Scripps Research Institute) for the Huh 7.5.1 cell line.
The present study was supported in part by the Program for the Training of Graduate Students in Regional Innovation, which was conducted by the Ministry of Commerce Industry and Energy of the Korean Government; grants MCBRG (M10501000022-05-N0100-02200) and SBD-NCRC (R15-2004-033-01001-0) from MOST; grants 02-PJ2-PG1-CH16-0002 and A050291 from KHIDI; grants KRF-2003-005-C0001 and FPR05B 1-310 of the 21C Frontier Functional Proteomics Project from KMST; and a grant from POSCO.
Footnotes
Published ahead of print on 30 May 2007.
REFERENCES
- 1.Alter, H. J., and L. B. Seeff. 2000. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin. Liver Dis. 20:17-35. [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., and V. Lohmann. 2001. Novel cell culture systems for the hepatitis C virus. Antiviral Res. 52:1-17. [DOI] [PubMed] [Google Scholar]
- 3.Bartenschlager, R., and T. Pietschmann. 2005. Efficient hepatitis C virus cell culture system: what a difference the host cell makes. Proc. Natl. Acad. Sci. USA 102:9739-9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becher, P., M. Orlich, and H. J. Thiel. 2000. Mutations in the 5′ nontranslated region of bovine viral diarrhea virus result in altered growth characteristics. J. Virol. 74:7884-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 6.Jaeckel, E., M. Cornberg, H. Wedemeyer, T. Santantonio, J. Mayer, M. Zankel, G. Pastore, M. Dietrich, C. Trautwein, and M. P. Manns. 2001. Treatment of acute hepatitis C with interferon alfa-2b. N. Engl. J. Med. 345:1452-1457. [DOI] [PubMed] [Google Scholar]
- 7.Kenny-Walsh, E. 2001. The natural history of hepatitis C virus infection. Clin. Liver Dis. 5:969-977. [DOI] [PubMed] [Google Scholar]
- 8.Kim, W. J., S. H. Back, V. Kim, I. Ryu, and S. K. Jang. 2005. Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol. Cell. Biol. 25:2450-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koutsoudakis, G., A. Kaul, E. Steinmann, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 80:5308-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, R. St George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 11.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 12.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 13.Moradpour, D., M. J. Evans, R. Gosert, Z. Yuan, H. E. Blum, S. P. Goff, B. D. Lindenbach, and C. M. Rice. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 78:7400-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rouille, Y., F. Helle, D. Delgrange, P. Roingeard, C. Voisset, E. Blanchard, S. Belouzard, J. McKeating, A. H. Patel, G. Maertens, T. Wakita, C. Wychowski, and J. Dubuisson. 2006. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J. Virol. 80:2832-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tscherne, D. M., C. T. Jones, M. J. Evans, B. D. Lindenbach, J. A. McKeating, and C. M. Rice. 2006. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 80:1734-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wasley, A., and M. J. Alter. 2000. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 20:1-16. [DOI] [PubMed] [Google Scholar]
- 18.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]