Abstract
Microviruses (canonical members are bacteriophages φX174, G4, and α3) are T=1 icosahedral virions with an assembly pathway mediated by two scaffolding proteins. The external scaffolding protein D plays a major role during morphogenesis, particularly in icosahedral shell formation. The results of previous studies, conducted with a cloned chimeric external scaffolding gene, suggest that the first α-helix acts as a substrate specificity domain, perhaps mediating the initial coat-external scaffolding protein interaction. However, the expression of a cloned gene could lead to protein concentrations higher than those found in typical infections. Moreover, its induction before infection could alter the timing of the protein's accumulation. Both of these factors could drive or facilitate reactions that may not occur under physiological conditions or before programmed cell lysis. In order to elucidate a more detailed mechanistic model, a chimeric external scaffolding gene was placed directly in the φX174 genome under wild-type transcriptional and translational control, and the chimeric virus, which was not viable on the level of plaque formation, was characterized. The results of the genetic and biochemical analyses indicate that α-helix 1 most likely mediates the nucleation reaction for the formation of the first assembly intermediate containing the external scaffolding protein. Mutants that can more efficiently use the chimeric scaffolding protein were isolated. These second-site mutations appear to act on a kinetic level, shortening the lag phase before virion production, perhaps lowering the critical concentration of the chimeric protein required for a nucleation reaction.
The assembly of viral proteins and nucleic acids into a biologically active virion involves diverse and numerous macromolecular interactions. Besides correct interactions among structural proteins, many viral systems require scaffolding protein support for proper morphogenesis. Scaffolding proteins temporarily associate with structural proteins, stimulating conformational changes that nucleate assembly and ensure morphogenetic fidelity (8, 12, 13).
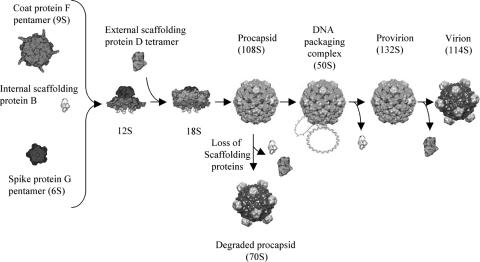
Microvirus morphogenesis is unique in that it depends on two scaffolding proteins, an external (D protein) and an internal (B protein) species. Together, these two proteins perform the analogous scaffolding functions found in systems with one scaffolding protein. The microvirus assembly pathway is illustrated in Fig. 1. The first detectable intermediates are the 9S and 6S particles, respective pentamers of the coat and major spike proteins (16). Five internal scaffolding proteins bind to the underside of the coat protein pentamers, inducing a conformational change that enables the pentamers to interact first with the major and minor spike proteins to form the 12S particle and then with the external scaffolding protein, creating the 18S particle (15, 16). Twelve of these particles then assemble into the procapsid (108S) most likely via twofold D-protein-related interactions (5, 6, 10, 16).
FIG. 1.
φX174 morphogenesis.
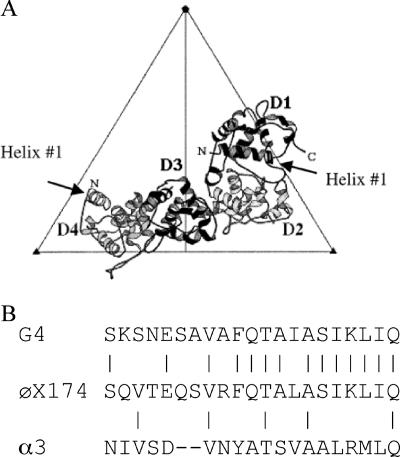
In the atomic structure of the φX174 procapsid (5, 6), there are four external scaffolding subunits per coat protein, arranged as two asymmetric dimers (D1D2 and D3D4), an arrangement that bears no resemblance to quasi-equivalence (Fig. 2A). Accordingly, each subunit makes a unique set of contacts with the underlying coat and neighboring D proteins. The external scaffolding protein consists of seven α-helices. The body of the protein, α-helices 2 to 6, which mediate the vast majority of intra- and interdimer contacts, is strongly conserved between all microviruses. However, considerable divergence occurs in α-helix 1 (Fig. 2B).
FIG. 2.
(A) The four external scaffolding protein subunits associated with each asymmetric unit. (B) Alignment of the first α-helices of the external scaffolding proteins of G4, α3, and φX174.
Previously, biochemical analyses were conducted with an α3/φX174 or G4/φX174 chimeric external scaffolding protein, in which α-helix 1 of φX174 was replaced by α-helix 1 of α3 or G4 phage, respectively. These results suggested that α-helix 1 acts as a coat protein substrate species specificity domain early in the morphogenetic pathway, interacting with α-helix 4 of the coat protein (3, 17). However, in those studies the chimeric protein was expressed from a high-copy-number plasmid that was induced before infection. This would result in a protein pool at the onset of infection. In typical positive-strand DNA virus life cycles, gene expression is dependent on negative-strand synthesis. Thus, protein concentrations higher than those found in typical infections and/or altering the timing of a protein's accumulation could drive or facilitate reactions that may not occur under physiological conditions (3). In an effort to elucidate a more mechanistic and physiologically relevant model, the chimeric gene was constructed directly in the phage genome. The results of biochemical, kinetic, and genetic analyses presented here suggest that the interactions between α-helix 1 of D protein and α-helix 4 of F protein are responsible for nucleation of the D-F protein association, which is the formation of the 18S particle.
MATERIALS AND METHODS
Phage plating, media, buffers, stock preparation, generation of single-stranded DNA, RF DNA, and DNA isolation.
The reagents, media, buffers, and protocols for single-stranded DNA and replicative form (RF) DNA isolation have been described previously (4, 7).
Bacterial strains, phage strains, and plasmids.
The Escherichia coli C strains C122 (sup0) and BAF30 (recA) have been described previously (4, 7). The C900 strain contains the host slyD mutation, which confers resistance to E-protein-mediated lysis (14). The RY7211 E. coli strain (1) that confers resistance to G4 phage E-protein-mediated lysis was kindly given by R. Young (Texas A&M University). The φX174 nullD mutant (2), φX174 cdah1 (3), and the plasmids pφXDJ (2) and pG4/φXD (17) have been described previously. In the naming of the plasmids encoding chimeric genes, the first phage mentioned indicates the origin of the DNA encoding α-helix 1. The name of the phage after the slash indicates the origin of the DNA encoding the rest of the gene.
To construct φX174 IPC (in-phage chimera), a downstream primer that encodes the complete 29 bp of φX174 sequence followed by the sequence of G4 α-helix 1 with an NheI site was designed. The φX174 genome served as a PCR template with the new downstream primer and an upstream primer that anneals to the unique XhoI site of φX174. The fragment was digested with XhoI and NheI and inserted into φX174 RF DNA after it was digested with the same enzymes. The ligated genome was transfected into cells harboring plasmid pφXDJ. Oligonucleotide-mediated mutagenesis (9) was used to move mutations between wild-type, nullD, and φX174 IPC backgrounds.
A G4 phage nullD mutant was constructed by site-directed mutagenesis by replacing the start codon with an ocher mutation. To construct pφX/G4 D, PCR was performed on the wild-type G4 genome using a downstream primer that introduces an NheI site immediately after α-helix 1 and an upstream primer that anneals to an indigenous SacII site. The PCR product and pφXDJ were digested with NheI and SacII before ligation.
Isolation of pfIPC mutants.
In order to isolate single IPC mutants capable of plaque formation (pfIPC), 106 φX174 IPC cells were plated on C122 and incubated at 37°C until plaques appeared. To isolate double pfIPC mutants, 106 pfIPC phage carrying the D333H mutation in gene F[pf(F)D333H] were plated on strain C122 and incubated at 28°C until plaques appeared.
Isolation of G4 mutants that can utilize the φX/G4 D protein.
G4 nullD was plated on cells expressing the φX/G4 D protein and incubated overnight at room temperature. Plaques were picked onto lawns seeded with C122 and BAF30 pφX/G4 and screened for a D protein complementation-dependent phenotype.
Detection of virion and intermediate particles from infected cells.
Protocols for the detection of large intermediates and end products from infected cells were previously described (17), except for the use of Nanosep (Pall Corp.) to concentrate proteins.
Determination of relative viral protein concentrations in infected cells.
To compare and approximate protein expression levels from cloned and genomic genes, 5.0 ml of lysis-resistant cells, with and without pG4φXD, were grown to a concentration of 1.0 × 108 cells/ml, infected as previously described (17), and incubated for 3 h. Cells were concentrated, and whole-cell samples were prepared for polyacrylamide gel electrophoresis. Gels were stained with Coomassie blue. Relative band intensities were calculated using one-dimensional image analysis software (Kodak Digital Science). The external scaffolding protein band was compared independently to the major coat, minor spike, and major spike proteins.
Growth curve kinetics.
Two milliliters of slyD cells, placed in a water bath at 37°C or 30°C, was infected with phage at time zero. At designated time points, 100-μl aliquots were removed, iced, and immediately lysed with T4 lysozyme and chloroform, and titers were determined on BAF30 pG4/φXD cells.
RESULTS
Biological activity of a φX174 strain carrying the chimeric G4/φX174 external scaffolding protein gene, φX174 IPC.
As previously reported, a cloned chimeric G4/φX174 external scaffolding gene was able to complement a φX174 nullD mutant (17). However, the expression of a cloned gene could lead to protein overexpression or alter the timing of its accumulation. To determine if the chimeric protein could support morphogenesis when expressed in a physiologically relevant manner, the chimeric gene was placed directly into the genome, yielding φX174 IPC, which replaced the wild-type gene. In contrast to the expressed cloned gene, the chimeric gene is most likely under the wild-type transcriptional and translational control, as it is regulated by the wild-type promoter and Shine-Delgarno sequence. Under these conditions, the chimeric protein failed to support growth at the level of plaque formation (Table 1), which is in contrast to the φX174 nullD complementation results (17). To determine whether the dependence on cloned gene expression was primarily a consequence of overexpression or altered timing of expression, protein levels were investigated in φX174 IPC-infected lysis-resistant cells with and without the cloned gene. The results of the analysis indicate that protein expression from the cloned gene was approximately 70% of the genome copy (data not shown). Thus, simple overexpression does not account for the ability of the cloned gene to complement the nullD mutant on the level of plaque formation. If a higher critical concentration of the chimeric protein is required to nucleate a certain step of assembly, it is more likely that the induction of the cloned gene before infection enables this concentration to be reached before programmed cell lysis.
TABLE 1.
φX174pfIPC plating efficienciesa
| Virus strain | Complementation under the indicated conditions:
|
Substitution(s) in pfIPCb | |||
|---|---|---|---|---|---|
| No plasmid
|
pG4/φX174 D
|
||||
| 28°C | 37°C | 28°C | 37°C | ||
| φX174 nullD | <10−4 | <10−4 | <10−4 | 1.0 | |
| φX174IPC | <10−4 | <10−4 | <10−4 | 1.0 | |
| φX174pfIPC mutant | |||||
| 1 | <10−4 | 1.0 | 1.0 | 1.0 | (F)D333H |
| 2 | 1.0 | 1.0 | 0.7 | 1.0 | (F)Q81R |
| 3 | <10−4 | 0.5 | 10−2 | 1.0 | (G)A106T |
| 4 | <10−4 | 1.0 | 10−3 | 1.0 | (G)A106V |
| 5 | <10−4 | 1.0 | <10−4 | 1.0 | (D)T46I |
| 6 | <10−4 | 1.0 | 10−3 | 1.0 | (D)V134P |
| 7 | 0.1 | 1.0 | 1.0 | 1.0 | (F)D333H/(F)D88G |
| 8 | 1.0 | 1.0 | 1.0 | 1.0 | (F)D333H/(F)T189I |
| 9 | 0.5 | <10−4 | 1.0 | 1.0 | (F)D333H/(F)A198T |
| 10 | 1.0 | 1.0 | 1.0 | 1.0 | (F)D333H/(F)A201V |
| 11 | 1.0 | 1.0 | 1.0 | 1.0 | (F)D333H//(F)V318P |
| 12 | 1.0 | 1.0 | 1.0 | 1.0 | (F)D333H/(F)T426A |
| 13 | 1.0 | 1.0 | 1.0 | 1.0 | (F)D333H/(G)A106V |
| 14 | 1.0 | 1.0 | 1.0 | 1.0 | (F)D333H/(D)V134P |
| 15 | 1.0 | 1.0 | 1.0 | 1.0 | (F)D333H/(D)D47N |
Complementation is reported as the plating efficiency, calculated as follows: titer under the designated condition/no. of PFU with cloned chimeric external scaffolding protein gene expression at 37°C.
Letters in parentheses indicate the gene where the mutation is located. The first letter and number indicate the wild-type amino acid found at that position; the letter after the number indicates the substituted amino acid.
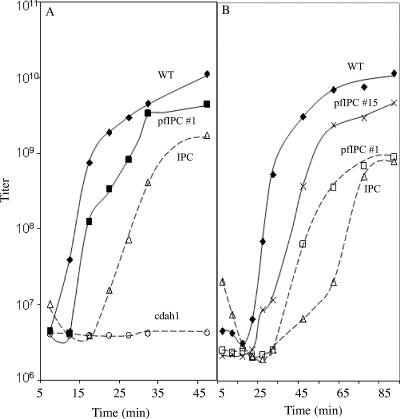
To test this hypothesis, the kinetics of phage formation was examined for wild type, φX174 IPC, and φX174 cdah1 in infected slyD cells. The slyD mutation blocks φX174 E-protein-mediated lysis (14); thus, delays in progeny production can be detected. The φX174 cdah1 strain also contains a chimeric D gene (3). In this strain, the first 22 codons of gene D, which encode α-helix 1, have been replaced by the analogous DNA sequence from bacteriophage α3. The chimeric protein expressed from cdah1 is known to have no activity, suggesting no interaction with the φX174 structural proteins, and it served as a negative control. Cells were infected at a multiplicity of infection of 3, and aliquots were removed at designated time points and chemically lysed, and progeny titer was determined. As can be seen in Fig. 3A, wild-type φX174 progeny production can be detected as quickly as 10 min postinfection. Progeny production in φX174 IPC-infected cells was delayed by 15 min while cdah1 failed to produce progeny even 90 min postinfection. The longer lag phase observed in φX174 IPC-infected cells supports the hypothesis that a higher critical concentration of the chimeric external scaffolding protein may be required for assembly. The inability of φX174 IPC to form plaques in lysis-sensitive cells is most likely due to the occurrence of cell lysis before progeny formation.
FIG. 3.
(A) Growth kinetics of at 37°C. (B) Growth kinetics at 30°C. The names of the φX174 strains are given on the graph. The lysis-resistant slyD host was used for all experiments. φX174 pfIPC 15 contains two plaque formation mutations. φX174 cdah1 has a chimeric external scaffolding gene; α-helix 1 is derived from bacteriophage α3.
Isolation of IPC mutants capable of plaque formation in wild-type (lysis-sensitive) cells.
To further elucidate the mechanism of α-helix 1 function, φX174 IPC mutants capable of plaque formation (pfIPC) were isolated by direct genetic selection at 37°C. A second-site mutation in the viral coat protein at the threefold axis of symmetry, pfIPC 1, was identified (Table 1), as in the previous study (17). The other mutations were novel, mapping to another region of the coat protein (pfIPC 2), the minor spike protein (pfIPC 3 and 4), and the external scaffolding protein pfIPC (mutants 5 and 6). However, the changes in the external scaffolding protein do not reside in the first α-helix.
Since most of the pfIPC mutants were not able to form plaques at 28°C, a second round of selection was performed with pfIPC 1 at 28°C in order to obtain double plaque formation mutations. Several double mutants that have overcome the cold-sensitive phenotype were isolated (Table 1, pfIPC 7 to 15). Most additional mutations were found in gene F at the threefold axis of symmetry as seen in our previous study (pfIPC 7 to 12). Other substitutions were found in the major spike (pfIPC 13) and external scaffolding proteins (pfIPC 5, 14, and 15). Mutations in the D gene ribosome binding site or promoter were not isolated (see Discussion). The complete genomes of the parental and of all the pfIPC mutants were determined, and no other mutations were found. Thus, the identified plaque formation mutations are most likely both necessary and sufficient to confer the observed phenotypes. Moreover, rescue experiments with mutagenic oligonucleotides encoding the plaque formation mutations (D)V134P and (G)A106V were performed to reconstruct the two strains. Phenotypes bred true.
The plaque formation mutations act on a kinetic level.
The chimeric D protein can support morphogenesis in lysis-deficient cells but not plaque formation in lysis-sensitive cells. In lysis-deficient cells, progeny production is delayed relative to wild type. These data suggest that the plaque formation mutations may act on a kinetic level. To test this hypothesis, the kinetics of pfIPC φX174 was examined. As can be seen in Fig. 3A, progeny production in pfIPC 1-infected cells at 37°C was observed 15 min postinfection, as opposed to 20 min postinfection in IPC-infected cells. The shorter lag phase in the pfIPC 1-infected cells suggests that the plaque formation substitution may lower the critical concentration of the chimeric protein needed to nucleate a stage of assembly. In order to see if double plaque formation mutations also acted by shortening lag phase in vivo, the kinetic growth curve of pfIPC 15 was determined at the temperature the mutant was isolated. As seen in Fig. 3B, progeny production of pfIPC 15 was observed 25 min postinfection at 30°C, 10 min earlier than the single plaque formation mutant. These results suggest that double plaque formation mutants also act at the kinetic level and are consistent with the observed plaque-forming phenotypes.
Further characterization of the plaque formation mutants.
The results of our previous study demonstrated that the chimeric external scaffolding proteins conferred two assembly defects (17). At lower temperatures, assembly is inhibited prior to procapsid formation. While virions were produced at higher temperatures, a significant amount of the degraded procapsids were also produced, suggesting that α-helix 1 can affect DNA packaging by stabilizing a pore at the threefold axis of symmetry through which DNA enters the procapsid.
In order to determine the assembly process in which the second-site mutations are acting, assembly intermediates synthesized in slyD cells were analyzed. After 4 h of incubation at 37°C, cells infected with wild-type φX174, IPC, and a single pfIPC mutant (pfIPC 1) were concentrated by centrifugation to remove unabsorbed virions. The double pfIPC mutant (pfIPC 15) infection was incubated at a lower temperature, the one at which the mutant was isolated. After chemical lysis, 1.3 to 1.4 g/cm3 of material containing soluble proteins and assembled particles was purified in CsCl gradients, concentrated, and analyzed by sucrose gradient sedimentation. After fractionation, infectious particles and noninfectious assembled particles were detected by spectroscopy (optical density at 280 nm). Infectious particle peaks were confirmed by direct plating assays.
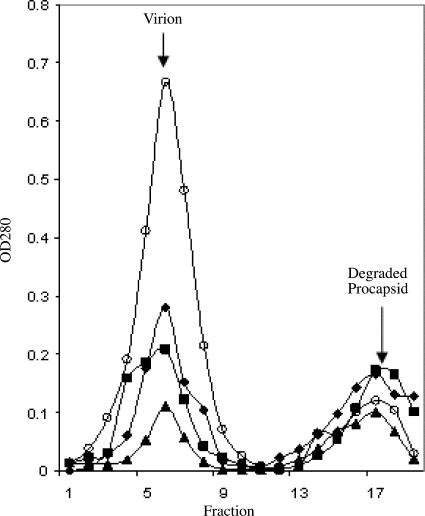
As can be seen in Fig. 4, all infections produced 114S virions (corresponding to fraction 6) and noninfectious 70S particles (fraction 17). The 70S particles are degraded procapsids, which have lost external scaffolding proteins during isolation. Thus, the accumulation of 70S particles indicates procapsid production (17). The wild-type infections produced significantly more virions than 70S particles. However, the φX174 IPC, pfIPC, and double pfIPC infections produced approximately the same ratio of virions and degraded procapsids. These results suggest that although IPC and pfIPC mutants can produce progeny over time, assembly integrity is very low, especially at the level of the DNA packaging process. Hence, these suppressors appear to work at the level of the assembly rate only prior to procapsid formation.
FIG. 4.
Particles produced in lysis-deficient slyD cells. Wild-type values were multiplied by 0.3 for purposes of graphing. Open circle, wild-type; diamond, φX174 IPC; square, φX174 pfIPC 1; triangle, φX174 pfIPC 15. OD280, optical density at 280 nm.
Biological activity of a φX174/G4 chimeric external scaffolding protein.
In order to determine if a similar biological phenomenon is observed in the G4 phage system, a cloned chimeric φX174/G4 external scaffolding gene was constructed and assayed for the ability to complement a G4 nullD mutant. Unlike the wild-type G4 D protein, which supported growth at all temperatures, the chimeric protein did not support plaque formation (Table 2). However, the G4 nullD mutant produced infectious progeny in lysis-deficient cells (data not shown). Extragenic mutations that allow plaque formation were isolated. A single and a double mutant were recovered. Both of these substitutions were observed in φX174 in this study or our previous study (17). The wild-type G4 protein does not complement the double mutant. Whether this indicates a switch in D protein substrate specificity remains to be determined. These results suggest that switching α-helix 1 between φX174 and G4 phage in the G4 system results in a similar phenomenon as that observed in phage φX174.
TABLE 2.
Complementation of nullD and utilizer mutation/nullD G4 mutants by wild-type and chimeric φX174/G4 external scaffolding proteinsa
| Strainb | External scaffolding protein complementation under the indicated conditions:
|
|||
|---|---|---|---|---|
| pφX/G4D
|
pG4D
|
|||
| 24°C | 30°C | 24°C | 30°C | |
| Wild type | 1.0 | 1.0 | 1.0 | 1.0 |
| nullD | 10−4 | 10−4 | 1.0 | 1.0 |
| ut(F)M212I/nullD | 1.4 | 0.4 | 0.2 | 1.0 |
| ut(F)M212I/(F)Q80R/nullD | 0.6 | 1.2 | 10−3 | 1.0 |
Complementation is reported as the plating efficiency (titer with the chimera at designated temperature/titer with the wild-type protein at 30°C).
The letter in parentheses indicates the gene in which the utilizer mutation resides. The first letter and number indicate the wild-type amino acid found at that position; the letter after the number indicates the substituted amino acid.
DISCUSSION
The results of previous studies with chimeric external scaffolding proteins suggest that α-helix 1 may be a substrate specificity domain vis-à-vis coat protein recognition (17). However, protein concentrations higher than those found in typical infections and/or altering the temporal expression of a viral gene by its preinduction from a plasmid could drive reactions that may not occur under physiological conditions (2, 3). In order to elucidate a more accurate mechanistic model, protein needs to be expressed under physiological conditions. To this end, the in-phage chimera φX174 IPC, in which the α-helix 1 of the external scaffolding gene was replaced by that of G4, was constructed. In contrast to the cloned gene, the chimeric gene is under the same transcriptional and translational control as the wild-type gene.
Chimeric external scaffolding protein may require a higher critical concentration for virion assembly.
φX174 IPC was not able to form plaques at any temperature without the coexpression of an exogenous external scaffolding protein, either the wild-type φX174 or the G4/φX chimeric protein. As previously reported, the cloned gene was able to complement a φX174 nullD mutant. Since protein expression from the cloned gene is roughly equal to the expression of the genomic copy, plaque formation is most likely a function of protein induction before infection. This could allow the chimeric protein to reach a critical concentration before programmed cell lysis. As demonstrated here (Fig. 3), fully infectious IPC virions are produced in lysis-deficient cells, but production is delayed relative to wild type. These observations are consistent with the hypothesis that a higher critical concentration of the chimeric external scaffolding protein is required for the formation of an assembly intermediate, most likely the 18S particle. Considering that the 18S particle is the first assembly intermediate in which the external scaffolding and coat protein pentamer come into contact, α-helix 1 most likely mediates the nucleation of 18S particle formation.
Second-site mutations act at the level of kinetics.
According to previous findings with the cloned gene, the chimeric external scaffolding protein confers two assembly defects (17). At lower temperatures, assembly is inhibited prior to procapsid formation. While virions were produced at higher temperatures, a significant amount of degraded procapsids was produced, suggesting that α-helix 1 can affect DNA packaging, perhaps by stabilizing a pore at the threefold axis of symmetry through which DNA enters the procapsid. Plaque-forming second-site mutations were isolated in the φX174 IPC background and characterized. While these mutations appear to act on a kinetic level, shortening the lag phase before virion production (Fig. 3), they appear to have little or no effect on the DNA packaging defect (Fig. 4). The φX174 IPC and pfIPC mutants produce approximately the same amount of infectious virions and degraded procapsids, while wild-type φX174 infections yield significantly more infectious virions.
Mechanisms conferred by second-site mutations.
Although 15 different pfIPC mutants have been isolated, mutations in either the D protein promoter or ribosome binding site were not recovered, suggesting that a single mutation in these genetic elements cannot increase protein levels to the required critical concentration to support plaque formation. Instead, the mutations most likely affect the affinities of interacting proteins or components.
Many of the second-site mutations are located in the viral coat protein at the threefold axis of symmetry, supporting a model in which α-helix 1 of the external scaffolding protein interacts with α-helix 4 of the coat protein (17). Structural studies of the φX174 closed procapsid, which is believed to be an off-pathway product, reveal very few interactions between the first α-helix of the D protein subunit and any structural proteins (5, 6). However, this off-pathway product has no pore at the threefold axis of symmetry, while the pore is observed in cryoelectron microscopy reconstruction (11). Thus, biologically important interactions were lost in the crystal structure. In the findings reported here, several mutations were also found in the major spike and external scaffolding proteins outside of α-helix 1. The suppressing D protein residues cluster in a loop in the D1 subunit and that extends toward the major spike protein (Fig. 5). The suppressing residue in the major spike protein is also in close proximity to α-helix 1 of both the D1 and D2 subunits. These mutations may act by creating an alternate assembly nucleation site for 18S particle assembly or may lead to a more stable 18S particle after formation.
FIG. 5.
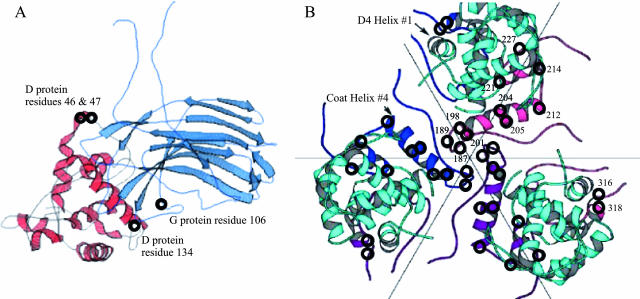
(A) Location of the second-site suppressor in the major spike and external scaffolding protein D1 subunit. Protein G and the D1 subunit are shown in light blue and red, respectively. The locations of the plaque formation mutations are depicted with circles and identified on the figure. (B) The clustering of the plaque formation mutation in the major coat protein at the threefold axis of symmetry. Coat proteins are depicted in blue, purple, and pink. The D4 external scaffolding protein subunit is depicted in cyan. Amino acid numbers are given on the pink coat protein subunit. Some of these second-site suppressors were previously isolated (17). Suppressors located at coat protein residues 12, 81, 138, 333, and 426 are not depicted.
Acknowledgments
This research was supported by National Science Foundation grant MCB 054297 to B.A.F.
Footnotes
Published ahead of print on 6 June 2007.
REFERENCES
- 1.Bernhardt, T. G., W. D. Roof, and R. Young. 2000. Genetic evidence that the bacteriophage phi X174 lysis protein inhibits cell wall synthesis. Proc. Natl. Acad. Sci. USA 97:4297-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burch, A. D., and B. A. Fane. 2000. Foreign and chimeric external scaffolding proteins as inhibitors of Microviridae morphogenesis. J. Virol. 74:9347-9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burch, A. D., and B. A. Fane. 2003. Genetic analyses of putative conformation switching and cross-species inhibitory domains in Microviridae external scaffolding proteins. Virology 310:64-71. [DOI] [PubMed] [Google Scholar]
- 4.Burch, A. D., J. Ta, and B. A. Fane. 1999. Cross-functional analysis of the Microviridae internal scaffolding protein. J. Mol. Biol. 286:95-104. [DOI] [PubMed] [Google Scholar]
- 5.Dokland, T., R. A. Bernal, A. Burch, S. Pletnev, B. A. Fane, and M. G. Rossmann. 1999. The role of scaffolding proteins in the assembly of the small, single-stranded DNA virus phiX174. J. Mol. Biol. 288:595-608. [DOI] [PubMed] [Google Scholar]
- 6.Dokland, T., R. McKenna, L. L. Ilag, B. R. Bowman, N. L. Incardona, B. A. Fane, and M. G. Rossmann. 1997. Structure of a viral procapsid with molecular scaffolding. Nature 389:308-313. [DOI] [PubMed] [Google Scholar]
- 7.Fane, B. A., and M. Hayashi. 1991. Second-site suppressors of a cold-sensitive prohead accessory protein of bacteriophage phi X174. Genetics 128:663-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fane, B. A., and P. E. Prevelige, Jr. 2003. Mechanism of scaffolding-assisted viral assembly. Adv. Protein Chem. 64:259-299. [DOI] [PubMed] [Google Scholar]
- 9.Fane, B. A., S. Shien, and M. Hayashi. 1993. Second-site suppressors of a cold-sensitive external scaffolding protein of bacteriophage phi X174. Genetics 134:1003-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujisawa, H., and M. Hayashi. 1977. Functions of gene C and gene D products of bacteriophage phi X 174. J. Virol. 21:506-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ilag, L. L., N. H. Olson, T. Dokland, C. L. Music, R. H. Cheng, Z. Bowen, R. McKenna, M. G. Rossmann, T. S. Baker, and N. L. Incardona. 1995. DNA packaging intermediates of bacteriophage phi X174. Structure 3:353-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King, J., and S. Casjens. 1974. Catalytic head assembling protein in virus morphogenesis. Nature 251:112-119. [DOI] [PubMed] [Google Scholar]
- 13.Prevelige, P. E., Jr., D. Thomas, and J. King. 1993. Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophys. J. 64:824-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roof, W. D., H. Q. Fang, K. D. Young, J. Sun, and R. Young. 1997. Mutational analysis of slyD, an Escherichia coli gene encoding a protein of the FKBP immunophilin family. Mol. Microbiol. 25:1031-1046. [DOI] [PubMed] [Google Scholar]
- 15.Siden, E. J., and M. Hayashi. 1974. Role of the gene beta-product in bacteriophage phi-X174 development. J. Mol. Biol. 89:1-16. [DOI] [PubMed] [Google Scholar]
- 16.Tonegawa, S., and M. Hayashi. 1970. Intermediates in the assembly of phi X174. J. Mol. Biol. 48:219-242. [DOI] [PubMed] [Google Scholar]
- 17.Uchiyama, A., and B. A. Fane. 2005. Identification of an interacting coat-external scaffolding protein domain required for both the initiation of phiX174 procapsid morphogenesis and the completion of DNA packaging. J. Virol. 79:6751-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]