Abstract
Although in vitro replication of the hepatitis C virus (HCV) JFH1 clone of genotype 2a (HCVcc) has been developed, a robust cell culture system for the 1a and 1b genotypes, which are the most prevalent viruses in the world and resistant to interferon therapy, has not yet been established. As a surrogate virus system, pseudotype viruses transiently bearing HCV envelope proteins based on the vesicular stomatitis virus (VSV) and retrovirus have been developed. Here, we have developed a replication-competent recombinant VSV with a genome encoding unmodified HCV E1 and E2 proteins in place of the VSV envelope protein (HCVrv) in human cell lines. HCVrv and a pseudotype VSV bearing the unmodified HCV envelope proteins (HCVpv) generated in 293T or Huh7 cells exhibited high infectivity in Huh7 cells. Generation of infectious HCVrv was limited in some cell lines examined. Furthermore, HCVrv but not HCVpv was able to propagate and form foci in Huh7 cells. The infection of Huh7 cells with HCVpv and HCVrv was neutralized by anti-hCD81 and anti-E2 antibodies and by sera from chronic HCV patients. The infectivity of HCVrv was inhibited by an endoplasmic reticulum α-glucosidase inhibitor, N-(n-nonyl) deoxynojirimycin (Nn-DNJ), but not by a Golgi mannosidase inhibitor, deoxymannojirimycin. Focus formation of HCVrv in Huh7 cells was impaired by Nn-DNJ treatment. These results indicate that the HCVrv developed in this study can be used to study HCV envelope proteins with respect to not only the biological functions in the entry process but also their maturation step.
Hepatitis C virus (HCV) is the major causative agent of blood-borne chronic non-A, non-B hepatitis, infecting at least 3% of the world's population. The majority of HCV-infected individuals develop chronic hepatitis that eventually progresses to liver cirrhosis and hepatocellular carcinoma (36). HCV is an enveloped single-stranded plus-sense RNA virus belonging to the genus Hepacivirus in the Flaviviridae family, which also includes members of the genus Flavivirus, such as yellow fever virus, dengue virus, and West Nile virus, and of the genus Pestivirus, such as bovine viral diarrhea virus and classical swine fever virus. The genome of HCV encodes a polyprotein of approximately 3,000 amino acids, which is subsequently processed into at least 10 viral proteins. The HCV envelope glycoproteins E1 and E2 are cleaved from the polyprotein by host signal peptidases and play a crucial role in the initiation of infection through interaction with cell surface receptor(s) in the HCV life cycle (17, 38).
A number of cellular components have been shown to participate in HCV adsorption and/or internalization, including human CD81 (hCD81) (52), low-density lipoprotein receptor (LDLr) (1), human scavenger receptor class B type I (SR-BI) (57), dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin (DC-SIGN), liver/lymph node-specific intercellular adhesion molecule-3-grabbing nonintegrin (L-SIGN or DC-SIGNR) (21, 34), glycosaminoglycans (2), and a tight junction component, claudin-1 (18). Recently, an in vitro cell culture system was developed for HCV of the genotype 2a JFH1 strain (HCVcc) isolated from a fulminant HCV patient (32, 63, 68). However, a robust cell culture system for HCV of the 1a and 1b genotypes, the most prevalent genotypes in the world, has not yet been successfully developed, except for the cell culture system of H77 or H77-S strain (1a genotype) (26, 65). Furthermore, it is currently not possible to obtain a sufficient amount of HCV particles for biological and physiochemical studies due to the low viral load in the sera of hepatitis C patients and the low yield of HCV particles in cell culture. Thus, the relative contribution of these receptor candidates in HCV attachment and entry remains unclear (44).
As surrogate systems for the investigation of HCV infection mechanisms, HCV-like particles (HCV-LP) produced in insect or mammalian cells by recombinant baculovirus vectors have been developed (7, 37). Although the binding of HCV-LP to the target cells has been well characterized, HCV-LP are not suitable for the analysis of the HCV entry steps due to the absence of a clear distinction between binding and internalization. On the other hand, both murine leukemia virus (MLV)- and human immunodeficiency virus-based pseudotype retroviral particles (HCVpp) bearing unmodified E1 and E2 proteins (5, 23) are capable of infecting human hepatoma cells, including Huh7 cells, and this infection can be inhibited by treatment with anti-hCD81 antibody and the soluble hCD81 protein or by a knockdown of hCD81 expression by small interfering RNAs (siRNAs) (67). Furthermore, the ectopic expression of hCD81 confers permissiveness to infection with HCVpp in normally nonpermissive HepG2 cells lacking expression of hCD81. These data suggest that expression of hCD81 is crucial for HCVpp infection (4). However, expression of this candidate receptor molecule is not sufficient to render nonhepatic cells permissive for HCVpp infection (6, 23). Indeed, it is also interesting to note that although neutralizing antibodies to HCVpp have been detected in the sera from persistently infected humans and chimpanzees (3, 23, 33, 66), these antibodies do not appear to play a significant role in the outcome of acute HCV infection (42). Therefore, further investigation is needed to assess the authenticity of the HCVpp as a surrogate system for HCV infection.
We and others have previously reported the generation of vesicular stomatitis virus (VSV)-based pseudotype viruses bearing chimeric or unmodified HCV E1 and E2 glycoproteins (HCVpv) in nonhepatic cell lines (27, 39, 60). Although HCVpv infected several cell lines, including human hepatoma cell lines (27, 39, 60), recombinant VSV bearing chimeric HCV E1 and E2 glycoproteins in place of VSV glycoprotein (G) was not infectious (9). This discrepancy in the cell tropism might be attributable to the differences in the constructs and strains of HCV envelope proteins or in the systems and cells in which the viruses were generated.
Human hepatocytes (Hc) are believed to be a main target for HCV replication, and it is reasonable to speculate that hepatocyte-specific host factors regulate the entry, replication, and assembly of HCV. Although HCVpp is an excellent system for examining the entry mechanisms of HCV, the system requires a high level of transfection of the expression plasmids, and thus production of HCVpp is limited to 293T cells due to their high transfectability. Furthermore, HCVpp are replication-defective and do not produce progeny virus in infected Hc, and thus reinfection with progeny viruses cannot be assessed. In this study, we generated replication-competent recombinant VSVs encoding the unmodified HCV E1 and E2 polyproteins of genotypes 1a and 1b in place of the G protein (HCVrv) in human cell lines. HCVrv was able to infect human hepatoma cell lines through an hCD81-dependent pathway and to form foci in Huh7 cells. Treatment with an ER α-glucosidase inhibitor was shown to inhibit not only infection but also focus formation of HCVrv, suggesting that modifications of envelope glycoproteins in the endoplasmic reticulum (ER) are required for infection with HCVrv.
MATERIALS AND METHODS
Plasmids and cells.
The cDNAs encoding the C-terminal 60 amino acids of the core to the last residue of p7 protein (c60-p7; nucleotides 735 to 2746) of the H77 (provided by Bukh) and Con1 (provided by Bartenschlager) (residues atg + 521 to 2773 bp) strains were generated by PCR amplification. All PCR products were cloned into pCAGGS/MCS-PM, carrying the puromycin gene for the establishment of the cell lines derived from pCAGGS (45) and designated pCAGc60-p7. The plasmid used for construction of HCVrv was pVSVΔG-P/M2.6, which has additional transcription units with two multiple cloning sites (MCS) located between the P and M genes (MCS-2) and the M and L genes (MCS-1). The c60-p7 gene was subcloned into pBluescript SK(+) from pCAGc60-p7 by digestion with EcoRI and EcoRV and designated pBSc60-p7. To construct pVSVΔG-c60-p7, the c60-p7 gene was excised from pBSc60-p7 with KpnI and XbaI and ligated into the KpnI and NheI sites of MCS-2 of pVSVΔG-P/M2.6. The cDNA of hCD81 was amplified by PCR from Huh7 cells and cloned into the BamHI and XbaI sites of the pcDNA3.1 plasmid, resulting in phCD81. The hepatic (Huh7, HepG2, Hep3B, and PLC/PRF/5) and nonhepatic (293T, HeLa, Vero, BHK, and CHOK1) cell lines were obtained from the American Type Culture Collection (Rockville, MD). The FLC4 cell line was established as described previously (37). The Huh7.5.1 cell line was kindly provided by F. Chisari. All cell lines were grown in Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich, St. Louis, MO) containing 10% fetal bovine serum (FBS). Human primary Hc were purchased from the Applied Cell Biology Research Institute (Kirkland, WA) and maintained using a CS-C serum-free medium kit (Applied Cell Biology Research Institute). To establish stable HepG2 or CHOK1 cell lines expressing hCD81, cells were transfected with phCD81 by the TransIT-LT1 (Mirus, Madison, WI) reagent, selected with DMEM containing 10% FBS and 2 mg/ml (HepG2) or 3.5 mg/ml (CHOK1) of G418 (PAA Laboratories GmbH, Linz, Austria), and sorted twice by FACSCalibur (Becton Dickinson, San Jose, CA) after staining with anti-hCD81 monoclonal antibody (JS-81; BD Biosciences Pharmingen, Mountain View, CA) to obtain high-expressing clones. Anti-E1 (BDI198; Biodesign International, Saco, ME) and anti-E2 (AP33) (13, 49) monoclonal antibodies or anti-VSVG polyclonal antibody (ab34774; Abcam Inc., Cambridge, MA) was used for detection of E1 and E2 of the H77 strain or VSVG by immunoblotting, respectively.
Reverse genetics of VSV.
Recombinant VSVs were generated as described previously (25, 30). Briefly, BHK cells were grown to 90% confluence on 35-mm tissue culture plates. The cells were infected with a recombinant vaccinia virus encoding T7 RNA polymerase (vTF7-3) (19) at a multiplicity of infection (MOI) of 5. After incubation at room temperature for 1 h, the cells were transfected with 3 μg of pBS-N, 5 μg of pBS-P, 1 μg of pBS-L, 8 μg of pBS-G, and 5 μg of pΔG-c60-p7 plasmids using a cationic liposome reagent (54). After 4 h, the supernatants were replaced with 10% FBS DMEM, and cells were incubated at 37°C for 48 h. The supernatants were then filtered through a 0.22-μm-pore-size filter (Millex-GS; Millipore) to remove vaccinia virus and were applied to BHK cells that had been transfected with pCAGVSVG (39) 24 h previously. Recovery of the virus was assessed by examining the cells for the cytopathic effects that are typical of a VSV infection after 24 to 36 h. Stocks of *G-complemented viruses, i.e., VSVΔG virus or recombinant viruses transiently bearing VSVG protein on the virion surface, were grown from single plaques on BHK cells transfected with pCAGVSVG and then stored at −80°C. The infectious titers of the recovered viruses were determined by a plaque assay.
Production and characterization of HCVpv, HCVrv, or HCVpp.
The construction of HCVpv and HCVrv is summarized in Fig. 1. To generate HCVpv in 293T or Huh7 cells transiently expressing E1 and E2 proteins, cells were transfected with pCAGc60-p7 (H77 or Con1 strain) using TransIT-LT1 (Mirus). After 24 h of incubation at 37°C, cells were infected at an MOI of 5 with the VSVΔG-GFP/G, in which the G envelope gene was replaced with the green fluorescent protein (GFP) gene and which was pseudotyped with the VSV G glycoprotein (39). The virus was adsorbed for 2 h at 37°C and then extensively washed four times with DMEM. After 24 h of incubation at 37°C, the culture supernatants were collected, centrifuged to remove cell debris, and stored at −80°C. HCVpp were produced as previously described from 293T cells cotransfected with an MLV Gag-Pol packaging construct, an MLV-based transfer vector encoding GFP, and the HCV envelope protein expression constructs (5). To generate HCVrv in various mammalian cell lines, cells were infected with the VSVG-complemented VSVΔG-c60-p7 at an MOI of 5 for 2 h at 37°C and then extensively washed four times with DMEM. After 48 h of incubation at 30°C, the culture supernatants were collected and stored at −80°C. The culture supernatants were pelleted through a 20% (wt/vol) sucrose cushion at 25,000 rpm for 2 h by using an SW28 rotor (Beckman Coulter, Tokyo, Japan). The pellets were resuspended in phosphate-buffered saline (PBS), mixed with 33% (wt/wt) cesium chloride, and centrifuged at 50,000 rpm for 48 h at 4°C by using an SW55Ti rotor (Beckman Coulter). After centrifugation, 12 fractions (0.5 ml each) were collected from the top and pelleted through a 20% (wt/vol) sucrose cushion by centrifugation at 50,000 rpm for 1 h at 4°C using an SW55Ti rotor. The pellets were resuspended in PBS and analyzed by immunoblotting to detect the incorporation of E1 or E2 proteins with anti-E1 (BDI198) or anti-E2 (AP33) monoclonal antibody, respectively. VSV N, P, and M were detected by anti-VSV polyclonal antibody, which was prepared by immunization of goats with purified VSVΔG. To determine the infectivities of HCVpv and HCVpp, infected cells were identified as GFP-positive cells under fluorescence microscopy or using FACSCalibur and expressed as infectious units (IU)/milliliter. The infectious titers of HCVrv were determined by a focus-forming assay as described below. To examine the effects of oligosaccharide modification of the E1 or E2 envelope proteins on the infectivity of the HCVpv and HCVrv, the cell lysates and the purified virions were digested with endoglycosidase H (Endo H) or peptide-N-glycosidase F (PNGase F) (Boehringer Mannheim, Mannheim, Germany), following a protocol provided by the manufacturer, and analyzed by immunoblotting. Pseudotype VSVs bearing VSVG (VSVpv) and MLV RD114 envelope protein (MLVpv) were produced in 293T cells transfected with pCAGVSVG and pFBASALF (provided by Miyazawa), respectively, and used as controls.
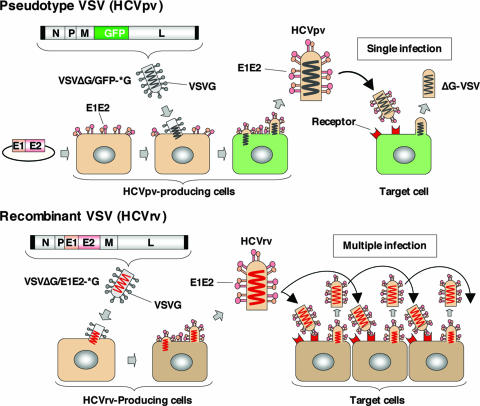
FIG. 1.
Schematic representation of the production of HCVpv and HCVrv. Pseudotype VSV (HCVpv): producer cells (Huh7 or 293T) were transfected with an expression plasmid encoding the HCV E1 and E2 genes and then infected with a VSVG-complemented pseudotype virus (VSVΔG/GFP-*G). The HCVpv released from the producer cells infected target cells but was not able to produce infectious progeny virus. Recombinant VSV (HCVrv): various mammalian producer cells were inoculated with a VSVG-complemented recombinant virus (VSVΔG/E1E2-*G) encoding the HCV E1 and E2 genes instead of VSVG. HCVrv was capable of undergoing a fully productive infection generating infectious progeny virus that could be passaged into naïve cells.
Immunofluorescence and focus-forming assay.
The cells infected with HCVpv, HCVrv, VSV, or HCVcc were cultured at 30°C with 0.8% methylcellulose in 10% FBS DMEM for the indicated periods and fixed with 4% paraformaldehyde solution for 1 h. Cells were washed once with PBS, treated with 0.5% Triton X-100 for 20 min for permeabilization, and then incubated with mouse monoclonal antibody to VSV N (10G4) (HCVpv, HCVrv, and VSV) or rabbit polyclonal antibody to NS5A (22) (HCVcc) for 1 h. Then, the cells were visualized by staining with Alexa 488-conjugated anti-mouse immunoglobulin G (IgG) or anti-rabbit IgG (Molecular Probes, Eugene, OR) for the immunofluorescence assay. The nuclei were counterstained with Hoechst 33258 (Molecular Probes). For the focus-forming assay, cells were treated with secondary antibodies and stained by using a VECTASTAIN Elite ABC anti-mouse IgG kit with a VIP substrate (Vector Laboratories, Burlingame, CA), following a protocol provided by the manufacturer. The infectious titers of the viruses were expressed as focus-forming units.
Inhibition of HCVpv or HCVrv infection by treatment with antibodies against hCD81, E1 and E2, HCV patient sera, and siRNA.
To determine the involvement of hCD81 in infection, Huh7 or HepCD81 cells were pretreated with 5 μg/ml of anti-hCD81 for 1 h at 37°C and inoculated with HCVpv or HCVrv. In addition, Huh7 cells on six-well plates were transfected with 80 nM of siRNAs targeted to hCD81 by using Nucleofector II (Amaxa GmbH, Cologne, Germany) according to the manufacturer's protocol. The hCD81 siRNAs (sc-35030) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). At 24 h posttransfection, cells were trypsinized, seeded at 8 × 103 cells/well into 96-well plates, and cultured for 48 h at 37°C. HCVpv or HCVrv was inoculated into the target cells, and infectivity was determined at 24 h postinfection. To characterize the infection with HCVpv and HCVrv, viruses were preincubated with 20 μg/ml of anti-E1 (AP21.010) (13) or anti-E2 (AP33) monoclonal antibodies, 1:50 diluted anti-E1 (R852) or anti-E2 (R646) polyclonal rabbit sera, and sera from chronic HCV patients or healthy donors for 1 h at 37°C and then inoculated into Huh7 cells. Informed consent was obtained from the patients and the donors. After 1 h of adsorption at 37°C, the cells were washed three times with DMEM containing 10% FBS, and infectivity was determined after 24 h of incubation at 37°C.
Effects of chemicals on HCVpv, HCVrv, and HCVcc infection.
To determine the entry pathways of the viruses, Huh7 cells were preincubated with various concentrations of bafilomycin A1 (Sigma) for 1 h at 37°C followed by infection with HCVpv, HCVrv, VSVpv, or MLVpv. The residual infectivity was determined as described above. N-(n-Nonyl) deoxynojirimycin (Nn-DNJ) and 1-deoxymannojirimycin hydrochloride (DMJ) were purchased from Toronto Research Chemicals Inc. (Downsview, ON, Canada). Nn-DNJ and DMJ were dissolved in ethanol and PBS, respectively, and diluted with medium before use. HCVcc was generated as previously described (47). Huh7 cells were inoculated with the viruses for 2 h at 37°C, replaced with medium containing either Nn-DNJ or DMJ, and cultured for 24 h (VSV), 72 h (HCVrv), or 96 h (HCVcc). The effects of Nn-DNJ or DMJ on the incorporation of the envelope proteins and generation of infectious particles were analyzed by immunoblotting and Coomassie staining. For the focus-forming assay, 0.8% methylcellulose in 10% FBS DMEM containing the reagents was overlaid on the cells. The infectious titers of VSV and HCVrv were determined by a focus-forming assay as described below. The infectious titers of HCVcc were evaluated by a quantitative core enzyme-linked immunosorbent assay as described previously (37).
RESULTS
Production and characterization of HCVrv.
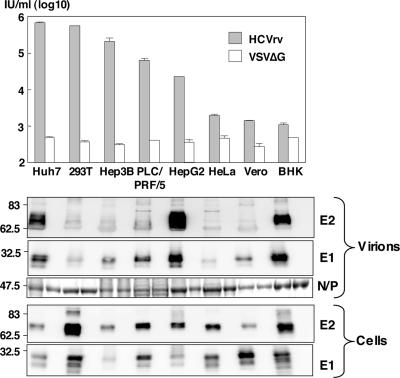
HCVrv was recovered from plasmids using established methods for the recovery of recombinant VSV in BHK cells. To ensure that infectious virus was produced, the recoveries were performed in cells transiently expressing VSV G protein. To determine if the HCV envelope proteins could mediate infection, the G-pseudotyped viruses were used to infect either Huh7 or 293T cells, and then the supernatants were titered on Huh7 cells. VSV lacking an envelope protein (VSVΔG) was used as a negative control. Infectivities of HCVrv generated in either 293T cells or Huh7 cells were dependent on the combination of incubation temperature and period (data not shown). The highest infectivity was constantly recovered in either cell line when cultured at 30°C for 48 h rather than when cultured at 37°C. Thus, HCVrv was prepared at 30°C for the remaining experiments. To determine whether the cell line used affected the generation of HCVrv, the infectivity in Huh7 cells and incorporation of HCV proteins into particles of HCVrv generated in various cell lines were examined (Fig. 2). HCVrv generated in Huh7 and 293T cells exhibited the highest infectivity in Huh7 cells, followed by Hep3B, PLC/PRF/5, and HepG2 cells. Significant infectivity was not observed for virus produced in HeLa, Vero, or BHK cells. Incorporation of the E1 and E2 proteins varied among the particles produced in the different cell lines. Although incorporation of E1 and E2 proteins into the particles was high in HCVrv generated in HepG2, BHK, and Huh7 cells, HCVrv produced in BHK cells exhibited the lowest infectivity to Huh7 cells. On the other hand, incorporation of the E1 and E2 proteins into HCVrv particles generated in 293T and Hep3B cells was low, whereas these viruses exhibited substantial infectivity to Huh7 cells. These results indicate that there is no clear correlation between the quantity of incorporation of HCV envelope proteins and the infectivity of HCVrv in Huh7 cells, although the producer cell type is important.
FIG. 2.
Production and characterization of HCVrv. (Top) The infectivity of HCVrv of the H77 strain produced in the indicated cell lines at 30°C for 48 h was determined in Huh7 cells at 37°C for 24 h postinfection by counting VSV N-positive cells. The results shown are from three independent assays, with the error bars representing the standard deviations. (Bottom) Expression and incorporation of the HCV E1 and E2 proteins in cells and purified viral particles. HCV E1 or E2 proteins were detected by immunoblotting with anti-E1 or anti-E2 monoclonal antibodies. An envelope-less pseudotype virus, VSVΔG, was used as a negative control.
Characterization of HCVrv and HCVpv.
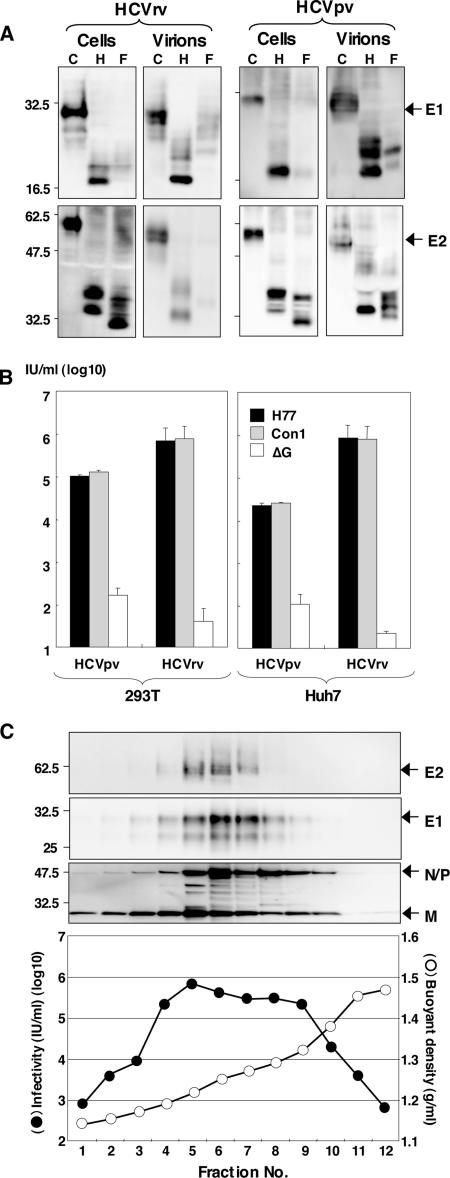
To examine the properties of the HCV envelope proteins incorporated into the recombinant and pseudotype VSV particles, E1 and E2 proteins of the H77 strain (genotype 1a) expressed in 293T cells and incorporated into the viral particles were examined by immunoblotting with anti-E1 (BDI198) and anti-E2 (AP33) monoclonal antibodies (Fig. 3A). The E1 and E2 proteins of the cell lysates and virions of HCVpv or HCVrv were sensitive to both Endo H and PNGase F, suggesting that both HCVrv and HCVpv possess E1 and E2 proteins with high-mannose glycans, as reported for the E1 and E2 proteins of HCVpp (48). Next, to examine the infectivity of HCVrv and HCVpv to the target cells, viruses bearing HCV envelope proteins of genotypes 1a (H77 strain) and 1b (Con1 strain) were generated in 293T or Huh7 cells and inoculated into Huh7 cells. The infectivities in Huh7 cells of HCVrv carrying E1 and E2 proteins of the H77 or Con1 strains were 10- to 20-fold higher (∼1 × 106 IU/ml) than those of HCVpv (∼1 × 105 IU/ml) (Fig. 3B). HCVpv generated in 293T cells exhibited higher infectivity than that generated in Huh7 cells. No difference in the infectivities of HCVpv and HCVrv between the 1a and 1b genotypes was observed. To determine the relationship between the incorporation of E1 and E2 proteins into HCVrv particles and their infectivities, the culture supernatants of 293T cells infected with HCVrv (H77 strain) were subjected to CsCl equilibrium gradient centrifugation, and each fraction was analyzed by immunoblotting and titration of infectivity in Huh7 cells (Fig. 3C). Immunoblot analyses revealed that incorporation of E1 and E2 proteins into HCVrv particles was detected in fractions 4 to 8 (Fig. 3C, top). These fractions exhibited the highest infectious titers (5 × 105 to 1 × 106 IU/ml), corresponding to buoyant densities of 1.2 to 1.3 g/ml (Fig. 3C, bottom).
FIG. 3.
Characterization of HCVrv and HCVpv. (A) The E1 and E2 proteins of the H77 strain expressed in 293T cells and incorporated into the particles of HCVrv and HCVpv were either untreated (C) or treated with endoglycosidase H (H) or peptide-N-glycosidase F (F). Following fractionation on sodium dodecyl sulfate-polyacrylamide gel gels, the glycoproteins were detected by immunoblotting with anti-E1 (BDI198) and anti-E2 (AP33) monoclonal antibodies. (B) The infectivities of HCVrv and HCVpv bearing HCV envelope proteins of genotypes 1a (H77 strain) and 1b (Con1 strain) generated in 293T or Huh7 cells were determined with Huh7 cells. The envelope-less VSV (ΔG) was used as a control. (C) (Top) CsCl gradient sedimentation of HCVrv produced in 293T cells. The supernatant was fractionated from the top of the gradient and analyzed by immunoblotting with anti-E2, anti-E1, and anti-VSV antibodies. (Bottom) The infectivity (filled circles) of each fraction was determined after the removal of CsCl with column purification. Fraction densities (open circles) are expressed in grams/milliliter.
Propagation of HCVrv.
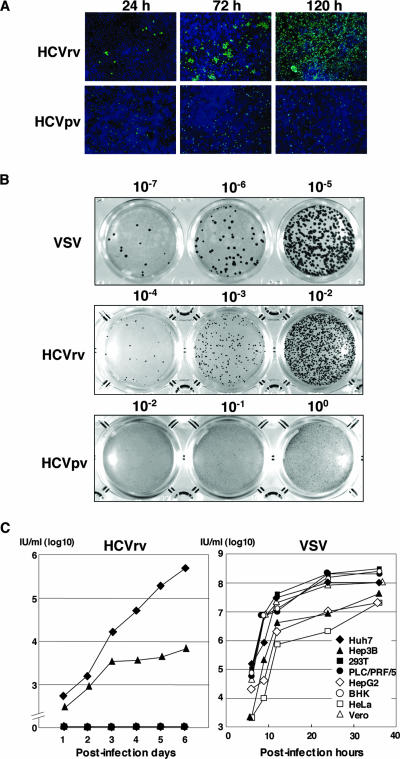
To examine the propagation of HCVrv in the target cells, Huh7 cells were infected with HCVrv at an MOI of 0.01 and incubated for up to 120 h. As a negative control, HCVpv was employed (Fig. 4A). A visible cytopathic effect was observed in Huh7 cells infected with HCVrv but not with HCVpv after 48 h incubation (data not shown). Immunofluorescence staining of Huh7 cells infected with HCVrv with antibody against VSV N revealed that VSV N protein was present from 24 h postinfection and had infected all cells at 120 h postinfection. In contrast, VSV N protein staining was decreased in cells infected with HCVpv at 120 h postinoculation. Focus formation of HCVrv in Huh7 cells was visualized by immunostaining under a methylcellulose overlay (Fig. 4B). Although the focus sizes of HCVrv were smaller than those of wild-type VSV, focus formation of HCVrv was clearly detected in a dose-dependent manner. In contrast, no focus formation was detected in cells infected with HCVpv. These results indicate that HCVrv is replication competent in Huh7 cells. To further determine the cell tropism for virus propagation, HCVrv was generated in various cell lines, and replication was assessed during incubation for up to 6 days (Fig. 4C, left). The growth kinetics of the wild-type VSV revealed an efficient replication of VSV in all the cell lines examined (Fig. 4C, right). Huh7 cells exhibited the highest susceptibility to propagation of HCVrv, followed by Hep3B cells, and no propagation was detected in the other cell lines. These results indicate that various human cell lines are capable of producing HCVrv that is infectious to Huh7 cells and that Huh7 cells are highly permissive to the propagation of HCVrv.
FIG. 4.
Propagation of HCVrv. (A) Detection of viral proteins in Huh7 cells infected with HCVpv or HCVrv. Huh7 cells were infected with HCVpv or HCVrv at an MOI of 0.01. Twenty-four, 72, and 120 h after infection, cells were fixed and stained with monoclonal antibody to VSV N protein and Alexa 488-conjugated secondary antibody. Cell nuclei were stained by Hoechst 33258. Pictures were taken using a fluorescence microscope by double exposure of the same fields with filters for Alexa 488 or Hoechst 33258. (B) Focus formation of HCVpv, HCVrv, or VSV in Huh7 cells. Huh7 cells were infected with serial 10-fold dilutions of HCVpv, HCVrv, or VSV and incubated at 30°C for 72 h for HCVpv and HCVrv or 24 h for VSV in a culture medium containing 0.8% methylcellulose. Foci of infected cells were detected by immunohistochemical staining. (C) Kinetics of HCVrv (left) and VSV (right) propagation in various cell lines. HCVrv and VSV generated in Huh7 cells were used to infect cells at an MOI of 0.01. The culture supernatant was collected at the indicated time points and titrated by a focus-formation assay. Infectious titers are expressed in IU/milliliter.
Involvement of hCD81 in the infection with HCVpv and HCVrv.
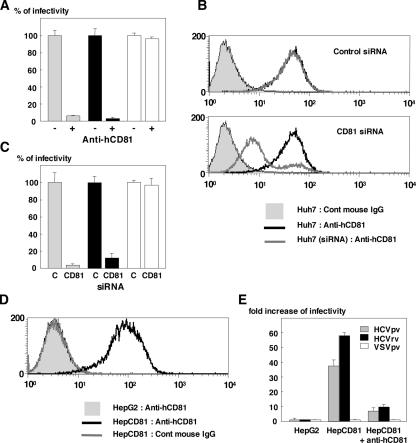
Among the candidates for entry receptor of HCV, hCD81 was shown to be most essential for the infection with HCVpp (5, 23) and HCVcc (27, 56, 60). The infection of Huh7 cells with HCVpv and HCVrv was inhibited by anti-hCD81 antibody, whereas no inhibition of VSVpv infection was observed (Fig. 5A). Treatment with siRNA targeted to hCD81 induced a reduction of hCD81 expression on the surface of Huh7 cells (Fig. 5B), and the susceptibility of hCD81-knockdown cells to infection with HCVpv and HCVrv, but not to that with VSVpv, was clearly reduced (Fig. 5C). To further determine the involvement of hCD81 in the infectivity of HCVpv and HCVrv, hCD81-negative HepG2 cells stably expressing hCD81 (HepCD81) were established, and fluorescence-activated cell sorter (FACS) analysis revealed that expression of hCD81 on the cell surface was higher than that of Huh7 cells (Fig. 5D). Although HCVpv and HCVrv are not infectious in HepG2 cells, HepCD81 cells were permissive to both HCVpv and HCVrv infection, and pretreatment with the anti-hCD81 antibody inhibited the infection of HepCD81 cells with HCVpv and HCVrv (Fig. 5E). These results indicate that hCD81 plays a crucial role in infection with HCVpv and HCVrv, as it has been reported to play in infection with HCVpp and HCVcc.
FIG. 5.
Involvement of hCD81 in the infection of HCVpv and HCVrv. (A) Effect of anti-hCD81 antibody on the infectivity of HCVpv (gray-filled bars), HCVrv (black-filled bars), or VSVpv (open bars) in Huh7 cells. (B) Cell surface expression of hCD81 on Huh7 cells transfected with siRNA targeted to hCD81 or control siRNA was examined by FACS analysis after staining with anti-hCD81 antibody. (C) Effect of knockdown of hCD81 in Huh7 cells by siRNA targeted to hCD81 on the infection of HCVpv, HCVrv, or VSVpv. (D) Cell surface expression of hCD81 on HepG2 and HepCD81 cells was examined by FACS analysis after staining with anti-hCD81 antibody. (E) Infectivity of HCVpv, HCVrv, or VSVpv to HepG2 or HepCD81 cells and the effect of anti-hCD81 antibody on the infection of the viruses to HepCD81 cells. The results shown are from three independent assays, with the error bars representing the standard deviations.
Infectivity of HCVpv and HCVrv in various cell lines.
To further examine the cell tropism of the viruses, HCVpv and HCVrv of the H77 and Con1 strains generated in 293T or Huh7 cells and HCVpp of the H77 strain generated in 293T cells were inoculated into various cell lines and primary Hc (Table 1). As expected, the control VSVΔG exhibited no infectivity in any of the cells examined (data not shown). The HCVpv and HCVrv derived from both genotypes were highly infectious in Huh7 cells, followed by HepCD81 and Hep3B cells, and weakly infectious in PLC/PRF/5, 293T, and Vero cells. No infectivity was detected in the other cell lines examined. The cell tropisms of the HCVpp were similar to those of HCVpv and HCVrv. Although the ectopic expression of hCD81 in Chinese hamster ovary cells (CHOCD81) did not confer susceptibility to HCVpv, HCVrv, or HCVpp infection, the expression of hCD81 in HepG2 cells (HepCD81) (Fig. 5A and D) rendered them permissive to infection with all of the viruses. Furthermore, Hc were not susceptible to the infection with HCVpv, HCVrv, or HCVpp, despite the expression of hCD81. These results suggest that expression of hCD81 is essential for the infection with HCVpv and HCVrv, as reported for infection with HCVpp and HCVcc, but conditions with a lack of hCD81 are insufficient for the infection with HCVpv, HCVrv, and HCVpp.
TABLE 1.
Infectivity of HCVpv, HCVrv, or HCVpp in various cells
| Target cells | Cell surface expression ofa:
|
Virus, producer cells, and strain (genotype)b
|
HCVpp virus produced in 293T cells and of strain H77 (genotype 1a)b | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HCVpv
|
HCVrv
|
||||||||||
| 293T
|
Huh7
|
293T
|
Huh7
|
||||||||
| hCD81 | SR-BI | H77 (1a) | Con1 (1b) | H77 (1a) | Con1 (1b) | H77 (1a) | Con1 (1b) | H77 (1a) | Con1 (1b) | ||
| Huh7 | ++ | ++ | +++ | +++ | ++ | ++ | +++ | +++ | +++ | +++ | +++ |
| HepG2 | − | ++ | − | − | − | − | − | − | − | − | − |
| HepCD81 | ++ | ++ | ++ | ++ | + | + | +++ | +++ | +++ | +++ | ++ |
| Hep3B | ++ | + | ++ | ++ | + | + | +++ | +++ | +++ | +++ | ++ |
| PLC/PRF/5 | ++ | + | + | + | − | − | + | + | + | + | − |
| FLC4 | − | ++ | − | − | − | − | − | − | − | − | − |
| Hc | ++ | − | − | − | − | − | − | − | − | − | − |
| HeLa | + | + | − | − | − | − | − | − | − | − | − |
| 293T | ++ | + | + | + | − | − | + | + | + | + | − |
| Vero | − | − | + | + | − | − | + | + | + | + | − |
| BHK | − | − | − | − | − | − | − | − | − | − | − |
| CHOK1 | − | − | − | − | − | − | − | − | − | − | − |
| CHOCD81 | ++ | − | − | − | − | − | − | − | − | − | − |
Cell surface expression of receptor candidates was examined by FACS analyses with specific antibodies. Mean fluorescence intensity shifts of less than 1, between 1 and 2, and between 2 and 3 are indicated as −, +, and ++, respectively.
Infectious titers higher than 5 × 104 IU/ml, between 5 × 103 and 5 × 104 IU/ml, between 5 × 102 and 5 × 103 IU/ml, and lower than 5 × 102 IU/ml are indicated as +++, ++, +, and −, respectively. The results were derived from at least three independent experiments, and the standard deviations did not exceed 30% of the mean values.
Neutralization of HCVpv and HCVrv infection by antibodies to HCV envelope proteins and sera of HCV patients.
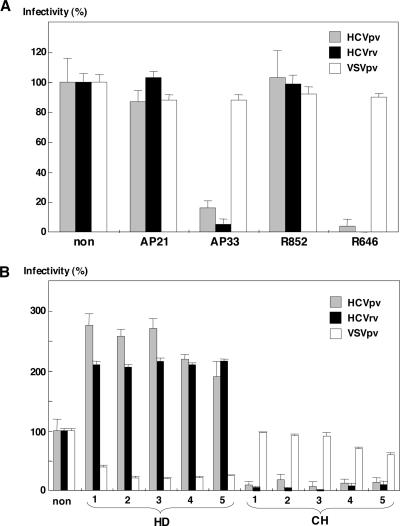
It has been reported that HCVpp can be neutralized by several well-characterized E2-specific monoclonal and polyclonal antibodies (5, 23, 49). The neutralization activity of anti-E1 (AP21.010) and anti-E2 (AP33) monoclonal antibodies (49) and anti-E1 (R852) and anti-E2 (R646) rabbit polyclonal antibodies raised against the E1 and E2 proteins of the H77 strain on the infection with HCVpv and HCVrv was determined (Fig. 6A). The infections with both HCVpv and HCVrv bearing E1 and E2 proteins of the H77 strain were clearly inhibited by anti-E2 (AP33) antibody or anti-E2 (R646) rabbit serum, consistent with a previous report on the effect of these antibodies on HCVpp infection (49), whereas no neutralization by AP21.010 and R852 antibodies was observed. The infections with HCVpv and HCVrv bearing E1 and E2 proteins of the Con1 strain were also inhibited by AP33 and R646 antibodies (data not shown), suggesting that the infectivity of HCVpv and HCVrv was cross-neutralized by anti-E2 antibody, as reported for HCVpp (49). These results indicate that the E2 protein plays a crucial role in the infectivity of both HCVpv and HCVrv. Although the addition of naïve human sera (HD) inhibited infection with VSVpv, infection with HCVpv or HCVrv was clearly enhanced, as reported for HCVpp infection of Huh7 cells (28, 42). To assess the neutralization ability of these antibodies in patients, HCVpv and HCVrv were incubated with a 2% concentration of the sera of chronic HCV patients infected with genotype 1b HCV (Fig. 6B). All of the sera of patients of genotype 1b showed high levels of neutralization activity against infection with HCVpv and HCVrv bearing envelope proteins of genotype 1a, whereas they had no effect on the infectivity of VSVpv, in contrast to the inhibition achieved by the naïve sera. These results indicate that HCV patients elicit high levels of antibodies that are likely to cross-neutralize the infectivity of HCVpv and HCVrv.
FIG. 6.
Neutralization of HCVpv and HCVrv infection by antibodies to HCV envelope proteins and sera of HCV patients. (A) Effect of anti-E1 (AP21.010) and anti-E2 (AP33) monoclonal antibodies and anti-E1 (R852) and anti-E2 (R646) rabbit sera on the infectivity of HCVpv (gray-filled bars), HCVrv (black-filled bars), or VSVpv (open bars) to Huh7 cells. The viruses were preincubated for 1 h at room temperature with the antibodies before infection of Huh7 cells. (B) Effects of human sera from healthy donors and HCV patients on the infection of HCVpv, HCVrv, or VSVpv. The viruses were preincubated for 1 h at room temperature with five different healthy human sera (HD) or chronic HCV patient sera (CH) diluted 1:50 before infection of Huh7 cells.
Inhibition of HCVpv and HCVrv infection by bafilomycin A1.
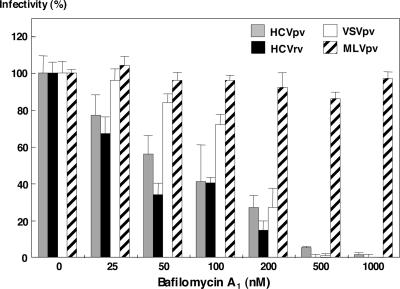
Enveloped viruses enter target cells through two different pathways: one is a pH-independent direct fusion at the plasma membrane, and the other is a pH-dependent receptor-mediated endocytosis (58). Previous studies have revealed that both HCVpp and HCVcc were sensitive to the inhibitors of vacuolar acidification, such as ammonium chloride, concanamycin A, or bafilomycin A1, suggesting that these viruses enter via a pH-dependent endocytosis into target cells (23, 61). To determine the entry pathway of HCVpv and HCVrv, Huh7 cells were pretreated with various concentrations of bafilomycin A1, and then the cells were inoculated with HCVpv, HCVrv, VSVpv, and MLVpv (Fig. 7). As expected, the treatment did not affect the infection with MLVpv bearing an envelope protein of MLV that enters cells via a pH-independent pathway. In contrast, infection with VSVpv bearing the G protein of VSV, which enters cells through pH-dependent endocytosis, was inhibited by the treatment with bafilomycin A1 in a dose-dependent manner. Infection with HCVpv and HCVrv was also clearly inhibited by the treatment with bafilomycin A1 in a dose-dependent manner, as with VSVpv. This suggests that low pH exposure is essential for the entry of HCVpv and HCVrv.
FIG. 7.
Inhibition of HCVpv and HCVrv infection by bafilomycin A1. HCVpv (gray-filled bars), HCVrv (black-filled bars), VSVpv (open bars), or MLVpv (striped bars) were inoculated to Huh7 cells after treatment with various concentrations of bafilomycin A1. The results shown are from three independent assays, with the error bars representing the standard deviations.
Effects of ER α-glucosidase inhibitors on HCVrv infection.
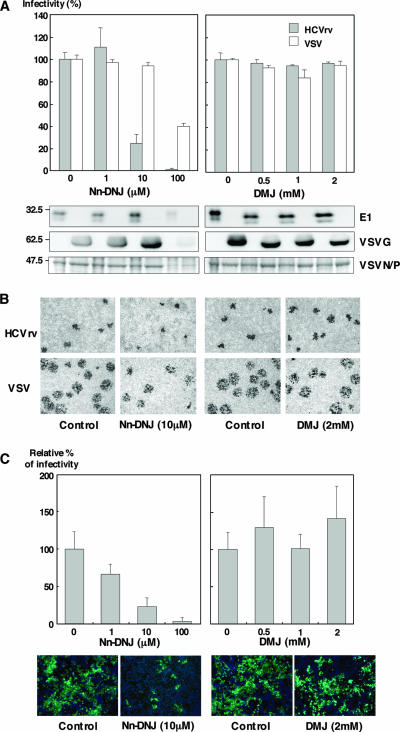
Previous studies have shown that deoxynojirimycin (DNJ) and Nn-DNJ, a long-alkyl-chain iminosugar derivative of DNJ, inhibit the infection of flaviviruses such as Japanese encephalitis virus (JEV) and dengue virus in a dose-dependent manner (15, 64). Although the effects of glycosylation inhibitors on the folding and assembly of HCV envelope proteins in the N-glycosylation steps and the binding properties of HCV-LP produced in insect cells have been reported (11, 12), glycobiological analyses of HCV envelope proteins involved in virus infectivity have not been reported yet. To determine the effects of the inhibitor of Golgi mannosidase (DMJ) and of ER α-glucosidase (Nn-DNJ) on the infectivity of HCVrv, Huh7 cells were treated with these inhibitors. Treatment of Huh7 cells with Nn-DNJ but not with DMJ reduced the infectivity of HCVrv in a dose-dependent manner, and this reduction was more efficient than that in the infectivity of VSV (Fig. 8A, top). Although immunoblotting and Coomassie staining of the particles revealed that incorporation of the envelope proteins and generation of HCVrv and VSV particles recovered from cells treated with 100 μM of Nn-DNJ were severely impaired by the cytotoxic effects of Nn-DNJ (Fig. 8A, bottom left), treatment with 10 μM of Nn-DNJ selectively reduced the infectivity of HCVrv but not of VSV without any cytotoxic effect (Fig. 8A, top left). In contrast, Huh7 cells treated with more than 0.5 mM of DMJ exhibited a slight reduction of molecular sizes of E1 or VSVG proteins incorporated into the particles (Fig. 8A, bottom right); no effect on the incorporation of the envelope proteins into the viral particles and the infectivity was observed (Fig. 8A, top right). Next, we assessed the effects of the inhibitors on the propagation of the viruses. Focus formation of HCVrv was also inhibited by the treatment with Nn-DNJ but not with DMJ (Fig. 8B). To further confirm the effect of modification of the envelope glycoproteins by ER α-glucosidase on the infectivity of HCV, Huh7.5.1 cells were treated with the inhibitors, and infectivity of HCVcc was determined (Fig. 8C, top). Treatment with Nn-DNJ clearly inhibited the infection with HCVcc in a dose-dependent manner, as it did the infection with HCVrv. Focus formation of HCVcc was also inhibited by the treatment of Huh7.5.1 cells with Nn-DNJ (Fig. 8C, bottom). These results indicate that modification of the glycans of HCV E1 and E2 proteins in the ER by α-glucosidase rather than that in the Golgi is crucial for the infectivity of both HCVrv and HCVcc.
FIG. 8.
Effects of ER α-glucosidase inhibitors on the infection with HCVrv and HCVcc. (A) (Top) Production of HCVrv and VSV in the presence of Nn-DNJ (left) or DMJ (right). Huh7 cells infected with HCVrv and VSV at MOIs of 0.1 and 0.01, respectively, were treated with various concentrations of Nn-DNJ or DMJ. Seventy-two hours (HCVrv) or 24 h (VSV) postinfection, culture supernatants were collected and titrated on Huh7 cells by a focus-forming assay. The results shown are from three independent assays, with the error bars representing the standard deviations. (Bottom) Purified viruses generated in Huh7 cells treated with Nn-DNJ or DMJ were analyzed by immunoblotting with anti-E1 (BDI198) and anti-VSVG (ab34774) or Coomassie staining. (B) Focus formation of HCVrv and VSV in the presence of Nn-DNJ or DMJ. Huh7 cells were infected with HCVrv or VSV, treated with Nn-DNJ (10 μM) or DMJ (2 mM) prior to an overlay of culture media containing 0.8% of methylcellulose, and stained with an anti-VSV N antibody after fixation at 72 h (HCVrv) and 24 h (VSV) postinfection. (C) (Top) Production of HCVcc in the presence of Nn-DNJ (left) or DMJ (right). Huh7.5.1 cells infected with HCVcc at an MOI of 0.01 were treated with various concentrations of Nn-DNJ or DMJ. Culture supernatants were collected and titrated by a quantitative core enzyme-linked immunosorbent assay at 96 h postinfection. (Bottom) Immunofluorescence assay of HCVcc infection in the presence of Nn-DNJ or DMJ. Huh7.5.1 cells were infected with HCVcc atan MOI of 0.01, treated with 10 μM of Nn-DNJ or 2 mM of DMJ prior to an overlay of culture media containing 0.8% of methylcellulose, and stained with an anti-NS5A antibody and Alexa 488-conjugated secondary antibody after fixation at 96 h postinfection. Cell nuclei were stained by Hoechst 33258. Pictures were taken using a fluorescence microscope by double exposure of the same fields with filters for Alexa 488 or Hoechst 33258.
DISCUSSION
In general, enveloped viruses attach to host target cells and enter into cells through the interaction between viral envelope proteins and cell surface receptors and coreceptors. Due to the lack of a robust cell culture system to support the replication of various HCV genotypes, surrogate systems have been developed to examine the mechanisms of HCV infection. Although in vitro binding assays have identified several candidate receptors for HCV (4), the final determination of a true entry receptor or coreceptor capable of internalizing HCV particles has to be made by an infection assay. Toward this end, pseudotype virus systems based on VSV (27, 39) and retroviruses (5, 23) have been established. Both VSV and retroviruses normally bud from the plasma membrane, and therefore foreign envelope proteins expressed on the cell surface have been believed to incorporate into the pseudotype particles. HCV E1 and E2 proteins form heterodimers that have static ER retention signals in their C-terminal transmembrane region (17) and pulse-chase experiments and endoglycanase treatment of the intracellular forms of the proteins or those incorporated into the HCVpp have revealed that only a small fraction of the HCV envelope proteins are translocated to the plasma membrane and modified to the complex-type glycans (48). In addition, it was demonstrated that recruitment of the foreign envelope proteins by MLV and the lentivirus core protein does not occur at the cell surface but takes place intracellularly in the endosomal pathway (55, 56). Production of pseudotype VSVs bearing unmodified envelope glycoproteins of bunyaviruses has also been reported, in spite of the static retention of the envelope glycoproteins in the intracellular compartment and the lack of translocation into the plasma membrane (46). Therefore, cell surface expression of HCV envelope glycoproteins may not necessarily be a prerequisite for generation of pseudotype particles based on VSV or retroviruses.
Recombinant VSV encoding foreign viral envelope proteins in place of the G protein has been shown to be a powerful tool for the investigation of viral infection and the development of vaccines for diseases caused by infection with viruses such as influenza virus, human immunodeficiency virus, respiratory syncytial virus, human papillomavirus, and filoviruses (20, 31). Although recombinant VSV encoding HCV envelope proteins has been generated as a surrogate model for HCV infection and a vaccine vector (9, 35), recombinant VSV generated in rodent cells possessing the chimeric E1 and/or E2 proteins has been shown to be noninfectious in a human hepatoma cell line that is susceptible to HCVpp infection (9). In this study, we successfully generated infectious recombinant and pseudotype VSVs incorporating unmodified E1 and E2 proteins in hepatic and nonhepatic human cell lines. The previously observed lack of infectivity of the recombinant VSV carrying the chimeric HCV envelope proteins might be attributable to the production of viral particles in rodent (BHK) cells (9), because in this study the HCVrv generated in BHK cells exhibited no infectivity in the target cells in spite of a sufficient amount of incorporation of the HCV envelope proteins. These results suggest that posttranslational modification or host factor(s) specific to human cells might be involved in the endowment of infectivity of recombinant VSVs. Furthermore, HCVrv can be produced in various cell lines upon infection with the G-complemented particles, which are known to exhibit infectivity in several cell lines, in contrast to the pseudotype viruses, infectious particles of which were recovered only in cells exhibiting a high competency of transfection, such as 293T cells. Therefore, generation of HCVrv in various human cells, including nonhepatic cells such as B cells, might be useful for investigating the cell-specific modification and/or factors determining the cell tropism of HCV infection.
Overwhelming evidence that hCD81 facilitates the entry of HCV into Hc via interaction with the E2 protein has been accumulated not only by surrogate models, such as purified E2 proteins, HCV-LP, and HCVpp, but also by authentic HCV particles and HCVcc of genotype 2a (4). In this study, both HCVpv and HCVrv were shown to be infectious in Huh7 cells, and this infectivity was shown to be mediated through the interaction with hCD81. Although overexpression of hCD81 in HepG2 cells which lack endogenous expression of hCD81 renders them susceptible to infection by surrogate viruses, primary human Hc and HeLa cells expressing hCD81 and the rodent CHO cells stably expressing hCD81 (CHOCD81 cells) were resistant to infection by HCVrv and HCVpv (Table 1) (5, 14, 67), suggesting that hCD81 is one of the important factors for HCV entry but is not sufficient for infectivity of HCV in target cells. Recently, it was shown that participation of hCD81 in the infection of HCVpp or HCVcc bearing HCV envelope proteins isolated during chronic HCV infection was reduced, suggesting that the affinity of HCV envelope proteins to hCD81 was reduced and HCV utilizes receptors other than hCD81 (62, 69). HCVrv is useful for studies of the generation of various genotypes of escape variants under pressure of neutralization antibody or antagonist against HCV receptor candidates. Further studies of the functional relevance of hCD81 and other receptor candidates in the entry steps of HCV, such as binding, endocytosis, and membrane fusion, are needed.
Bafilomycin A1, an H+-ATPase inhibitor, was shown to reduce the infectivities of both HCVpv and HCVrv in a dose-dependent manner, as it did for the infectivities of both HCVpp and HCVcc (6, 23, 29, 61), suggesting that these viruses require low-pH-induced conformational changes of the envelope proteins upon entry. Furthermore, as with HCVcc (40, 61), preexposure of HCVpv and HCVrv to acidic pH did not reduce their infectivity (data not shown), indicating that additional factors are required for the internalization of the viruses. Recently, entry of HCVpp was shown to depend on the clathrin-mediated endocytosis through the knockdown of clathrin heavy chain by siRNA or chlorpromazine (8, 40), and dominant-negative mutants of Rab5 or Rab7, which are involved in the transport of clathrin-coated vesicles, revealed that entry of HCVpp requires delivery to early but not to late endosomes (40). N-linked glycosylation processing events in the ER are important for the secretion of several enveloped viruses. ER α-glucosidase I and II are involved in the trimming of terminal glucose on the core oligosaccharides, and the resulting monoglucosylated glycoproteins are able to bind to the ER chaperones calnexin (CNX) and/or calreticulin (CRT). ER α-glucosidase inhibitors, DNJ or castanospermine, which block the trimming step of N-linked glycosylation, have been shown to prevent the interaction of CNX and/or CRT with the folding glycoproteins, and the production of many enveloped viruses is inhibited by these inhibitors (41). In this study, we found that infection with both HCVrv and HCVcc was inhibited in a dose-dependent manner by treatment with Nn-DNJ, which is an N-alkylated derivative of DNJ exhibiting a stronger effect than DNJ. HCV E1 and E2 proteins were shown to interact with CNX and CRT, and these interactions were inhibited by the treatment with ER α-glucosidase inhibitors (12). One possible function of the HCV p7 protein, the formation of ion channels, has also been shown to be inhibited by the treatment with long-alkyl-chain iminosugar derivatives (50). Recently, it was reported that HCV-LPs produced in the presence of ER α-glucosidase inhibitors incorporated unprocessed, triglucosylated N-glycans and misfolded E1 and E2 proteins and lost their ability to bind hepatoma cell lines (11). Our results demonstrate that the modification of E1 and E2 proteins in the glycosylation steps in the ER is required to confer infectivity to HCVrv and HCVcc. The presence of E1 and E2 proteins on the surrogate viruses and HCVcc possessing high-mannose glycans indicate that these viruses are not released through the trans-Golgi network. In the case of West Nile virus, mature particles propagated in mammalian cells possess complex types of carbohydrates, in contrast to those generated in insect cells, which have high-mannose glycans (16). We still do not know the exact nature of modifications of the mature envelope proteins on authentic HCV particles. Further studies of the relationship between the modification of HCV envelope proteins and their infectivity are needed to clarify the life cycle of HCV. The neutralizing activity of antibodies against HCV have been assessed in the past using HCVpv (10, 43), HCVpp (3, 33, 42), and HCVcc (63, 65), as well as by the inhibition of binding of purified E2 protein to hCD81 (24, 53) and of HCV-LP to target cells (59). Sera from patients chronically infected with HCV and experimentally infected chimpanzees were shown to specifically neutralize HCVpp infection (3, 33, 42). In the present study, sera from patients infected with genotype 1b of HCV and anti-E2 monoclonal antibodies exhibited high levels of neutralization activity against infection with both HCVpv and HCVrv bearing HCV envelope proteins of genotypes 1a and 1b. One of the characteristics of HCV infection is the establishment of a persistent infection. Therefore, the high prevalence of neutralizing antibodies to the surrogate viruses and HCVcc suggests that HCV particles exhibiting similar phenotypes to surrogate viruses and HCVcc would be easily eliminated by neutralizing antibodies and thus not be able to participate in the establishment of a persistent infection. Recently, it was reported that HCV escapes from neutralizing antibody and T-cell responses by the continuous generation of escape variants during chronic infection (51, 62). However, it was demonstrated that viral clearance in acute HCV infection was not correlated with the presence of neutralizing antibodies against HCVpp (33, 42), and 75% of HCVpp bearing HCV envelope proteins of various genotypes are not infectious (29). Therefore, it is reasonable to speculate that HCV particles exhibiting characteristics similar to those of the surrogate viruses are produced in large numbers and act as decoys in HCV patients, eliciting strong neutralizing antibodies against the viruses, and that a small portion of HCV particles exhibiting characteristics different from those of the surrogate viruses may participate in the establishment of persistent infection by escaping from the host immune surveillance system. The authenticity of the surrogate virus systems for the study of HCV infection remains controversial, and further studies are needed to clarify their profiles.
In conclusion, we generated replication-incompetent HCVpv and replication-competent HCVrv possessing HCV envelope proteins as novel surrogate models for the study of HCV. HCVpv and HCVrv were shown to have infection mechanisms similar to those of HCVpp and HCVcc. HCVrv has the following advantages compared to HCVcc: (i) infectious particles bearing HCV envelope proteins of various genotypes are capable of generating in various cell lines or primary cells, in contrast to the strict restriction of generating the infectious HCVcc in the Huh7-derived cell lines; (ii) isolation of escape mutants carrying mutations in the envelope proteins under various pressures may be easily obtained due to the higher replication efficiency than that of HCVcc; and (iii) in vivo investigation of the HCV envelope proteins for entry using humanized mice with human Hc and for immunogenicity for a future vaccine development are possible. Therefore, replication-competent HCVrv established in this study may provide valuable tools not only for understanding the entry mechanisms of HCV in a manner that is cell type and species dependent but also for developing novel therapeutics and vaccines.
Acknowledgments
We thank H. Murase for secretarial work and T. Ohtsubaki for excellent technical assistance. We also thank F. Cosset and F. Chisari for provision of the HCVpp system and Huh7.5.1 cells.
This research was supported in part by grants-in-aid from the Ministry of Health, Labor, and Welfare; the Ministry of Education, Culture, Sports, Science, and Technology; and the 21st Century Center of Excellence Program of Japan and by the Foundation for Biomedical Research and Innovation, Japan. H.T. was supported by research fellowships of the Japan Society for the Promotion of Science for Young Scientists.
Footnotes
Published ahead of print on 6 June 2007.
REFERENCES
- 1.Agnello, V., G. Abel, M. Elfahal, G. B. Knight, and Q. X. Zhang. 1999. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 96:12766-12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barth, H., C. Schafer, M. I. Adah, F. Zhang, R. J. Linhardt, H. Toyoda, A. Kinoshita-Toyoda, T. Toida, T. H. Van Kuppevelt, E. Depla, F. Von Weizsacker, H. E. Blum, and T. F. Baumert. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 278:41003-41012. [DOI] [PubMed] [Google Scholar]
- 3.Bartosch, B., J. Bukh, J. C. Meunier, C. Granier, R. E. Engle, W. C. Blackwelder, S. U. Emerson, F. L. Cosset, and R. H. Purcell. 2003. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc. Natl. Acad. Sci. USA 100:14199-14204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartosch, B., and F. L. Cosset. 2006. Cell entry of hepatitis C virus. Virology 348:1-12. [DOI] [PubMed] [Google Scholar]
- 5.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartosch, B., A. Vitelli, C. Granier, C. Goujon, J. Dubuisson, S. Pascale, E. Scarselli, R. Cortese, A. Nicosia, and F. L. Cosset. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624-41630. [DOI] [PubMed] [Google Scholar]
- 7.Baumert, T. F., S. Ito, D. T. Wong, and T. J. Liang. 1998. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J. Virol. 72:3827-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchard, E., S. Belouzard, L. Goueslain, T. Wakita, J. Dubuisson, C. Wychowski, and Y. Rouille. 2006. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 80:6964-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buonocore, L., K. J. Blight, C. M. Rice, and J. K. Rose. 2002. Characterization of vesicular stomatitis virus recombinants that express and incorporate high levels of hepatitis C virus glycoproteins. J. Virol. 76:6865-6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burioni, R., Y. Matsuura, N. Mancini, H. Tani, T. Miyamura, P. E. Varaldo, and M. Clementi. 2002. Diverging effects of human recombinant anti-hepatitis C virus (HCV) antibody fragments derived from a single patient on the infectivity of a vesicular stomatitis virus/HCV pseudotype. J. Virol. 76:11775-11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapel, C., C. Garcia, P. Roingeard, N. Zitzmann, J. Dubuisson, R. A. Dwek, C. Trepo, F. Zoulim, and D. Durantel. 2006. Antiviral effect of alpha-glucosidase inhibitors on viral morphogenesis and binding properties of hepatitis C virus-like particles. J. Gen. Virol. 87:861-871. [DOI] [PubMed] [Google Scholar]
- 12.Choukhi, A., S. Ung, C. Wychowski, and J. Dubuisson. 1998. Involvement of endoplasmic reticulum chaperones in the folding of hepatitis C virus glycoproteins. J. Virol. 72:3851-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clayton, R. F., A. Owsianka, J. Aitken, S. Graham, D. Bhella, and A. H. Patel. 2002. Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles. J. Virol. 76:7672-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cormier, E. G., F. Tsamis, F. Kajumo, R. J. Durso, J. P. Gardner, and T. Dragic. 2004. CD81 is an entry coreceptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 101:7270-7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Courageot, M. P., M. P. Frenkiel, C. D. Dos Santos, V. Deubel, and P. Despres. 2000. Alpha-glucosidase inhibitors reduce dengue virus production by affecting the initial steps of virion morphogenesis in the endoplasmic reticulum. J. Virol. 74:564-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis, C. W., H. Y. Nguyen, S. L. Hanna, M. D. Sanchez, R. W. Doms, and T. C. Pierson. 2006. West Nile virus discriminates between DC-SIGN and DC-SIGNR for cellular attachment and infection. J. Virol. 80:1290-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubuisson, J. 2000. Folding, assembly and subcellular localization of hepatitis C virus glycoproteins. Curr. Top. Microbiol. Immunol. 242:135-148. [DOI] [PubMed] [Google Scholar]
- 18.Evans, M. J., T. von Hahn, D. M. Tscherne, A. J. Syder, M. Panis, B. Wolk, T. Hatziioannou, J. A. McKeating, P. D. Bieniasz, and C. M. Rice. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801-805. [DOI] [PubMed] [Google Scholar]
- 19.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garbutt, M., R. Liebscher, V. Wahl-Jensen, S. Jones, P. Moller, R. Wagner, V. Volchkov, H. D. Klenk, H. Feldmann, and U. Stroher. 2004. Properties of replication-competent vesicular stomatitis virus vectors expressing glycoproteins of filoviruses and arenaviruses. J. Virol. 78:5458-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardner, J. P., R. J. Durso, R. R. Arrigale, G. P. Donovan, P. J. Maddon, T. Dragic, and W. C. Olson. 2003. L-SIGN (CD 209L) is a liver-specific capture receptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 100:4498-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamamoto, I., Y. Nishimura, T. Okamoto, H. Aizaki, M. Liu, Y. Mori, T. Abe, T. Suzuki, M. M. Lai, T. Miyamura, K. Moriishi, and Y. Matsuura. 2005. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J. Virol. 79:13473-13482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishii, K., D. Rosa, Y. Watanabe, T. Katayama, H. Harada, C. Wyatt, K. Kiyosawa, H. Aizaki, Y. Matsuura, M. Houghton, S. Abrignani, and T. Miyamura. 1998. High titers of antibodies inhibiting the binding of envelope to human cells correlate with natural resolution of chronic hepatitis C. Hepatology 28:1117-1120. [DOI] [PubMed] [Google Scholar]
- 25.Jeetendra, E., K. Ghosh, D. Odell, J. Li, H. P. Ghosh, and M. A. Whitt. 2003. The membrane-proximal region of vesicular stomatitis virus glycoprotein G ectodomain is critical for fusion and virus infectivity. J. Virol. 77:12807-12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanda, T., A. Basu, R. Steele, T. Wakita, J. S. Ryerse, R. Ray, and R. B. Ray. 2006. Generation of infectious hepatitis C virus in immortalized human hepatocytes. J. Virol. 80:4633-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lagging, L. M., K. Meyer, R. J. Owens, and R. Ray. 1998. Functional role of hepatitis C virus chimeric glycoproteins in the infectivity of pseudotyped virus. J. Virol. 72:3539-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavillette, D., Y. Morice, G. Germanidis, P. Donot, A. Soulier, E. Pagkalos, G. Sakellariou, L. Intrator, B. Bartosch, J. M. Pawlotsky, and F. L. Cosset. 2005. Human serum facilitates hepatitis C virus infection, and neutralizing responses inversely correlate with viral replication kinetics at the acute phase of hepatitis C virus infection. J. Virol. 79:6023-6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavillette, D., A. W. Tarr, C. Voisset, P. Donot, B. Bartosch, C. Bain, A. H. Patel, J. Dubuisson, J. K. Ball, and F. L. Cosset. 2005. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology 41:265-274. [DOI] [PubMed] [Google Scholar]
- 30.Lawson, N. D., E. A. Stillman, M. A. Whitt, and J. K. Rose. 1995. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. USA 92:4477-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lichty, B. D., A. T. Power, D. F. Stojdl, and J. C. Bell. 2004. Vesicular stomatitis virus: re-inventing the bullet. Trends Mol. Med. 10:210-216. [DOI] [PubMed] [Google Scholar]
- 32.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 33.Logvinoff, C., M. E. Major, D. Oldach, S. Heyward, A. Talal, P. Balfe, S. M. Feinstone, H. Alter, C. M. Rice, and J. A. McKeating. 2004. Neutralizing antibody response during acute and chronic hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 101:10149-10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lozach, P. Y., H. Lortat-Jacob, A. de Lacroix de Lavalette, I. Staropoli, S. Foung, A. Amara, C. Houles, F. Fieschi, O. Schwartz, J. L. Virelizier, F. Arenzana-Seisdedos, and R. Altmeyer. 2003. DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J. Biol. Chem. 278:20358-20366. [DOI] [PubMed] [Google Scholar]
- 35.Majid, A. M., H. Ezelle, S. Shah, and G. N. Barber. 2006. Evaluating replication-defective vesicular stomatitis virus as a vaccine vehicle. J. Virol. 80:6993-7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Major, M. E., B. Rehermann, and S. M. Feinstone. 2001. Hepatitis C viruses, p. 1127-1162. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 37.Matsuo, E., H. Tani, C. K. Lim, Y. Komoda, T. Okamoto, H. Miyamoto, K. Moriishi, S. Yagi, A. H. Patel, T. Miyamura, and Y. Matsuura. 2006. Characterization of HCV-like particles produced in a human hepatoma cell line by a recombinant baculovirus. Biochem. Biophys. Res. Commun. 340:200-208. [DOI] [PubMed] [Google Scholar]
- 38.Matsuura, Y., T. Suzuki, R. Suzuki, M. Sato, H. Aizaki, I. Saito, and T. Miyamura. 1994. Processing of E1 and E2 glycoproteins of hepatitis C virus expressed in mammalian and insect cells. Virology 205:141-150. [DOI] [PubMed] [Google Scholar]
- 39.Matsuura, Y., H. Tani, K. Suzuki, T. Kimura-Someya, R. Suzuki, H. Aizaki, K. Ishii, K. Moriishi, C. S. Robison, M. A. Whitt, and T. Miyamura. 2001. Characterization of pseudotype VSV possessing HCV envelope proteins. Virology 286:263-275. [DOI] [PubMed] [Google Scholar]
- 40.Meertens, L., C. Bertaux, and T. Dragic. 2006. Hepatitis C virus entry requires a critical postinternalization step and delivery to early endosomes via clathrin-coated vesicles. J. Virol. 80:11571-11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mehta, A., N. Zitzmann, P. M. Rudd, T. M. Block, and R. A. Dwek. 1998. Alpha-glucosidase inhibitors as potential broad based anti-viral agents. FEBS Lett. 430:17-22. [DOI] [PubMed] [Google Scholar]
- 42.Meunier, J. C., R. E. Engle, K. Faulk, M. Zhao, B. Bartosch, H. Alter, S. U. Emerson, F. L. Cosset, R. H. Purcell, and J. Bukh. 2005. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc. Natl. Acad. Sci. USA 102:4560-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer, K., A. Basu, C. T. Przysiecki, L. M. Lagging, A. M. Di Bisceglie, A. J. Conley, and R. Ray. 2002. Complement-mediated enhancement of antibody function for neutralization of pseudotype virus containing hepatitis C virus E2 chimeric glycoprotein. J. Virol. 76:2150-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moriishi, K., and Y. Matsuura. 2003. Mechanisms of hepatitis C virus infection. Antivir. Chem. Chemother. 14:285-297. [DOI] [PubMed] [Google Scholar]
- 45.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 46.Ogino, M., H. Ebihara, B. H. Lee, K. Araki, A. Lundkvist, Y. Kawaoka, K. Yoshimatsu, and J. Arikawa. 2003. Use of vesicular stomatitis virus pseudotypes bearing hantaan or seoul virus envelope proteins in a rapid and safe neutralization test. Clin. Diagn. Lab. Immunol. 10:154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okamoto, T., Y. Nishimura, T. Ichimura, K. Suzuki, T. Miyamura, T. Suzuki, K. Moriishi, and Y. Matsuura. 2006. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 25:5015-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Op De Beeck, A., C. Voisset, B. Bartosch, Y. Ciczora, L. Cocquerel, Z. Keck, S. Foung, F. L. Cosset, and J. Dubuisson. 2004. Characterization of functional hepatitis C virus envelope glycoproteins. J. Virol. 78:2994-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Owsianka, A., A. W. Tarr, V. S. Juttla, D. Lavillette, B. Bartosch, F. L. Cosset, J. K. Ball, and A. H. Patel. 2005. Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J. Virol. 79:11095-11104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pavlovic, D., D. C. Neville, O. Argaud, B. Blumberg, R. A. Dwek, W. B. Fischer, and N. Zitzmann. 2003. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc. Natl. Acad. Sci. USA 100:6104-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pestka, J. M., M. B. Zeisel, E. Blaser, P. Schurmann, B. Bartosch, F. L. Cosset, A. H. Patel, H. Meisel, J. Baumert, S. Viazov, K. Rispeter, H. E. Blum, M. Roggendorf, and T. F. Baumert. 2007. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc. Natl. Acad. Sci. USA 104:6025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 53.Rosa, D., S. Campagnoli, C. Moretto, E. Guenzi, L. Cousens, M. Chin, C. Dong, A. J. Weiner, J. Y. Lau, Q. L. Choo, D. Chien, P. Pileri, M. Houghton, and S. Abrignani. 1996. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc. Natl. Acad. Sci. USA 93:1759-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rose, J. K., L. Buonocore, and M. A. Whitt. 1991. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques 10:520-525. [PubMed] [Google Scholar]
- 55.Sandrin, V., P. Boulanger, F. Penin, C. Granier, F. L. Cosset, and B. Bartosch. 2005. Assembly of functional hepatitis C virus glycoproteins on infectious pseudoparticles occurs intracellularly and requires concomitant incorporation of E1 and E2 glycoproteins. J. Gen. Virol. 86:3189-3199. [DOI] [PubMed] [Google Scholar]
- 56.Sandrin, V., D. Muriaux, J. L. Darlix, and F. L. Cosset. 2004. Intracellular trafficking of Gag and Env proteins and their interactions modulate pseudotyping of retroviruses. J. Virol. 78:7153-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scarselli, E., H. Ansuini, R. Cerino, R. M. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneider-Schaulies, J. 2000. Cellular receptors for viruses: links to tropism and pathogenesis. J. Gen. Virol. 81:1413-1429. [DOI] [PubMed] [Google Scholar]
- 59.Steinmann, D., H. Barth, B. Gissler, P. Schurmann, M. I. Adah, J. T. Gerlach, G. R. Pape, E. Depla, D. Jacobs, G. Maertens, A. H. Patel, G. Inchauspe, T. J. Liang, H. E. Blum, and T. F. Baumert. 2004. Inhibition of hepatitis C virus-like particle binding to target cells by antiviral antibodies in acute and chronic hepatitis C. J. Virol. 78:9030-9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tamura, K., A. Oue, A. Tanaka, N. Shimizu, H. Takagi, N. Kato, A. Morikawa, and H. Hoshino. 2005. Efficient formation of vesicular stomatitis virus pseudotypes bearing the native forms of hepatitis C virus envelope proteins detected after sonication. Microbes Infect. 7:29-40. [DOI] [PubMed] [Google Scholar]
- 61.Tscherne, D. M., C. T. Jones, M. J. Evans, B. D. Lindenbach, J. A. McKeating, and C. M. Rice. 2006. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 80:1734-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.von Hahn, T., J. C. Yoon, H. Alter, C. M. Rice, B. Rehermann, P. Balfe, and J. A. McKeating. 2007. Hepatitis C virus continuously escapes from neutralizing antibody and T-cell responses during chronic infection in vivo. Gastroenterology 132:667-678. [DOI] [PubMed] [Google Scholar]
- 63.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu, S. F., C. J. Lee, C. L. Liao, R. A. Dwek, N. Zitzmann, and Y. L. Lin. 2002. Antiviral effects of an iminosugar derivative on flavivirus infections. J. Virol. 76:3596-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yi, M., R. A. Villanueva, D. L. Thomas, T. Wakita, and S. M. Lemon. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. USA 103:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu, M. Y., B. Bartosch, P. Zhang, Z. P. Guo, P. M. Renzi, L. M. Shen, C. Granier, S. M. Feinstone, F. L. Cosset, and R. H. Purcell. 2004. Neutralizing antibodies to hepatitis C virus (HCV) in immune globulins derived from anti-HCV-positive plasma. Proc. Natl. Acad. Sci. USA 101:7705-7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, J., G. Randall, A. Higginbottom, P. Monk, C. M. Rice, and J. A. McKeating. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 78:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhong, J., P. Gastaminza, J. Chung, Z. Stamataki, M. Isogawa, G. Cheng, J. A. McKeating, and F. V. Chisari. 2006. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J. Virol. 80:11082-11093. [DOI] [PMC free article] [PubMed] [Google Scholar]