Abstract
The molecular chaperone heat shock protein 90 (Hsp90) is involved in multiple cellular processes including protein maturation, complex assembly and disassembly, and intracellular transport. We have recently shown that a disruption of Hsp90 activity in cultured Drosophila melanogaster cells suppresses Flock House virus (FHV) replication and the accumulation of protein A, the FHV RNA-dependent RNA polymerase. In the present study, we investigated whether the defect in FHV RNA polymerase accumulation induced by Hsp90 suppression was secondary to an effect on protein A synthesis, degradation, or intracellular membrane association. Treatment with the Hsp90-specific inhibitor geldanamycin selectively reduced FHV RNA polymerase synthesis by 80% in Drosophila S2 cells stably transfected with an inducible protein A expression plasmid. The suppressive effect of geldanamycin on protein A synthesis was not attenuated by proteasome inhibition, nor was it sensitive to changes in either the mRNA untranslated regions or protein A intracellular membrane localization. Furthermore, geldanamycin did not promote premature protein A degradation, nor did it alter the extremely rapid kinetics of protein A membrane association. These results identify a novel role for Hsp90 in facilitating viral RNA polymerase synthesis in Drosophila cells and suggest that FHV subverts normal cellular pathways to assemble functional replication complexes.
The small genome of viruses relative to other organisms requires that they appropriate cellular machinery to complete their replication cycle. For example, no virus encodes the complete set of nucleic acid and protein constituents necessary for the autonomous translation of viral mRNAs, and therefore, viruses utilize diverse and often elaborate mechanisms to subvert the cellular translation apparatus to their benefit (7, 10, 34). Many seminal discoveries in the field of translation research have come from studies with viral mRNAs, such as the description of internal ribosome entry sites (IRES), the realization that efficient translation initiation occurs through the formation of a closed loop structure, and the identification of unusual translation events that expand genetic repertoires through ribosomal frameshifting, read-through translation, shunting, and leaky scanning (reviewed in reference 10). The use of alternative translation mechanisms by viral pathogens can be crucial for effective countermeasures against cellular innate antiviral responses, such as bypassing or inhibiting the global translation suppression mediated by protein kinase R activation (34). The important link between virus replication and cellular translation is particularly evident with viruses that contain a positive-strand RNA genome. These viruses, with the notable exception of retroviruses, generally do not encapsidate RNA replication proteins, and therefore, an essential early step in the viral life cycle after entry is viral mRNA translation. Thus, studies that investigate the molecular mechanisms of viral mRNA translation and its impact on replication may reveal novel antiviral drug targets.
To study virus replication and mRNA translation, we use Flock House virus (FHV), a versatile model pathogen that replicates robustly in Saccharomyces cerevisiae (24, 26, 32), Caenorhabditis elegans (22), and Drosophila melanogaster (19, 25, 41). The FHV genome is bipartite, with two positive-sense RNA segments copackaged into a nonenveloped icosahedral virion (2). The larger 3.1-kb RNA segment, RNA1, encodes protein A, the FHV RNA-dependent RNA polymerase, whereas the smaller 1.4-kb segment, RNA2, encodes the structural capsid protein precursor. During viral RNA replication, FHV produces a subgenomic 0.4-kb RNA, RNA3, which encodes the RNA interference suppressor protein B2 (21). FHV assembles its viral RNA replication complexes in association with intracellular membranes (25), consistent with all characterized positive-strand RNA viruses (1). FHV RNA replication complexes are targeted and anchored to the mitochondrial outer membrane by protein A via an amino-proximal transmembrane domain (24) that resembles the signal-anchor sequences of cellular mitochondrial outer membrane proteins (40). However, FHV RNA replication complexes can be retargeted to alternative intracellular membranes such as the endoplasmic reticulum by modification of the protein A amino-proximal targeting domain (26).
We hypothesize that FHV uses cellular chaperone pathways to assemble viral RNA replication complexes based on the previously observed connections between virus replication and cellular chaperones (35) and the demonstrated role of cellular chaperones in endogenous mitochondrial protein targeting and transport (48). We have previously demonstrated that the inhibition of the heat shock protein 90 (Hsp90) chaperone using both pharmacologic and genetic approaches suppresses FHV replication in cultured Drosophila S2 cells (19). Hsp90 inhibition reduces protein A accumulation but does not affect the activity of preformed FHV RNA replication complexes, suggesting that Hsp90 activity is important for an early step in the FHV life cycle, such as during the initial stages of viral RNA replication complex assembly. However, these experiments could not distinguish between specific effects of Hsp90 inhibition on protein A synthesis, degradation, intracellular trafficking, and membrane association.
In this report, we further examine the role of Hsp90 in FHV RNA replication and demonstrate that geldanamycin, a specific Hsp90 inhibitor, selectively suppressed protein A synthesis in Drosophila S2 cells independent of its intracellular membrane localization. Furthermore, we demonstrate that Hsp90 inhibition neither accelerated protein A degradation nor altered its rapid association with intracellular membranes.
MATERIALS AND METHODS
Plasmids.
We used standard molecular biology procedures for all cloning steps and sequenced all plasmid regions generated by PCR. The metallothionein (MT) promoter-driven FHV RNA1 and protein A expression plasmids pS2F1 and pS2FA, and the control β-galactosidase expression plasmid pS2LacZ have been previously described (19). To generate pS2FB, a Drosophila Cu2+-inducible FHV protein B2 expression plasmid, we initially constructed the in vitro expression vector pIVT-FB by PCR amplifying the protein B2 open reading frame from pS2F1 and inserting the product as a MluI/SalI fragment into pCMV-TnT (Promega, Madison, WI). Primer sequences used for amplification are available upon request. We subsequently inserted the EcoRI/NotI fragment from pIVT-FB into pMT-V5/HisA (Invitrogen, Carlsbad, CA) to generate pS2FB. To generate pS2FA-Gal L, a protein A expression plasmid with a 5′ GAL1 leader sequence and a C-terminal hemagglutinin (HA) epitope tag, we inserted the PstI (blunt)/XhoI fragment from pFA-C/HA (24) into the EcoRV/XhoI site of pMT-V5/HisA. To generate pS2FA-5′vUTR, a protein A expression plasmid with a C-terminal HA tag and the FHV 5′ untranslated region (UTR), we inserted the AlwNI/BlpI fragment from pS2FA into pS2F1. To generate pS2FA-3′vUTR, a protein A expression plasmid with a 5′ GAL1 leader sequence, an FHV 3′ UTR, and a C terminus without an HA tag, we inserted the BglII/XhoI fragment from pS2F1 into pS2FA-Gal L. To generate the endoplasmic reticulum-retargeted Drosophila protein A expression vectors pS2FA-P450 and pS2FA-HCV, we initially inserted the PstI (blunt)/AlwNI fragments from pFA-P450 and pFA-HCV (26) into the EcoRV/XhoI site of pMT-V5/HisA and subsequently exchanged the BlpI/AlwNI fragment with the same region from pS2FA (19) to insert C-terminal HA epitope tags.
Antibodies and inhibitors.
We obtained rabbit polyclonal antibodies against HA from Santa Cruz Biotechnology (Santa Cruz, CA), rabbit polyclonal antibodies against Hsp83 and Hsp60 from Stressgen Biotechnologies (Victoria, British Columbia, Canada), rabbit polyclonal antibodies against the voltage-dependent anion channel (VDAC) from Affinity Bioreagents (Golden, CO), and mouse monoclonal antibodies against tubulin from the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA). We obtained agarose-conjugated Staphylococcus aureus protein A or goat antibodies against mouse immunoglobulin G from Sigma (St. Louis, MO). Mouse monoclonal antibodies against FHV protein A have been previously described (26). Rabbit polyclonal antibodies against FHV protein B2 were generously provided by Paul Ahlquist (University of Wisconsin—Madison). We obtained secondary antibodies for immunoblotting from Jackson Immunoresearch (West Grove, PA). Hippuristanol, a eukaryotic translation initiation inhibitor (6), was generously provided by Junichi Tanaka (University of the Ryukyus, Japan). We obtained geldanamycin, lactacystin, cycloheximide, and MG132 from Sigma, and we stored all inhibitors at −20°C as concentrated stock solutions in single-use aliquots.
Cell culture and induction protocols.
We cultured Drosophila S2 cells and generated cells stably transfected with Cu2+-inducible expression plasmids as previously described (19). Amino acid-deficient Schneider's Drosophila medium (SDM) is not commercially available, and therefore, we used the modified recipe of Schneider and Blumenthal that excluded bacteriological peptone (11) to generate complete SDM (cSDM) supplemented with 10 units penicillin per ml, 10 μg streptomycin per ml, and 10% heat-inactivated fetal bovine serum. We used the same recipe to generate Met-Cys-free SDM that also excluded yeast extract and that was supplemented with 1% dialyzed fetal bovine serum and the antibiotics listed above.
Metabolic labeling and immunoprecipitation analysis.
We induced stably transfected S2 cells in cSDM with 1 mM Cu2+ for 2 h, washed cells twice with Met-Cys-deficient SDM, and incubated cells for 1 h in Met-Cys-deficient SDM with 1 mM Cu2+ to deplete intracellular amino acid pools. We labeled cells with 100 to 500 μCi per ml PRO-MIX 35S cell labeling mix (Amersham) for 15 to 90 min and stopped translation and respiration by adding 100 μg cycloheximide per ml and 0.1% sodium azide. Specific label concentrations and time periods are given for individual experiments in Results.
For routine immunoprecipitation experiments, we washed cells once with Tris-buffered saline (100 mM sodium chloride, 50 mM Tris [pH 7.2]) and lysed cells on ice in radioimmunoprecipitation assay (RIPA) buffer (Tris-buffered saline with 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], and 1 mM phenylmethylsulfonyl fluoride). We centrifuged lysates at 10,000 × g for 5 min to remove nuclei and insoluble debris, precleared lysates with goat immunoglobulin conjugated to agarose, and formed immune complexes by incubation with primary antibodies for 2 h. Immune complexes were precipitated by incubation with Staphylococcus aureus protein A or goat antibodies against mouse immunoglobulin G conjugated to agarose overnight at 4°C with gentle rotation, washed extensively with RIPA buffer, eluted with reducing SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer (62.5 mM Tris [pH 6.8], 2% SDS, 5% glycerol, 14.4 mM 2-mercaptoethanol, 0.02% bromophenol blue), and separated in 8% or 15% gels. After electrophoresis, gels were fixed in a solution containing 25% methanol and 7% acetic acid, impregnated with 1 M sodium salicylate, dried under a vacuum, and exposed to Blue Light AutoRad film (ISC BioExpress, Kaysville, UT) at −80°C. Digital film images were obtained using an Alpha Innotech Fluorchem 8900 apparatus, and band intensities were quantitated with AlphaEaseFC software.
Polysome analysis.
We induced stably transfected S2 cells in cSDM with 1 mM Cu2+ for 6 h in the presence of inhibitors. We used inhibitor concentrations that were 200-fold above the 50% inhibitory concentration (IC50) for the suppression of FHV replication or total protein synthesis in Drosophila S2 cells, which corresponded to 3 μM for geldanamycin (19) and 5 μM for hippuristanol (K. M. Castorena and D. J. Miller, unpublished data). Approximately 5 × 108 cells per group were washed twice with ice-cold phosphate-buffered saline (PBS) (100 mM sodium chloride, 50 mM sodium phosphate [pH 7.4]) containing 100 μg per ml cycloheximide, lysed in 0.5 ml ice-cold polysome fractionation buffer (20 mM HEPES [pH 7.4], 100 mM potassium chloride, 5 mM magnesium chloride, 1% Triton X-100, 0.5% sodium deoxycholate, 2 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 100 μg per ml cycloheximide, 0.1 U per ml RNasin), and centrifuged at 10,000 × g for 5 min to remove nuclei and insoluble debris. We loaded postnuclear lysates onto linear 10 to 50% sucrose gradients prepared in polysome fractionation buffer, centrifuged them at 4°C and 100,000 × g for 2 h with a Beckman MLS-50 rotor, and harvested eight equal-volume (0.5-ml) fractions from the top of the gradient. We extracted fractions with phenol-chloroform, precipitated total RNA with 2 volumes of ethanol, and analyzed samples by denaturing formaldehyde-agarose gel electrophoresis and Northern blotting for protein A mRNA as previously described (19, 25).
Cell fractionation.
To rapidly separate cells into cytosolic and membrane fractions, we washed 35S-labeled cells once with ice-cold PBS containing 100 μg per ml cycloheximide and 0.1% sodium azide, incubated cells at 4°C with fractionation buffer (PBS with 100 μg per ml cycloheximide, 0.1% sodium azide, and 0.01% saponin) for 10 min, centrifuged samples at 10,000 × g for 5 min to pellet cell remnants, and recovered supernatants as cytosolic fractions. Pelleted samples were washed once with fractionation buffer and lysed in RIPA buffer as described above to obtain membrane fractions. Fractions were analyzed by either immunoprecipitation as described above or by immunoblotting as previously described (19).
Statistical analyses.
We used a two-tailed Student's t test assuming unequal variances for statistical comparisons, and we considered a P value of <0.05 to be statistically significant.
RESULTS
Hsp90 inhibition and FHV protein A synthesis in Drosophila S2 cells.
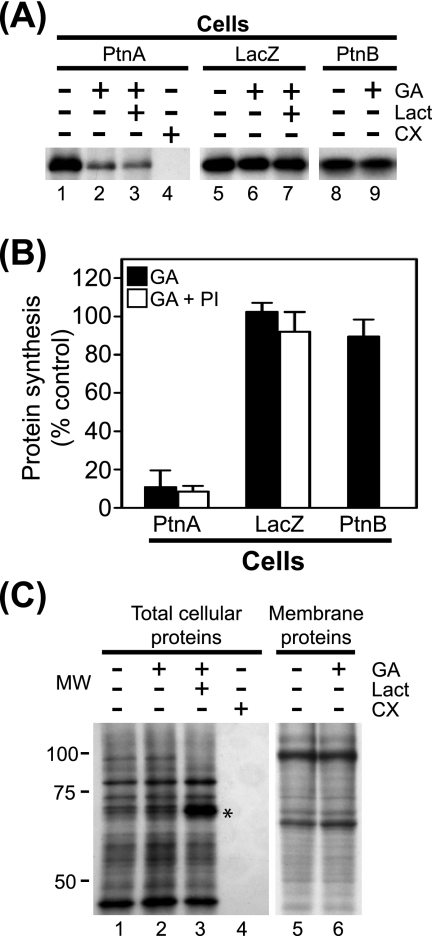
We previously demonstrated that Hsp90 inhibition suppresses FHV protein A accumulation in Drosophila S2 cells stably transfected with pS2FA, an MT promoter-driven C-terminal HA-tagged protein A expression plasmid (19). We initially determined that treatment with geldanamycin, a selective Hsp90 inhibitor (29, 33), did not suppress protein A mRNA accumulation in S2 cells stably transfected with pS2FA when measured 12 h after Cu2+ induction (data not shown). Thus, to investigate whether suppressed protein A accumulation was due in part to decreased synthesis, we examined the effects of geldanamycin in 35S metabolic labeling experiments (Fig. 1). We induced Drosophila S2 cells stably transfected with pS2FA with Cu2+ for 2 h, incubated cells with [35S]Met-Cys for 90 min in the presence of inhibitors, and subsequently immunoprecipitated full-length protein A from cell lysates with HA-specific antibodies (Fig. 1A, lanes 1 to 4). Treatment with geldanamycin suppressed protein A synthesis by >80% compared to controls (Fig. 1A, lane 2, and B) but had negligible effects on the synthesis of either total cellular proteins (Fig. 1C, lane 2) or membrane-associated cellular proteins (Fig. 1C, lane 6). In contrast, the translation elongation inhibitor cycloheximide completely suppressed the synthesis of both protein A (Fig. 1A, lane 4) and total cellular protein (Fig. 1C, lane 4). The inhibitory activity of geldanamycin was not due to promoter suppression, as it had a minimal effect on Cu2+-induced β-galactosidase production (Fig. 1A, lane 6, and B), consistent with reporter protein accumulation, and activity results in S2 cells (19).
FIG. 1.
Hsp90 inhibition suppresses FHV protein A synthesis. (A) Drosophila S2 cells stably transfected with pS2FA (lanes 1 to 4), pS2LacZ (lanes 5 to 7), or pS2FB (lanes 8 and 9) were induced with Cu2+ and labeled with 100 μCi per ml [35S]Met-Cys in the presence of 1 μM geldanamycin (GA), 10 μM lactacystin (Lact), or 100 μg per ml cycloheximide (CX) for 90 min. Radiolabeled proteins were immunoprecipitated with either HA-specific antibodies for FHV protein A (PtnA) and β-galactosidase or FHV protein B2 (PtnB)-specific antibodies, separated by SDS-PAGE, and analyzed by fluorography. (B) Quantitative data for the effect of geldanamycin and proteasome inhibitors (PI) on FHV protein A, protein B2, and β-galactosidase synthesis compared to no-inhibitor controls. Proteasome inhibitor results are composite data from experiments using either lactacystin or MG132. (C) Total lysates (lanes 1 to 4) or carbonate-resistant membrane fractions (lanes 5 and 6) from cells metabolically labeled with [35S]Met-Cys in the presence of the inhibitors listed above the lanes. Lanes 1 to 4 correspond to similarly numbered lanes in A. Samples for lanes 5 and 6 were prepared by differential centrifugation as previously described (25) and represent cellular proteins tightly associated with intracellular membranes (5). The asterisk indicates a cellular protein whose degradation is inhibited by lactacystin. Results are representative of three independent experiments, and for B, results represent the means ± standard errors of the means relative to vehicle-only controls. MW, molecular weight (in thousands).
We also examined whether lactacystin, an irreversible proteasome inhibitor (20) that was shown to partially attenuate the protein A accumulation defect mediated by Hsp90 inhibition during a 12-h incubation (19), also attenuated the effect of geldanamycin on protein A synthesis. Lactacystin had no significant impact on the geldanamycin-mediated suppression of full-length protein A synthesis (Fig. 1A, lane 3, and B) but did augment the recovery of some 35S-labeled cellular proteins (Fig. 1C, lane 3; see also Fig. 4B). Similar results were obtained with the reversible proteasome inhibitor MG132 (data not shown), suggesting that geldanamycin did not promote rapid proteasome-mediated degradation of newly synthesized protein A.
FIG. 4.
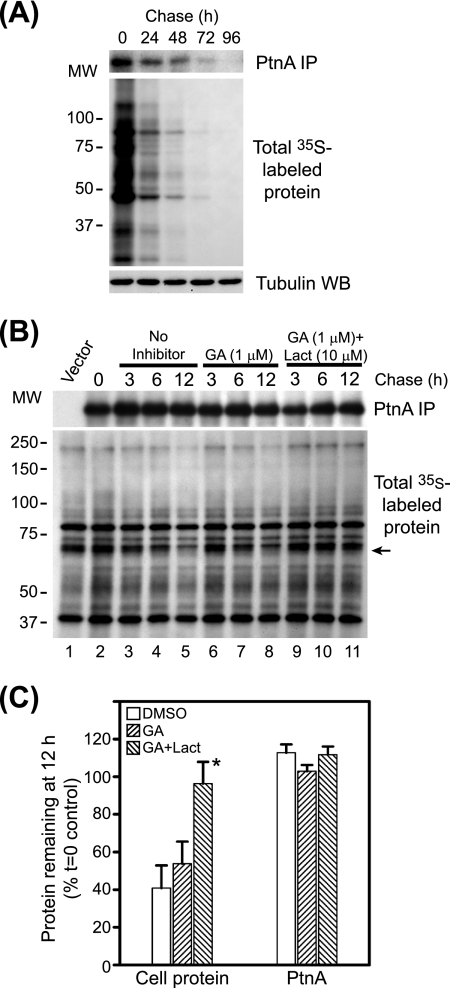
Hsp90 suppression does not accelerate FHV protein A degradation. (A) Basal FHV protein A stability in transfected Drosophila S2 cells. Cells stably transfected with pS2FA were induced with Cu2+ and labeled with 125 μCi per ml [35S]Met-Cys for 90 min, washed extensively, and cultured with cSDM in the absence of Cu2+ for 4 days with daily passages to maintain normal cell proliferation. We harvested equivalent numbers of cells at 24-h intervals and analyzed FHV protein A (PtnA) by immunoprecipitation (IP) and fluorography (top), total 35S-labeled protein by fluorography (middle), and total cellular protein by immunoblotting (WB) for tubulin (bottom). (B) Cells stably transfected with pS2FA were induced with Cu2+ and labeled with 250 μCi per ml [35S]Met-Cys for 90 min; cultured in cSDM in the presence of no inhibitor (lanes 2 to 5), geldanamycin (GA) (lanes 6 to 8), or geldanamycin plus lactacystin (Lact) (lanes 9 to 11); and harvested at 3, 6, and 12 h, and immunoprecipitated protein A (top) and total 35S-labeled protein (bottom) were analyzed by fluorography. MW, molecular weight (in thousands) (C) Quantitative analysis of protein recovery at 12 h (Fig. 4B, lanes 5, 8, and 12) compared to the time-zero (t=0) control (see B, lane 2). For cellular protein quantitation, we used the radiolabeled band indicated by the arrow in B. *, the P value was <0.02 compared to the vehicle-only control. Results are representative of at least three independent experiments, and for C, results represent the means ± standard errors of the means relative to untreated controls at time zero. DMSO, dimethyl sulfoxide.
To further examine the relationship between Hsp90 inhibition and FHV protein A synthesis, we conducted a dose-response analysis. Geldanamycin suppressed protein A synthesis in a dose-dependent manner, with a calculated IC50 of 92 nM, which was approximately twofold higher that the IC50 value calculated for geldanamycin-mediated suppression of protein A accumulation in Drosophila S2 cells (19). The different experimental conditions under which IC50 values were determined, for example, synthesis over 90 min versus accumulation over 12 h, make a direct quantitative comparison difficult. Nevertheless, these results indicated that geldanamycin potently suppressed FHV protein A synthesis without disrupting general cellular translation.
To determine whether Hsp90 inhibition also impacted the synthesis of other viral proteins, we examined the effect of geldanamycin on the synthesis of FHV protein B2, which is translated from subgenomic RNA3 (2) and functions as an RNA interference suppressor in plants (21), nematodes (22), and insects (21, 41). We generated Drosophila S2 cells stably transfected with pS2FB, a Cu2+-inducible FHV protein B2 expression plasmid, and examined protein B2 synthesis by 35S metabolic labeling and immunoprecipitation. Geldanamycin had a minimal effect on FHV protein B2 synthesis (Fig. 1A, lane 9, and B), in contrast to results with protein A (Fig. 1A, lane 2). We obtained similar results with protein B2 synthesis in S2 cells using a fivefold-higher concentration of geldanamycin (data not shown). These results further emphasized the selectivity of Hsp90 inhibition on FHV RNA polymerase synthesis.
Hsp90 inhibition and polysomal protein A mRNA distribution.
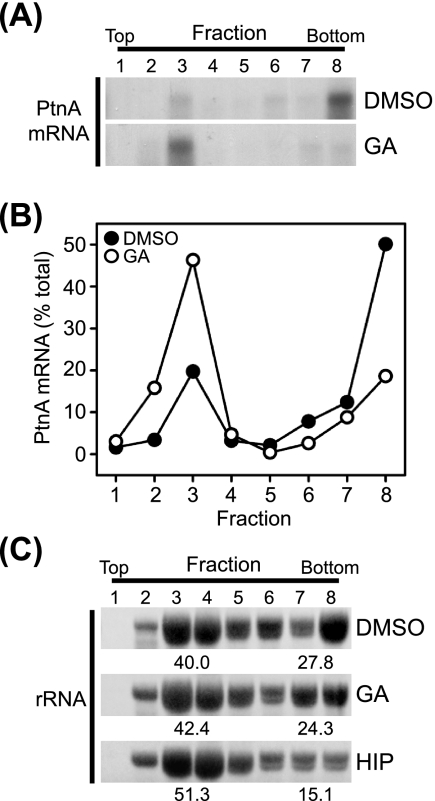
To further examine the impact of Hsp90 inhibition on FHV protein A synthesis, we analyzed the effect of geldanamycin treatment on the association of protein A mRNA with polysomes (Fig. 2). The association of mRNAs with multiple ribosomes, termed polysomes, frequently indicates active translation, and polysomal mRNAs can be identified and isolated from cell lysates by gradient centrifugation due to their increased density (23). We induced Drosophila S2 cells stably transfected with pS2FA with Cu2+ for 6 h in the presence of inhibitors, fractionated cell lysates in linear sucrose density gradients, and analyzed fractions for total RNA and protein A mRNA by ethidium bromide staining and Northern blotting, respectively. Treatment with geldanamycin resulted in a shift of protein A mRNA from the high-density polysome fractions to lower-density fractions (Fig. 2A and B). The redistribution of protein A mRNA with geldanamycin treatment occurred without a dramatic change in rRNA distribution within the sucrose gradient (Fig. 2C, middle), consistent with the minimal impact of geldanamycin on total protein synthesis (Fig. 1C). In contrast, treatment with hippuristanol, a general eukaryotic translation initiation inhibitor (6), resulted in the redistribution of rRNA from high- to low-density fractions, consistent with an overall reduction in polysomal mRNA (Fig. 2C, bottom). These results were consistent with the 35S metabolic labeling experiments (Fig. 1) and further supported the conclusion that geldanamycin suppressed FHV protein A synthesis.
FIG. 2.
Hsp90 inhibition reduces FHV protein A mRNA association with polysomes. (A) Drosophila S2 cells stably transfected with pS2FA were induced with Cu2+ in the presence of dimethyl sulfoxide (DMSO) or geldanamycin (GA), cell lysates were separated by linear sucrose gradient density centrifugation, equal-volume fractions were recovered from the top (low density) to the bottom (high density) of the gradient, total RNA was recovered by phenol-chloroform extraction and ethanol precipitation, and FHV protein A (PtnA) mRNA content was analyzed by Northern blotting with a 32P-labeled riboprobe (25). (B) Quantitative data for the effect of geldanamycin on protein A mRNA polysome association. The total protein A mRNA recovered was the sum of the densitometry analysis of lanes 1 to 8 (see above [A]). (C) Ethidium bromide-stained rRNA in sucrose gradient fractions from cells treated with dimethyl sulfoxide, geldanamycin, or the general translation initiation inhibitor hippuristanol (HIP). The quantitative values represent the percentages of total rRNA present in fractions 3 and 4 or fractions 7 and 8. Results are representative of two independent experiments, and for B and quantitative values in C, results represent the means from those experiments.
Hsp90 inhibition and FHV protein A mRNA 5′ and 3′ UTRs.
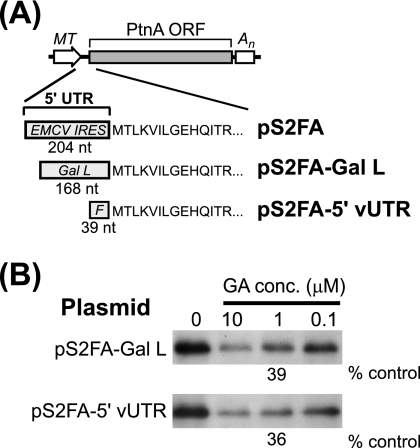
The protein A expression plasmid pS2FA contains a modified 5′ UTR with an encephalomyocarditis virus (EMCV) IRES inserted to disrupt its function as a replication template and enhance translation efficiency in animal cells (19). To examine whether the mRNA 5′ UTR influenced geldanamycin-mediated suppression of protein A synthesis, we used S2 cells stably transfected with pS2FA-Gal L, a modified protein A expression plasmid whose 5′ UTR contained the yeast GAL1 leader (Fig. 3A). Protein A synthesis in S2 cells stably transfected with pS2FA-Gal L was more efficient than that in cells transfected with pS2FA (comparative data not shown), consistent with the observation that the EMCV IRES is not highly active in insect cells (12). Nevertheless, geldanamycin also suppressed protein A synthesis in pS2FA-Gal L-transfected S2 cells in a dose-dependent manner, where 1 μM decreased synthesis by 61% (Fig. 3B). We further examined the impact of mRNA sequence on protein A synthesis using pS2FA-5′vUTR, a protein A expression plasmid with an authentic FHV 5′ UTR (Fig. 3A). Similar to results with pS2FA-Gal L, 1 μM geldanamycin decreased protein A synthesis in S2 cells stably transfected with pS2FA-5′vUTR by 64% (Fig. 3B).
FIG. 3.
mRNA 5′ UTR does not affect geldanamycin-mediated suppression of FHV protein A synthesis. (A) Schematics of pS2FA and derivatives with modified 5′ UTRs. All plasmids contain an MT promoter and a simian virus 40 polyadenylation signal (An). The 5′ UTR of pS2FA contains 204 nucleotides (nt) with an EMCV IRES, whereas pS2FA-Gal L contains 168 nt with the yeast GAL1 leader (Gal L). The 5′ UTR of pS2FA-5′vUTR contains 39 nt with the authentic FHV 5′ UTR sequence (F). ORF, open reading frame. (B) Drosophila S2 cells stably transfected with pS2FA-Gal L or pS2FA-5′vUTR were induced with Cu2+ and labeled with 100 μCi per ml [35S]Met-Cys for 90 min in the presence of vehicle only or decreasing geldanamycin (GA) concentrations (conc.). Radiolabeled protein A was immunoprecipitated with HA-specific antibodies, separated by SDS-PAGE, and analyzed by fluorography. Results are representative of three or four independent experiments, and the quantitative values are the means from those experiments relative to vehicle-only controls.
We also examined whether the mRNA 3′ UTR influenced geldanamycin-mediated suppression of protein A synthesis. To facilitate these experiments, we generated pS2FA-3′vUTR, a protein A expression plasmid with a 5′ GAL1 leader but an authentic FHV C terminus and 3′ UTR and hence no C-terminal HA tag or polyadenylation sequence. The absence of a C-terminal HA tag in pS2FA-3′vUTR necessitated the use of monoclonal antibodies against FHV protein A (26) for selective immunoprecipitation of radiolabeled proteins. Similar to results with pS2FA-Gal L and pS2FA-5′vUTR (Fig. 3B), 1 μM geldanamycin decreased protein A synthesis in S2 cells stably transfected with pS2FA-3′vUTR by 65% (data not shown). These results indicated that geldanamycin-mediated suppression of protein A synthesis was independent of the mRNA 5′ and 3′ UTRs.
Hsp90 inhibition and FHV protein A degradation.
The inability of proteasome inhibitors to attenuate geldanamycin-mediated suppression of protein A synthesis (Fig. 1) suggested that Hsp90 inhibition did not promote rapid protein A degradation. However, to directly examine protein turnover, we conducted pulse-chase experiments (Fig. 4). The normal half-life of FHV protein A is unknown, although results with Drosophila cells infected with black beetle virus, a closely related alphanodavirus (2), suggest that nodavirus RNA polymerases are stable for at least 36 h after synthesis (13). We initially examined the baseline kinetics of FHV protein A degradation in Drosophila S2 cells (Fig. 4A). We induced S2 cells stably transfected with pS2FA with Cu2+ for 2 h, pulse labeled the cells with [35S]Met-Cys for 90 min, and immunoprecipitated full-length protein A with HA-specific antibodies at 24-h intervals up to 96 h. We detected FHV protein A in cells for at least 72 h after synthesis and calculated a half-life of approximately 38 h (Fig. 4A). The doubling time of S2 cells is less than 24 h under the growth conditions used in these experiments (19). Thus, the majority of the signal loss that we observed may have been due to dilution rather than degradation if protein A was divided equally between daughter cells during cytokinesis and mitochondrial partitioning (42). Nevertheless, although these results may have underestimated the actual half-life of FHV protein A in Drosophila S2 cells, they demonstrated that it was normally a highly stable protein after synthesis.
We previously examined the effects of Hsp90 inhibition on FHV replication and protein A accumulation at 12 h after Cu2+ induction due in part to the antiproliferative effects of geldanamycin on S2 cells during longer incubations (19). Thus, we limited our examination of protein A degradation to 12 h after inhibitor addition (Fig. 4B). We pulse labeled S2 cells stably transfected with pS2FA as described above, chased cells in cSDM in the presence of geldanamycin with or without the proteasome inhibitor lactacystin, and immunoprecipitated full-length protein A from cell lysates 3, 6, and 12 h later. Neither geldanamycin nor lactacystin had a significant effect on protein A degradation over a 12-h time period, whereas lactacystin did delay the degradation of some cellular proteins (Fig. 4B and C). These pulse-chase results, in conjunction with the 35S metabolic labeling and polysome analysis data presented above (Fig. 1 to 3), suggested that the suppressive effects of geldanamycin on FHV protein A accumulation and RNA replication (19) were due primarily to viral RNA polymerase synthesis suppression rather than degradation enhancement.
Hsp90 inhibition and FHV protein A membrane association.
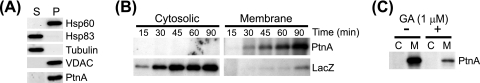
The association of protein A with intracellular membranes in Drosophila cells has important functional consequences for FHV RNA replication complex activity (45, 46). Thus, we investigated whether protein A membrane association was also disrupted by inhibiting Hsp90 activity. Since protein translation and membrane association may be linked processes within cells, we examined the effects of geldanamycin on protein A membrane association in the context of protein synthesis (Fig. 5). To rapidly separate small-volume cultures into cytosolic and membrane fractions, we developed a lysis and differential centrifugation technique using the detergent saponin. Low concentrations of saponin, a natural amphiphilic detergent that forms complexes with cholesterol to disrupt lipid bilayers (3), selectively permeabilize the plasma membrane and leave organelle membranes intact (43). We treated S2 cells with 0.01% saponin in PBS for 10 min on ice and separated lysates into soluble and pellet fractions by differential centrifugation. We subsequently extracted pellet fractions with RIPA buffer to recover cellular proteins either bound to membranes or enclosed within organelles and to remove nuclei and insoluble debris from the final samples and analyzed supernatant and extracted pellet fractions by immunoblotting. Cytosolic proteins such as Hsp83, the sole Drosophila Hsp90 family chaperone (8), and monomeric tubulin partitioned into the supernatant fraction, whereas the membrane protein VDAC and the mitochondrial matrix protein Hsp60 partitioned into the pellet fraction (Fig. 5A). Under these saponin-based lysis and fractionation conditions, FHV protein A partitioned almost exclusively into the pellet fraction, indicating that at steady-state levels it is predominantly membrane associated, consistent with previous observations (24, 25). We used this protocol of differential solubilization and centrifugation for subsequent fractionation experiments, and we refer to supernatant and pellet fractions as cytosolic and membrane fractions, respectively.
FIG. 5.
Newly synthesized FHV protein A rapidly associates with cellular membranes in Drosophila S2 cells. (A) Cells were incubated with PBS and 0.01% saponin on ice for 10 min and centrifuged at 10,000 × g for 5 min to recover the supernatant fraction (S) and the resultant pellet, which was extracted in an equal volume of RIPA buffer and centrifuged at 10,000 × g for 5 min to remove nuclei and insoluble cellular debris and to recover a final pellet (P) fraction. Equal-volume fractions were separated by SDS-PAGE and immunoblotted with antibodies against the cytosolic proteins Hsp83 and tubulin, the mitochondrial membrane protein VDAC, the mitochondrial matrix protein Hsp60, and FHV protein A (PtnA). (B) Cells stably transfected with pS2FA or pS2LacZ were induced with Cu2+, incubated with 125 μCi per ml [35S]Met-Cys, and harvested at 15, 30, 45, 60, and 90 min. Cells were separated into cytosolic and membrane fractions as described above, and FHV protein A and β-galactosidase were immunoprecipitated with HA-specific antibodies and analyzed by fluorography. (C) Cells stably transfected with pS2FA were induced and labeled as described above for 90 min in the presence or absence of 1 μM geldanamycin (GA) and separated into cytosolic (C) and membrane (M) fractions. Full-length FHV protein A was immunoprecipitated and analyzed by fluorography. Results are representative of at least four independent experiments.
We initially examined the baseline kinetics of full-length protein A association with intracellular membranes by 35S metabolic labeling and fractionation (Fig. 5B). We induced S2 cells stably transfected with pS2FA or pS2LacZ, incubated cells with 100 μCi per ml [35S]Met-Cys, harvested cultures at 15- or 30-min intervals for up to 90 min, fractionated cells using saponin as described above, and immunoprecipitated radiolabeled protein A or β-galactosidase with HA-specific antibodies. We detected full-length protein A at 15 min in membrane fractions, and recovery increased throughout the labeling period (Fig. 5B, upper right). However, we were unable to recover detectable full-length protein A from cytosolic fractions at any time point (Fig. 5B, upper left), suggesting that protein A rapidly associated with intracellular membranes, perhaps even prior to the complete translation of the entire polypeptide chain. In contrast to results with protein A, full-length β-galactosidase partitioned predominantly into cytosolic fractions, except for a small quantity recovered from membrane fractions at later time points (Fig. 5B, bottom). We also examined whether the residual protein A synthesized in the presence of 1 μM geldanamycin (Fig. 1A) partitioned into cytosolic or membrane fractions. Total recovery was reduced in lysates from cells treated with geldanamycin, but full-length protein A remained almost exclusively in the membrane fraction (Fig. 5C).
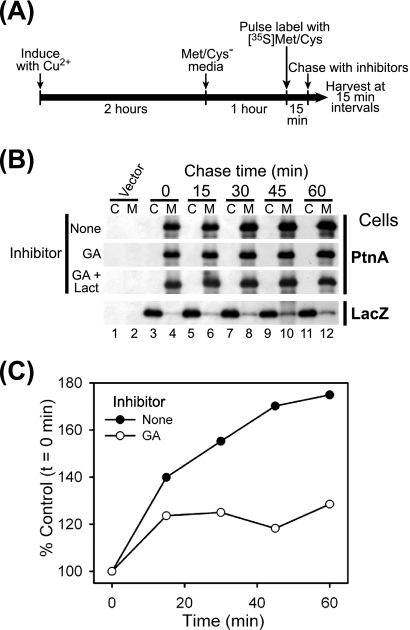
The inclusion of geldanamycin during the labeling period prevented drawing definitive conclusions regarding potential different effects of Hsp90 inhibition on protein A synthesis versus membrane association. Thus, we conducted additional pulse-chase fractionation experiments (Fig. 6). We pulse labeled cells for 15 min with 500 μCi per ml [35S]Met-Cys to increase labeling density and enhance detection sensitivity, chased cells in cSDM in the presence of inhibitors, and harvested samples at 15-min intervals (Fig. 6A). We immediately fractionated and immunoprecipitated full-length protein A or β-galactosidase with HA-specific antibodies (Fig. 6B). We did not recover detectable full-length protein A in cytosolic fractions at any time point in control cells without inhibitors but observed a time-dependent increase in full-length protein A recovery from membrane fractions (Fig. 6B, top, and C). The temporal increase in membrane-associated protein A despite the absence of extracellular radioactive label was likely secondary to residual intracellular [35S]Met-Cys pools or the completion of polypeptide synthesis initiated during the 15-min pulse-labeling period. In cells treated with geldanamycin, there was only a marginal increase in full-length protein A recovered from the membrane fraction (Fig. 6B and C), and the addition of lactacystin had no significant effect (Fig. 6B). Furthermore, we did not recover full-length protein A from cytosolic fractions in cells treated with geldanamycin alone or with proteasome inhibitors at any time point. In contrast, radiolabeled β-galactosidase was recovered predominantly from cytosolic fractions throughout the chase period (Fig. 6B, bottom). These results indicated that geldanamycin did not dislodge membrane-associated protein A and further suggested an intimate temporal link between protein A synthesis and membrane association.
FIG. 6.
Hsp90 inhibition does not alter FHV protein A membrane association in Drosophila S2 cells. (A) Schematic of pulse-chase fractionation protocol. (B) Cells stably transfected with pS2FA or pS2LacZ were induced with Cu2+; pulse labeled with 500 μCi per ml [35S]Met-Cys for 15 min; chased with cSDM and no inhibitor, 1 μM geldanamycin (GA), or 1 μM geldanamycin plus 10 μM lactacystin (Lact); harvested at 15-min intervals; separated into cytosolic (C) and membrane (M) fractions; and analyzed by immunoprecipitation as described in the legend of Fig. 5. (C) Quantitative temporal analysis of full-length FHV protein A recovery from the membrane fraction after pulse labeling and treatment with vehicle only (closed circles) or 1 μM geldanamycin (GA) (open circles) compared to the time-zero (t=0) control (Fig. 5B, lane 4). Results are representative of two independent experiments, and results for C represent the means from those experiments.
Hsp90 inhibition and retargeted FHV protein A synthesis.
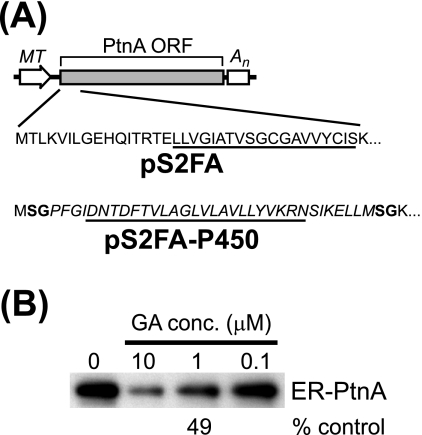
The amino-proximal transmembrane domain and adjacent residues of protein A are an important region for membrane association and mitochondrial targeting (24). In S. cerevisiae, this region can be replaced with several alternative targeting domains to redirect protein A to the endoplasmic reticulum (26). We examined whether we could also redirect protein A to the endoplasmic reticulum in Drosophila S2 cells and whether geldanamycin suppressed the synthesis of retargeted protein A (Fig. 7). We used the yeast cytochrome P450 oxidoreductase endoplasmic reticulum targeting sequence (37) to replace the endogenous protein A mitochondrial targeting signal and generate the Drosophila expression plasmid pS2FA-P450 (Fig. 7A). Chimeric protein A in S2 cells stably transfected with pS2FA-P450 colocalized with endogenous Drosophila endoplasmic reticulum but not Golgi apparatus markers by immunofluorescence microscopy (data not shown). We subsequently examined the effects of geldanamycin on the synthesis of endoplasmic reticulum-retargeted protein A by 35S metabolic labeling and immunoprecipitation (Fig. 7B). Geldanamycin suppressed retargeted protein A synthesis in a dose-dependent manner, where 1 μM decreased synthesis by 51%. Similar results were obtained when we used a chimeric protein A retargeted to the endoplasmic reticulum via the insertion of an inverted targeting signal from hepatitis C virus (HCV) NS5B (data not shown). Although the potency of geldanamycin in suppressing the synthesis of retargeted protein A was reduced compared to that of wild-type protein A (Fig. 1A), these results indicated that Hsp90 inhibition suppressed FHV RNA polymerase synthesis independent of its intracellular localization.
FIG. 7.
Hsp90 inhibition suppresses the synthesis of retargeted FHV protein A. (A) Schematics of pS2FA, which encodes wild-type mitochondrial targeted protein A, and pS2FA-P450, which encodes a chimeric protein A retargeted to the endoplasmic reticulum. The amino-terminal coding sequences are shown, and the transmembrane domains are underlined. The endoplasmic reticulum targeting signal from the yeast cytochrome P450 oxidoreductase is italicized, and the residues in boldface type are encoded by the unique BspEI sites used to generate the chimeric junctions (26). ORF, open reading frame. (B) Drosophila S2 cells stably transfected with pS2FA-P450 were induced with Cu2+ and labeled with 100 μCi per ml [35S]Met-Cys for 90 min in the presence of decreasing geldanamycin (GA) concentrations (conc.). Radiolabeled endoplasmic reticulum-targeted protein A (ER-PtnA) was immunoprecipitated with HA-specific antibodies, separated by SDS-PAGE, and analyzed by fluorography. Results are representative of at least three independent experiments, and the quantitative value is the mean from those experiments relative to the vehicle-only control.
DISCUSSION
In this study, we further investigated the role of the molecular chaperone Hsp90 in FHV RNA replication complex assembly in Drosophila cells. Pharmacologic inhibition of Hsp90 activity with geldanamycin selectively suppressed protein A synthesis independent of its intracellular localization but neither enhanced protein A degradation nor directly altered its membrane association. These results suggest that one important function of Hsp90 in the FHV life cycle is to facilitate replication complex assembly by promoting efficient synthesis of the viral RNA polymerase. Previous studies have demonstrated that Hsp90 also plays an important role in the replication of other viruses that either contain an RNA genome or utilize RNA as a genomic intermediate during replication, including hepatitis B virus (HBV) (16-18), HCV (30, 44), influenza virus (27, 28), and reovirus (14). However, Hsp90 appears to have virus-specific functions at unique steps in the viral life cycle, as it facilitates the interactions between HBV reverse transcriptase and pregenomic viral RNA (18), modulates the activity of both HCV NS2/3 protease (44) and NS5A (30), promotes the assembly and nuclear import of influenza virus RNA polymerase (28), and supports the oligomerization of the reovirus cell attachment protein σ1 (14). These observations highlight the complexity and diversity of the mechanisms employed by viruses to appropriate cellular pathways for their own purposes.
One consistent observation in the studies that examined the importance of Hsp90 in the replication of HBV, HCV, and influenza virus was the demonstration of a physical interaction between Hsp90 and a particular virus-specific protein involved in genome replication (16, 27, 28, 30, 44). We previously hypothesized that Hsp90 functioned via a similar physical interaction with FHV protein A to promote viral RNA polymerase posttranslational maturation and intracellular targeting (19), similar to the role of Hsp90 in cellular client protein maturation (31) and endogenous mitochondrial protein trafficking (48). The results presented in this report are more consistent with a model whereby Hsp90 functions at an earlier stage of FHV RNA replication complex assembly, such as during viral RNA polymerase synthesis. Furthermore, despite repeated attempts under diverse experimental conditions, we have been unable to convincingly demonstrate a direct physical interaction between full-length FHV protein A and Hsp90 chaperones from either Drosophila cells or rabbit reticulocyte lysates (S. A. Weeks, K. A. Stapleford, and D. J. Miller, unpublished data). However, we cannot exclude a transient low-affinity interaction between protein A and Hsp90, particularly one that occurs prior to complete polypeptide synthesis. We are currently using affinity purification approaches to further investigate potential FHV protein A-Hsp90 physical interactions.
The abundant cytosolic chaperone Hsp90 is generally thought to be involved in the posttranslation folding, trafficking, and turnover of cellular proteins (31), but it has also been directly implicated in cellular translation via its role in the maturation of the alpha subunit of eukaryotic translation initiation factor 2 (9, 36, 47). However, the observations that Hsp90 inhibition did not disrupt Cu2+-induced β-galactosidase, FHV protein B2, or cellular protein synthesis (Fig. 1) suggest that a direct suppression of a ubiquitous translation factor involved in either general protein production or the synthesis of membrane-associated cellular proteins could not account for the inhibitory effect of geldanamycin on FHV protein A synthesis. An alternative hypothesis to the direct mechanism described above is that a cellular protein whose maturation or stability within cells is dependent upon a functional Hsp90 chaperone complex selectively facilitates FHV protein A synthesis. Consistent with this hypothesis, geldanamycin did not inhibit the in vitro translation of protein A in rabbit reticulocyte lysates (K. A. Stapleford and D. J. Miller, unpublished data). This discrepancy with results in Drosophila S2 cells was not due to a species-specific effect, as Hsp90 inhibition with geldanamycin or radicicol potently suppressed FHV protein A accumulation in cultured rodent cells (Castorena and Miller, unpublished). Discordant results on the effects of Hsp90 inhibition on protein synthesis in cells and in vitro have also been described for the Src kinase p56lck (4, 15). However, Hsp90 facilitates the posttranslational membrane association and subsequent stabilization of p56lck in cells (4), whereas we saw a minimal effect of geldanamycin on either protein A stability (Fig. 4) or membrane association (Fig. 5 and 6), suggesting a unique role for Hsp90 in protein A synthesis.
The inability to recover newly synthesized full-length protein A from the cytosol of Drosophila S2 cells (Fig. 5) was unanticipated and suggests that FHV RNA polymerase translation and membrane association are linked processes. However, we cannot exclude the potential presence of a small cytosolic full-length protein A fraction that was not detected with our experimental conditions. We attempted to examine the temporal link between protein A translation and membrane association using 35S metabolic labeling and fractionation experiments with an amino-terminal HA-tagged protein A expression plasmid. Surprisingly, the presence of an amino-terminal epitope tag altered membrane association in Drosophila S2 cells such that approximately 50% of full-length protein A was recovered in the cytosolic fraction (D. J. Miller, unpublished data). Although these results precluded drawing definitive conclusions regarding the temporal pattern of protein A membrane association in relation to synthesis, they indicated that the amino-terminal coding region of FHV protein A was a key determinant in this process. The potential link between protein A translation, membrane association, and subsequent RNA replication complex assembly is consistent with an emerging model for FHV replication where processes such as viral RNA synthesis, translation, capsid assembly, and genome packaging are linked temporally and spatially within cells (38, 39). Studies are in progress to further examine the mechanisms whereby FHV utilizes cellular chaperones to assemble functional RNA replication complexes and potentially highlight additional mechanisms that viruses employ to subvert cellular pathways to efficiently complete their replication cycles.
Acknowledgments
We thank Donna Gschwend and Bethany Sprader for assistance and all laboratory members for their helpful comments on the research and manuscript. We thank Paul Ahlquist and Junichi Tanaka for generously providing reagents.
This work was supported by National Institutes of Health grant R01-AI062749. S.A.W. and K.A.S. were funded by training grants T32-AI007528 and T32-GM007315, respectively.
Footnotes
Published ahead of print on 23 May 2007.
REFERENCES
- 1.Ahlquist, P., A. O. Noueiry, W. M. Lee, D. B. Kushner, and B. T. Dye. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 77:8181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball, L. A., and K. L. Johnson. 1998. Nodaviruses of insects, p. 225-267. In L. K. Miller and L. A. Ball (ed.), The insect viruses. Plenum Publishing Corporation, New York, NY.
- 3.Bangham, A. D., R. W. Horne, A. M. Glauert, J. T. Dingle, and J. A. Lucy. 1962. Action of saponin on biological cell membranes. Nature 196:952-955. [DOI] [PubMed] [Google Scholar]
- 4.Bijlmakers, M. J., and M. Marsh. 2000. Hsp90 is essential for the synthesis and subsequent membrane association, but not the maintenance, of the Src-kinase p56lck. Mol. Biol. Cell 11:1585-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonifacino, J. S. 2000. Characterization of cellular proteins, p. 5.0.1-5.5.11. In K. S. Morgan (ed.), Current protocols in cell biology. John Wiley & Sons, Inc., New York, NY.
- 6.Bordeleau, M. E., A. Mori, M. Oberer, L. Lindqvist, L. S. Chard, T. Higa, G. J. Belsham, G. Wagner, J. Tanaka, and J. Pelletier. 2006. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2:213-220. [DOI] [PubMed] [Google Scholar]
- 7.Bushell, M., and P. Sarnow. 2002. Hijacking the translation apparatus by RNA viruses. J. Cell Biol. 158:395-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cutforth, T., and G. M. Rubin. 1994. Mutations in Hsp83 and cdc37 impair signaling by the sevenless receptor tyrosine kinase in Drosophila. Cell 77:1027-1036. [DOI] [PubMed] [Google Scholar]
- 9.Donze, O., and D. Picard. 1999. Hsp90 binds and regulates Gcn2, the ligand-inducible kinase of the alpha subunit of eukaryotic translation initiation factor 2. Mol. Cell. Biol. 19:8422-8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dreher, T. W., and W. A. Miller. 2006. Translational control in positive strand RNA plant viruses. Virology 344:185-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Echalier, G. 1997. Composition of the body fluid of Drosophila and the design of culture media for Drosophila cells, p. 1-67. In G. Echalier (ed.), Drosophila cells in culture. Academic Press, London, United Kingdom.
- 12.Finkelstein, Y., O. Faktor, O. Elroy-Stein, and B. Z. Levi. 1999. The use of bi-cistronic transfer vectors for the baculovirus expression system. J. Biotechnol. 75:33-44. [DOI] [PubMed] [Google Scholar]
- 13.Friesen, P. D., and R. R. Rueckert. 1981. Synthesis of black beetle virus proteins in cultured cells: differential expression of RNAs 1 and 2. J. Virol. 37:876-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilmore, R., M. C. Coffey, and P. W. Lee. 1998. Active participation of Hsp90 in the biogenesis of the trimeric reovirus cell attachment protein σ1. J. Biol. Chem. 273:15227-15233. [DOI] [PubMed] [Google Scholar]
- 15.Hartson, S. D., E. A. Ottinger, W. Huang, G. Barany, P. Burn, and R. L. Matts. 1998. Modular folding and evidence for phosphorylation-induced stabilization of an hsp90-dependent kinase. J. Biol. Chem. 273:8475-8482. [DOI] [PubMed] [Google Scholar]
- 16.Hu, J., and C. Seeger. 1996. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc. Natl. Acad. Sci. USA 93:1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, J., D. Toft, D. Anselmo, and X. Wang. 2002. In vitro reconstitution of functional hepadnavirus reverse transcriptase with cellular chaperone proteins. J. Virol. 76:269-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu, J., D. O. Toft, and C. Seeger. 1997. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 16:59-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kampmueller, K. M., and D. J. Miller. 2005. The cellular chaperone heat shock protein 90 facilitates Flock House virus RNA replication in Drosophila cells. J. Virol. 79:6827-6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, D. H., and A. L. Goldberg. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397-403. [DOI] [PubMed] [Google Scholar]
- 21.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 22.Lu, R., M. Maduro, F. Li, H. W. Li, G. Broitman-Maduro, W. X. Li, and S. W. Ding. 2005. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature 436:1040-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merrick, W. C., and J. O. Hensold. 2000. Analysis of eukaryotic translation in purified and semipurified systems, p. 11.9.1-11.9.26. In K. S. Morgan (ed.), Current protocols in cell biology. John Wiley & Sons, Inc., New York, NY. [DOI] [PubMed]
- 24.Miller, D. J., and P. Ahlquist. 2002. Flock house [sic] virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J. Virol. 76:9856-9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, D. J., M. D. Schwartz, and P. Ahlquist. 2001. Flock house [sic] virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J. Virol. 75:11664-11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, D. J., M. D. Schwartz, B. T. Dye, and P. Ahlquist. 2003. Engineered retargeting of viral RNA replication complexes to an alternative intracellular membrane. J. Virol. 77:12193-12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Momose, F., T. Naito, K. Yano, S. Sugimoto, Y. Morikawa, and K. Nagata. 2002. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J. Biol. Chem. 277:45306-45314. [DOI] [PubMed] [Google Scholar]
- 28.Naito, T., F. Momose, A. Kawaguchi, and K. Nagata. 2007. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J. Virol. 81:1339-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ochel, H. J., K. Eichhorn, and G. Gademann. 2001. Geldanamycin: the prototype of a class of antitumor drugs targeting the heat shock protein 90 family of molecular chaperones. Cell Stress Chaperones 6:105-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okamoto, T., Y. Nishimura, T. Ichimura, K. Suzuki, T. Miyamura, T. Suzuki, K. Moriishi, and Y. Matsuura. 2006. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 25:5015-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pratt, W. B., and D. O. Toft. 2003. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 228:111-133. [DOI] [PubMed] [Google Scholar]
- 32.Price, B. D., R. R. Rueckert, and P. Ahlquist. 1996. Complete replication of an animal virus and maintenance of expression vectors derived from it in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:9465-9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roe, S. M., C. Prodromou, R. O'Brien, J. E. Ladbury, P. W. Piper, and L. H. Pearl. 1999. Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J. Med. Chem. 42:260-266. [DOI] [PubMed] [Google Scholar]
- 34.Schneider, R. J., and I. Mohr. 2003. Translation initiation and viral tricks. Trends Biochem. Sci. 28:130-136. [DOI] [PubMed] [Google Scholar]
- 35.Sullivan, C. S., and J. M. Pipas. 2001. The virus-chaperone connection. Virology 287:1-8. [DOI] [PubMed] [Google Scholar]
- 36.Uma, S., S. D. Hartson, J. J. Chen, and R. L. Matts. 1997. Hsp90 is obligatory for the heme-regulated eIF-2α kinase to acquire and maintain an activable conformation. J. Biol. Chem. 272:11648-11656. [DOI] [PubMed] [Google Scholar]
- 37.Venkateswarlu, K., D. C. Lamb, D. E. Kelly, N. J. Manning, and S. L. Kelly. 1998. The N-terminal membrane domain of yeast NADPH-cytochrome P450 (CYP) oxidoreductase is not required for catalytic activity in sterol biosynthesis or in reconstitution of CYP activity. J. Biol. Chem. 273:4492-4496. [DOI] [PubMed] [Google Scholar]
- 38.Venter, P. A., N. K. Krishna, and A. Schneemann. 2005. Capsid protein synthesis from replicating RNA directs specific packaging of the genome of a multipartite, positive-strand RNA virus. J. Virol. 79:6239-6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venter, P. A., and A. Schneemann. 2007. Assembly of two independent populations of Flock House virus particles with distinct RNA packaging characteristics in the same cell. J. Virol. 81:613-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waizenegger, T., T. Stan, W. Neupert, and D. Rapaport. 2003. Signal-anchor domains of proteins of the outer membrane of mitochondria: structural and functional characteristics. J. Biol. Chem. 278:42064-42071. [DOI] [PubMed] [Google Scholar]
- 41.Wang, X. H., R. Aliyari, W. X. Li, H. W. Li, K. Kim, R. Carthew, P. Atkinson, and S. W. Ding. 2006. RNA interference directs innate immunity against viruses in adult Drosophila. Science 312:452-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warren, G. 1993. Membrane partitioning during cell division. Annu. Rev. Biochem. 62:323-348. [DOI] [PubMed] [Google Scholar]
- 43.Wassler, M., I. Jonasson, R. Persson, and E. Fries. 1987. Differential permeabilization of membranes by saponin treatment of isolated rat hepatocytes. Release of secretory proteins. Biochem. J. 247:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waxman, L., M. Whitney, B. A. Pollok, L. C. Kuo, and P. L. Darke. 2001. Host cell factor requirement for hepatitis C virus enzyme maturation. Proc. Natl. Acad. Sci. USA 98:13931-13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, S. X., P. Ahlquist, and P. Kaesberg. 1992. Active complete in vitro replication of nodavirus RNA requires glycerophospholipid. Proc. Natl. Acad. Sci. USA 89:11136-11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, S. X., and P. Kaesberg. 1991. Synthesis of template-sense, single-strand Flockhouse [sic] virus RNA in a cell-free replication system. Virology 183:392-396. [DOI] [PubMed] [Google Scholar]
- 47.Yang, J., J. M. Yang, M. Iannone, W. J. Shih, Y. Lin, and W. N. Hait. 2001. Disruption of the EF-2 kinase/Hsp90 protein complex: a possible mechanism to inhibit glioblastoma by geldanamycin. Cancer Res. 61:4010-4016. [PubMed] [Google Scholar]
- 48.Young, J. C., N. J. Hoogenraad, and F. U. Hartl. 2003. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112:41-50. [DOI] [PubMed] [Google Scholar]