Abstract
The type I interferons (IFNs) are potent mediators of antiviral immunity, and many viruses have developed means to block their expression or their effects. Semliki Forest virus (SFV) infection induces rapid and profound silencing of host cell gene expression, a process believed to be important for the inhibition of the IFN response. In SFV-infected cells, a large proportion of the nonstructural protein nsp2 is found in the nucleus, but a role for this localization has not been described. In this work we demonstrate that a viral mutant, SFV4-RDR, in which the nuclear localization sequence of nsp2 has been rendered inactive, induces a significantly more robust IFN response in infected cells. This mutant virus replicates at a rate similar to that of the parental SFV4 strain and also shuts off host cell gene expression to similar levels, indicating that the general cellular shutoff is not responsible for the inhibition of IFN expression. Further, the rate of virus-induced nuclear translocation of early IFN transcription factors was not found to differ between the wild-type and mutant viruses, indicating that the effect of nsp2 is at a later stage. These results provide novel information about the mode of action of this viral IFN antagonist.
Type I interferons (type I IFNs) are central players in initiation of innate immune defenses to virus infections. By upregulating many antiviral genes, their actions establish an antiviral state. Induction of type I IFN in most cell types is triggered by recognition of viral replication intermediates, such as double-stranded RNA in the cytoplasm. Detection of double-stranded RNA is mediated by retinoic acid-inducible gene I and melanoma differentiation-associated factor 5 (21, 22, 42), which activate latent type I IFN transcription factors, namely IFN regulatory factor 3 (IRF-3), nuclear factor kappa B (NF-κB), and activating protein 1. IRF-3 is activated by phosphorylation and dimerization, while NF-κB is activated by degradation of its inhibitory protein, IκB. The activated transcription factors relocate to the nucleus, where they promote the transcription of genes for early type I IFNs, beta interferon (IFN-β), and alpha 4 interferon (IFN-α4). Expression and secretion of these early type I IFNs lead to autocrine and paracrine stimulation of the cell surface alpha/beta interferon (IFN-α/β) receptor (IFNAR1), resulting in expression of the late type I IFN subsets. Many genes are upregulated upon type I IFN stimulation, and together these constitute activities which block the spread of many diverse virus infections. The coevolution of viruses with their hosts has ensured that many viral strategies exist for antagonizing the activation of these transcription factors to promote viral replication and spread (18). IRF-3 activity is targeted by many viruses, including hepatitis C virus and rotavirus, in order to suppress the expression of type I IFN. Similarly, several viral NF-κB antagonists are known. Most, including for example vaccinia virus K1L, hepatitis C virus Core, and HIV Vpu, inhibit degradation of IκB, maintaining NF-κB in its inactive cytoplasmic form. Another viral antagonist of this pathway, the A238L protein of African swine fever virus, inhibits NF-κB activity at a later stage (38); the A238L protein is an IκB homologue and associates with cytoplasmic p65 after proteasome-mediated degradation of endogenous IκB. In another strategy, the 3C protease of poliovirus cleaves and inactivates NF-κB (31).
Semliki Forest virus (SFV) is an RNA virus of the alphavirus genus of the Togaviridae family. The virus particles are enveloped and contain a single-stranded positive-sense 42S RNA genome. The 5′ two-thirds area of the genome codes for the viral nonstructural proteins 1 to 4 (nsp1 to -4), which are translated directly from this genomic RNA. The structural proteins are encoded within the 3′ one-third of the genome and are translated from a subgenomic 26S mRNA. The nonstructural proteins together with cellular proteins form the viral replicase complex, which catalyzes the production of new viral 42S and 26S RNAs via a negative-strand intermediate. Infection of cells in culture or in vivo with SFV results in the induction of type I IFNs; indeed, many studies have used SFV as a positive control for the induction of IFN.
A major event in the infection of cells with SFV and related viruses is the profound inhibition of cellular transcription and translation, which favors the production of viral RNA and proteins. Very soon after infection, the profile of total cellular macromolecular synthesis changes from cellular RNAs and proteins to predominantly viral gene products. Various mechanisms have been proposed for this (12, 17, 27, 30, 37, 41), but the exact series of events remains obscure. The virus benefits from this shutoff of host molecular synthesis as it frees components of the cellular gene expression apparatus for the production of virus products. It has been proposed that the shutoff also benefits the virus by reducing the capacity of the infected cell to signal, via type I IFNs and other cytokines, to surrounding cells, thus expediting virus propagation (13, 16).
Each of the nsp's has discrete activities within the infected cell. nsp1 is involved in modification of intracellular membranes. nsp3 has an important but yet-undefined function. nsp4 is the RNA-dependent RNA polymerase. nsp2, the most well studied of the SFV nsp's, is a papain-like protease responsible for processing the viral nonstructural polypeptide to release each nsp in a temporally regulated manner (40). It also has NTPase, RNA helicase, and RNA triphosphatase activities (15, 33, 34). In infected cells, a large proportion of nsp2 is found in association with the nuclear matrix, although its function there is unknown (32). The nuclear localization sequence of nsp2 consist of a core nonapeptide (A645LPRRRVTWN) (35); however, in fusion constructs, a larger fragment is required to direct nuclear translocation of β-galactosidase (35). Mutation of the second R residue of the nsp2 nuclear localization sequence to D results in a virus termed SFV4-RDR, in which nsp2 does not translocate to the nucleus (35). Apart from a short lag period, SFV4-RDR has replication kinetics similar to that of parental SFV4 in BHK cells (33). The short lag phase is most likely due to a slower processing of the nsp12 precursor. Although cellular DNA synthesis is not inhibited by SFV4-RDR as much as by SFV4 infection, levels of cellular transcription and translation inhibition are similar (33). Despite the similarities in tissue culture, SFV4 is neurovirulent in BALB/c mice, whereas SFV4-RDR is not (33). SFV4-RDR infection results in only limited focal central nervous system infection and limited cell damage (10). However, in IFNAR1-defective mice, which have no functional type I IFN system, SFV4-RDR and SFV4 are equally virulent, indicating that the differences observed relate to interaction of these viruses with the type I IFN system (10).
In the present work, we have analyzed the role of type I IFN in the control of SFV4-RDR virus replication. Using primary mouse embryo fibroblast (MEF) cells competent for production of and reaction to type I IFNs, we show that despite efficient inhibition of cellular transcription and translation, SFV4-RDR infection induces significantly more type I IFN than does SFV4 infection. Our results therefore indicate that nuclear localization of nsp2 or a function within the region of nsp2 containing the nuclear localization signal normally suppresses type I IFN production in SFV-infected cells.
MATERIALS AND METHODS
Cells and viruses.
BHK-21 cells (ATCC) were cultured in Glasgow minimal essential medium supplemented with 5% fetal calf serum (FCS) (Sigma), 20 mM HEPES, 10% tryptose phosphate broth, 2 mM l-glutamine, and penicillin-streptomycin (Invitrogen). Wild-type (wt) B6, wt sv129, and IFNAR−/− sv129 MEFs (obtained from 12- to 13-day-old fetuses) and L929 cells were cultured in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% FCS, 4 mM l-glutamine, and penicillin-streptomycin. wt SFV4 and SFV4-RDR were both derived from the SFV4 infectious clone (pSP6-SFV4) as described previously (25). Virus titration was performed by quantification of plaque numbers on wt MEF cells. SFV4 stocks were used for infection as follows: cell monolayers were washed with phosphate-buffered saline (PBS), and virus was added in 300 μl minimal essential medium (Invitrogen) supplemented with 0.2% bovine serum albumin, 2 mM l-glutamine, and 20 mM HEPES with periodic shaking for 1 h at 37°C. Virus solutions were then removed and cells washed with PBS before addition of prewarmed complete medium.
RNA extraction and quantitative PCR.
At various times postinfection, cells incubated with virus were trypsinized, removed from the flask, and pelleted by gentle centrifugation. Cell pellets were immediately snap-frozen on dry ice and stored at −70°C. RNA was extracted from 40 mg of cell pellet using the QIAGEN RNeasy Mini kit following the manufacturer's protocol. RNA quality and quantity were assessed on an Agilent bioanalyzer (Agilent technologies) using the RNA 6000 Nano assay. High-quality RNA samples were reverse transcribed as described previously (28). To minimize variability, samples to be directly compared were reverse transcribed at the same time using the same master mix (all reagents were from Invitrogen). Standards and test samples were always assayed in triplicate. Briefly, in a total volume of 20 μl (made up in RNase-free water), reaction mixes contained the following: 50 pM of each primer, 40 mM deoxynucleoside triphosphates, 25 mM MgCl2, 1:20,000 SYBR Green (Biogene Ltd.), 5 U Fast Start Taq (Roche Applied Science), and 2 μl cDNA. Tubes were heated to 94°C for 5 min, and the PCR was then cycled through 94°C for 20 s, 62°C for 20 s, and 72°C for 20 s for 40 cycles on a RotorGene 3000 instrument (Corbett Research). Cell culture-derived samples were normalized to total RNA determined on an Agilent bioanalyzer. Sequences of the primers used in the assay were as follows: SFV4, 5′-CGCATCACCTTCTTTTGTG-3′ and 5′-CCAGACCACCCGAGATTTT-3′; IFN-β, 5′-CACAGCCCTCTCCATCAACT-3′ and 5′-GCATCTTCTCCGTCATCTCC-3′; β-actin, 5′-CGTTGACATCCGTAAAGACC-3′ and 5′-CTGGAAGGTGGACAGTGAG-3′; glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-AACTCCCACTCTTCCACCTT-3′ and 5′-GCCCCTCCTGTTATTATGG-3′.
Type I IFN bioassay.
A bioassay was employed to determine the total amount of active IFN-α/β (20). Briefly, flat-bottomed 96-well plates were seeded with 1.5 × 104 L929 cells/well. The following day, twofold serially diluted samples or an IFN-α/β standard (Gu02-901-511; National Institute for Allergy and Infectious Diseases) was added to the cells. After overnight incubation, the cells were infected with SFV4 (350 IU/well). Mitochondrial dehydrogenase activity was assayed 2 days after infection using 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide according to the instructions of Sigma kit CGD-1. The 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide was dissolved in OPTIMEM reduced-serum medium lacking phenol red (Invitrogen) supplemented with 2% FCS, 2 mM l-glutamine, and penicillin-streptomycin. The absorbance was read at 570 nm and normalized against the absorbance at 630 nm in a microplate reader (Elx 800 UV; BIO-TEK Instruments, Winooski, Vermont). The data were converted to international units using murine IFN-α/β as a standard (National Institute for Allergy and Infectious Diseases, NIH) in all assays. Samples were considered below the level of detection if the signal did not reach three-quarters of the plateau level of the standard. All samples analyzed in the bioassay were UV inactivated to abolish any residual infectivity of the viral particles used for stimulation. Levels of tumor necrosis factor alpha (TNF-α) were quantified by enzyme-linked immunosorbent assay (ELISA) (R&D Systems) according to the manufacturer's instructions.
Western blotting.
For analysis of protein by Western immunoblotting, cells were lysed on ice in lysis buffer (1% NP-40, 50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 2 mM EDTA) and clarified by centrifugation at 6,000 × g for 5 min in a microcentrifuge at 4°C. The protein concentration was determined with the Bio-Rad DC protein assay, and equal amounts were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to Hybond-P membranes (Amersham), and blotted with rabbit polyclonal antibodies specific for either phospho-eIF2α (phosphorylated on serine 51; Biosource) or total eIF2α (Santa Cruz). The secondary antibody was horseradish peroxidase-conjugated goat antirabbit serum (BD Biosciences). Chemiluminescence was detected using ECL reagents (Amersham). Membranes were stripped by washing in 100 mM 2-mercaptoethanol, 2% SDS, and 62.5 mM Tris-HCl (pH 6.7) for 45 min at 50°C, washed, blocked, and probed again as described above.
Protein expression assays.
For metabolic labeling of virus-infected cultures, cells were incubated first in methionine-free minimum essential medium supplemented with 2 mM l-glutamine and 20 mM HEPES (starvation medium) for 20 min. Newly produced proteins were then labeled by incubation in starvation medium supplemented with 50 μCi/ml [35S]methionine (Amersham) for 10 min (Pulse medium). Cells were then washed in cold PBS, lysed on ice in lysis buffer, and clarified by centrifugation at 6,000 × g for 5 min in a microcentrifuge at 4°C. The protein concentration was determined using the Bio-Rad DC protein assay, and equal quantities were then analyzed on 10% SDS-PAGE gels, which were subsequently dried and subjected to autoradiography. Densitometry analysis was performed using Bio-Rad Quantity One software.
RNA quantitation by [14C]uridine labeling.
Equal numbers of MEF cells were infected as described above with SFV4 or SFV4-RDR. For labeling, the medium was removed and replaced with complete medium containing 1 μCi/ml [14C]uridine (Amersham). After the labeling period, total cell RNA was isolated using the GenElute kit (Sigma) according to the manufacturer's instructions and quantified by spectrophotometry. Equal quantities of each sample were analyzed by formaldehyde agarose gel electrophoresis. The gel was then soaked for 30 min in 1 M salicylic acid (Sigma), dried, and exposed to X-ray film (Fujifilm). Densitometry analysis was performed using Bio-Rad Quantity One software.
Immunofluorescence.
Antibodies used included murine anti-SFV4 E2, murine anti-SFV4 replicase, goat anti-IRF-3 (Santa Cruz), and goat anti-NF-κB p65 (Santa Cruz). Secondary antibodies were Cy3-conjugated donkey antigoat sera and Cy2-conjugated donkey antimouse serum (both ML grade from Jackson Immunoresearch). Immunofluorescence was performed as previously described (27). Briefly, cells grown on coverslips were fixed by incubation in 4% paraformaldehyde (in PBS) for 8 to 10 min at room temperature, followed by incubation in methanol for 8 to 10 min at −20°C. The coverslips were blocked, and primary antibodies were diluted in blocking buffer and incubated with the cells for 1 to 3 h. Secondary antibodies were diluted in blocking buffer containing 0.5 μg/ml Hoechst 33258 (Molecular Probes) for identification of cell nuclei. Washed coverslips were then mounted in vinol mounting medium, and images were captured using a Leitz DM RB fluorescence microscope.
RESULTS
SFV4-RDR is attenuated for growth in type I IFN-competent cells.
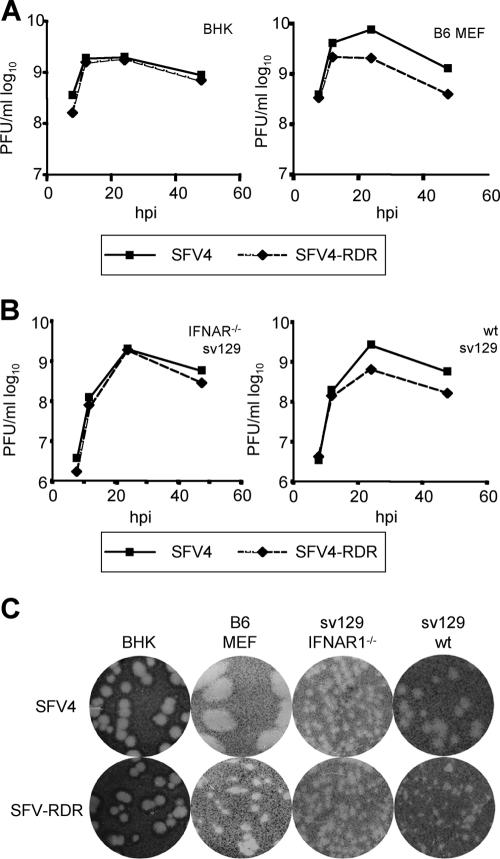
Previous experiments comparing SFV4 and SFV4-RDR showed that the replication kinetics of these two viruses in BHK cells were similar (33). These experiments, performed at a high multiplicity of infection (MOI), indicate that the nuclear localization signal of nsp2 is not important for virus growth, at least in BHK cells. To compare the abilities of these two viruses to spread in cells with or without an intact IFN system, BHK cells and MEFs from IFN-competent B6 mice were infected with SFV4 and SFV4-RDR at a low MOI (0.05), and the amount of infectious virus released into the media was quantified by plaque assay (Fig. 1A). Upon low-MOI infection of type I IFN-incompetent BHK cells, SFV4 and SFV4-RDR replicated with similar kinetics. Both infections produced high titers by 12 h and led to the death of all cells in the cultures by 24 h. This is consistent with results in the previous study (33) and shows that the mutant virus had no significant defect in any step of the viral life cycle in BHK cells. However, when similar low-MOI infections were performed with type I IFN-competent B6 MEFs, relative to those of SFV4, SFV4-RDR titers were reduced. Again, by 12 h, both viruses had grown to high titers. However, the titer of SFV4 continued to rise until 24 h, whereas the titer of SFV4-RDR did not, and while all cells were killed by the SFV4 infection, the SFV4-RDR infection did not result in complete death of the culture; a significant fraction of the cells survived. In repeat experiments, these findings were highly reproducible and indicate that the SFV4-RDR virus is defective in its ability to spread in a culture of type I IFN-competent cells.
FIG. 1.
(A) BHK (left panel) or wt B6 MEF (right panel) cells were infected with SFV4 or SFV4-RDR at a MOI of 0.05, samples were taken at the indicated time points, and infectious virus was quantified by plaque assay on BHK cells. Data points are averages for three experiments. (B) sv129 IFNAR1−/− (left panel) or wt sv129 (right panel) MEFs were infected with SFV4 or SFV4-RDR at a MOI of 0.05, samples were taken at the indicated time points, and infectious virus was quantified by plaque assay on BHK cells. Data points are averages for two experiments. (C) Comparison of viral plaque sizes of SFV4 and SFV4-RDR on BHK cells, wt B6 MEFs, sv129 IFNAR1−/− MEFS, and wt sv129 MEFs.
To confirm that this was not a phenomenon related to other properties of these two cell types, the studies were repeated with a second set of interferon-competent and -incompetent cells, MEFs derived from sv129 wt or sv129-IFNAR1−/− mice. The results are presented in Fig. 1B. Again, in cells lacking a type I IFN signaling system (the IFNAR1−/− cells), the growth curves of SFV4 and SFV4-RDR were very similar, while in cells with an intact IFN system, SFV4-RDR replicated less efficiently than SFV4. In repeat experiments, the final titer of SFV4-RDR was consistently >3.5-fold lower than that of SFV4. As with wt B6 MEFs, many of the SFV4-RDR-infected wt sv129 MEFs survived, whereas none of the IFNAR1−/− cells infected by either virus were viable after 48 h. These experiments show that although SFV4-RDR replicates well in IFN-incompetent cells, it is clearly attenuated in wt MEFs (both B6 and sv129), which are competent for production of and responses to type I IFNs.
To further investigate this phenomenon, we examined the viral plaque sizes of SFV4 and SFV4-RDR grown on all four cell types. The results clearly show that on BHK and IFNAR−/− cell monolayers, the plaque sizes of the two viruses were identical, whereas on both strains of wt MEFs, the SFV4-RDR plaques were smaller than the SFV4 plaques (Fig. 1C). In particular, relative to SFV4, SFV4-RDR gave rise to very small plaques on wt B6 MEFs, indicating that this mutant virus is severely defective in its ability to spread in these type I IFN-competent MEFs. The mutant also grew less well than SFV4 on the sv129 MEFs. Relative to SFV4, SFV-RDR exhibited a reduced-plaque-size phenotype only in wt MEFs of either background (the small plaques shown on the IFNAR−/− MEFs are due to earlier fixing of cells because of crowding on the plates). Taken together with the in vivo data on these viruses in IFNAR−/− mice (10), these results demonstrate that relative to SFV4, growth of SFV4-RDR is suppressed by the action of type I IFN. This could result from differences in IFN induction by these two viruses or from differences in the susceptibility of the two viruses to IFN-mediated antiviral activities.
Infection of MEFs with SFV4-RDR leads to significantly higher levels of type I IFN.
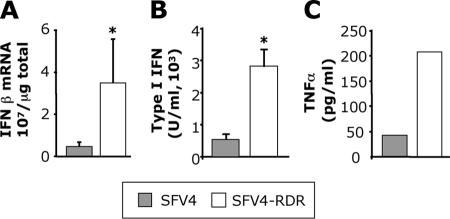
The levels of type I IFN induced in MEFs infected with SFV4 and SFV4-RDR were determined using two different assays. First, we measured the amount of IFN-β mRNA in infected cells. At 12 h postinfection (hpi), total cellular RNA was extracted from triplicate cultures of sv129 MEFs infected at MOI 50 with SFV4 or SFV4-RDR. With both viruses, this MOI was observed by immunostaining to result in infection of >98% of cells in the culture (data not shown). Levels of IFN-β mRNA transcripts were measured by quantitative reverse transcriptase PCR (QPCR). Approximately seven times more IFN-β mRNA was produced by SFV4-RDR-infected cells than by SFV4-infected cells (Fig. 2A). In a second approach, we quantified the levels of functional IFN activity produced by infected cells. Sv129 MEFs were infected with SFV4 or SFV4-RDR at a MOI of 50. Supernatants were sampled at 24 hpi, and type I IFN levels were quantified using a bioassay (20). The results are shown in Fig. 2B. SFV4-RDR-infected MEFs produced significantly more functional type I IFN than SFV4-infected cells. Supernatants were also analyzed by ELISA for levels of the proinflammatory cytokine TNF-α. Levels were dramatically higher with SFV4-RDR than with SFV4 (Fig. 2C), indicating that this effect was not specific to type I IFNs but was also observed for other virus-induced cytokines. All of the experiments presented in Fig. 2 were repeated in murine L929 cells, with similar or even greater differences between the two viruses (data not shown).
FIG. 2.
(A) Total cellular RNA was taken at 12 hpi from MEFs infected with SFV4 or SFV4-RDR and analyzed by QPCR for IFN-β mRNA levels. Bars represent the mean of triplicate measurements from a representative experiment, error bars are standard deviations, and the asterisk indicates a P value of <0.05 (Student's t test). (B) Supernatants from SFV4- or SFV4-RDR-infected MEFs were taken at 24 hpi and analyzed by bioassay for levels of functional IFN. Bars represent the mean of triplicate measurements from a representative experiment, error bars are standard deviations, and the asterisk indicates a P value of <0.05 (Student's t test). (C) Supernatants from infected MEFs were taken at 24 hpi and analyzed by ELISA for levels of TNF-α. Bars represent the mean of triplicate measurements from one representative experiment of three.
SFV4-RDR induces inhibition of host RNA production as efficiently as SFV4.
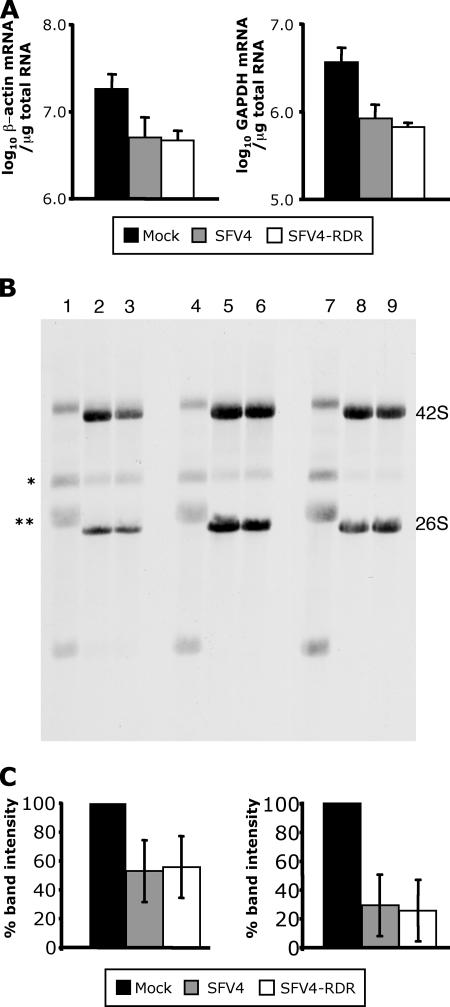
The levels of type I IFN produced during SFV4-RDR infection were much higher than those produced by SFV4 infection. A similar observation has been made with nsp2 mutants of the related Sindbis virus (SIN), and here it was proposed that the increased IFN resulted from an incomplete inhibition of cellular gene expression; wt SIN completely suppresses host cell gene expression, whereas nsp2 mutants do not (13, 14, 16). To determine whether there were differences in the shutoff of host cell gene transcription in SFV4- and SFV4-RDR-infected cells, total cellular RNA was purified from SFV4-, SFV4-RDR-, or mock-infected MEFs at 12 hpi, and as representative markers of cellular transcripts, levels of β-actin and GAPDH mRNA were determined by QPCR (Fig. 3A). Relative to those in the mock-infected control cultures, levels of these cellular mRNAs were equally reduced in the two virus infections.
FIG. 3.
(A) Total cellular RNA was taken at 12 hpi from mock-infected MEFs (black bars) or MEFs infected with SFV4 (gray bars) or SFV4-RDR (white bars) and analyzed by QPCR for β-actin (left panel) and GAPDH (right panel) mRNA levels. Each bar represents the mean of triplicate measurements from a representative experiment, and error bars are standard deviations. (B) Total cellular and viral RNA synthesis was visualized by [14C]uridine labeling between 1 and 3 hpi (lanes 1 to 3), 3 and 5 hpi (lanes 4 to 6), or 5 and 7 hpi (lanes 7 to 9) in wt B6 MEFs mock infected (lanes 1, 4, and 7), infected at an MOI of 50 with SFV4 (lanes 2, 5, and 8), or infected at a MOI of 50 with SFV4-RDR (lanes 3, 6, and 9). Positions of the viral genomic 42S and subgenomic 26S RNAs are indicated on the right. (C) Densitometry analysis was performed on the intensities of cellular RNA bands labeled with an asterisk (left panel) or two asterisks (right panel) in panel B between 2 and 5 hpi. Data are averages from four experiments, and error bars are standard deviations.
Levels of newly produced RNA in the early stages of virus infection were also investigated by metabolic labeling. B6 MEFs were infected at a MOI of 50 with SFV4 or SFV4-RDR or mock infected and labeled for 2-h periods with [14C]uridine. Total RNA was extracted and analyzed by agarose gel electrophoresis. Three labeling periods were used, 1 to 3 hpi, 3 to 5 hpi, and 5 to 7 hpi (Fig. 3B). Several major RNA species were present in the mock-infected cells; these represent the rRNAs and their precursors. In cells infected with SFV4 or SFV4-RDR, even at the earliest 1- to 3-hpi labeling time period, the major RNA species were the viral genomic 42S RNA and the subgenomic 26S mRNA, with few rRNAs. This experiment also demonstrates that the RNA replication kinetics of SFV4 and SFV4-RDR were indistinguishable. In order to quantify the transcriptional shutoff caused by these two viral infections, densitometry analysis was performed on two prominent bands, corresponding to cellular RNAs. Figure 3C shows that while these two cellular RNAs differ in the extent to which each is shut off, in neither case is there a difference between cells infected with SFV4 or SFV4-RDR. It is evident that some variation of the extent of shutoff exists between individual experiments, but in each case the values for the two viruses were very similar. These data indicate that a profound inhibition of cellular RNA production occurs rapidly in both SFV4- and SFV4-RDR-infected cells and that this is not different between these two viruses.
SFV4-RDR induces inhibition of host protein production as efficiently as SFV4.
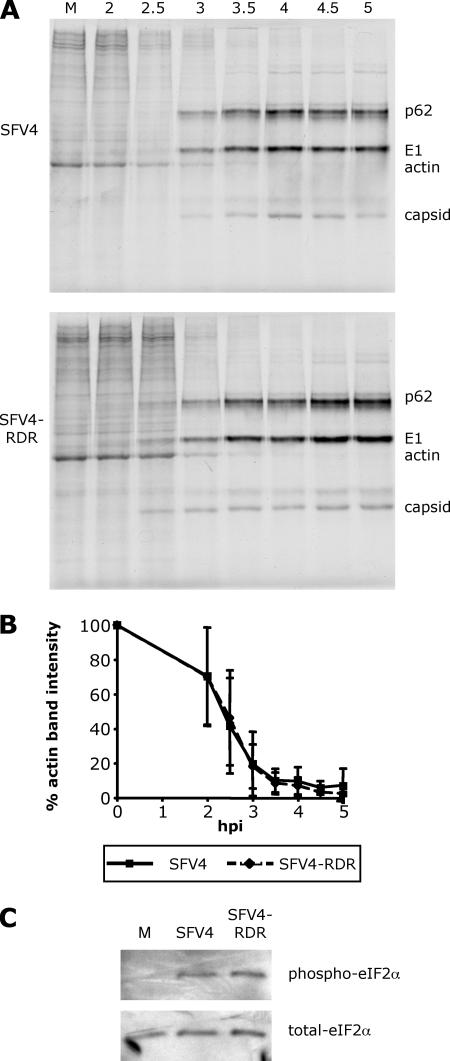
We also investigated virus-induced inhibition of cellular translation in SFV4- and SFV4-RDR-infected cells. Replicate cultures of B6 MEFs were infected at a MOI of 10, and at various times postinfection, cultures were metabolically pulse-labeled with [35S]methionine. Analysis of cell lysates by SDS-PAGE demonstrated that the protein production profile of SFV4- and SFV4-RDR-infected MEFs changed from cellular to predominantly viral at similar times (Fig. 4A). Between 2 and 3 hpi, cellular protein synthesis declined sharply to negligible levels. From 3 hpi onwards, protein synthesis was mainly devoted to virus structural proteins. In the experiment shown and in some but not all repeat experiments, viral protein synthesis in the SFV-RDR-infected cells appeared to begin at a low level at the 2.5-hpi time point. The cause of this is unknown, although our analysis of viral RNA production (Fig. 3) suggested that it is likely not due to increased 26S mRNA production at these times. In any event, it does not appear to alter the kinetics of shutoff of host protein synthesis. When densitometric analysis was carried out on the intensity of the bands corresponding to the cellular protein actin (Fig. 4B), it was found to be reduced at similar rates in cells infected with either virus. As with the quantification analysis of transcription shutoff, there was variation between each experiment, but the means and the standard deviations for each virus were almost identical at each time point. Thus, these data show that both viruses induced shutoff of host cell translation to similar extents and with similar kinetics. In a separate approach to studying translation, we analyzed the extent of phosphorylation of the α subunit of the translation initiation factor eIF2. Lysates from cells infected with SFV4 or SFV4-RDR were prepared after 5 h of infection and analyzed by immunoblotting for the phosphorylated and nonphosphorylated forms of this protein. A representative experiment shown in Fig. 4C shows that the levels of phosphorylation did not differ between SFV4- and SFV4-RDR-infected cells. Taken together, the results in Fig. 3 and 4 show that the SFV4-RDR virus does not have a defect in its ability to interfere with the gene expression apparatus in infected cells. Thus, the higher levels of antiviral and proinflammatory cytokines measured during SFV4-RDR infection cannot be attributed to a reduced global suppression of host cell transcription or translation by this virus.
FIG. 4.
(A) The kinetics of protein synthesis at early times postinfection in SFV4-infected (upper panel) or SFV4-RDR-infected (lower panel) MEFs (each at a MOI of 10) was analyzed by [35S]methionine labeling. Positions of cellular actin and viral p62, E1, and capsid proteins are indicated on the right. (B) Densitometry was performed on the intensity of the band corresponding to actin. Data are averages from three experiments, and error bars are standard deviations. (C) The phosphorylation state of eIF2α in these cells 5 h postinfection or mock infection (M) was determined by immunoblotting for phospho-eIF2α (upper panel) and related to total eIF2α by stripping and reprobing the membrane (lower panel).
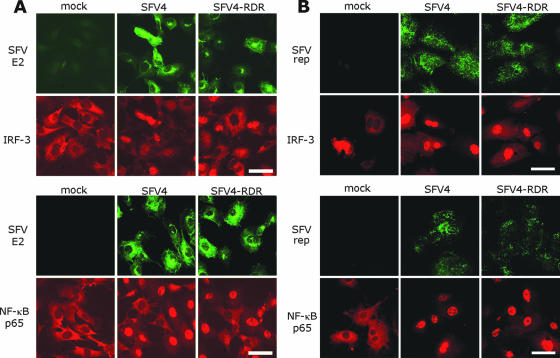
SFV4-RDR does not block the nuclear entry of type I IFN transcription factors.
As discussed above, many viruses have the ability to specifically block the type I IFN system. One strategy is to block nuclear translocation of IFN-inducing transcription factors (18). To investigate whether nuclear translocation of IRF-3 or NF-κB was suppressed in SFV4- or SFV4-RDR infected cells, MEFs were infected at both low and high MOI and fixed at various times after infection, and the locations of these two transcription factors were investigated by immunofluorescence. Infected cultures were stained using antibody for either IRF-3 or NF-κB and, in order to visualize virus-infected cells, a monoclonal antibody to the SFV E2 spike protein or to the SFV replicase complex. At 5 hpi in the low-MOI-infected cells, complete translocation of both transcription factors was seen in every cell which expressed the E2 spike protein in cultures infected with either virus (Fig. 5A). Analysis at earlier times was made difficult by low viral protein expression and by the asynchronicity of low-MOI infections in MEFs. However, it could be seen that the numbers of cells exhibiting nuclear localization of either transcription factor did not differ between the two viruses at any time during infection (data not shown). In order to ensure complete and synchronous infection, this analysis was also performed at a high MOI. At 3 hpi in cells infected at a high MOI with either virus, all cells were positive for the SFV replicase and had exhibited nuclear translocation of both IRF-3 and NF-κB (Fig. 5B). These early components of the type I IFN induction pathway were therefore clearly activated early in infection by both viruses.
FIG. 5.
B6 MEFs were infected at a MOI of 1 (A) or 50 (B) with SFV4 or SFV4-RDR or were mock infected. Cells were fixed at 5 hpi (A) or 3 hpi (B) and immunostained with antibodies to SFV E2 or replicase (green fluorescence) and either IRF-3 (upper panels, red fluorescence) or NF-κB p65 (lower panels, red fluorescence). Bar, 50 μm.
DISCUSSION
The discoveries of many viral suppressors of the type I IFN response (reviewed in reference 18) illustrate the potency of this pathway in the control of virus infections. Pathogenic viruses have evolved in many ways to inhibit the host's ability to respond to infection, and these suppressive effects are found in most stages of the pathway. Suppressors include viral proteins that can block the initial activation of the upstream sensors of viral infection (retinoic acid-inducible gene I and melanoma differentiation-associated factor 5) (6, 8, 11), block the activation and nuclear entry of type I IFN transcription factors (4, 5, 24, 36, 38), inhibit general transcription (1, 7, 9, 39), act as decoy type I IFN receptors (2), and block the downstream type I IFN effector molecules (19, 23).
In this study we have analyzed the interactions between SFV and the type I interferon response in mouse embryo fibroblasts. Our tool was the virus mutant SFV4-RDR, which differs from SFV4 by a single point mutation in the RRR (wt) nuclear localization signal within nsp2. Early work with this virus in mice showed that an intact type I IFN interferon system could attenuate the neurovirulence of SFV4-RDR but not the neurovirulence of SFV4 (10, 33). Here we show that relative to SFV4, infection of MEFs with SFV4-RDR resulted in increased expression of type I IFN genes and increased levels of functional interferon and the proinflammatory cytokine TNF-α.
Several RNA viruses block the cell's ability to produce large amounts of type I IFN by blocking general cellular gene expression. In these viral infections, constitutively active transcription factors are inactivated and total cellular transcription is blocked, including that of type I IFN genes and other infection-stimulated genes. In many cases, inhibition of cellular RNA synthesis is accompanied by translational shutoff, possibly aiding the immediate cessation of cellular gene expression (1, 9). Recent work with nsp2 mutants of the related Sindbis alphavirus suggests that this virus also causes a general inhibition of cellular RNA synthesis, resulting in inhibition of the type I IFN system (13, 16). The SIN nsp2 mutant SIN/G, which is incapable of inhibiting cellular protein and RNA synthesis in infected murine cells, induces a significant increase in type I IFN relative to that with the parental virus. The SIN2V mutant also induces high levels of IFN relative to those with the parental virus but causes only a partial shutoff of cellular transcription. In contrast, our SFV4-RDR mutant replicates with kinetics comparable to those of SFV4 and induces similar levels of global transcription and translation shutoff but nevertheless induces significantly more type I IFN. This indicates that the differences in cytokine responses are not due to an inability of SFV4-RDR to globally suppress host cell macromolecular synthesis.
Two explanations must be considered for these differences; first, that SFV4-RDR activates the IFN system more efficiently than SFV4 does, and second, that SFV4 has the ability to specifically suppress IFN induction, while the ability of SFV4-RDR to do this has been reduced or lost. It is difficult to separate these possibilities, but since nuclear translocation of the key interferon-inducing transcription factors IRF-3 and NF-κB occurred with equal kinetics in all virus-infected cells, in both SFV4- and SFV4-RDR-infected cultures, we conclude that both viruses efficiently activate early events in the induction of type I IFN gene expression. nsp2 of SFV must therefore have the ability to suppress the induction of type I IFN at some point in the gene induction pathway beyond the nuclear translocation of these transcription factors. SFV4 and SFV4-RDR differ in a single amino acid in the nuclear localization signal of nsp2; while it is tempting to speculate that translocation of nsp2 to the nucleus in SFV4-infected cells is required for suppression of the IFN system, the possibility that the RDR mutation affects a function of nsp2 manifest in the cytoplasm cannot be ruled out.
The observation that the levels of another virus-induced cytokine, TNF-α, were also increased in SFV4-RDR-infected cells points to a non-IFN-specific effect on host cell gene expression. It is possible that a larger group of genes is inhibited by nsp2 via inhibition of the activity of a particular transcription factor, such as, for example, NF-κB. Alternatively, it is possible that the nuclear fraction of nsp2 may inhibit transcription on a more general level in a manner similar to those discussed for other RNA viruses. Either way, an nsp2-dependent mechanism exists to inhibit the production of cellular transcripts which are strongly promoted soon after virus infection, for example, host response cytokines. However, as we have shown that both SFV4 and SFV4-RDR lead to inhibition of host transcription by a common global mechanism but differ in their levels of type I IFN and TNF-α induction, the relative contributions of specific and general transcription inhibition mechanisms to these differences cannot be readily determined.
The SFV nsp2 protein has a number of activities which could be involved in the inhibition of host responses (15, 26, 29, 33, 34). Notably, it contains a papain-like protease domain which is primarily responsible for the processing of the nsp1234 polyprotein and maturation of the viral replicase complex (29, 40). Other RNA viruses encode proteases which are transported to the nucleus and result in the cleavage of cellular transcription factors (3). It is possible that SFV4 nsp2 protease activity is maintained upon nuclear entry and that this is responsible for the inhibition of the type I IFN response. The observation that cellular DNA synthesis is inhibited by SFV4 but not by SFV4-RDR (33) suggests that nuclear entry of nsp2 has clear effects on the metabolism of the host cell. How nsp2 antagonizes transcription of cellular innate immune signaling genes remains to be elucidated.
Acknowledgments
We thank Åsa Hidmark for preparation of primary MEFs.
This work was supported by the European Union 5th Framework program (Project SFVectors) and by a Eurogendis grant to L.B.
Footnotes
Published ahead of print on 6 June 2007.
REFERENCES
- 1.Ahmed, M., M. O. McKenzie, S. Puckett, M. Hojnacki, L. Poliquin, and D. S. Lyles. 2003. Ability of the matrix protein of vesicular stomatitis virus to suppress beta interferon gene expression is genetically correlated with the inhibition of host RNA and protein synthesis. J. Virol. 77:4646-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcami, A., J. A. Symons, and G. L. Smith. 2000. The vaccinia virus soluble alpha/beta interferon (IFN) receptor binds to the cell surface and protects cells from the antiviral effects of IFN. J. Virol. 74:11230-11239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amineva, S. P., A. G. Aminev, A. C. Palmenberg, and J. E. Gern. 2004. Rhinovirus 3C protease precursors 3CD and 3CD′ localize to the nuclei of infected cells. J. Gen. Virol. 85:2969-2979. [DOI] [PubMed] [Google Scholar]
- 4.Baigent, S. J., G. Zhang, M. D. Fray, H. Flick-Smith, S. Goodbourn, and J. W. McCauley. 2002. Inhibition of beta interferon transcription by noncytopathogenic bovine viral diarrhea virus is through an interferon regulatory factor 3-dependent mechanism. J. Virol. 76:8979-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barro, M., and J. T. Patton. 2005. Rotavirus nonstructural protein 1 subverts innate immune response by inducing degradation of IFN regulatory factor 3. Proc. Natl. Acad. Sci. USA 102:4114-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Billecocq, A., M. Spiegel, P. Vialat, A. Kohl, F. Weber, M. Bouloy, and O. Haller. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 78:9798-9806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breiman, A., N. Grandvaux, R. Lin, C. Ottone, S. Akira, M. Yoneyama, T. Fujita, J. Hiscott, and E. F. Meurs. 2005. Inhibition of RIG-I-dependent signaling to the interferon pathway during hepatitis C virus expression and restoration of signaling by IKKɛ. J. Virol. 79:3969-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, M. E., P. M. Lieberman, A. J. Berk, and A. Dasgupta. 1993. Direct cleavage of human TATA-binding protein by poliovirus protease 3C in vivo and in vitro. Mol. Cell. Biol. 13:1232-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fazakerley, J. K., A. Boyd, M. L. Mikkola, and L. Kaariainen. 2002. A single amino acid change in the nuclear localization sequence of the nsP2 protein affects the neurovirulence of Semliki Forest virus. J. Virol. 76:392-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 12.Frolov, I. 2004. Persistent infection and suppression of host response by alphaviruses. Arch. Virol. Suppl. 2004:139-147. [DOI] [PubMed] [Google Scholar]
- 13.Frolova, E. I., R. Z. Fayzulin, S. H. Cook, D. E. Griffin, C. M. Rice, and I. Frolov. 2002. Roles of nonstructural protein nsP2 and alpha/beta interferons in determining the outcome of Sindbis virus infection. J. Virol. 76:11254-11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garmashova, N., R. Gorchakov, E. Frolova, and I. Frolov. 2006. Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription. J. Virol. 80:5686-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomez de Cedron, M., N. Ehsani, M. L. Mikkola, J. A. Garcia, and L. Kaariainen. 1999. RNA helicase activity of Semliki Forest virus replicase protein NSP2. FEBS Lett. 448:19-22. [DOI] [PubMed] [Google Scholar]
- 16.Gorchakov, R., E. Frolova, and I. Frolov. 2005. Inhibition of transcription and translation in Sindbis virus-infected cells. J. Virol. 79:9397-9409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorchakov, R., E. Frolova, B. R. G. Williams, C. M. Rice, and I. Frolov. 2004. PKR-dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J. Virol. 78:8455-8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haller, O., G. Kochs, and F. Weber. 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, B., M. Gross, and B. Roizman. 1997. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hidmark, A. S., G. M. McInerney, E. K. Nordstrom, I. Douagi, K. M. Werner, P. Liljestrom, and G. B. Karlsson Hedestam. 2005. Early alpha/beta interferon production by myeloid dendritic cells in response to UV-inactivated virus requires viral entry and interferon regulatory factor 3 but not MyD88. J. Virol. 79:10376-10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato, H., S. Sato, M. Yoneyama, M. Yamamoto, S. Uematsu, K. Matsui, T. Tsujimura, K. Takeda, T. Fujita, O. Takeuchi, and S. Akira. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23:19-28. [DOI] [PubMed] [Google Scholar]
- 22.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101-105. [DOI] [PubMed] [Google Scholar]
- 23.Kitajewski, J., R. J. Schneider, B. Safer, S. M. Munemitsu, C. E. Samuel, B. Thimmappaya, and T. Shenk. 1986. Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2 alpha kinase. Cell 45:195-200. [DOI] [PubMed] [Google Scholar]
- 24.La Rocca, S. A., R. J. Herbert, H. Crooke, T. W. Drew, T. E. Wileman, and P. P. Powell. 2005. Loss of interferon regulatory factor 3 in cells infected with classical swine fever virus involves the N-terminal protease, Npro. J. Virol. 79:7239-7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liljestrom, P., S. Lusa, D. Huylebroeck, and H. Garoff. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lulla, A., V. Lulla, K. Tints, T. Ahola, and A. Merits. 2006. Molecular determinants of substrate specificity for Semliki Forest virus nonstructural protease. J. Virol. 80:5413-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McInerney, G. M., N. L. Kedersha, R. J. Kaufman, P. Anderson, and P. Liljeström. 2005. Importance of eIF2α phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell. 16:3753-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKimmie, C. S., and J. K. Fazakerley. 2005. In response to pathogens, glial cells dynamically and differentially regulate Toll-like receptor gene expression. J. Neuroimmunol. 169:116-125. [DOI] [PubMed] [Google Scholar]
- 29.Merits, A., L. Vasiljeva, T. Ahola, L. Kaariainen, and P. Auvinen. 2001. Proteolytic processing of Semliki Forest virus-specific non-structural polyprotein by nsP2 protease. J. Gen. Virol. 82:765-773. [DOI] [PubMed] [Google Scholar]
- 30.Montgomery, S. A., P. Berglund, C. W. Beard, and R. E. Johnston. 2006. Ribosomal protein S6 associates with alphavirus nonstructural protein 2 and mediates expression from alphavirus messages. J. Virol. 80:7729-7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neznanov, N., K. M. Chumakov, L. Neznanova, A. Almasan, A. K. Banerjee, and A. V. Gudkov. 2005. Proteolytic cleavage of the p65-RelA subunit of NF-kappaB during poliovirus infection. J. Biol. Chem. 280:24153-24158. [DOI] [PubMed] [Google Scholar]
- 32.Peranen, J., M. Rikkonen, P. Liljestrom, and L. Kaariainen. 1990. Nuclear localization of Semliki Forest virus-specific nonstructural protein nsP2. J. Virol. 64:1888-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rikkonen, M. 1996. Functional significance of the nuclear-targeting and NTP-binding motifs of Semliki Forest virus nonstructural protein nsP2. Virology 218:352-361. [DOI] [PubMed] [Google Scholar]
- 34.Rikkonen, M., J. Peranen, and L. Kaariainen. 1994. ATPase and GTPase activities associated with Semliki Forest virus nonstructural protein nsP2. J. Virol. 68:5804-5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rikkonen, M., J. Peranen, and L. Kaariainen. 1992. Nuclear and nucleolar targeting signals of Semliki Forest virus nonstructural protein nsP2. Virology 189:462-473. [DOI] [PubMed] [Google Scholar]
- 36.Ronco, L. V., A. Y. Karpova, M. Vidal, and P. M. Howley. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 12:2061-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tait, S. W., E. B. Reid, D. R. Greaves, T. E. Wileman, and P. P. Powell. 2000. Mechanism of inactivation of NF-kappa B by a viral homologue of I kappa b alpha. Signal-induced release of I kappa b alpha results in binding of the viral homologue to NF-kappa B. J. Biol. Chem. 275:34656-34664. [DOI] [PubMed] [Google Scholar]
- 39.Thomas, D., G. Blakqori, V. Wagner, M. Banholzer, N. Kessler, R. M. Elliott, O. Haller, and F. Weber. 2004. Inhibition of RNA polymerase II phosphorylation by a viral interferon antagonist. J. Biol. Chem. 279:31471-31477. [DOI] [PubMed] [Google Scholar]
- 40.Vasiljeva, L., A. Merits, A. Golubtsov, V. Sizemskaja, L. Kaariainen, and T. Ahola. 2003. Regulation of the sequential processing of Semliki Forest virus replicase polyprotein. J. Biol. Chem. 278:41636-41645. [DOI] [PubMed] [Google Scholar]
- 41.Ventoso, I., M. A. Sanz, S. Molina, J. J. Berlanga, L. Carrasco, and M. Esteban. 2006. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 20:87-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]