Abstract
The basis for the switch from CCR5 to CXCR4 coreceptor usage seen in ∼50% of human immunodeficiency virus type 1 (HIV-1) subtype B-infected individuals as disease advances is not well understood. Among the reasons proposed are target cell limitation and better immune recognition of the CXCR4 (X4)-tropic compared to the CCR5 (R5)-tropic virus. We document here X4 virus emergence in a rhesus macaque (RM) infected with R5-tropic simian/human immunodeficiency virus, demonstrating that coreceptor switch can happen in a nonhuman primate model of HIV/AIDS. The switch to CXCR4 usage in RM requires envelope sequence changes in the V3 loop that are similar to those found in humans, suggesting that the R5-to-X4 evolution pathways in the two hosts overlap. Interestingly, compared to the inoculating R5 virus, the emerging CXCR4-using virus is highly neutralization sensitive. This finding, coupled with the observation of X4 evolution and appearance in an animal with undetectable circulating virus-specific antibody and low cellular immune responses, lends further support to an inhibitory role of antiviral immunity in HIV-1 coreceptor switch.
The human immunodeficiency virus (HIV) enters target cells via interaction of the viral glycoprotein with the cellular receptor CD4 and two principal coreceptors, CCR5 (R5 viruses) and CXCR4 (X4 viruses) (2). Most HIV type 1 (HIV-1) transmission results in a predominantly R5 virus infection. With time, X4 variants arise and coexist with R5 virus variants in ∼50% of subtype B-infected individuals, and this event is associated with rapid CD4+ T-cell loss and disease progression (22, 37). The determinant(s) of phenotypic change from R5 to X4 maps largely to the V3 loop of the envelope gp120 (6, 18, 39) and can be inferred by analysis of the amino acid sequence of this region (11). Although the underlying basis for virus coreceptor switch late in infection remains ill defined, several hypotheses that include changes in target cell populations during the course of infection and/or differential immune recognition of X4 and R5 viruses have been proposed (31, 34). Furthermore, it is unclear whether X4 viruses evolve during the course of infection or are transmitted but selected against early in infection.
We have used infection of rhesus macaques (RM) with simian/human immunodeficiency viruses (SHIV) expressing the envelopes of X4 and R5 HIV-1 isolates to study the impact of coreceptor usage in virus transmission and pathogenesis. We previously reported that both X4 and R5 SHIVs can be transmitted intravenously or intravaginally but showed that the basis for the immunodeficiencies caused by these viruses is different. Whereas primary infection with X4 SHIV caused severe and sustained peripheral blood and secondary lymphoid tissue CD4+ T-cell loss, infection with R5 SHIV resulted in transient loss of CD4+ T cells at these sites (15, 17). Thus, infection with SHIVs of different coreceptor usage recapitulates the different stages of HIV infection in humans: R5 SHIV provides a model of early infection with gradual peripheral CD4+ T cell effects, while X4 SHIV infection reproduces the precipitous CD4+ T-cell decline observed in late-stage disease concomitant with the emergence of X4 virus. The R5 SHIV used in these studies, designated SHIVSF162P3, was generated through successive rapid transfer in RM of the molecular clone SHIVSF162, recovered from passage 3 macaque T353 after 20 weeks of infection (14). Although SHIVSF162P3 can replicate to high titers and induce simian AIDS (SAIDS) in ∼50% of infected RM by intravenous or intravaginal inoculations (reference 13 and unpublished data), no expansion or switch to CXCR4 usage has been observed with progression to disease. In the present work, we hypothesized that infection with an isolate recovered at a later stage of infection in macaque T353 may provide a greater chance of observing coreceptor switch. We reasoned this late isolate would be more diverse and divergent, allowing for rapid evolution of the viral population which may lead to a change in coreceptor preference. We tested this hypothesis by infection of RM with SHIVSF162P3N, an R5 isolate recovered at the time of euthanasia of T353 after 66 weeks of infection.
MATERIALS AND METHODS
Cells.
Human and rhesus peripheral blood mononuclear cells (PBMC) were obtained by Ficoll-Hypaque gradient purification followed by stimulation with 3 μg/ml of phytohemagglutinin A (PHA; Sigma-Aldrich, St. Louis, MO) for human PBMC (huPBMC) and 5 μg/ml staphylococcus enterotoxin B (SEB; Sigma-Aldrich) for rhesus PBMC (rhPBMC) in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 20 U/ml of interleukin-2 (Novartis, Emeryville, CA). 293T cells and TZM-bl cells expressing CD4, CCR5, and CXCR4 (41) and containing integrated reporter genes for firefly luciferase and β-galactosidase under control of the HIV-1 long terminal repeat were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FBS, penicillin, streptomycin, and l-glutamine. U87.CD4 cells stably expressing CD4 and one of the chemokine receptors (10) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FBS, antibiotics, 1 μg/ml puromycin (Sigma-Aldrich), and 300 μg/ml G418 (Geneticin; Invitrogen, Carlsbad, CA).
Virus isolation.
SHIVSF162P3N was recovered by coculturing of week 66 PBMC from macaque T353 with PHA-stimulated huPBMC as previously described (26). The amplified virus was propagated in huPBMC, and the titer was determined in SEB-stimulated rhPBMC. SHIVBR24N was recovered by culturing of PHA-stimulated huPBMC with week 28 plasma from macaque BR24. Recovered virus as well as the control X4 SHIVSF33A and R5 SHIVSF162P3 viruses were amplified and titers were determined in SEB-stimulated rhPBMCs.
Animal infection.
All infections were carried out in adult RM (Macaca mulatta) individually housed at the Tulane National Primate Research Center in compliance with its Guide for the Care and Use of Laboratory Animals. Animals were confirmed to be serologically negative for simian type D retrovirus, SIV, and simian T-cell lymphotropic virus prior to infection. Intravenous inoculations were carried out with 2.4 × 103 and 4.2 × 103 50% tissue culture infectious doses of SHIVSF162P3N and SHIVBR24N, respectively. Whole blood was sampled at designated time intervals. Plasma viremia was quantified by branched DNA analysis (Siemens Medical Solutions Diagnostic Clinical Lab, Emeryville, CA), and absolute CD4+ and CD8+ cell counts were monitored by TruCount (BD Biosciences, Palo Alto, CA). Animals with clinical signs of SAIDS were euthanized by intravenous administration of ketamine-HCl followed by an overdose of sodium pentobarbital. Tissue samples were obtained by surgery or at the time of necropsy. The percentages of CD4+ T cells in the tissue samples were analyzed by flow cytometry (FACScalibur) using CD3-fluorescein isothiocyanate, CD4-phycoerythrin, and CD8-peridinin chlorophyll protein antibodies. Further phenotyping of peripheral CD4+ T cells was performed by using CD28-allophycocyanin and CD95-phycoerythrin. Except for CD3-fluorescein isothiocyanate (BioSource, Camarillo, CA), all antibodies were obtained from BD Biosciences.
Determination of coreceptor usage.
Blocking experiments in rhPBMC and TZM-bl cells were performed to determine coreceptor usage of replication-competent SHIVs. For rhPBMC, 1 × 106 SEB-stimulated cells were infected with 3 ng p27gag antigen equivalent of the indicated SHIV in the absence or presence of 1 μM of the CXCR4 inhibitor AMD3100 or CCR5 inhibitor TAK-779. After incubation for 3 h at 37°C, cells were washed and cultured in 200 μl interleukin-2 and appropriate inhibitor-supplemented RPMI medium in 96-well plates. Culture supernatants were collected over time, and p27gag antigen content was quantified according to the manufacturer's instructions (Coulter Corp., Miami, FL). Percentage blocking at day 7 to 10 postinfection was determined by calculating the amount of p27gag production in the presence relative to that in the absence of inhibitor. For TZM-bl cells, 7 × 103 cells per well of a 96-well plate were inoculated, in triplicate, with 1 ng p27gag antigen equivalent of the indicated SHIV in the absence or presence of inhibitors. The cells were lysed after 72 h of incubation at 37°C and processed for β-galactosidase activity according to the manufacturer's instructions (Applied Biosystems, Foster City, CA). The percentage of blocking was then determined. For assessment of coreceptor usage of reporter viruses, 7 × 103 U87.CD4.CCR5 or U87.CD4.CXCR4 cells were seeded in 96-well plates 24 h before use and infected, in triplicate, with 10 ng p24gag equivalent of the indicated pseudotyped viruses, followed by incubation for 72 h at 37°C. At the end of the incubation period, cells were harvested, lysed, and processed according to the manufacturer's instructions (Promega, Madison, WI). Entry, as quantified by luciferase activity, was measured with an MLX microtiter plate luminometer (Dynex Technologies, Inc., Chantilly, VA). Infection of coreceptor-bearing cells with NL4-3 virus generated in the absence of Env served as a negative control.
Neutralization assay and antibodies.
Virus neutralization was assessed using TZM-bl cells in 96-well plates. Briefly, an equal volume (50 μl) of SHIV (1 ng p27gag equivalent) and serial dilutions of heat-inactivated macaque serum or monoclonal antibodies (MAbs) was incubated for 1 h at 37°C and then added to cells, in triplicate wells, for an additional 2 h at 37°C. A 100-μl aliquot of medium was then added to each well, and the virus-serum cultures were maintained for 72 h. Control cultures received virus in the absence of antibodies. At the end of the culture period, the cells were lysed and processed for β-galactosidase activity. A neutralization curve was generated by plotting the percentage of neutralization versus serum or MAb dilution. Results shown are representative of two independent experiments. The human MAb immunoglobulin G1b12 (IgG1b12) was obtained from D. Burton (Scripps Research Institute, La Jolla, CA), and CD4-IgG2 (PRO 542) was generously provided by Progenics Pharmaceuticals (Tarrytown, NY).
Plasmid constructs and pseudotype virus production.
For expression of envelope glycoproteins, full-length gp160 coding sequences of SHIVSF162P3N and SHIVBR24N were subcloned into the pCAGGS expression plasmid to generate EnvP3N and EnvBR24N, respectively. Briefly, viral RNA was prepared from 500 μl virus supernatant using a commercially available RNA extraction kit (QIAGEN, Chatsworth, CA) and reverse transcribed with SuperScript III reverse transcriptase (Invitrogen) and random hexamer primers (Amersham Pharmacia, Piscataway, NJ). Full-length gp160 was amplified from cDNA of SHIVSF162P3N by primers SH43 (5′-AAGACAGAATTCATGAGAGTGAAGGGGATCAGGAAG-3′) and SH44 (5′-AGAGAGGGATCCTTATAGCAAAGCCCTTTCAAAGCCCT-3′) and from SHIVBR24N with primers SH52 (5′-TAGATCGAATTCTAGAGCCCTGGAAGCATCCAGGAAGTCAGCCTA-3′) and SH53 (5′-AGAGAGGGATCCTCCAGTCCCCCCTTTTCTTTTAAAAA-3′). PCR-based overlapping extension methodology was then employed to replace the V3 loop of EnvP3N with that of BR24N to generate the P3N(V3) Env expression plasmid. With EnvP3N serving as template for PCR amplification, the outer primers used were SH43 and SH44 and the inner primers were SH56 (5′-TATAATATAAGAAAAAGTATACGTATACATAGAGGACCGGGGAGAGCATTTTATGC-3′) and SH57 (5′-TCTATGTATACGTATACTTTTTCTTATATTATAGTTAGGTCTTGTACAATTAATTTCTAC-3′), which encompassed the amino acid changes found in the V3 loop of EnvBR24N (underlined) flanked by adjacent sequence. The resulting amplified fragment was verified by sequencing and subcloned back into pCAGGS. To generate luciferase reporter viruses capable of only a single round of replication, an envelope trans-complementation assay was used as previously described (7). P3N, BR24N, or P3N(V3) Env expression plasmid and the NL4.3LucE-R- vector were cotransfected with polyethylenimine (Polyscience, Warrington, PA) into 2.5 × 106 293T cells plated in a 100-mm plate. Cell culture supernatants were harvested 72 h later, filtered through 0.45-μm filters, and stored at −70°C in 1-ml aliquots. Pseudotype viruses were quantified for p24gag content (Beckman Coulter, Fullerton, CA).
DNA, RNA extraction, sequencing, and analysis.
Proviral DNA was extracted from 1 × 106 to 3 × 106 PBMC with a DNA extraction kit (QIAGEN), and viral RNA was prepared from 500 μl plasma or virus supernatant as described above. The V1-to-V5 region of gp120 was amplified from the reverse transcription products by using Taq DNA polymerase (QIAGEN) with primers ED5 and ED12 or ES7 and ES8 as previously described (9). PCR products were cloned with the TOPO TA cloning kit (Invitrogen) per the manufacturer's instructions followed by direct automated sequencing of cloned gp120 amplicons (SeqWright, Fisher Scientific, Houston, TX). Nucleotide sequences were aligned using the CLUSTALX 1.81 program and further adjusted manually (19). Pair-wise genetic distances were calculated using Kimura's two-parameter model of molecular evolution (20). A phylogenetic tree was constructed using the maximum likelihood method, and bootstrap values were generated with 1,000 repetitions.
Sequence-specific PCR for HR insertion.
Proviral and cDNA products from various time points postinfection were subjected to PCR using Taq DNA polymerase and primers ES7/ES8 as described elsewhere (9). Detection of HR sequence in amplified ES7/ES8 products was achieved by using primers V3-S (5′-CTGTTAAATGGCAGTCTAGC-3′) and SH30 (5′-GCTCTCCCCGGTCCTCTATG-3′) in PCRs. The cycling profile was 95°C for 15 min followed by 40 cycles of 95°C for 15 s and 72°C for 70 s. Amplified products were visualized by electrophoresis in ethidium bromide-stained 2% agarose gels. Appropriate mixing and titration experiments with R5 and X4 variant target sequences that differ in the HR region were performed. The sensitivity of the detection assay was shown to be one X4 variant copy in 106 R5 targets.
RESULTS
High levels of virus replication in macaques infected with the late isolate SHIVSF162P3N.
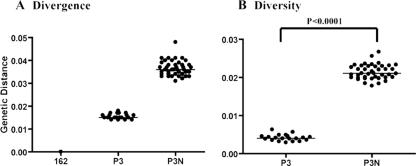
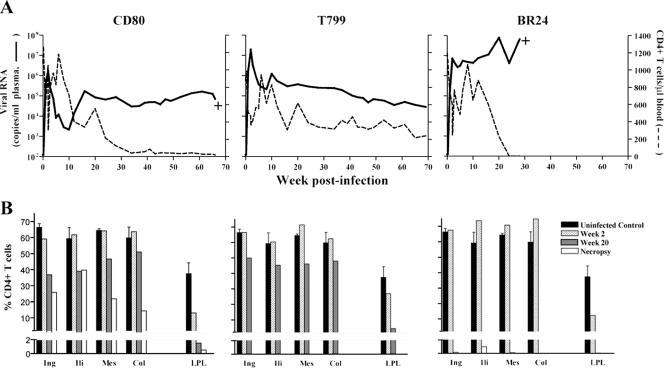
We hypothesized that a late-stage SAIDS-associated CCR5-tropic SHIV isolate, being more diverse and divergent, replicates better and evolves more rapidly than an earlier isolate, providing a greater chance of observing coreceptor switch. To test this hypothesis, we first determined the genetic composition of SHIVSF162P3N, an R5 isolate recovered after the onset of SAIDS (week 66), and compared it to the parental clone, SHIVSF162, as well as the earlier isolate, SHIVSF162P3. Sequence analysis revealed that SHIVSF162P3N is indeed more divergent from the parental clone than is the week 20 SHIVSF162P3 virus (Fig. 1A) and is more diverse (Fig. 1B). The average genetic distances among the Env clones sequenced are greater for the week 66 isolate than the week 20 isolate (P < 0.0001). Three RM were then inoculated intravenously with SHIVSF162P3N. Infection was established in all three macaques, with peak plasma viremia of 106 to 107 RNA copies/ml by 2 weeks postinfection (wpi) (Fig. 2A). Viral replication declined thereafter in two macaques (CD80 and T799) but rebounded by 10 wpi, reaching set points of 105 to 106 RNA copies/ml plasma. A transient drop in peripheral CD4+ T cells, characteristic of R5 SHIV infection (17), accompanied peak infection but recovered close to preinfection values by 8 wpi. With the rise in viremia at 10 wpi, progressive CD4+ T-cell decline followed, which was less protracted in CD80 than T799. Both macaques seroconverted by 6 wpi (Table 1), and peripheral SIV Gag-specific gamma interferon-secreting CD8+ T cells could be detected by intracellular cytokine staining (data not shown). CD80 developed clinical signs consistent with SAIDS and was euthanized at 66 wpi. At the time of death, CD80 had viral loads of >105 RNA copies/ml of plasma and a CD4+ T-cell count of 19 cells/μl of blood. Histopathological findings included multifocal, lymphoid hyperplasia of peripheral and iliac lymph nodes, thymic atrophy, and multiple opportunistic infections (Cryptosporidium, Helicobacter pylori, and Microsporidia).
FIG. 1.
Divergence and diversity of the late week 66 isolate SHIVSF162P3N. (A) Viral divergence from the founder clone SHIVSF162 (162) of SHIVSF162P3 (P3) and SHIVSF162P3N (P3N). The V1-V5 Env gp120 sequence for 20 and 41 clones of P3 and P3N, respectively, was determined. Nucleotide sequences were aligned and the means and standard deviations for all pair-wise comparisons between the sequences of the two isolates and the molecular clone 162 were then calculated using Kimura's two-parameter model of molecular evolution (20). (B) Viral population diversity of P3 and P3N. Genetic distances between all possible pairs of Env nucleotide sequences for viral RNA in the two virus inocula are shown. Bars indicate median values. The P value is from an unpaired t test.
FIG. 2.
Virologic and immunologic measurements in R5 SHIVSF162P3N-infected RM CD80, T799, and BR24. (A) Viral load and absolute CD4+ T-cell counts. (B) Percentages of CD4+ T cells in the inguinal (Ing), iliac (Ili), mesenteric (Mes), and colonic (Col) lymph nodes and LPLs from the jejunum during peak (week 2) and chronic (week 20) stages of infection and at time of necropsy. Baseline values generated from three uninfected macaques are shown for reference. +, death due to euthanasia. Surgery was not performed on BR24 at week 20 due to the development of clinical signs consistent with SAIDS.
TABLE 1.
Humoral immune response in SHIVSF162P3N-infected macaques
| RM | Protein | Antibody response at wpia:
|
|||
|---|---|---|---|---|---|
| 0 | 6 | 12 | 28 | ||
| BR24 | gp120 | − | − | − | − |
| gp41 | − | − | − | − | |
| p31 | − | − | − | − | |
| p27 | − | − | − | − | |
| CD80 | gp120 | − | ++++ | ++++ | ++++ |
| gp41 | − | ++++ | ++++ | +++ | |
| p31 | − | − | ++ | ++ | |
| p27 | − | ++++ | ++++ | +++ | |
| T799 | gp120 | − | ++++ | ++++ | ++++ |
| gp41 | − | ++++ | ++++ | +++ | |
| p31 | − | + | +++ | ++ | |
| p27 | − | ++++ | ++++ | ++ | |
SHIV-specific antibody responses were detected using the strip immunoblot assay in accordance with the manufacturer's instructions (Novartis, Emeryville, CA). Macaque sera reactivity was determined by comparison with two internal IgG strip controls. Scoring relative to level I and II control bands was as follows: −, an absent band; ±, intensity lower than the level I band; +, intensity equal to the level I band; ++, intensity greater than the level I but less than the level II band; +++, intensity equal to the level II band; ++++, intensity greater than the level II band.
The third infected RM, BR24, demonstrated a rapid progressor phenotype. This macaque had sustained viremia that was 2 logs higher (∼107 RNA copies/ml plasma) and failed to develop strong antiviral immune responses. Circulating virus-specific antibody was undetectable (Table 1), and the response of peripheral CD8+ T cells to stimulation with SIV Gag peptides was low and intermittent (data not shown). After a period of transient acute loss, peripheral CD4+ T-cell counts in BR24 also recovered to within the normal range up to 16 wpi but dropped precipitously thereafter. Concomitant with the onset of rapid CD4+ T-cell loss was a spike in viremia to >108 RNA copies/ml of plasma at 20 wpi. BR24 was euthanized at week 28 with clinical signs of SAIDS. At necropsy, BR24 showed histopathological lesions that included severe lymphoid depletion of peripheral and alimentary lymph nodes along with generalized inflammation, thymic dysinvolution, and opportunistic infections (Crytosporidium and Pneumocystis carinii). The circulating CD4+ T-cell count in this animal at the time of death was 1 cell/μl of blood.
Analyses of the percentages of CD4+ T cells in tissue compartments during peak (2 wpi) and chronic (20 wpi) infection in CD80 and T799 revealed moderate CD4+ T-cell loss (<30%) in peripheral lymph nodes (LN) but close to a 90% drop in CD4+ T lymphocytes at effector sites such as the lamina propria (LP) of the gut by 20 wpi (Fig. 2B). At necropsy, >99% of CD4+ T lymphocytes in the gut of CD80 were depleted, but with preservation of 15 to 40% CD4+ T cells in all secondary lymphoid tissues examined. A similar pattern of peak CD4+ T cells loss was observed in macaque BR24, with depletion only in the gut at 2 wpi (Fig. 2B). At necropsy, however, there was massive depletion of LN CD4+ T cells as well as LP lymphocytes (LPL). This pattern of severe CD4+ T-cell loss differs from CD80 and is reminiscent of that seen in X4 SHIV-infected macaques (17), raising the possibility of X4 emergence in BR24. To investigate this possibility, virus was recovered from BR24 plasma obtained at necropsy. The coreceptor usage of the recovered virus, designated SHIVBR24N, was then determined.
Emergence of a CXCR4-using and neutralization-sensitive variant in macaque BR24.
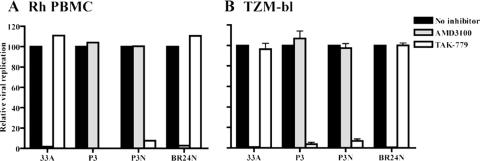
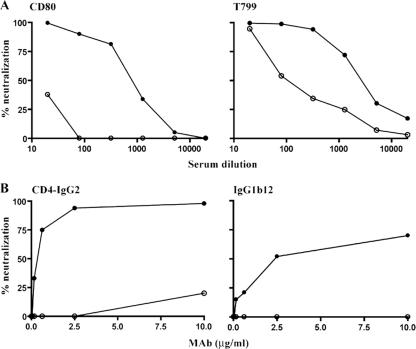
We confirmed CCR5 usage of SHIVSF162P3 and SHIVSF162P3N by demonstrating that the CCR5 inhibitor TAK-779 and not the CXCR4 inhibitor AMD3100 blocked replication of these viruses in rhPBMC (Fig. 3A). In contrast, replication of SHIVBR24N, similar to the X4 SHIVSF33A virus, was not inhibited by TAK-779 but was completely blocked by AMD3100. Similar results were obtained using TZM-bl cells (Fig. 3B), showing that SHIVBR24N uses CXCR4 for entry in vitro. Because a recent report indicated that increased neutralization sensitivity of CXCR4-using HIV-1 strains emerged during the transition from CCR5 to CXCR4 usage (4), the susceptibility of SHIVBR24N to antibody neutralization was also examined. Compared to the inoculating R5 SHIVSF162P3N virus, we found that the X4 SHIVBR24N isolate is highly sensitive to neutralization with sera from macaques CD80 and T799 (Fig. 4A). A 90% neutralization of SHIVBR24N was achieved at dilutions of 1:160 and 1:1,280 of CD80 and T799 sera, respectively. In contrast, the R5 SHIVSF162P3N inoculating virus was resistant to neutralization with serum from macaque CD80, while a similar degree of neutralization (90%) with T799 serum required a 32-fold-higher titer (1:40). SHIVBR24N is also more sensitive than SHIVSF162P3N to neutralization with the broadly cross-reactive CD4 binding site monoclonal antibodies CD4-IgG2 and IgG1b12 (Fig. 4B). CD4-IgG2 potently neutralized SHIVBR24N at concentrations of 2 μg/ml, while IgG1b12 achieved 70% neutralization at the highest concentration tested (10 μg/ml). These CD4 binding site MAbs, however, were unable to neutralize SHIVSF162P3N. Collectively, the data showed emergence of a neutralization-sensitive CXCR4-utilizing variant in macaque BR24.
FIG. 3.
Coreceptor usage of SHIVBR24N. CXCR4 and CCR5 usage of the inoculating virus SHIVSF162P3N (P3N) and SHIVBR24N (BR24N), the virus recovered from BR24 at end-stage disease, was determined by blocking entry into rhPBMC (A) and TZM-bl (B) cells with 1 μM CXCR4-specific (AMD3100) or CCR5-specific (TAK-779) inhibitor. X4 SHIVSF33A (33A) and R5 SHIVSF162P3 (P3) served as controls. For TZM-bl cells, error bars indicate standard errors of data in triplicate wells. Results shown are representative of at least two independent experiments, and for rhPBMC cells from two different animals were used.
FIG. 4.
Neutralization sensitivity of SHIVSF162P3N and SHIVBR24. The neutralization susceptibilities of the inoculating R5 SHIVSF162P3N (○) and emerging X4 SHIVBR24 (•) viruses to antibodies in sera from macaques CD80 and T799 (A) and CD4 binding site MAbs CD4-IgG2 and IgG1b12 (B) were determined as described in Materials and Methods. Data shown are representative of at least two independent neutralization experiments.
Sequence of the SHIVBR24N variant envelope gp120.
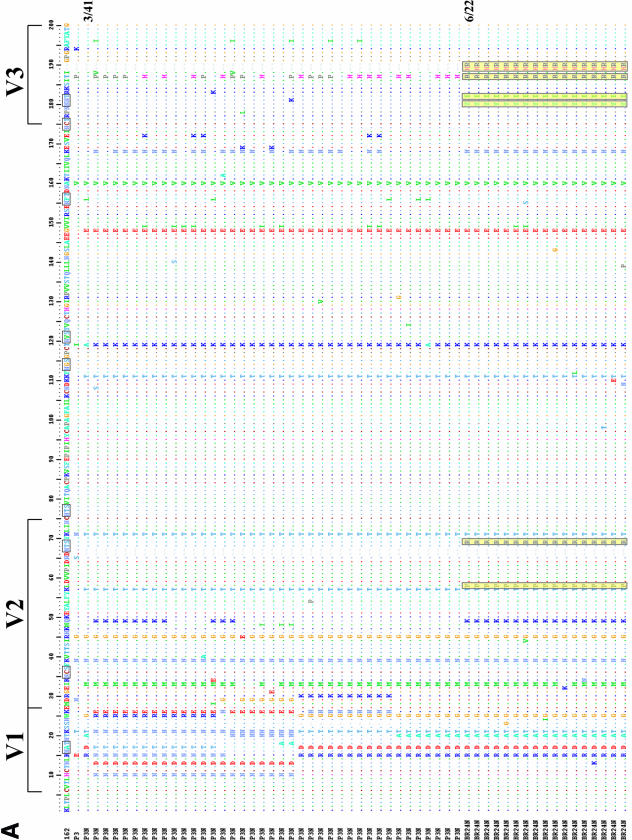
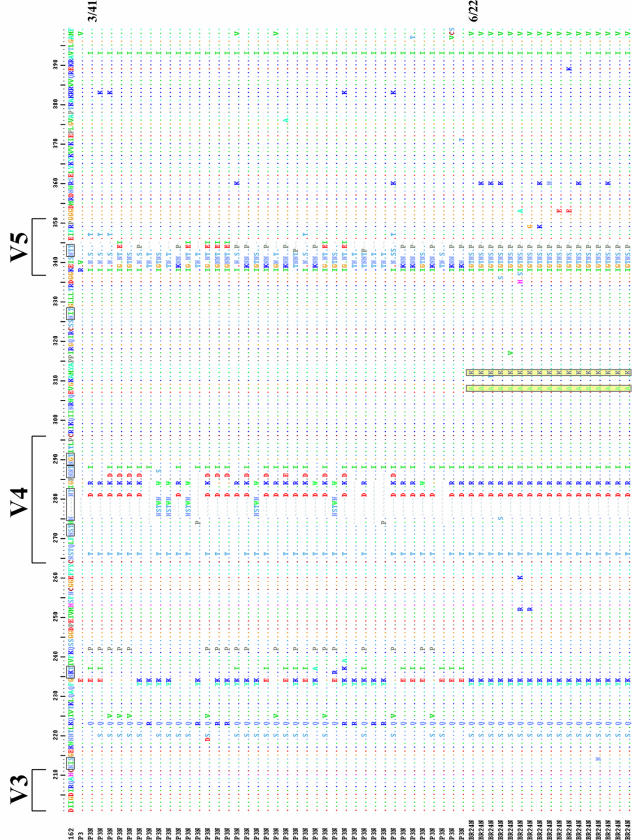
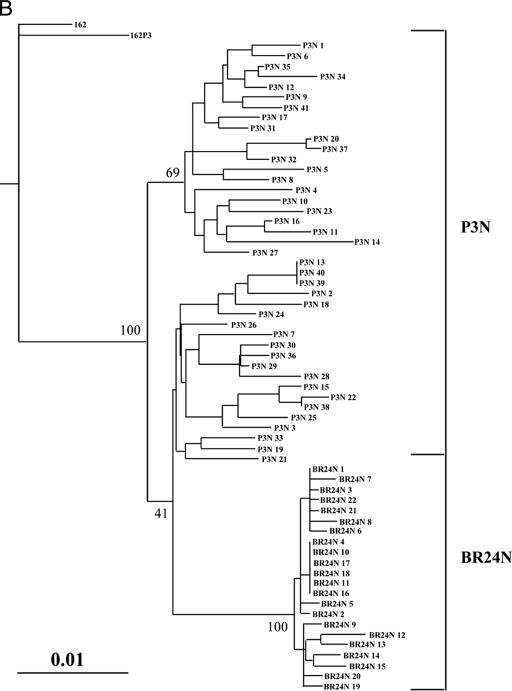
We sequenced and compared the envelope gp120 V1-to-V5 region of the inoculating R5 SHIVSF162P3N virus and the X4 SHIVBR24N variant to understand their relationship (Fig. 5A). All SHIVBR24N Env clones sequenced (22 in total) harbored the same nine unique amino acid changes compared to the inoculating strain. Two of the changes are in the V2 region and two are in the C4 region, with five clustering in the V3 domain. The nonsynonymous serine-to-arginine change in V2 as well as the asparagine-to-tyrosine and threonine-to-isoleucine changes in V3 are predicted to result in loss of potential N-linked glycosylation sites in these regions of the SHIVBR24N envelope gp120. Furthermore, insertion of two positively charged amino acids, histidine and arginine immediately upstream of the GPGR crown V3 loop sequence, increases the overall charge of this domain, and the two amino acid changes in C4 are within a conserved gp120 structure implicated in CCR5 binding (35). The conservation of this particular set of changes in regions known to influence envelope functions, including coreceptor usage (23, 36), points to their importance in determining the phenotypic characteristics of the SHIVBR24N variant. Phylogenetic tree analysis of Env V1-V5 sequences showed two loosely clustered viral populations within the inoculating SHIVSF162P3 virus (Fig. 5B). The SHIVBR24N sequences, although limited in diversity, are markedly divergent from those of the inoculum, with the branching point of variant-associated sequences having its origin within one of the inoculating R5-associated subpopulations (100% bootstrap value). Collectively, the data support derivation of the X4 SHIVBR24N variant from the inoculating R5 virus.
FIG. 5.
(A) Env V1-V5 amino acid sequence comparison of SHIVSF162P3N and SHIVBR24N. Sequences of 41 SHIVSF612P3N and 22 SHIVBR24N Env clones were aligned with the reference SHIVSF162 (GenBank accession number M38428) and the consensus SHIVSF162P3 (GenBank accession number AF536757) sequences. Gaps are indicated by wave lines and dots denote identical residues in the sequence. Potential N-linked glycosylation sites are bracketed, and the nine unique amino acid changes shared by all the BR24N sequences are shaded in yellow. The numbers shown on the right of a sequence represent the number of clones containing this sequence out of the total number of clones sequenced. Those lacking numbers on the right represent unique sequences. (B) Phylogenetic tree showing the relationship between the SHIV162P3N and SHIVBR24N sequences. A maximum likelihood tree rooted on the sequence of the molecular clone SHIVSF162 was generated. The scale bar indicates the genetic distance along the horizontal branches, and the numbers at the nodes are bootstrap values.
The V3 loop of SHIVBR24N dictates CXCR4 usage.
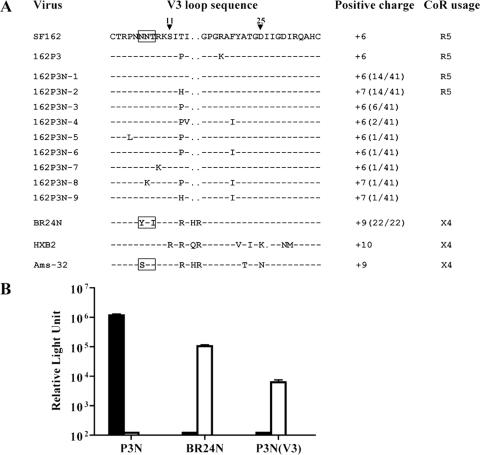
The V3 domain of HIV-1 gp120 is known to contribute to coreceptor use. A closer examination of this region revealed the presence of two major V3 loop species each representing 14 of 41 Env clones sequenced in the R5 SHIVSF162P3N virus inoculum (Fig. 6A) but homogeneity in the recovered X4 SHIVBR24N virus. As predicted, the overall positive charge of this region of gp120 is higher for SHIVBR24N (+9) than SHIVSF162P3N (+6 or +7), but basic amino acids at V3 positions 11 and 25 that frequently distinguish primary X4 and R5 viruses (11) were absent in SHIVBR24N. Of note, insertion of the two basic amino acids (histidine and arginine) at the crown of the loop was at the same position as those found in the prototypic X4 HIV-1 strains HXB2 and Ams-32. Furthermore, the N-terminal glycan, which has been shown to modulate antibody neutralization susceptibility and interaction of HIV-1 Env with chemokine receptors (1, 27, 33), is lost in both SHIVBR24N and Ams-32. The lack of the N-terminal V3 loop glycan in SHIVBR24N envelope gp120 likely contributes to increased sensitivity of the virus to serum as well as CD4-IgG2 and IgG1b12 neutralization.
FIG. 6.
(A) Comparison of V3 loop sequences of SHIV162P3N, SHIVBR24N, and well-characterized X4 and R5 viruses. Gaps are indicated as dots, dashes denote similarities in sequence, and the total positive charge of this region was calculated and is shown. Positions 11 and 25 within the V3 loop are indicated, and the N-terminal glycan site is bracketed. The numbers in parentheses represent the number of clones with the indicated V3 loop sequence per the total number sequenced. (B) Relative entry into U87.T4.CCR5 cells (black bars) and U87.T4.CXCR4 cells (white bars) of luciferase reporter viruses expressing the envelope of SHIVSF162P3N (EnvP3N), SHIVBR24N (EnvBR24N), and V3 loop recombinant EnvP3N(V3). All data are the means of triplicates + standard errors of the means.
To determine whether the V3 loop of SHIVBR24N determines CXCR4 usage, the 37-amino-acid V3 domain of this virus was used to replace the corresponding region of SHIVSF162P3N gp120 (EnvP3N) to generate the recombinant EnvP3N(V3). In single-round infectivity assays, we found that, contrary to EnvP3N, the recombinant EnvP3N(V3) was not able to mediate entry into U87.T4 cells expressing CCR5 (Fig. 6B). Instead, similar to the envelope of SHIVBR24N (EnvBR24N), EnvP3N(V3) used CXCR4 as the coreceptor. Taken together, the data demonstrate that the V3 loop of SHIVBR24N is a major determinant of coreceptor preference.
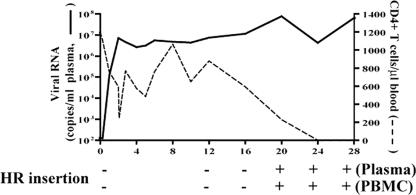
Tracking X4 evolution and emergence in macaque BR24.
The presence of the two amino acid insertions in the V3 loop of SHIVBR24N allows for the design of PCR primers that can distinguish between the inoculating R5 virus and the X4 variant to track evolution of the latter in macaque BR24. Signature X4 variant “HR” sequence could not be detected in the SHIVSF162P3N inoculum (data not shown) or at early times in the infection of macaque BR24 (Fig. 7). It was first detected in both plasma and PBMC of BR24 at 20 wpi, coinciding with the spike in viremia. Interestingly, although X4 emergence was associated with loss of peripheral CD4+ T cells and disease progression, it appeared to occur after the onset of precipitous peripheral CD4+ T-cell decline seen at weeks 12 to 16.
FIG. 7.
Evolution of X4 variants in macaque BR24 as tracked by the presence of the signature V3 HR insertion sequence, using sequence-specific PCR. + and − indicate presence and absence of the insertion.
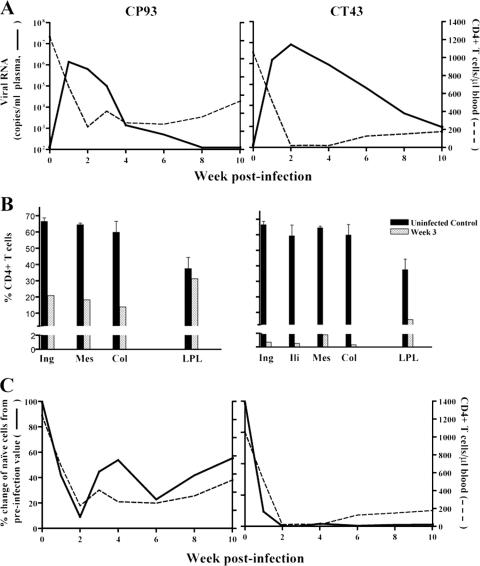
SHIVBR24N uses CXCR4 in vivo.
To establish coreceptor usage of SHIVBR24N in vivo, two macaques were inoculated intravenously with cell-free virus. Both animals were infected, and peak viremia in CP93 (106 RNA copies/ml plasma) was 1 log lower than in CT43 (Fig. 8A). A rapid and sustained drop in peripheral CD4+ T cells that was more dramatic for CT43 than CP93 and accompanied virus replication. Examination of the percentages of tissue CD4+ T cells during peak infection (3 wpi) showed >70% loss in all LN of CP93 examined but little or no loss in the gut (Fig. 8B). In CT43, the macaque with a 1-log-higher peak viremia, massive depletion (>98%) of CD4+ T cells was seen in the LN, with substantial depletion in the gut as well. Although the extent of CD4+ T-cell loss among the two SHIVBR24N-infected macaques varied due to a difference in viral load, the pattern and sites of CD4+ T-cell depletion are similar to those found in X4-infected and not R5 SHIV-infected macaques (Fig. 2B) (17). Analyses of the circulating T-cell subpopulations showed early targeting and depletion of naïve CD4+ T cells, where the rate of loss of this T-cell subset paralleled the CD4+ T-cell drop (Fig. 8C). Naïve CD4+ T lymphocytes express high levels of CXCR4 but not CCR5 (3, 17). Thus, the in vivo findings corroborated the in vitro data, showing CXCR4 usage of SHIVBR24N.
FIG. 8.
Virologic and immunologic measurements in SHIVBR24N-infected RM. (A) Viral load and absolute CD4+ T-cell counts. (B) Percentage of CD4+ T cells in various lymphoid compartments and the gut LPL after 3 weeks of infection. Baseline values generated from three uninfected macaques are shown for reference. (C) Percent change of naïve CD4+ T cells as identified by the CD95low CD28high phenotype in peripheral blood of BR24. The baseline naïve CD4+ T-cell subset value for each macaque was set at 100%.
DISCUSSION
This report documents phenotypic coreceptor switch in an animal model of HIV/AIDS. Functional, sequence, and in vivo infectivity studies showed that contrary to the inoculating strain, R5 SHIVSF162P3N, the virus recovered at end-stage disease of macaque BR24 uses CXCR4 for entry. Although it is formally possible that the X4 variant is present in the inoculum at an extremely low level, the temporal correlation between the appearance of the X4 variant, the spike in viremia, and progression to disease, coupled with the phylogenetic relationship between the variant and the inoculating R5 SHIVSF162P3N virus as well as the inability to detect the X4 variant signature sequence in the inoculum with sequence-specific PCR that has a sensitivity of detection of 1 in 106 copies, strongly suggests that evolution to CXCR4 usage occurred in macaque BR24.
The mechanism(s) for X4 emergence late in infection has been debated for many years. Various frequencies of X4 viruses have been reported throughout the course of HIV-1 natural infection (38), but their dominance is seen only towards the end stage of disease. We and others previously proposed an immunological restriction on coreceptor switch that is based on the assumption that the X4 virus is more susceptible to immune control (13, 29). The appearance of X4 variants late in infection, therefore, is presumed to be the consequence rather than the cause of the collapse of the host immune system. In this regard, our findings of evolution and emergence of neutralization-sensitive X4 variants in an R5 SHIV-infected macaque with undetectable antiviral antibody responses are of interest. Increased neutralization sensitivity of recently emerged CXCR4-using HIV-1 variants has also been reported (4). This, together with data indicating similar V3 sequence changes associated with CXCR4 usage in SHIVBR24N and the HIV-1 isolate Ams-32, raises the possibility that similar mechanistic pathways underlie phenotypic conversion in some HIV-1-infected patients and macaques. We suggest that adaptation to efficient usage of CXCR4 requires conformational changes in the envelope glycoprotein, such as the loss of the N-terminal V3 loop glycan (1, 27, 33), that may render the virus more susceptible to antibody neutralization. In the absence of such immune selection pressures in the rapid progressor BR24, or in patients with end-stage disease, the X4 variants that evolved during the course of infection can now emerge and subsequently dominate. Further studies on the impact of amino acid changes in the V2, V3, and C4 regions of SHIVBR24N envelope gp120 on virus entry, receptor usage efficiency, and neutralization susceptibility should further our understanding of the basis for the late appearance of X4 viruses.
Surprisingly, emergence of the X4 variant in macaque BR24 appears to follow rather than precede or coincide with the onset of precipitous peripheral CD4+ T-cell loss. Similar observations have also been made in HIV-1-infected infants and adults (5, 32). It is conceivable that X4 viruses are present at the time of accelerated CD4+ T-cell decline in macaque BR24 but are below the limit of detection or in anatomical sites, such as the lymph nodes and thymus, that were not sampled. This scenario supports the idea that X4 emergence is the cause of the rapid loss of total CD4+ T cells. Alternatively, even though circulating virus-specific antibody and the cytokine-producing CD8+ T-cell response is either absent or at low levels, the presence of antiviral immunity at lymphoid and/or mucosal tissue compartments early in infection cannot be excluded. Thus, it is tempting to speculate that the onset of the rapid drop in peripheral CD4+ T cells seen in macaque BR24 at 16 wpi is due to further impairment of the suppressive capacity of the immune system as a consequence of sustained high-level viral replication. The final collapse of the immune system removes any selection pressures acting against X4 viruses, allowing for their emergence at 20 wpi. This latter interpretation of the observation is consistent with the notion that X4 appearance is the result, rather than the cause of, immune failure. Although we cannot formally exclude a contribution of target cell availability to the phenotypic switch in BR24, we deem this possibility less likely given that CCR5-expressing cells were also limiting in macaque CD80 at end-stage disease, but X4 virus failed to emerge in this seropositive animal. Target cell availability has also been suggested not to be a driving force for the R5-to-X4 switch in HIV-1-infected individuals (40).
In vitro, two amino acid residue changes in the V3 domain of HIV-1 strain SF162 are sufficient to expand to CXCR4 usage (8, 16, 21). The same two V3 mutations are seen for multiple CXCR4-using variants selected either by target cell limitation or CCR5 inhibition, leading to the conclusion that SF162 has a single pathway for acquiring CXCR4 use (21). We show here that coreceptor switch of a SHIV molecular clone expressing the SF162 envelope in RM requires amino acid residue changes in the V3 loop that are different from those reported in tissue culture systems. The need to escape evolving immune selective pressures while at the same time maintaining viral fitness and efficient coreceptor usage on diverse tissue cells of the macaque host likely explains the differences in sequence change required for adaptation of the SF162 envelope to function with CXCR4 in vivo and in vitro. Whether intermediates between CCR5 and CXCR4 use are involved in the R5-to-X4 evolutionary process in vivo, as well as the costs and benefits in expanding or switching coreceptor usage on virus receptor binding and replication fitness, merits further investigation.
The identification of an R5 SHIV that can switch to CXCR4 usage in infected RM through genetic adaptations similar to those seen in humans holds promise in providing the experimental tool critically needed to fully elucidate the mechanistic basis for and obstacles to coreceptor switch in HIV-1 infection. The coreceptor switch we observed was in an animal with rapid disease progression, a clinical course that is rarely seen in HIV-1-infected individuals (12, 25, 28, 30). Accordingly, it will be important to demonstrate that coreceptor switch occurs in additional SHIVSF162P3N-infected macaques, especially in conventional progressors. Nevertheless, the experimental system can be employed to examine whether administration of antibodies to CD8 T cells or B cells that perturb the immune system to artificially promote a rapid progressor condition, or CCR5 blockers (24) to alter target cell availability at or during the course of SHIVSF162P3N infection of RM, hastens the process of X4 virus evolution and emergence, which inevitably heralds an unfavorable clinical outcome.
Acknowledgments
We thank Lisa Chakrabarti and Jay Levy for discussions and critical comments, Zhiwei Chen for help with phylogenetic tree analysis, and Lily Tsai for help with the strip immunoblot analysis. AMD3100, TAK779, and TZM-bl cells and the U87.CD4 indicator cell line were obtained through the NIH AIDS Research and Reference Reagent Program.
This work was supported by National Institutes of Health grants R01AI46980 and R37AI41945.
Footnotes
Published ahead of print on 30 May 2007.
REFERENCES
- 1.Back, N. K., L. Smit, J. J. De Jong, W. Keulen, M. Schutten, J. Goudsmit, and M. Tersmette. 1994. An N-glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology 199:431-438. [DOI] [PubMed] [Google Scholar]
- 2.Berger, E. A. 1997. HIV entry and tropism: the chemokine receptor connection. AIDS 11(Suppl. A):S3-S16. [PubMed] [Google Scholar]
- 3.Bleul, C. C., L. Wu, J. A. Hoxie, T. A. Springer, and C. R. Mackay. 1997. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc. Natl. Acad. Sci. USA 94:1925-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bunnik, E. M., E. D. Quakkelaar, A. C. van Nuenen, B. Boeser-Nunnink, and H. Schuitemaker. 2007. Increased neutralization sensitivity of recently emerged CXCR4-using human immunodeficiency virus type 1 strains compared to coexisting CCR5-using variants from the same patient. J. Virol. 81:525-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casper, C., L. Naver, P. Clevestig, E. Belfrage, T. Leitner, J. Albert, S. Lindgren, C. Ottenblad, A. B. Bohlin, E. M. Fenyo, and A. Ehrnst. 2002. Coreceptor change appears after immune deficiency is established in children infected with different HIV-1 subtypes. AIDS Res. Hum Retrovir. 18:343-352. [DOI] [PubMed] [Google Scholar]
- 6.Cocchi, F., A. L. DeVico, A. Garzino-Demo, A. Cara, R. C. Gallo, and P. Lusso. 1996. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat. Med. 2:1244-1247. [DOI] [PubMed] [Google Scholar]
- 7.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 8.Dejucq, N., G. Simmons, and P. R. Clapham. 2000. T-cell line adaptation of human immunodeficiency virus type 1 strain SF162: effects on envelope, vpu and macrophage-tropism. J. Gen. Virol. 81:2899-2904. [DOI] [PubMed] [Google Scholar]
- 9.Delwart, E. L., and C. J. Gordon. 1997. Tracking changes in HIV-1 envelope quasispecies using DNA heteroduplex analysis. Methods 12:348-354. [DOI] [PubMed] [Google Scholar]
- 10.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 11.Fouchier, R. A., M. Groenink, N. A. Kootstra, M. Tersmette, H. G. Huisman, F. Miedema, and H. Schuitemaker. 1992. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J. Virol. 66:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganeshan, S., R. E. Dickover, B. T. Korber, Y. J. Bryson, and S. M. Wolinsky. 1997. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. J. Virol. 71:663-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harouse, J. M., C. Buckner, A. Gettie, R. Fuller, R. Bohm, J. Blanchard, and C. Cheng-Mayer. 2003. CD8+ T cell-mediated CXC chemokine receptor 4-simian/human immunodeficiency virus suppression in dually infected rhesus macaques. Proc. Natl. Acad. Sci. USA 100:10977-10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harouse, J. M., A. Gettie, T. Eshetu, R. C. Tan, R. Bohm, J. Blanchard, G. Baskin, and C. Cheng-Mayer. 2001. Mucosal transmission and induction of simian AIDS by CCR5-specific simian/human immunodeficiency virus SHIV(SF162P3). J. Virol. 75:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harouse, J. M., A. Gettie, R. C. Tan, J. Blanchard, and C. Cheng-Mayer. 1999. Distinct pathogenic sequela in rhesus macaques infected with CCR5 or CXCR4 utilizing SHIVs. Science 284:816-819. [DOI] [PubMed] [Google Scholar]
- 16.Harrowe, G., and C. Cheng-Mayer. 1995. Amino acid substitutions in the V3 loop are responsible for adaptation to growth in transformed T-cell lines of a primary human immunodeficiency virus type 1. Virology 210:490-494. [DOI] [PubMed] [Google Scholar]
- 17.Ho, S. H., L. Shek, A. Gettie, J. Blanchard, and C. Cheng-Mayer. 2005. V3 loop-determined coreceptor preference dictates the dynamics of CD4+-T-cell loss in simian-human immunodeficiency virus-infected macaques. J. Virol. 79:12296-12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang, S. S., T. J. Boyle, H. K. Lyerly, and B. R. Cullen. 1991. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science 253:71-74. [DOI] [PubMed] [Google Scholar]
- 19.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 20.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol 16:111-120. [DOI] [PubMed] [Google Scholar]
- 21.Kiselyeva, Y., R. Nedellec, A. Ramos, C. Pastore, L. B. Margolis, and D. E. Mosier. 2007. Evolution of CXCR4-using human immunodeficiency virus type 1 SF162 is associated with two unique envelope mutations. J. Virol. 81:3657-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koot, M., I. P. Keet, A. H. Vos, R. E. de Goede, M. T. Roos, R. A. Coutinho, F. Miedema, P. T. Schellekens, and M. Tersmette. 1993. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann. Intern. Med. 118:681-688. [DOI] [PubMed] [Google Scholar]
- 23.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lederman, M. M., A. Penn-Nicholson, M. Cho, and D. Mosier. 2006. Biology of CCR5 and its role in HIV infection and treatment. JAMA 296:815-826. [DOI] [PubMed] [Google Scholar]
- 25.Liu, S. L., T. Schacker, L. Musey, D. Shriner, M. J. McElrath, L. Corey, and J. I. Mullins. 1997. Divergent patterns of progression to AIDS after infection from the same source: human immunodeficiency virus type 1 evolution and antiviral responses. J. Virol. 71:4284-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luciw, P. A., E. Pratt-Lowe, K. E. Shaw, J. A. Levy, and C. Cheng-Mayer. 1995. Persistent infection of rhesus macaques with T-cell-line-tropic and macrophage-tropic clones of simian/human immunodeficiency viruses (SHIV). Proc. Natl. Acad. Sci. USA 92:7490-7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malenbaum, S. E., D. Yang, L. Cavacini, M. Posner, J. Robinson, and C. Cheng-Mayer. 2000. The N-terminal V3 loop glycan modulates the interaction of clade A and B human immunodeficiency virus type 1 envelopes with CD4 and chemokine receptors. J. Virol. 74:11008-11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michael, N. L., A. E. Brown, R. F. Voigt, S. S. Frankel, J. R. Mascola, K. S. Brothers, M. Louder, D. L. Birx, and S. A. Cassol. 1997. Rapid disease progression without seroconversion following primary human immunodeficiency virus type 1 infection—evidence for highly susceptible human hosts. J. Infect. Dis. 175:1352-1359. [DOI] [PubMed] [Google Scholar]
- 29.Miedema, F., M. Tersmette, and R. A. van Lier. 1990. AIDS pathogenesis: a dynamic interaction between HIV and the immune system. Immunol. Today 11:293-297. [DOI] [PubMed] [Google Scholar]
- 30.Montagnier, L., C. Brenner, S. Chamaret, D. Guetard, A. Blanchard, J. de Saint Martin, J. D. Poveda, G. Pialoux, and M. L. Gougeon. 1997. Human immunodeficiency virus infection and AIDS in a person with negative serology. J. Infect. Dis. 175:955-959. [DOI] [PubMed] [Google Scholar]
- 31.Moore, J. P., S. G. Kitchen, P. Pugach, and J. A. Zack. 2004. The CCR5 and CXCR4 coreceptors—central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Res. Hum Retrovir. 20:111-126. [DOI] [PubMed] [Google Scholar]
- 32.Nelson, J., T. Riddle, N. Shire, M. Sherman, K. Franco, and H. Sheppard. 2007. Sequential turnover of env variants and co-receptor switching during HIV-1 chronic infection, abstr 252. 14th Conf. Retrovir. Opportunistic Infect., Los Angeles, CA.
- 33.Polzer, S., M. T. Dittmar, H. Schmitz, and M. Schreiber. 2002. The N-linked glycan g15 within the V3 loop of the HIV-1 external glycoprotein gp120 affects coreceptor usage, cellular tropism, and neutralization. Virology 304:70-80. [DOI] [PubMed] [Google Scholar]
- 34.Regoes, R. R., and S. Bonhoeffer. 2005. The HIV coreceptor switch: a population dynamical perspective. Trends Microbiol. 13:269-277. [DOI] [PubMed] [Google Scholar]
- 35.Rizzuto, C., and J. Sodroski. 2000. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res. Hum Retrovir. 16:741-749. [DOI] [PubMed] [Google Scholar]
- 36.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 37.Rowland-Jones, S. L. 2003. Timeline: AIDS pathogenesis: what have two decades of HIV research taught us? Nat. Rev. Immunol. 3:343-348. [DOI] [PubMed] [Google Scholar]
- 38.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shioda, T., J. A. Levy, and C. Cheng-Mayer. 1991. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature 349:167-169. [DOI] [PubMed] [Google Scholar]
- 40.van Rij, R. P., M. D. Hazenberg, B. H. van Benthem, S. A. Otto, M. Prins, F. Miedema, and H. Schuitemaker. 2003. Early viral load and CD4+ T cell count, but not percentage of CCR5+ or CXCR4+ CD4+ T cells, are associated with R5-to-X4 HIV type 1 virus evolution. AIDS Res. Hum Retrovir. 19:389-398. [DOI] [PubMed] [Google Scholar]
- 41.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]