Abstract
The flavivirus capsid protein not only is a component of nucleocapsids but also plays a role in viral replication. In this study, we found a small capsid protein in cells infected with Japanese encephalitis virus (JEV) but not in the viral particles. The small capsid protein was shown to be generated by processing with host cysteine protease cathepsin L. An in vitro cleavage assay revealed that cathepsin L cleaves the capsid protein between amino acid residues Lys18 and Arg19, which are well conserved among the mosquito-borne flaviviruses. A mutant JEV resistant to the cleavage of the capsid protein by cathepsin L was generated from an infectious cDNA clone of JEV by introducing a substitution in the cleavage site. The mutant JEV exhibited growth kinetics similar to those of the wild-type JEV in monkey (Vero), mosquito (C6/36), and porcine (PK15) cell lines, whereas replication of the mutant JEV in mouse macrophage (RAW264.7) and neuroblastoma (N18) cells was impaired. Furthermore, the neurovirulence and neuroinvasiveness of the mutant JEV to mice were lower than those of the wild-type JEV. These results suggest that the processing of the JEV capsid protein by cathepsin L plays a crucial role in the replication of JEV in neural and macrophage cells, which leads to the pathogenesis of JEV infection.
The genus Flavivirus within the family Flaviviridae comprises over 70 viruses, many of which are predominantly arthropod-borne viruses, such as Japanese encephalitis virus (JEV), West Nile virus (WNV), Murray Valley encephalitis virus (MVE), dengue virus (DEN), yellow fever virus (YFV), and tick-borne encephalitis virus (TBEV). They frequently cause significant morbidity and mortality in mammals and birds (5). JEV is distributed in the south and southeast regions of Asia and is kept in a zoonotic transmission cycle between pigs or birds and mosquitoes (5, 42, 45). JEV spreads to dead-end hosts, including humans, through the bite of JEV-infected mosquitoes and causes infection of the central nervous system with a high mortality rate (5, 45). JEV has a single-stranded positive-strand RNA genome of approximately 11 kb, which is capped at the 5′ end but lacks a 3′ polyadenine tail (24). The ability of the flaviviral genomic RNA to cyclize is crucial for viral replication (1, 14). Among mosquito-borne flaviviruses, two complementary cyclization sequences, mapped in the capsid protein-coding region and 3′ untranslated region (UTR), mediated the cyclization by RNA-RNA base pairing, together with a second pair of complementary sequences, named 5′ and 3′ upstream AUG regions (1, 10, 14, 19, 25). The genomic RNA includes a single large open reading frame, and a polyprotein translated at the endoplasmic reticulum (ER) membrane is cleaved co- and posttranslationally by host and viral proteases to yield three structural proteins, the capsid, precursor membrane (prM), and envelope (E) proteins, and at least seven nonstructural proteins, NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 (24).
Although the capsid protein has very little amino acid homology among flaviviruses—for example, the homologies of the capsid protein of JEV to those of WNV, DEN type 2 (DEN2), and TBEV were only 67%, 33%, and 25%, respectively—the structural properties, such as the hydrophobicity profile, abundance of basic amino acid residues, and secondary and tertiary structures, are well conserved (11, 18, 27). The flavivirus capsid protein commonly contains two hydrophobic sequences in the center and the carboxyl terminus. The latter serves as a signal sequence of prM. The signal/anchor sequence is cleaved off by the viral protease NS2B/3, and this cleavage is required for the subsequent liberation of the amino terminus of prM by the host signal peptidase (26, 43, 49). The mature capsid protein may be associated with the ER membrane through the central hydrophobic region (23, 29). Because the capsid protein has RNA-binding ability via the basic amino acid clusters at its amino and carboxyl termini, it is believed to bind to the genomic RNA to form a nucleocapsid (20). Unlike other envelope viruses, the nucleocapsid structures are rarely found in cells infected with flaviviruses (48), although the nucleocapsid of TBEV can assemble in vitro (21). Therefore, viral assembly is thought to be a coordinated process between the membrane-associated capsid protein and two envelope glycoproteins, prM and E, in the ER membrane.
In conflict with their roles as structural proteins, the capsid proteins of some flaviviruses are localized not only in the cytoplasm but also in the nuclei of the infected cells (4, 28, 32, 44, 46-48). We previously reported that the JEV capsid protein has also been detected in both the nucleoli and cytoplasm and that the mutant virus defective in the nuclear localization of capsid protein exhibited impaired viral growth in mammalian cells and neuroinvasiveness in mice (32). Furthermore, we have also reported that the nuclear and cytoplasmic localizations of the JEV capsid protein are dependent on binding to the host nucleolar protein B23 (46). It has been reported that, in addition to the JEV capsid protein, the WNV and DEN capsid proteins bind to several host proteins, such as Jab1, a component of the COP9 signalosome complex (34), the chaperone protein HSP70 (35), and the heterogenous nuclear ribonucleoprotein K (8), to regulate these functions. Recently, Clyde and Harris have shown that the small capsid protein isoform translated from the second AUG codon of the DEN genome by leaky scanning is important for viral replication (9). In this context, these properties of the flaviviral capsid proteins raised the possibility that they play some roles in viral growth as “nonstructural” proteins.
In this study, we detected a small capsid protein in JEV-infected cells, but not in the released viral particles. The small capsid protein has been shown to be generated by host protease cathepsin L. Cathepsin L was capable of cleaving the capsid protein between amino acid residues Lys18 and Arg19. Furthermore, we have generated a mutant JEV carrying a capsid protein resistant to cleavage by cathepsin L. The characterization of this mutant JEV indicated that cleavage of the capsid protein by cathepsin L plays important roles in viral replication in mouse neuroblastoma and macrophage cells and in the pathogenesis of encephalitis in vivo. These results suggest a novel mechanism for JEV to adapt host cells by the processing of the capsid protein.
MATERIALS AND METHODS
Cells.
The mammalian cell lines Vero (monkey kidney), 293T (human kidney), PK15 (pig kidney), RAW264.7 (mouse macrophage), and N18 (mouse neuroblastoma) were maintained in Dulbecco's modified Eagle's minimal essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Mosquito cell line C6/36 (Aedes albopictus) was grown in Eagle's minimal essential medium supplemented with 10% FBS. Vero cell lines Vero/siNC and Vero/siCTSL, stably expressing the hairpin small interfering RNAs (siRNA) for the nonsense sequence and cathepsin L, respectively, were established by transfection with plasmids pSilencer/NC and pSilencer/CTSL (see below), respectively, and selected with DMEM containing 10% FBS and 50 μg/ml hygromycin B (Sigma, St. Louis, MO).
Plasmids.
The cDNA for the capsid protein of JEV AT31 (amino acid residues 2 to 105) was amplified from pMWATG1 (54) by PCR using Ex-Taq (Takara, Shiga, Japan) and cloned between the FLAG and hemagglutinin (HA) tags in pcDNA3.1FlagHA (36). From this plasmid, the capsid cDNAs with or without FLAG and/or HA tags were amplified by PCR and subcloned into a mammalian expression vector pCAGPM (31) and designated pCAG/FLAG-JEC-HA, pCAG/FLAG-JEC, pCAG/JEC-HA, and pCAG/JEC. By the same procedure, the plasmids encoding FLAG- and HA-tagged DEN2 and DEN4 capsid proteins, pCAG/FLAG-DEN2C-HA and pCAG/FLAG-DEN4C-HA, were generated from the plasmids encoding the capsid proteins of DEN2 and DEN4, respectively (the kind gifts from F. Hasebe and M. Tadano, respectively). For mutational analyses of the amino acid residues from 14 to 23 (based on the JEV capsid protein sequence), a series of point mutants of the FLAG- and HA-tagged JEV capsid proteins were synthesized by PCR-based mutagenesis (17). All of the mutant genes, as well as the wild-type gene, were cloned into pCAGPM. The JEV capsid gene was cloned into pcDNA 3.1/myc-His (Invitrogen, Carlsbad, CA), and the cDNA encoding the JEV capsid protein fused with myc and His tags was amplified and cloned into bacterial expression vector pET32a (Merck Novagen, Darmstadt, Germany). The resulting plasmid was designated pET32/JECmycHis. The cDNAs of human cathepsins B and L were amplified from 293T cells by reverse transcription-PCR and cloned into pcDNA 3.1/myc-His. An enzymatically inactive mutation of cathepsin L in which Cys138 was replaced with Ala was generated by PCR-based mutagenesis. Expression vector pSilencer/CTSL, for a hairpin siRNA for African green monkey cathepsin L, was generated by annealing with synthesized nucleotides (sense, GAT CCG GCG ATG CAC AAC AGA TTA TTC AAG AGA TAA TCT GTT GTG CAT CGC CTT TTT TGG AAA; antisense, AGC TTT TCC AAA AAA GGC GAT GCA CAA CAG ATT ATC TCT TGA ATA ATC TGT TGT GCA TCG CCG) and insertion into the BamHI and HindIII sites of pSilencer 2.1 U6 hygro (Ambion Inc., Austin, TX). pSilencer/NC, encoding an siRNA with no homology to mammalian genes, was used as a negative control. pMWAT/L17A carrying replacements of cytosine at nucleotide 144 and thymine at nucleotide 145 with guanine and cytosine, respectively, in pMWATG1, an infectious cDNA clone of JEV, was constructed by PCR-based mutagenesis which results in the replacement of Leu17 in the capsid protein with Ala (see Fig. 5A). In addition, adenine-to-guanine and guanine-to-cytosine mutations were introduced into pMWATG1 and pMWAT/L17A at nucleotides 10865 and 10866 of the JEV gene, respectively. The resulting plasmids were named pMWAT/CSmt and pMWAT/L17ACSmt, respectively.
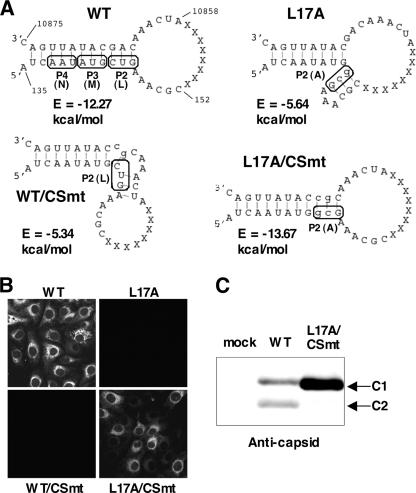
FIG. 5.
Construction of a mutant JEV carrying the capsid protein resistant to cleavage by cathepsin L. (A) Predicted RNA secondary structures of the wild-type (WT) and mutant viral genomes. Nucleotides 135 to 152 and bases 10858 to 10875 in the 5′ and 3′ termini, respectively, connected by 8 nonsense nucleotides (X) alternative to bases 153 to 10857, were applied to the computer program GENETYX-MAC, version 12, to calculate free energies (E). The secondary RNA structures with minimum free energies are illustrated. RNA sequences encoding Asp15 (P4), Met16 (P3), and Leu17 (P2) in the 5′ cyclization sequences of the WT JEV, Leu17 (P2) for WT/CSmt, and Ala17 (P2) for L17A and L17A/CSmt are boxed. The mutated nucleotides are shown by lowercase letters. (B) Vero cells (5 × 106) were electroporated with 10 μg of in vitro-transcribed genomic RNA of WT, L17A, WT/CSmt, or L17A/CSmt virus and immunostained with an anti-E antibody at 4 days posttransfection. (C) Expression of capsid proteins in cells infected with WT or L17A/CSmt JEV. Vero cells were inoculated with the JEVs at an MOI of 10 and analyzed by immunoblotting with anticapsid antibody at 1 day postinfection.
Viruses.
The wild-type and L17A/CSmt JEVs were generated from plasmids pMWATG1 and pMWAT/L17ACSmt, respectively, by a method described previously (54). The infectivity of the viruses was determined by an immunostaining focus assay as described previously (32) and expressed in focus-forming units (FFU). The JEV particles were purified from the supernatant of the infected Vero cells as described previously with some modifications (32). Briefly, the virions were clarified by centrifugation at 6,000 × g for 30 min and precipitated with 10% polyethylene glycol (molecular mass, approximately 6,000 kDa). The precipitates were collected by centrifugation at 10,000 × g for 45 min and centrifuged at 147,000 × g for 20 h on a 20 to 60% sucrose gradient. The fractions ranging from 1.16 to 1.19 g/ml in gravity were used as the purified virion.
Antibodies.
Anti-JEV capsid protein rabbit polyclonal antibody (PAb) was prepared as described previously (32). Monoclonal antibodies (MAbs) to JEV E (10B4) and NS3 proteins (34A1) were generous gifts from E. Konishi and K. Yasui, respectively. Anti-FLAG tag (M2) and anti-β-actin MAbs were purchased from Sigma. Anti-HA (HA11) and anti-myc tag (9E10) MAbs were purchased from Covance (Richmond, CA). An antinucleolin MAb (MS-3) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-PA28-alpha and anti-cathepsin L rabbit PAbs were purchased from Affinity Bioreagents (Golden, CO) and Merck Calbiochem (Darmstadt, Germany), respectively.
Infection, transfection, immunoblotting, and cell fractionation.
A monolayer of Vero or N18 cells was infected at multiplicities of infection (MOI) of 5 and 10 with the wild-type and L17A/CSmt JEVs. Plasmids were transfected by TransIT LT-1 (Mirus, Madison, WI) and Lipofectamine 2000 (Invitrogen) for Vero and 293T cells, respectively, according to the manufacturers' instructions. At 24 h after inoculation or transfection, cells were lysed on ice by Triton lysis buffer (20 mM Tris-HCl [pH 7.4], 135 mM NaCl, 1% Triton X-100, 10% glycerol) supplemented with a protease inhibitor cocktail (Biovision, Mountain View, CA) and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting as previously described (36, 46). JEV-infected cells were fractionated using a Nuclear/Cytosol Fractionation kit (Biovision).
Inhibition of capsid protein processing.
E64d and CA074Me were purchased from the Peptide Institute (Osaka, Japan). Z-Phe-Tyr-(tert-butyl)-diazomethyl ketone (DMK) (Z-FY-DMK), Z-Val-Ala-Asp-fluoromethyl ketone (FMK) (Z-VAD-FMK), PD150606, and bafilomycin A1 were purchased from Merck Calbiochem. Chloroquine and ammonium chloride were obtained from Sigma and Nacalai Tesque (Kyoto, Japan), respectively. Chloroquine and ammonium chloride were dissolved in distilled water, and bafilomycin A1 was dissolved in ethanol. The other reagents were dissolved in dimethyl sulfoxide (DMSO). At 24 h after inoculation or transfection, cells were incubated with the culture medium containing each reagent or solvent for 8 h at 37°C and examined by immunoblotting. To determine the effects of CA074Me or FY-DMK on the cleavage of the capsid protein, cells transfected with pCAG/FLAG-JEC-HA were treated with the inhibitor for 8 h at 37°C. The ratios of the densities of the slower- and faster-migrating capsid proteins (C1 and C2, respectively) detected by immunoblotting were calculated by Multi Gauge software (Fujifilm, Tokyo, Japan). The relative cleavage values were determined as the C2 to C1 ratio in the presence of inhibitor/the C2 to C1 ratio in the absence of inhibitor. The inhibitory effects of CA074Me or Z-FY-DMK to cathepsins B and L were determined as described previously (7, 13) with some modifications. Briefly, Vero cells (2 × 105) were treated with CA074Me or Z-FY-DMK for 4 h at 37°C and lysed with 25 μl of acidic lysis buffer consisting of 100 mM sodium acetate (pH 5.0), 1 mM EDTA, 0.5% Triton X-100, 2 mM AEBSF [4-(2-aminoethyl)benzenesulfonyl fluoride] (Merck Calbiochem), 5 μg/ml aprotinin (Nacalai Tesque), 100 μM bestatin (Sigma), and 15 μM pepstatin (Peptide Institute). Insoluble materials were sedimented in a microcentrifuge at 4°C. Ten microliters of each lysate was mixed with 90 μl of reaction buffer (100 mM sodium acetate [pH 5.0], 1 mM EDTA, 4 mM dithiothreitol, 2 mM AEBSF, 5 μg/ml aprotinin, 100 μM bestatin, 15 μM pepstatin). The resulting samples were mixed with 100 μl of cathepsin B-specific (100 μM Z-Arg-Arg-MCA [4-methylcoumaryl-7-amide; Peptide Institute], 0.1% Brij 35) (3) or cathepsin L-specific (100 μM [Z-Phe-Arg]2-R110 [Molecular Probes, Eugene, OR], 0.1% Brij 35) (2) substrate solutions in a black 96-well plate (Corning, Corning, NY). After incubation for 30 min at room temperature, fluorescence was measured using a fluorescence multiwell plate reader (CytoFluor 4000 LX1; Applied Biosystems, Foster City, CA) with an excitation of 360 nm and an emission of 460 nm for cathepsin B and with an excitation of 485 nm and an emission of 460 nm for cathepsin L. The relative cleavage value in the absence of each inhibitor was defined as 1.
In vitro processing of the JEV capsid protein.
The JEV capsid protein fused with thioredoxin and myc-His tags in the N and C termini, respectively, was purified using TALON metal affinity resin (Clontech, Mountain View, CA) from the lysate of Escherichia coli transformed by pET32/JECmycHis. The purified protein was dialyzed with acidic dialysis buffer (50 mM sodium acetate [pH 5.5], 1 mM EDTA) for 24 h at 4°C. The recombinant JEV capsid protein (33 μg [1 nmol]/100 μl) was incubated with 0.01 units (170 ng) of human cathepsin L (Merck Calbiochem) for 2 h at room temperature. According to the manufacturer's instructions, one unit is defined as an amount of the enzyme capable of hydrolyzing 1.0 μmol of Z-Phe-Arg-AMC (7-amino-4-methylcoumarin) per minute at 37°C. The resulting samples were subjected to SDS-PAGE and Western blotting using anti-myc MAb. The N-terminal peptide sequences of the cleaved capsid proteins were determined by the Edman degradation method at the APRO Life Science Institute (Tokushima, Japan).
Computer analyses of the flavivirus capsid genes.
The amino acid sequences of the flavivirus capsid proteins were aligned with the software package GENETYX- MAC, version 12 (GENETYX, Tokyo, Japan). The GenBank accession numbers of the analyzed sequences are as follows: JEV AT31 strain, AB196923; MVE 1-51 strain, AF161266; WNV IS-98 STD1 strain, AF481864; DEN1 Singapore S275/90 strain, M87512; DEN2 New Guinea C strain, M29095; DEN3 H87 strain, M93130; DEN4 814669 strain, AF326573; YFV 17D strain, X03700. Nucleotides 135 to 152 and bases 10858 to 10875 in the 5′ and 3′ termini, respectively, connected by 8 X nucleotides alternative to bases 153 to 10857, of the wild-type and mutant JEV genomes were applied to GENETYX-MAC to predict RNA secondary structures with minimum free energy.
Growth kinetics of JEVs in vitro.
Vero, C6/36, PK15, N18, RAW264.7, Vero/siNC, and Vero/siCTSL cells in 24-well plates (2 × 105) were infected with the wild-type or L17A/CSmt virus at an MOI of 5 for 1 h, washed three times with a medium to remove unbound viruses, and incubated with a medium supplemented with 5% FBS for a total duration of 72 h. To examine the effect of the cathepsin L inhibitor on virus growth, DMSO or 1 μM Z-FY-DMK was added to the culture medium over the incubation period (24 h). The culture supernatants were used for titration of infectious virus.
Mouse experiments.
The pathogenicity of JEV to mice was determined as described previously (32). Briefly, 3-week-old female ICR mice were purchased from CLEA Japan (Osaka, Japan) and kept in special pathogen-free environments. Groups of 10 mice were intracerebrally inoculated with 30 μl of 10-fold-diluted solutions of wild-type or L17A/CSmt virus. The virus-diluting solution (DMEM) was administered to two mice as a control. The mice were observed for 2 weeks after inoculation to determine survival rates. The value of the 50% lethal dose (LD50) of each virus was determined by the method by Reed and Müench (39). To examine viral growth in the brain, 100 FFU of the viruses were intracerebrally administered to the mice. At 3 and 5 days after inoculation, the mice were euthanized, and the brains were collected. The infectious titers in the homogenates of the brains were determined in Vero cells as described above. Groups of 10 mice were inoculated intraperitoneally with 1 × 106 FFU (100 μl) of the viruses. The mice were observed for 3 weeks after inoculation to determine survival rates.
RESULTS
JEV-infected cells contained a small capsid protein.
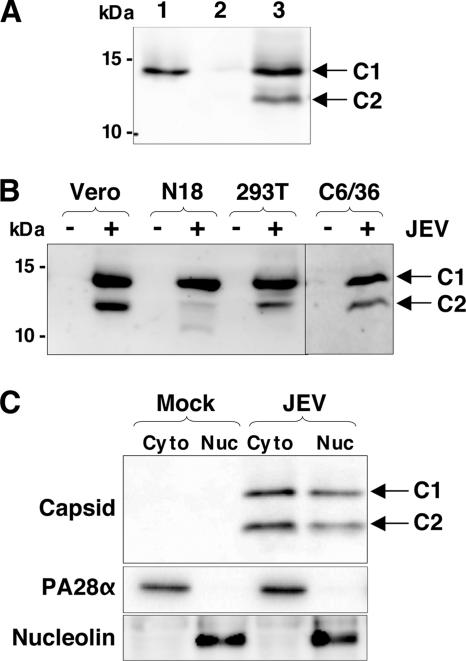
Western blotting analyses of Vero cells infected with JEV revealed capsid proteins of 14 and 12 kDa, which were designated C1 and C2, respectively, in contrast to the purified viral particles, in which only C1 was detected (Fig. 1A), indicating that C1 is a mature capsid protein missing a signal sequence of the prM protein. The C2 protein was also detected in the other cell lines examined, and a further processed capsid protein was detected in N18 cells infected with JEV (Fig. 1B). It was shown that the JEV capsid protein is localized in the nuclei as well as in the cytoplasm of the infected cells (32). The C1 and C2 proteins were also detected in both the cytoplasmic and nuclear fractions (Fig. 1C). These results indicate that two forms of the capsid proteins, C1 and C2, are generated in cells infected with JEV, and the larger capsid (C1) is selectively incorporated into the viral particles.
FIG. 1.
Detection of C2 protein in cells infected with JEV. (A) Detection of the capsid proteins from the purified viral particles and cells infected with JEV. Lane 1, purified JEV particles produced in Vero cells; lanes 2 and 3, mock- and JEV-infected Vero cells, respectively. Arrows indicate a mature capsid protein (C1) and a further-processed capsid protein (C2). (B) Detection of the capsid protein from various cell lines infected with JEV. (C) Detection of the C1 and C2 proteins in the cytoplasmic (Cyto) and nuclear (Nuc) fractions of Vero cells infected with JEV. PA28-α and nucleolin are control proteins of the cytoplasmic and nuclear fractions, respectively.
The C2 protein lacks the amino terminus.
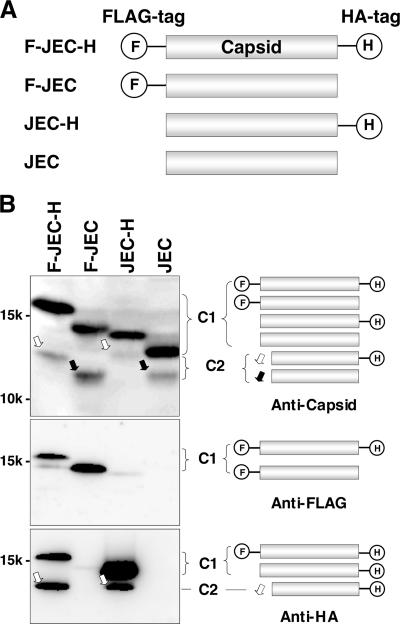
To determine which terminus is missing in the C2 protein, expression plasmids encoding a series of capsid proteins with or without amino-terminal FLAG and carboxyl-terminal HA tags (F-JEC-H, F-JEC, JEC-H, and JEC) were generated (Fig. 2A). Both the C1 and C2 isoforms were detected in Vero cells transfected with each of the expression plasmids by immunoblotting with anti-JEV capsid PAb (Fig. 2B). The size of the C2 proteins in cells transfected with JEC was similar to that of F-JEC, which has the amino-terminal FLAG tag, whereas larger products were detected in the cells transfected with F-JEC-H and JEC-H, which have the carboxyl-terminal HA tag. Consistent with this observation, anti-HA antibody recognized both isoforms in cells expressing F-JEC-H and JEC-H, whereas anti-FLAG antibody detected only C1 in cells expressing F-JEC-H and F-JEC. These results indicate that the C2 protein lacks the amino-terminal region of the JEV capsid protein.
FIG. 2.
The C2 protein lacks the amino terminus. (A) Series of the capsid protein constructs with or without FLAG and HA tags in the amino and carboxyl termini, respectively. (B) Expression of a series of the capsid proteins in Vero cells. The cell lysates expressing F-JEC-H, F-JEC, JEC-H, and JEC were examined by immunoblotting using anti-capsid, anti-FLAG, and anti-HA antibodies. The molecules detected by the immunoblotting are indicated on the right. White and black arrows indicate the C2 proteins with and without HA tags in the carboxyl terminus, respectively.
The JEV capsid protein is processed by cathepsin L.
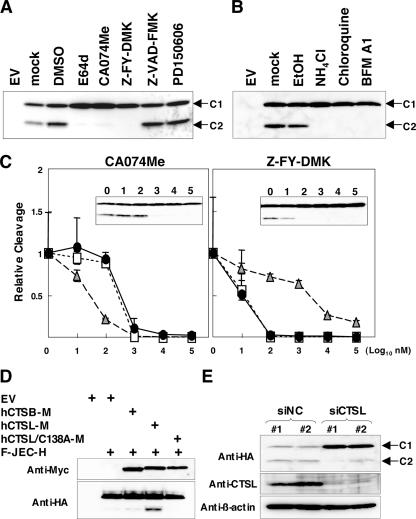
The C2 protein missing the amino-terminal region of the JEV capsid protein may be generated through cleavage by a host cell protease(s) or translation from the second start codon by leaky scanning, as reported in the case of DEN2 (9). To assess these possibilities, cells expressing F-JEC-H were treated with various protease inhibitors. C2 production was completely abrogated by treatment with broad-spectrum cysteine protease inhibitor E64d at the concentration of 50 μM, along with an increase in C1 expression (Fig. 3A), indicating that the JEV C2 protein was generated via cleavage of the C1 protein by a cysteine protease(s) but not leaky scanning. To identify the cysteine protease responsible for the processing of the JEV capsid protein, specific inhibitors for individual cysteine proteases were examined in cells expressing F-JEC-H. The inhibitors for cathepsins B and L, CA074Me (10 μM) (6) and Z-FY-DMK (10 μM) (40), impaired the processing, while an inhibitor of caspases, Z-VAD-FMK (20 μM), and an inhibitor of calpains, PD150606 (20 μM), exhibited no effect (Fig. 3A). Cathepsins B and L are known to be present in the late endosome and lysosome. The treatments with inhibitors of these acidic compartments, ammonium chloride (10 mM), chloroquine (50 μM), and bafilomycin A1 (100 nM), also blocked the processing of the capsid protein (Fig. 3B). To determine whether cathepsin B or L is a dominant protease for cleavage of the JEV capsid protein, the dose dependency of the effects of cathepsin inhibitors CA074Me and Z-FY-DMK on the cleavage of F-JEC-H was examined. The processing of the JEV capsid protein was inhibited in a manner that correlated closely with the inactivation of cathepsin L rather than that of cathepsin B (Fig. 3C). Furthermore, overexpression of cathepsin L, but not cathepsin B and inactive cathepsin L (C138A), resulted in an increase of C2 production in 293T cells (Fig. 3D). In addition, production of C2 from F-JEC-H was significantly decreased in two independent clones of Vero cells stably expressing siRNA for cathepsin L (Fig. 3E). These results indicate that cathepsin L is responsible for the processing of the JEV capsid protein to generate the C2 protein.
FIG. 3.
JEV capsid protein is processed by cathepsin L. (A) Effects of cysteine protease inhibitors on the processing of the JEV capsid protein. Vero cells expressing F-JEC-H were treated with 50 μM E64d, 10 μM CA074Me, 10 μM Z-FY-DMK, 20 μM Z-VAD-FMK, or 20 μM PD150606 for 8 h at 37°C and examined by immunoblotting using an anti-HA antibody. EV, empty vector. (B) Effects of anti-acidic compartment reagents on the processing of the JEV capsid protein. Vero cells expressing F-JEC-H were treated with 10 mM ammonium chloride, 50 μM chloroquine, or 100 nM bafilomycin A1 (BFM A1) for 8 h at 37°C and examined by immunoblotting using an anti-HA antibody. EtOH, ethanol. (C) Dose-dependent effects of two cathepsin inhibitors, CA074Me and Z-FY-DMK, on F-JEC-H processing. Vero cells expressing F-JEC-H were treated with CA074Me or Z-FY-DMK at the indicated concentrations for 8 h at 37°C and examined by immunoblotting using an anti-HA antibody. The relative cleavage values for the capsid protein (solid circles) were calculated as the intensity of C2 compared to that of C1 in three independent experiments. A representative image of the immunoblotting is indicated in each graph panel. The relative levels of cleavage of the substrates specific to cathepsin B (gray triangles) and cathepsin L (open squares) were determined as described in Materials and Methods. The value for the control sample without treatment of each inhibitor was taken as 1. (D) Effects of the overexpression of cathepsins on the processing of the JEV capsid protein. 293T cells were cotransfected with plasmids encoding myc-tagged human cathepsin B (hCTSB-M), cathepsin L (hCTSL-M), or inactive cathepsin L (hCTSL/C138A-M) with F-JEC-H. Immunoblot analysis was carried out using the antibodies shown at the left. (E) Processing of F-JEC-H in Vero cells stably expressing hairpin siRNA corresponding to the negative control (siNC) or cathepsin L (siCTSL). Immunoblot analysis was carried out using the antibodies shown at the left.
Identification of the site of the cleavage of the JEV capsid protein by cathepsin L.
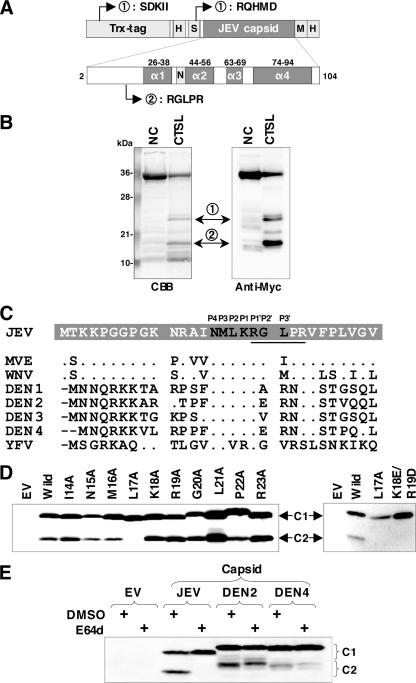
To determine the site of the cleavage of the JEV capsid protein by cathepsin L, a recombinant capsid protein possessing amino-terminal thioredoxin, His, and S tags and carboxyl-terminal myc and His tags was prepared (Fig. 4A). The in vitro incubation of the purified capsid protein with cathepsin L at room temperature for 60 min generated two major cleaved products, detectable by anti-myc antibody (Fig. 4B). The amino-terminal amino acid sequencing revealed that the mass of cleaved product 1 contained two peptides beginning with the residues Ser-Asp-Lys-Ile-Ile (a minor peptide) and Arg-Gln-His-Met-Asp (a major peptide), corresponding to a region of the thioredoxin and S tags, respectively (Fig. 4A and B). On the other hand, cleaved product 2 contained a single peptide beginning with Arg-Gly-Leu-Pro-Arg, corresponding to amino acid residues 19 to 23 of the JEV capsid protein. This result indicates that the JEV capsid protein is cleaved between Lys18 and Arg19 by cathepsin L in vitro (Fig. 4C). To further confirm the cleavage of the capsid protein in mammalian cells, a series of F-JEC-H proteins with alanine substitutions in each residue around the cleavage site (Ile14 to Arg23) was expressed in Vero cells (Fig. 4D). As indicated in the reports that a hydrophobic amino acid residue at position P2 is responsible for the substrate specificity of cathepsin L (37, 38), the replacement of Leu17 (P2) with alanine was crucial for capsid protein processing. In addition, although the single replacements at the cleavage site of Lys18 (P1) and Arg19 (P1′) with alanine had no effect on cleavage, the double substitution of acidic amino acids (Lys18 to Glu and Arg19 to Asp) resulted in impairment of C2 production (Fig. 4D). These results indicate that the JEV capsid protein is cleaved between Lys18 and Arg19 by cathepsin L in vitro and in vivo.
FIG. 4.
Identification of the site of cleavage of JEV capsid protein by cathepsin L. (A) Schematic diagram of the recombinant JEV capsid protein. The His, S, and myc tags are indicated as H, S, and M, respectively. Four α-helices (α1 to 4) of the JEV capsid protein were predicted by Ma et al. (27). The nuclear localization signal (N) was mapped to residues Gly42 and Pro43 (32). Products 1 and 2 of in vitro cleavage by cathepsin L began at the indicated positions. Trx, thioredoxin. (B) The purified capsid protein (33 μg [1 nmol]/100 μl) was treated with 0.01 units of recombinant human cathepsin L (CTSL) at room temperature for 60 min and analyzed by Coomassie brilliant blue (CBB) staining and immunoblotting using an anti-myc antibody after SDS-PAGE. The amino-terminal amino acid sequences of cleavage products 1 and 2 were determined by the Edman degradation method. (C) Alignment of the amino-terminal amino acid sequences of the mosquito-borne flaviviral capsid proteins. Positions P4 to P3′ of the site of cleavage of the JEV capsid protein by cathepsin L are shown at the top of the sequences. The amino-terminal amino acid sequences of cleavage product 2 generated by cathepsin L in vitro are underlined. Identical and deleted residues compared with the JEV capsid protein are indicated as dots and bars, respectively. (D) Identification of crucial residues for capsid protein processing by cathepsin L in vivo. A series of the mutant constructs derived from F-JEC-H were expressed in Vero cells and analyzed by immunoblotting using an anti-HA antibody. (E) Effect of a cysteine protease inhibitor E64d on the processing of the DEN capsid proteins. Vero cells expressing the FLAG- and HA-tagged capsid proteins of JEV, DEN2, and DEN4 were treated with DMSO or 50 μM E64d for 8 h at 37°C and examined by immunoblotting using an anti-HA antibody.
Production of the C2 proteins of DENs.
The P4 to P1′ region of the cathepsin L cleavage site is conserved among many mosquito-borne flaviviruses, including MVE, WNV, and DENs (Fig. 4C), and the 5′-complementary cyclization sequences are overlapped through the P4 to P2 sites (1, 19) (Fig. 5A). The C2 proteins were also detected in cells expressing the capsid proteins of DEN2 and DEN4 (Fig. 4E). To determine whether the C2 proteins of DEN are generated in the same manner as the C2 proteins of JEV, we examined the effect of the cysteine protease inhibitor E64d on the productions of the DEN C2 proteins. When cells were treated with E64d at a concentration of 50 μM, the C2 protein was diminished in cells expressing the capsid protein of JEV, but not in those expressing DEN2 and DEN4. However, it should be noted that treatment with the inhibitor induced a slight delay in migration of the C2 proteins of DENs. These results suggest that cysteine proteases do not play a major role in the production of the C2 proteins of DENs but play some roles in their processing.
Construction of a mutant JEV carrying the capsid protein resistant to cleavage by cathepsin L.
To assess the biological significance of the cleavage of the JEV capsid protein by cathepsin L, a mutant JEV with Leu17 replaced by Ala (L17A) was generated (Fig. 5A). However, the electroporation of the mutant RNA did not result in the production of the viral antigen (Fig. 5B) and infectious particles (data not shown). The coding region for Leu17 slightly overlaps the 5′ cyclization sequences, suggesting that the lack of replication of the L17A mutant is caused by the unstable and inappropriate secondary structure of viral RNA (Fig. 5A). To examine this possibility, L17A/CSmt, carrying additional complementary mutations in the 3′ UTR, which was predicted to have a stable secondary structure, and WT/CSmt, carrying a mutation only in the 3′ UTR as a control, were generated (Fig. 5A). Upon electroporation of the genomic RNAs into Vero cells, RNA of L17A/CSmt but not of WT/CSmt exhibited replication (Fig. 5B). As we expected, the C2 protein was not detected in Vero cells infected with L17A/CSmt (Fig. 5C). These results further confirm that RNA-RNA base pairing mediated by the two complementary cyclization sequences in the capsid coding region and 3′ UTR is required for replication of JEV.
Involvement of capsid protein cleavage on the cell type-specific replication of JEV.
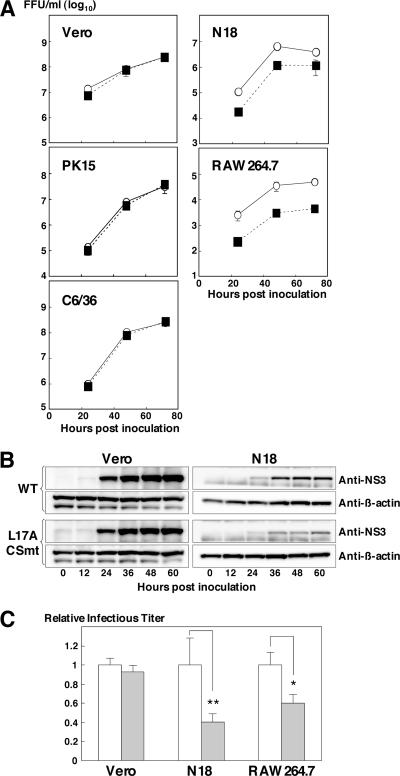
To examine the biological function of the C2 protein, the growth kinetics of the mutant L17A/CSmt was examined in several cell lines. As shown in Fig. 6A, L17A/CSmt was comparably replicated in Vero, C6/36, and PK15 cells compared with wild-type JEV, whereas growth of L17A/CSmt was 3.3- to 6.1-fold lower and 10.8- to 11.8-fold lower than that of wild-type JEV in N18 and RAW264.7 cells, respectively. In addition, L17A/CSmt exhibited reduced synthesis of the viral protein in N18 cells but not in Vero cells (Fig. 6B), suggesting that impairment of L17A/CSmt replication in N18 cells might be attributable to the reduction of viral protein synthesis. To further confirm the involvement of capsid protein cleavage in the cell type-specific restriction of L17A/CSmt replication, we examine the effect of the cathepsin L inhibitor on JEV replication. The cathepsin L inhibitor suppressed the growth of the wild-type virus in N18 and RAW264.7 cells, but not in Vero cells (Fig. 6C). Furthermore, the wild-type virus replicated equally in Vero/siNC and Vero/siCTSL cells (data not shown). These results suggest that generation of the C2 protein is required for the efficient replication of JEV in murine macrophage and neural cells.
FIG. 6.
Growth kinetics of L17A/CSmt in various cell lines. (A) The wild-type (WT; open circles) and mutant L17A/CSmt (solid squares) JEVs were inoculated into Vero, C6/36, PK15, N18, and RAW264.7 cells at an MOI of 10. After the indicated times, the infective titers in the culture supernatants on Vero cells were determined. (B) Viral protein synthesis in Vero and N18 cells infected with the WT or L17/CSmt virus. The NS3 and β-actin proteins were detected by immunoblotting with anti-JEV NS3 and anti-β-actin MAbs, respectively. (C) The WT JEV was inoculated into Vero, N18, and RAW264.7 cells at an MOI of 10 and incubated in the presence of DMSO (white bars) or 1 μM Z-FY-DMK (gray bars). At 24 h after inoculation, the infectious titers in the culture supernatants on Vero cells were determined. Asterisks showed significant differences by t test (**, P < 0.01; *, P < 0.05).
Neurovirulence and neuroinvasiveness of L17A/CSmt in mice.
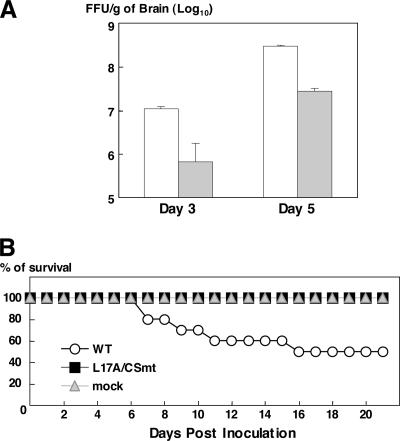
To compare the levels of neurovirulence of the wild-type and mutant viruses, we determined the LD50 values by intracerebral inoculation of the viruses in 3-week-old ICR mice. The LD50 value of L17A/CSmt (12.3 FFU) was approximately five times higher than that of the wild-type JEV (2.7 FFU). Although no significant difference in symptoms was observed between mice inoculated with 100 FFU of the wild-type and the mutant viruses, L17A/CSmt required longer periods than the wild-type JEV to kill mice (wild type versus L17A/CSmt: 6.8 ± 0.9 versus 8.4 ± 1.4 days postinoculation). To examine the growth kinetics of the viruses in the mouse brain, 100 FFU of each virus were intracerebrally injected and the progeny viruses in the brain were determined. The growth of L17A/CSmt was 16.3 and 11.0 times lower than that of the wild-type virus at 3 and 5 days after inoculation, respectively (Fig. 7A). Next, to compare the levels of neuroinvasiveness of the wild-type and mutant viruses, ICR mice were intraperitoneally inoculated with 1 × 106 FFU of each virus. All of the 10 mice inoculated with L17A/CSmt survived, whereas one-half of the mice inoculated with the wild-type JEV died by 10.0 days postinoculation on average (Fig. 7B). These results indicated that the L17A/CSmt mutant resistant to the cleavage by cathepsin L exhibits impaired neurovirulence and neuroinvasiveness in mice.
FIG. 7.
Neurovirulence and neuroinvasiveness of L17A/CSmt in mice. (A) Growth of the wild-type (WT; white bars) and mutant L17A/CSmt (gray bars) virus in mouse brain. One hundred FFU of each virus were intracerebrally injected into 3-week-old ICR mice, and the progeny viruses in the brain at 3 or 5 days after inoculation on Vero cells were determined. (B) Neuroinvasiveness of the WT and mutant JEVs to mice. Ten ICR mice were intraperitoneally inoculated with 1 × 106 FFU of each virus, and the survival rates of the mice were determined for 21 days.
DISCUSSION
Posttranslational modifications, including proteolysis, glycosylation, and phosphorylation, play a key role in regulating the functions of various proteins. Flavivirus proteins are translated as a single large precursor polyprotein, and proteolysis by host and viral proteases, such as signal peptidase, NS2B/3, and furin, is crucial for viral propagation (24). In this study, we demonstrate that some fraction of the mature JEV capsid proteins (C1) are further processed into a small form of capsid protein (C2) by cathepsin L, a papain-like cysteine protease. Furthermore, the C2 protein was shown to play a role in the replication of JEV in neural and macrophage cells and pathogenicity in mice. It is well established that cathepsins, a large group of lysosomal proteases, are involved in the bulk degradation of proteins in the lysosome. On the other hand, limited proteolysis by cathepsins has also been shown to convert a hormone (12), a neurotransmitter (51), and transactivators (15, 16, 33) from inactive precursors to the active forms and facilitate entry of several viruses (7, 13, 41).
It has been shown that the JEV C2 protein can be generated by the cleavage of the amino-terminal 18 amino acids from the C1 capsid protein by cathepsin L. However, the amino-terminal part of the cleavage product was not detected even though a FLAG tag was added (Fig. 2B). Therefore, the fate of the N-termimal 18 residues is currently unknown. The C2 protein was detected only in the cells, not in the viral particles, in contrast to the C1 protein, which was detected in both. The amino-terminal 32 amino acids and carboxyl-terminal 26 amino acids of the capsid protein of Kunjin virus (KUN), an Australian subtype of WNV, are essential for binding to the genomic RNA (20). The amino-terminal region of the capsid protein is well conserved between JEV and WNV. Therefore, it is possible that the JEV C2 protein is not incorporated into viral particles due to lack of the amino-terminal region of the capsid protein, required for binding to the viral RNA. Three-dimensional structural analyses revealed that the DEN and KUN capsid proteins contain four α-helixes and form a homodimer and a homotetramer (11, 27), and the amino-terminal 20 amino acids of the DEN capsid protein were shown to be flexible and not resolvable by nuclear magnetic resonance assay (27). In addition, a deletion mutant of the capsid protein of KUN lacking the amino-terminal 22 amino acids was used to determine the crystal structure (11). Therefore, the amino-terminal region of the flaviviral capsid proteins might not be involved in the self-assembly of the capsid proteins.
The capsid proteins in the fraction that are degraded rather than secreted as virions are likely to come in contact with cathepsin L in the acidic compartments such as the lysosome. Furthermore, subcellular fractionation indicated that the C2 protein had also migrated into the nucleus after processing. Our previous studies have shown that nuclear localization of the capsid protein and binding with the host nucleolar protein B23 are important for JEV replication (32, 46). These data suggest that the JEV capsid protein is translocated from the cytoplasm to the nucleus through the acidic compartment. Although the trafficking mechanisms of the capsid protein remain unknown, the C2 protein is able to migrate into the nucleus through the nuclear localization signal and B23-binding domain at Gly42 and Pro43 (32, 46). The C2 protein of a mutant JEV in which Gly42 and Pro43 were replaced with alanines (32) was impaired in nuclear localization, and the mutant capsid protein missing the amino-terminal amino acids was detected in the nucleus, especially in the nucleolus, when it was expressed by plasmid transfection (data not shown). On the other hand, it has been reported that cathepsin L or a cathepsin L-like protease is expressed in the nucleus and cleaves some host proteins, such as CDP/Cux (16), RB, and SP-1 (15, 33). Therefore, the JEV capsid protein might be alternatively processed in the nucleus by the proteases. In the case of WNV, the export of the capsid protein from the nucleus was facilitated in a Jab1-binding manner (34), and the Jab1-binding motif (Pro-Gly-Gly-Pro; residues 5 to 8) was also conserved in the JEV capsid protein. Therefore, the C2 protein lacking the Jab1-binding motif due to cleavage with cathepsin L might be able to escape from Jab1-dependent nuclear export and accumulate in the nucleus.
It has been established that the primary determinants of the specificity for cathepsin L are the S2 subsite (as shown in other papain-like proteases) and the hydrophobic residues at the P2 position of the substrates (37, 38). In addition, basic residues show a preference for the P1 position of substrates (38). These properties are in good agreement with our results that the cleavage site of the JEV capsid protein by cathepsin L is between Lys18 and Arg19 and that Leu17 at the P2 site was crucial for the cleavage. The residues P4 to P1′ are well conserved among mosquito-borne flaviviruses except for YFV, and the amino acid changes of the YFV capsid protein occur only within hydrophobic (Leu to Val at the P2 site) and basic (Lys to Arg at the P1 site) residues, respectively (Fig. 4C). Therefore, the capsid protein of YFV may also be cleaved by cathepsin L.
On the other hand, it has been reported that a small capsid protein of DEN2 was generated by leaky scanning (9). Due to the lack of a Kozak consensus sequence around the first start codon in many mosquito-borne flaviviruses, including DEN2, the smaller capsid protein of DEN2 is translated from the second or third AUG codon (9). In this context, two independent mechanisms of leaky scanning and processing by cathepsin L might be involved in the production of the small capsid protein. If both mechanisms were involved in the processing of the capsid protein of DEN, the C2 products that were generated by leaky scanning that started at residue Met15 and then were processed by cathepsin L at Arg18 should be present. This hypothesis is supported by the detection of the slowly migrating C2 proteins of DEN2 and -4 by SDS-PAGE due to treatment with E64d (Fig. 4E). The fast-migrating forms of the C2 proteins of DEN2 and -4 may be generated by cleavage by cathepsin L, while the slowly migrating forms detected in the presence of the inhibitor may be generated by leaky scanning. In contrast, the JEV genome possesses the ideal Kozak consensus sequence around the first AUG codon (9), and thus leaky scanning should not be involved in the production of the C2 protein.
Generation of the L17A capsid mutant in combination with the changes in the CSmt region (L17A/CSmt mutant) was necessary to ensure that altering the Leu17 codon did not also affect the 3′ cyclization sequence essential for viral replication. The RNA-RNA interaction between the 5′ cyclization sequences, in which the conserved amino acids required for cathepsin L cleavage are partially encoded, and the 3′ cyclization sequences was predicted for the flaviviruses (19), and the importance of the interaction for replication has been demonstrated in many flaviviruses (1, 10, 14, 19, 25). In this study we further confirmed the crucial role of the interaction of both the 5′ and 3′ ends of the viral RNA for JEV replication. Replication is a prerequisite for the viral life cycle; therefore, the capacity for the processing of the capsid protein, which is partially encoded in the 5′ cyclization sequences, by cathepsin L should be acquired during the viral adaptation to the hosts.
The growth kinetics of the L17A/CSmt JEV was reduced in RAW264.7 and N18 cells, but not in Vero, PK15, and C6/36 cells. It is noteworthy that the neural cells and the cells of monocyte/macrophage lineage are known to support JEV replication in vivo (22, 30, 50). The present study could not completely exclude the possibility that the complementary mutations in the cyclization sequences and/or the structure of the mutant capsid protein may be responsible for the reduced replication of the mutant virus in specific cells in culture or in vivo. It has been previously reported that DEN RNA with complementary mutations in the cyclization sequences recovered its direct interaction and self-primed RNA synthesis to the same level as seen in the wild-type RNA in a cell-free system (52, 53), whereas similar mutations significantly delayed RNA replication of the KUN replicon (19). Suppression of viral replication in N18 and RAW264.7 cells by treatment with the cathepsin L inhibitor further supports the possibility that the cleavage of capsid protein rather than RNA alteration in the cyclization sequences plays a crucial role in viral replication. Generation of the C2 protein is not a prerequisite for the cell-specific replication of JEV, because the processing of the capsid protein by cathepsin L was observed in all of the cells examined. However, we do not know the reason why the cell lines that showed the lowest production of the C2 protein exhibited the lowest viral production and the largest difference in growth of wild-type and L17A/Csmt viruses at the moment. Interaction of the C2 protein with a host factor(s) may be required for efficient replication of JEV in neural and macrophage cells, in which virus replicates at a low level, whereas the C2 protein may be unnecessary for replication in highly replication-competent cells, such as Vero, C6/36, and PK15 cells. The importance of the small capsid protein for viral replication has been shown in a study of DEN2, but a cell tropism for viral replication has not been reported (9).
Consistent with the data obtained in vitro, the L17A/CSmt mutant exhibited slow growth in the mouse brain. In addition, the limited growth of the mutant JEV in RAW264.7 and N18 cells may be a reflection of its reduced neuroinvasiveness. The symptoms of mice intracerebrally inoculated with the L17A/CSmt mutant were indistinguishable from those inoculated with the wild type, although disease induction required more time and a larger amount of virus than that due to inoculation with the wild type. These results suggest that the C2 protein is involved in viral replication in vivo but does not directly participate in virulence. This is in clear contrast to the mutant JEV defective in the nuclear localization of the capsid protein, which exhibited neurovirulence comparable to that of the wild type in spite of severe impairment of growth in the brain (32).
The present study demonstrated that cleavage of the capsid protein by cathepsin L and the resulting C2 protein missing the amino-terminal 18 amino acids plays a role in JEV replication in the nerve and macrophage cell lines, suggesting that the capsid protein has additional functions other than nucleocapsid formation. The limited genomic information of flaviviruses may constrain the multiassignment strategies of the viral proteins during the evolutional adaptation of the viruses to their hosts.
Acknowledgments
We thank H. Murase for her secretarial work. We also thank T. Wakita for providing the JEV infectious clone plasmids, E. Konishi and K. Yasui for the gifts of the anti-E and NS3 antibodies, respectively, and F. Hasebe and M. Tadano for the plasmids encoding the DEN2 and DEN4 capsid proteins, respectively.
This research was supported in part by grants-in-aid from the Ministry of Health, Labor, and Welfare; the Ministry of Education, Culture, Sports, Science, and Technology; the 21st Century Center of Excellence Program; the Foundation for Biomedical Research and Innovation; and the Zoonoses Control Project of the Ministry of Agriculture, Forestry and Fisheries of Japan.
Footnotes
Published ahead of print on 6 June 2007.
REFERENCES
- 1.Alvarez, D. E., M. F. Lodeiro, S. J. Luduena, L. I. Pietrasanta, and A. V. Gamarnik. 2005. Long-range RNA-RNA interactions circularize the dengue virus genome. J. Virol. 79:6631-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assfalg-Machleidt, I., G. Rothe, S. Klingel, R. Banati, W. F. Mangel, G. Valet, and W. Machleidt. 1992. Membrane permeable fluorogenic rhodamine substrates for selective determination of cathepsin L. Biol. Chem. Hoppe-Seyler. 373:433-440. [DOI] [PubMed] [Google Scholar]
- 3.Barrett, A. J., and H. Kirschke. 1981. Cathepsin B, cathepsin H, and cathepsin L. Methods Enzymol. 80:535-661. [DOI] [PubMed] [Google Scholar]
- 4.Bulich, R., and J. G. Aaskov. 1992. Nuclear localization of dengue 2 virus core protein detected with monoclonal antibodies. J. Gen. Virol. 73:2999-3003. [DOI] [PubMed] [Google Scholar]
- 5.Burke, D. S., and T. P. Monath. 2001. Flaviviruses, p. 1043-1125. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 6.Buttle, D. J., M. Murata, C. G. Knight, and A. J. Barrett. 1992. CA074 methyl ester: a proinhibitor for intracellular cathepsin B. Arch. Biochem. Biophys. 299:377-380. [DOI] [PubMed] [Google Scholar]
- 7.Chandran, K., N. J. Sullivan, U. Felbor, S. P. Whelan, and J. M. Cunningham. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308:1643-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, C. J., H. W. Luh, S. H. Wang, H. J. Lin, S. C. Lee, and S. T. Hu. 2001. The heterogeneous nuclear ribonucleoprotein K (hnRNP K) interacts with dengue virus core protein. DNA Cell Biol. 20:569-577. [DOI] [PubMed] [Google Scholar]
- 9.Clyde, K., and E. Harris. 2006. RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. J. Virol. 80:2170-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corver, J., E. Lenches, K. Smith, R. A. Robison, T. Sando, E. G. Strauss, and J. H. Strauss. 2003. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J. Virol. 77:2265-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dokland, T., M. Walsh, J. M. Mackenzie, A. A. Khromykh, K. H. Ee, and S. Wang. 2004. West nile virus core protein; tetramer structure and ribbon formation. Structure 12:1157-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunn, A. D., H. E. Crutchfield, and J. T. Dunn. 1991. Thyroglobulin processing by thyroidal proteases. Major sites of cleavage by cathepsins B, D, and L. J. Biol. Chem. 266:20198-20204. [PubMed] [Google Scholar]
- 13.Ebert, D. H., J. Deussing, C. Peters, and T. S. Dermody. 2002. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 277:24609-24617. [DOI] [PubMed] [Google Scholar]
- 14.Filomatori, C. V., M. F. Lodeiro, D. E. Alvarez, M. M. Samsa, L. Pietrasanta, and A. V. Gamarnik. 2006. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 20:2238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu, Y. H., T. Nishinaka, K. Yokoyama, and R. Chiu. 1998. A retinoblastoma susceptibility gene product, RB, targeting protease is regulated through the cell cycle. FEBS Lett. 421:89-93. [DOI] [PubMed] [Google Scholar]
- 16.Goulet, B., A. Baruch, N. S. Moon, M. Poirier, L. L. Sansregret, A. Erickson, M. Bogyo, and A. Nepveu. 2004. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol. Cell 14:207-219. [DOI] [PubMed] [Google Scholar]
- 17.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, C. T., L. Ma, J. W. Burgner, T. D. Groesch, C. B. Post, and R. J. Kuhn. 2003. Flavivirus capsid is a dimeric alpha-helical protein. J. Virol. 77:7143-7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75:6719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khromykh, A. A., and E. G. Westaway. 1996. RNA binding properties of core protein of the flavivirus Kunjin. Arch. Virol. 141:685-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiermayr, S., R. M. Kofler, C. W. Mandl, P. Messner, and F. X. Heinz. 2004. Isolation of capsid protein dimers from the tick-borne encephalitis flavivirus and in vitro assembly of capsid-like particles. J. Virol. 78:8078-8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura-Kuroda, J., M. Ichikawa, A. Ogata, K. Nagashima, and K. Yasui. 1993. Specific tropism of Japanese encephalitis virus for developing neurons in primary rat brain culture. Arch. Virol. 130:477-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kofler, R. M., F. X. Heinz, and C. W. Mandl. 2002. Capsid protein C of tick-borne encephalitis virus tolerates large internal deletions and is a favorable target for attenuation of virulence. J. Virol. 76:3534-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.). Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 25.Lo, M. K., M. Tilgner, K. A. Bernard, and P. Y. Shi. 2003. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 77:10004-10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lobigs, M., and E. Lee. 2004. Inefficient signalase cleavage promotes efficient nucleocapsid incorporation into budding flavivirus membranes. J. Virol. 78:178-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma, L., C. T. Jones, T. D. Groesch, R. J. Kuhn, and C. B. Post. 2004. Solution structure of dengue virus capsid protein reveals another fold. Proc. Natl. Acad. Sci. USA 101:3414-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makino, Y., M. Tadano, T. Anzai, S. P. Ma, S. Yasuda, and T. Fukunaga. 1989. Detection of dengue 4 virus core protein in the nucleus. II. Antibody against dengue 4 core protein produced by a recombinant baculovirus reacts with the antigen in the nucleus. J. Gen. Virol. 70:1417-1425. [DOI] [PubMed] [Google Scholar]
- 29.Markoff, L., B. Falgout, and A. Chang. 1997. A conserved internal hydrophobic domain mediates the stable membrane integration of the dengue virus capsid protein. Virology 233:105-117. [DOI] [PubMed] [Google Scholar]
- 30.Mathur, A., M. Bharadwaj, R. Kulshreshtha, S. Rawat, A. Jain, and U. C. Chaturvedi. 1988. Immunopathological study of spleen during Japanese encephalitis virus infection in mice. Br. J. Exp. Pathol. 69:423-432. [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuo, E., H. Tani, C. Lim, Y. Komoda, T. Okamoto, H. Miyamoto, K. Moriishi, S. Yagi, A. H. Patel, T. Miyamura, and Y. Matsuura. 2006. Characterization of HCV-like particles produced in a human hepatoma cell line by a recombinant baculovirus. Biochem. Biophys. Res. Commun. 340:200-208. [DOI] [PubMed] [Google Scholar]
- 32.Mori, Y., T. Okabayashi, T. Yamashita, Z. Zhao, T. Wakita, K. Yasui, F. Hasebe, M. Tadano, E. Konishi, K. Moriishi, and Y. Matsuura. 2005. Nuclear localization of Japanese encephalitis virus core protein enhances viral replication. J. Virol. 79:3448-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishinaka, T., Y. H. Fu, L. I. Chen, K. Yokoyama, and R. Chiu. 1997. A unique cathepsin-like protease isolated from CV-1 cells is involved in rapid degradation of retinoblastoma susceptibility gene product, RB, and transcription factor SP1. Biochim. Biophys. Acta 1351:274-286. [DOI] [PubMed] [Google Scholar]
- 34.Oh, W., M. R. Yang, E. W. Lee, K. M. Park, S. Pyo, J. S. Yang, H. W. Lee, and J. Song. 2006. Jab1 mediates cytoplasmic localization and degradation of West Nile virus capsid protein. J. Biol. Chem. 281:30166-30174. [DOI] [PubMed] [Google Scholar]
- 35.Oh, W. K., and J. Song. 2006. Hsp70 functions as a negative regulator of West Nile virus capsid protein through direct interaction. Biochem. Biophys. Res. Commun. 347:994-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okamoto, K., K. Moriishi, T. Miyamura, and Y. Matsuura. 2004. Intramembrane proteolysis and endoplasmic reticulum retention of hepatitis C virus core protein. J. Virol. 78:6370-6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Portaro, F. C., A. B. Santos, M. H. Cezari, M. A. Juliano, L. Juliano, and E. Carmona. 2000. Probing the specificity of cysteine proteinases at subsites remote from the active site: analysis of P4, P3, P2′ and P3′ variations in extended substrates. Biochem. J. 347:123-129. [PMC free article] [PubMed] [Google Scholar]
- 38.Puzer, L., S. S. Cotrin, M. F. Alves, T. Egborge, M. S. Araujo, M. A. Juliano, L. Juliano, D. Bromme, and A. K. Carmona. 2004. Comparative substrate specificity analysis of recombinant human cathepsin V and cathepsin L. Arch. Biochem. Biophys. 430:274-283. [DOI] [PubMed] [Google Scholar]
- 39.Reed, L. J., and H. Müench. 1938. A simple method of estimationg fifty per cent endpoints. Am. J. Hyg. 27:493. [Google Scholar]
- 40.Shaw, E., S. Mohanty, A. Colic, V. Stoka, and V. Turk. 1993. The affinity-labelling of cathepsin S with peptidyl diazomethyl ketones. Comparison with the inhibition of cathepsin L and calpain. FEBS Lett. 334:340-342. [DOI] [PubMed] [Google Scholar]
- 41.Simmons, G., D. N. Gosalia, A. J. Rennekamp, J. D. Reeves, S. L. Diamond, and P. Bates. 2005. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA 102:11876-11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solomon, T., H. Ni, D. W. Beasley, M. Ekkelenkamp, M. J. Cardosa, and A. D. Barrett. 2003. Origin and evolution of Japanese encephalitis virus in southeast Asia. J. Virol. 77:3091-3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stocks, C. E., and M. Lobigs. 1998. Signal peptidase cleavage at the flavivirus C-prM junction: dependence on the viral NS2B-3 protease for efficient processing requires determinants in C, the signal peptide, and prM. J. Virol. 72:2141-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tadano, M., Y. Makino, T. Fukunaga, Y. Okuno, and K. Fukai. 1989. Detection of dengue 4 virus core protein in the nucleus. I. A monoclonal antibody to dengue 4 virus reacts with the antigen in the nucleus and cytoplasm. J. Gen. Virol. 70:1409-1415. [DOI] [PubMed] [Google Scholar]
- 45.Tsai, T. F. 2000. New initiatives for the control of Japanese encephalitis by vaccination: minutes of a WHO/CVI meeting, Bangkok, Thailand, 13-15 October 1998. Vaccine 18:1-25. [DOI] [PubMed] [Google Scholar]
- 46.Tsuda, Y., Y. Mori, T. Abe, T. Yamashita, T. Okamoto, T. Ichimura, K. Moriishi, and Y. Matsuura. 2006. Nucleolar protein b23 interacts with Japanese encephalitis virus core protein and participates in viral replication. Microbiol. Immunol. 50:225-234. [DOI] [PubMed] [Google Scholar]
- 47.Wang, S. H., W. J. Syu, K. J. Huang, H. Y. Lei, C. W. Yao, C. C. King, and S. T. Hu. 2002. Intracellular localization and determination of a nuclear localization signal of the core protein of dengue virus. J. Gen. Virol. 83:3093-3102. [DOI] [PubMed] [Google Scholar]
- 48.Westaway, E. G., A. A. Khromykh, M. T. Kenney, J. M. Mackenzie, and M. K. Jones. 1997. Proteins C and NS4B of the flavivirus Kunjin translocate independently into the nucleus. Virology 234:31-41. [DOI] [PubMed] [Google Scholar]
- 49.Yamshchikov, V. F., and R. W. Compans. 1994. Processing of the intracellular form of the west Nile virus capsid protein by the viral NS2B-NS3 protease: an in vitro study. J. Virol. 68:5765-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, K. D., W. T. Yeh, R. F. Chen, H. L. Chuon, H. P. Tsai, C. W. Yao, and M. F. Shaio. 2004. A model to study neurotropism and persistency of Japanese encephalitis virus infection in human neuroblastoma cells and leukocytes. J. Gen. Virol. 85:635-642. [DOI] [PubMed] [Google Scholar]
- 51.Yasothornsrikul, S., D. Greenbaum, K. F. Medzihradszky, T. Toneff, R. Bundey, R. Miller, B. Schilling, I. Petermann, J. Dehnert, A. Logvinova, P. Goldsmith, J. M. Neveu, W. S. Lane, B. Gibson, T. Reinheckel, C. Peters, M. Bogyo, and V. Hook. 2003. Cathepsin L in secretory vesicles functions as a prohormone-processing enzyme for production of the enkephalin peptide neurotransmitter. Proc. Natl. Acad. Sci. USA 100:9590-9595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.You, S., B. Falgout, L. Markoff, and R. Padmanabhan. 2001. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 276:15581-15591. [DOI] [PubMed] [Google Scholar]
- 53.You, S., and R. Padmanabhan. 1999. A novel in vitro replication system for dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 274:33714-33722. [DOI] [PubMed] [Google Scholar]
- 54.Zhao, Z., T. Date, Y. Li, T. Kato, M. Miyamoto, K. Yasui, and T. Wakita. 2005. Characterization of the E-138 (Glu/Lys) mutation in Japanese encephalitis virus by using a stable, full-length, infectious cDNA clone. J. Gen. Virol. 86:2209-2220. [DOI] [PubMed] [Google Scholar]