Abstract
Both untranslated regions (UTRs) of plus-strand RNA virus genomes jointly control translation and replication of viral genomes. In the case of the Enterovirus genus of the Picornaviridae family, the 5′UTR consists of a cloverleaf-like terminus preceding the internal ribosomal entry site (IRES) and the 3′ terminus is composed of a structured 3′UTR and poly(A). The IRES and poly(A) have been implicated in translation control, and all UTR structures, in addition to cis-acting genetic elements mapping to the open reading frame, have been assigned roles in RNA replication. Viral UTRs are recognized by viral and host cell RNA-binding proteins that may codetermine genome stability, translation, plus- and minus-strand RNA replication, and scaffolding of viral replication complexes within host cell substructures. In this report, we describe experiments with coxsackie B viruses with a cell type-specific propagation deficit in Sk-N-Mc neuroblastoma cells conferred by the combination of a heterologous IRES and altered 3′UTR. Serial passage of these constructs in Sk-N-Mc cells yielded genetic adaptation by mutations within the viral nonstructural proteins 3A and 3C. Our data implicate 3A and/or 3C or their precursors 3AB and/or 3CD in a functional complex with the IRES and 3′UTR that drives viral propagation. Adaptation to neuroblastoma cells suggests an involvement of cell type-specific host factors or the host cell cytoplasmic milieu in this phenomenon.
Enterovirus (EV) plus-strand RNA genome translation and replication are controlled by cis-acting sequence elements mapping to the open reading frame and both 5′ and 3′ untranslated regions (UTRs) as well as nonstructural viral gene products. Their ∼7,400-nucleotide (nt) genomes feature complex structured 5′ and 3′UTRs of ∼750- and ∼80- to 110-nt length, respectively, flanking the open reading frame for a single polyprotein (24). The EV 5′UTR consists of a ∼90-nt “cloverleaf” structure serving as a cis-acting replication signal (4) preceding a ∼450-nt internal ribosomal entry site (IRES) involved in cap-independent translation initiation (23, 39). The 3′UTRs of EVs are structurally highly conserved, except coxsackie B viruses (CBVs), whose 3′UTRs contain a stem-loop domain (SLD) added to the conventional EV arrangement (42) (see Fig. 1A).
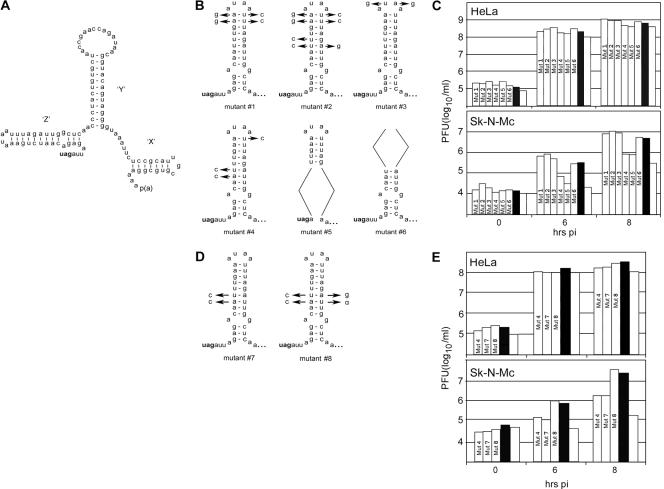
FIG. 1.
CBV-RICO variants with mutant 3′UTRs display differential growth in Sk-N-Mc cells. (A) Proposed secondary structure of the CBV 3′UTR (42). Predicted SLDs are marked X, Y, and Z; all EVs share a SLD X/Y arrangement, but SLD Z is unique to CBVs (42). The stop codon of the polyprotein is shown in bold type. p(a), poly(A) tail. (B) Sequences of mutant Z SLDs 1 to 6. Arrows indicate nucleotide substitutions; the stop codon of the polyprotein is shown in bold type. (C) Growth kinetics of mutants 1 to 6 (shown as Mut 1 to Mut 6 in the bars) in HeLa and Sk-N-Mc cells. For comparison, CBV-RICO (solid black bars) and RICO-ΔZ (open bars) were analyzed in parallel. (D) Sequences of mutants 7 and 8. Mutant 4 was altered by converting a mutation in the distal stem to the wt sequence (mutant 7) and restoring base pairing in the lower stem (mutant 8). (E) Growth kinetics in HeLa and Sk-N-Mc cells of mutants 4, 7, and 8 (Mut 4, Mut 7, and Mut 8), CBV-RICO (solid black bars), and RICO-ΔZ (open bars). Viral titers at test intervals represent the averages of two independent experiments.
The 5′ cloverleaf and 3′UTR are recognized by the same viral proteins with RNA-binding properties and have been implicated in genome replication functions. Viral proteins 3AB (the precursor of 3A, a small membrane-anchoring and RNA-binding protein and 3B, the genome-linked protein VPg) and 3CD (the precursor of the 3C proteinase and the 3D RNA-dependent RNA polymerase) interact with these terminal RNA structures (3, 15, 21, 37). Both the cloverleaf and the 3′UTR are required for efficient minus-strand synthesis (3, 5, 27, 28, 43, 50), although viable EVs with 3′UTR deletions have been derived (52).
In addition to these cis-acting genetic elements and viral RNA-binding proteins, host factors may play a role in the control of EV genome replication and translation. In addition to 3CD and 3AB, the cloverleaf attracts the poly(rC)-binding protein 2 (PCBP2) (15, 35). PCBP2 has been proposed to be involved in a switch from translation to replication at plus-strand RNA templates (16). Also, an interaction of PCBP2 with the poly(A)-binding protein has been speculated to mediate template circularization by linking the cloverleaf and poly(A) (22).
The complexity of EV translation and RNA synthesis has obstructed unraveling the mechanistic details of their regulation. Several multifunctional viral proteins interact with cis-acting signals on both ends on a potentially circular template. Often, the functions of precursor polypeptides are disparate from those of processed viral proteins. For example, although they differ only by the removal of the carboxy-terminal 22 amino acids of 3B, 3AB is believed to costimulate RNA replication by supporting the function of 3CD and 3D, while 3A does not (32, 36). cis-acting signals recognized by viral RNA-binding proteins simultaneously contribute to template stability, translation, and/or RNA replication. Moreover, the viral UTRs are recognized by host RNA-binding proteins that may either support or interfere with proper ribonucleoprotein (RNP) formation. The distribution or functional state of such host proteins may differ in a tissue type-specific manner, leading to various effects on viral functions according to host cell origin (30, 31). Both UTRs may assume complex configurations that are difficult to predict empirically, or they may engage in long-range interactions. RNA conformation, long-range interactions (e.g., circularization [22]), and the functional roles of UTR signals are likely to be influenced by interaction with viral or host RNA-binding factors and may fluctuate during the virus life cycle.
The roles of viral UTRs in genome translation and replication most commonly are assessed through genetic manipulation. Frequently, these studies are limited by unintended effects on higher-order structure and, consequently, on virus viability. We investigated the role of viral cis-acting genetic elements and their interaction with host or viral trans-acting gene products in viral propagation by constructing recombinant EVs with heterologous UTRs.
Insertion of a heterologous human rhinovirus type 2 (HRV2) IRES into the poliovirus (PV) genome causes a cell type-specific propagation deficit in neuron-like cell lines, e.g., Sk-N-Mc neuroblastoma (19, 20) or HEK-293 neuroblasts (8) and attenuates neurovirulence in mice transgenic for the human PV receptor (19) and nonhuman primates (20). Furthermore, the HRV2 IRES exhibits neuron-specific deficits that reduce neurovirulence when inserted to drive expression of a critical neurovirulence gene of herpes simplex virus type 1 (7). We reported that the heterologous HRV2 IRES mediates incorporation of viral RNA into a nonperforming RNP that is excluded from polysomes in neurons, but not in permissive cancer cell lines (31). Polysomal exclusion correlates with interaction of a neuron-specific heterodimer composed of the double-stranded RNA-binding protein 76 (DRBP76) and nuclear factor 45 (NF45) with the HRV2 IRES (30, 31).
We observed that cell type-specific performance of the HRV2 IRES within the context of EV genomes codepends on the conformation of the CBV3 3′UTR (see Fig. 1A) (10). More specifically, the unique SLD Z of CBVs is able to partially compensate for functional deficits of the HRV2 IRES in Sk-N-Mc neuroblastoma cells, suggesting that IRES function and its cell type-specific restrictions are influenced by cis-acting sequences in the 3′UTR (10). Here we report experiments designed to unravel the roles of 5′ and 3′UTRs in cell type-specific viral propagation in cells of neuronal lineage. We constructed a series of variant 3′UTRs to delineate the genetic basis of Z SLD-mediated stimulation of viral growth. Serial passaging of CBV3s replicating under control of the HRV2 IRES and 3′UTRs with deficient Z SLDs in neuroblastoma cells yielded adaptation mutations in the nonstructural viral proteins 3A/3AB either isolated or in conjunction with mutations in 3C/3CD. We observed mixed populations composed of viruses with a range of mutations in confined regions of 3A and/or 3C rather than homogeneous adaptation variants with mutations at defined sites.
MATERIALS AND METHODS
Construction of recombinant viruses.
Generation of a CBV3 chimera containing the HRV2 IRES (CBV-RICO) and CBV-RICO variants containing the 3′UTR of poliovirus (RICO-PV) or the 3′UTR of CBV3 carrying a deletion of stem-loop Z (RICO-ΔZ) have been described before (10). CBV-RICO mutants 1 to 8 contain modified 3′UTRs that were inserted into a cassette containing an XbaI restriction site spanning the termination codon of the CBV3 polyprotein via silent mutagenesis. Mutant 3′UTRs were PCR generated with reverse primer (primer 0) and one of the forward primers (primers 1 to 8; primer numbers correspond to mutant numbers) (primer sequences are listed in Table 1). The PCR products were digested with XbaI and ClaI and ligated with the CBV-RICO cassette vector.
TABLE 1.
Oligonucleotide primers used in this work
| Primer | Sequence (5′-3′) |
|---|---|
| 0 | GGATCGATGGGT18CCGCACCGAATGC |
| 1 | CTTCTAGATTAGAGACAATTTGGGATAACCTAGATTGGCT |
| 2 | CTTCTAGATTAGAGACAATCCGAAATAATTTGGATTGGCT |
| 3 | CTTCTAGATTAGAGACAATTTGAAAGGATTTAGATTGGCT |
| 4 | CTTCTAGATTAGAGACACCTTGAAATAACTTAGATTGGCT |
| 5 | CTTCTAGATTGTTGAAATAATTTAGAACCCTACTGTGCTAACC |
| 6 | CTTCTAGATTAGAGACAATATTGGCTCAACCCTACTGTGC |
| 7 | CCTCTAGATTAGAGACACCTTGAAATAATTTAGATTGGCTCAACCCTAC |
| 8 | CCTCTAGATTAGAGACACCTTGAAATAATTTAGGGTGGCTCAACCCTAC |
| 9 | CCAGATCACTTCGACGGATAC |
| 10 | CCACTGCTTCCAGCATGTA |
| 11 | GCACCAGAGACATCACCACCGCCCG |
| 12 | CGGGCGGTGGTGATGTCTCTGGTGC |
| 13 | CGGACCTGCTCAAAACGGTAGATAGTGA |
| 14 | TCACTATCTACCGTTTTGAGCAGGTCCG |
| 15 | GTGAGGCTGTGACGGAGTACTGCAAAG |
| 16 | CTTTGCAGTACTCCGTCACAGCCTCAC |
| 17 | GTGAGGCTGTGATGGAGTACTGCAAAG |
| 18 | CTTTGCAGTACTCCATCACAGCCTCAC |
| 19 | CCATCTTGATGAATGGTCAAGAGGTTGGT |
| 20 | ACCAACCTCTTGACCATTCATCAAGATGG |
| 21 | GACATCACCACCGCCCGCCATTGCGGACCTGCTCAAATCGGTAGATAGTGAGGCTGTGACGGAGTACTGCAAAG |
| 22 | CTCCGTCACAGCCTCACTATCTACCGATTTGAGCAGGTCCGCAATGGCGGGCGGTGGTGATGTCTCTGGTGCAAC |
| 23 | CCATCGATCCGCACCGAATGCGGAGA |
| 24 | GGATCGATGCT12CTCCGGAATTA |
| 25 | CTCTTGTGTGTGGGAAGGC |
| 26 | GATACCCAGTACCTTGCCG |
Adaptive mutations acquired through serial passage of mutants 4, 5, RICO-PV, and RICO-ΔZ (see Fig. 2B) were cloned into CBV-RICO, RICO-ΔZ, CBV-ΔZ, or CBV3 cDNA using PCR fragments corresponding to the regions coding for 3A to 3C of the viral genome. First, two overlapping PCR products were generated with one of the flanking primers (primer 9 or 10) and corresponding internal primers containing the desired sequence alteration. Second, corresponding fragments were combined through PCR performed with both flanking primers using the overlapping PCR products as the template. The internal primers and the mutations (see Fig. 2B) they introduce are as follows: primers 11 and 12, mutation [P17S]; primers 13 and 14, [S28T]; primers 15 and 16, [R35T]; primers 17 and 18, [R35M]; and primers 19 and 20, [D51G]. Double mutation [R35M:D51G] was generated by fusion of three PCR fragments produced in the first round with primer pairs (primers 9 and 18, primers 17 and 20, and primers 10 and 19). The resulting PCR products were digested with BsmBI and ligated into CBV-RICO, RICO-ΔZ, CBV-ΔZ, or CBV3 full-length cDNA digested with BsmBI.
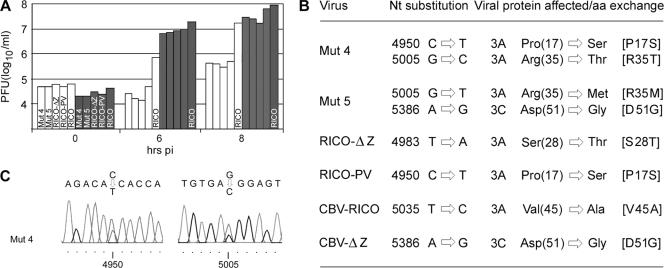
FIG. 2.
Adaptation of mutants 4 and 5 (Mut 4 and Mut 5), RICO-ΔZ, RICO-PV, CBV-RICO, and CBV-ΔZ through serial passage in Sk-N-Mc cells. (A) One-step growth curves of prepassage viruses (open bars) and viruses subjected to eight serial passages (gray bars). Individual constructs are indicated for the 0-h time point and are shown in the same order for the 6- and 8-h time points; CBV-RICO (RICO) is indicated in each set of bars. (B) Nucleotide (Nt) substitutions identified in the indicated viruses through sequencing of bulk PCR product and their effect on the amino acid sequence of the affected viral protein. Nucleotide numbering refers to the genome of CBV-RICO. Amino acid (aa) numbering refers to the residue within CBV3 3A or 3C. Adaptation mutations are shown as numbers and letters in brackets. (C) Chromatograms depicting sequencing results for mutant 4. Double peaks indicate the presence of adaptation mutations (the wt sequence is shown above the chromatograms); nucleotide numbering corresponds to that in panel B.
Derivation of viruses from infectious cDNA.
All recombinant plasmids were linearized with restriction endonuclease ClaI for in vitro transcription with T7 RNA polymerase as described previously (19). Approximately 5 μg of in vitro-transcribed RNA was transfected into 2.5 × 106 HeLa cells with DMRIE-C transfection reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. The transfected cells were incubated for 48 h at 37°C and subjected to two consecutive freeze-thaw cycles, and the resulting lysate was analyzed by plaque assay. Viruses derived from in vitro-transcribed RNA were propagated in HeLa cells to generate large-scale virus stocks. To this end, 500 μl of the transfected cell lysate was added to 8 × 106 HeLa cells in a 10-cm culture dish, which was gently rocked for 30 min at room temperature. Afterwards, 5 ml of culture medium was added, and the cells were incubated at 37°C overnight. After two freeze-thaw cycles, the infection procedure was repeated to infect 108 HeLa cells to generate a large-scale virus preparation. To determine the full-length genome sequences of mutants 4 and 5, RICO-ΔZ, RICO-PV, and CBV-RICO, aliquots from virus stocks were used to infect HeLa cells in a 3.5-cm dish using the procedure described above. Upon the onset of cytopathic effects 8 hours postinfection (p.i.), the cells were subjected to two freeze-thaw cycles and total RNA was extracted from the resulting lysate as described below.
One-step growth curves and serial passaging of viruses.
One-step growth curves were established as follows. HeLa R19, Sk-N-Mc, or HEK-293 cell monolayers in 60-mm dishes were overlaid with 500 μl of virus suspension at a multiplicity of infection of 10 and gently agitated for 30 min at room temperature. After three rinses with 2 ml of serum-free Dulbecco's minimal essential medium (DMEM; Invitrogen, Carlsbad, CA), cells were overlaid with 1.5 ml of DMEM containing 2% fetal bovine serum (Invitrogen, Carlsbad, CA) and incubated at 37°C for the indicated intervals. At chosen time points, the cultures were subjected to two freeze-thaw cycles, and the resulting lysates were used to determine the viral titer by plaque assay on HeLa cells as described elsewhere (14). Mutants 4, 5, RICO-ΔZ, RICO-PV, CBV-RICO, and CBV-ΔZ were serially passaged in Sk-N-Mc cells for eight consecutive rounds. Infection of Sk-N-Mc cells for passaging was essentially performed as described above, except the rinsing step was omitted and infected cultures were incubated at 37°C for 12 h. Subsequently, the cells were lysed by two freeze-thaw cycles, and the lysate was used to infect a new culture.
Population sequencing of viral genomes.
Viral sequences were determined from total RNA isolated from HeLa cells infected with prepassage virus stocks or from infected Sk-N-Mc cells after the indicated passage using TRIzol reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. Amplification of viral genomes by reverse transcription-PCR (RT-PCR) was performed as described before (8). RT was performed using primer 23 (for all viruses containing CBV3 3′UTR variants) or 24 (for RICO-PV), complementary to the 3′ terminus of viral RNA. Sections of the resulting cDNAs were amplified with different sets of primers to generate overlapping PCR fragments covering the entire viral genome and sequenced.
Clonal analysis.
To determine the sequences of mixed populations resulting from adaptation of viruses to neuroblastoma cells, we performed the following assays. Total RNA extracted from Sk-N-Mc cells infected with mutant 4 and 5 viruses after eight passages was subjected to RT-PCR with primers 25 and 26 to amplify the region coding for nonstructural viral proteins 3A to 3C. This region had been identified previously through whole-genome sequencing of passaged viruses to contain adaptive mutations (see Fig. 2B). The resulting PCR fragments were phosphorylated using T4 polynucleotide kinase (NEB, Beverly, MA) and cloned into the EcoRV site of pBluescript II KS+ (Stratagene, La Jolla, CA). Individual clones were isolated, and 39 clones (16 for mutant 4 and 23 for mutant 5) were sequenced in both directions with primers 25 and 26.
Northern blots.
Northern blot riboprobes were produced from a plasmid generated by engineering a BglII site into the multiple cloning site of pBluescript II KS+. CBV-RICO cDNA was digested with EcoRI and BglII and a 721-base-pair portion of the P1 coding region was cloned into the EcoRI-BglII site of the modified pBS vector. In vitro transcription with T3 polymerase generated sense probe to detect plus-strand viral RNA. In vitro transcription reactions were conducted in the presence of [α-32P]UTP to radioactively label the probes and treated with RQ1 DNase (Promega, Madison, WI) for 45 min at 37°C. RNA probes were purified over G-50 Sephadex columns (GE Healthcare, Little Chalfont, United Kingdom). Northern blotting was performed according to previously established procedures (57). Total RNA from infected Sk-N-Mc cells was extracted with TRIzol-LS (Invitrogen, Carlsbad, CA), and quantified via UV spectroscopy. Ten micrograms of total cellular RNA was used for Northern blot analysis of each sample (57).
RESULTS
The lower portion of SLD Z is critical for chimeric CBV3 propagation in a cell type-specific manner.
We previously observed cooperative effects on cell type-specific propagation of CBV3 mediated by heterologous UTRs (10). In neuroblastoma cells, virus containing an HRV2 IRES and cognate 3′UTR (CBV-RICO) grows to titers ∼10- to 50-fold higher than the titers of variants lacking SLD Z (RICO-ΔZ; Fig. 1) (10). SLD Z alleviates a neuron-specific deficit associated with the foreign IRES; it is irrelevant for proper virus propagation in HeLa cells (10).
To unravel the influence of 3′UTR structure on cell type-specific viral propagation, we constructed a series of CBV-RICO mutants with altered Z SLDs (Fig. 1B and D). These were designed to disrupt primary sequence motifs and/or predicted stable secondary structure. Mutants 1 and 2 disrupt AU-rich elements without altering base pairing in the distal stem, mutant 3 contains a terminal loop with altered primary sequence, mutant 4 contains an enlarged terminal loop combined with disrupted stability of the lower stem, and mutants 5 and 6 feature deletions of the proximal stem and distal stem/terminal loop, respectively (Fig. 1B).
The viruses were derived via transfection of in vitro-transcribed viral RNA into HeLa cells as described in Materials and Methods. All variants readily emerged in transfected cells. Plaque assay of lysates obtained 48 h after transfection revealed virus yields comparable to that of wild-type (wt) CBV3 derived in parallel. This suggests that none of the constructs had overt growth defects in HeLa cells. The viruses were subjected to at least six passages in HeLa cells to generate large-scale virus preparations used in growth assays (see Materials and Methods). To exclude selection of adaptation mutants in this process, we sequenced the entire genomes of mutants 4, 5, RICO-ΔZ, RICO-PV (CBV-RICO containing a poliovirus 3′UTR), and CBV-RICO from total RNA isolated from infected HeLa cells (CBV-ΔZ and CBV3 were not sequenced) (see Materials and Methods). None of the viruses analyzed displayed sequence variation from the cloned cDNA constructs.
Analysis of propagation kinetics of mutants 1 to 6 in HeLa and Sk-N-Mc neuroblastoma cells revealed two distinct growth phenotypes (Fig. 1C). All mutants exhibited comparable growth in HeLa cells, but mutants 4 and 5 displayed significant growth inhibition in the neuroblastoma cell line (Fig. 1C). Growth of mutants 4 and 5 was more efficient than with outright SLD Z deletion (RICO-ΔZ) but was clearly impaired compared to all other mutants, which propagated to levels equal to that of CBV-RICO. Since mutants 4 and 5 contain an altered proximal portion of SLD Z, these findings implicate this substructure in the observed phenotype.
To further analyze the role of SLD Z structure in viral growth, we generated two variants of mutant 4 (mutants 7 and 8 [Fig. 1D]). We converted the substitution in the distal stem to wt (mutant 7) and restored the proximal stem in mutant 7 by substituting the pairing nucleotides (mutant 8). Mutants 7 and 8 were tested for propagation in HeLa and Sk-N-Mc cells in parallel with mutant 4, CBV-RICO, and RICO-ΔZ (Fig. 1E). While all viruses propagated to the same levels in HeLa cells, mutants 4 and 7 exhibited significant growth deficits in Sk-N-Mc cells (Fig. 1E). Restoration of the proximal stem in SLD Z rescued mutant 8, which propagated as well as CBV-RICO did in Sk-N-Mc cells (Fig. 1E).
Serial passaging of viruses with defective growth in neuroblastoma cells yields adaptation mutations in viral nonstructural gene products.
What is the mechanism determining the effect of UTR structure on cell type-specific propagation of CBV-RICO? We addressed this issue by testing for the emergence of adaptation variants with improved growth in Sk-N-Mc cells (Fig. 2). To this end, we subjected mutants 4, 5, RICO-ΔZ, and RICO-PV to eight serial passages in Sk-N-Mc cells. In addition, to probe for adaptation to the HRV2 IRES or ΔZ alone, we passaged CBV-RICO and CBV-ΔZ (wt CBV3 with ΔZ). Finally, to exclude cell type-specific adaptation of CBV3, we performed identical passaging procedures with the wt parent. We then performed one-step growth curves in Sk-N-Mc cells with prepassaged and passaged viruses in parallel (Fig. 2A). Mutants 4, 5, RICO-ΔZ, RICO-PV, and CBV-RICO adapted to significantly improved growth in SK-N-Mc cells compared to their unpassaged counterparts (Fig. 2A). Only the wt CBV3 growth rate did not differ after eight serial passages (data not shown). The effect of adaptation on growth 6 h p.i. was far more pronounced for variants with absent or defective Z SLDs than on CBV-RICO (Fig. 2A). Adapted SLD Z variants grew to ∼10-fold-higher titers 6 h p.i. than unpassaged CBV-RICO (containing the wt CBV3 3′UTR) (Fig. 2A).
Next, we sequenced the entire genomes of mutants 4, 5, RICO-ΔZ, RICO-PV, and CBV-RICO by RT-PCR from viral RNA isolated from infected Sk-N-Mc cells after eight passages (see Materials and Methods; only the coding region for 3A-3C of the adapted CBV-ΔZ genome was sequenced). Notably, none of the viruses had acquired sequence changes mapping to the heterologous 5′ and 3′UTR sequences. However, sequencing revealed variation in the coding regions for the viral nonstructural proteins 3A and/or 3C in all recombinants (Fig. 2B). Mutations [R35T] and [R35M] affecting the same residue of 3A occurred separately after passage of mutants 4 and 5. [P17S] in 3A and [D51G] in 3C emerged in two separate incidences after passaging of mutant 4/RICO-PV and mutant 5/CBV-ΔZ, respectively. Adaptation mutations occurred with the HRV2 IRES:ΔZ combination (mutant 5, RICO-ΔZ/PV), with only the HRV2 IRES (CBV-RICO) as well as with ΔZ alone (CBV-ΔZ) (Fig. 2B). However, the average growth benefit from adaptation of SLD Z variants and CBV-RICO was ∼280-fold and ∼80-fold versus ∼20-fold and ∼5-fold at 6 and 8 hours pi, respectively (Fig. 2A). This shows that, although adaptation occurs with the intact CBV3 3′UTR, the combination of HRV2 IRES:ΔZ derives far higher growth benefits than the foreign IRES alone.
Adapted SLD Z variants consist of swarms of genetically heterogeneous viruses.
Our sequencing approach (i.e., analysis of bulk PCR product) does not permit analysis of individual variants within a mixed population. However, the sequence chromatograms revealed double peaks at mutation sites (Fig. 2C), indicating the presence of genetically diverse viruses in the population. Therefore, we performed clonal analysis by subcloning PCR product generated from adapted mutants 4 and 5 and sequencing the coding region spanning 3A-3C of individual clones (see Materials and Methods for details) (Table 2 and 3).
TABLE 2.
Sequencing of adapted mutant 4 virus populations
| Clone | Adaptation mutationa
|
||
|---|---|---|---|
| [P17S] | [R35T] | Other | |
| 1 | − | − | [S28T] |
| 2 | + | − | − |
| 3 | − | [R35M]b | [L26F] |
| 4 | − | − | − |
| 5 | − | + | − |
| 6 | − | + | − |
| 7 | − | − | − |
| 8 | + | − | − |
| 9 | − | + | − |
| 10 | − | + | − |
| 11 | + | − | − |
| 12 | + | − | − |
| 13 | − | + | − |
| 14 | − | − | − |
| 15 | − | + | − |
| 16 | − | + | − |
The presence (+) or absence (−) of the indicated mutation in the clones is shown. All adaptation mutations are in 3A (see Fig. 3).
[R35M] is the predominant mutation in the coding region for 3A in the population of adapted mutant 5 viruses.
TABLE 3.
Sequencing of adapted mutant 5 virus populations
| Clone | Adaptation mutationa
|
|||
|---|---|---|---|---|
| [R35M]b | [D51G]c | Otherb | Otherc | |
| 1 | + | + | [L44S] | [F4S] |
| 2 | + | + | − | − |
| 3 | − | + | − | − |
| 4 | − | + | − | − |
| 5 | − | + | [E32G] | − |
| 6 | + | − | − | − |
| 7 | − | + | − | − |
| 8 | − | + | − | − |
| 9 | − | + | − | − |
| 10 | + | + | [D30G] | − |
| 11 | + | − | − | − |
| 12 | − | + | − | [M49T] |
| 13 | − | + | − | − |
| 14 | + | − | − | − |
| 15 | + | + | − | [V54A] |
| 16 | + | + | − | − |
| 17 | + | + | − | − |
| 18 | + | + | − | − |
| 19 | − | + | − | − |
| 20 | + | + | − | − |
| 21 | + | − | − | − |
| 22 | + | + | − | − |
| 23 | + | − | − | − |
The presence (+) or absence (−) of the indicated viral construct in the clones is shown.
Adaptation mutations in 3A (see Fig. 3).
Adaptation mutations in 3C.
This method revealed the composition of mixed populations of adapted mutants 4 and 5 (Tables 2 and 3). Adapted mutant 4 populations were composed of parental virus and variants with mutations only in 3A (Table 2). The ratio of parental virus and viruses containing adaptation mutations [P17S] and [R35T] was ∼1:1:2. None of the clones contained both mutations combined. One clone had [R35M] (a mutation in the majority of adapted mutant 5 viruses) instead of [R35T], and two clones displayed point mutations that were not apparent from sequencing of the bulk PCR product. This is explained by the low incidence of these variants (1 out of 16). Both minor mutations, like the major changes in adapted mutant 4 identified before, mapped to the amino-terminal portion of 3A ([L26F] and [S28T]) (Fig. 3).
FIG. 3.
Aligned amino acid sequences of the amino-terminal portion of 3A. The sequences of PV type 1 (Mahoney) (GenBank accession no. V01149), HRV2 (GenBank accession no. X02316), and CBV3 (20) (GenBank accession no. AY752946) are shown. Mutated residues are marked by asterisks. Gaps introduced to maximize alignment are indicated by dashes. Numbering refers to that of 3A of CBV3.
The composition of adapted mutant 5 population was more complex and revealed mutations in 3A and 3C (Table 3). Out of 23 clones, 5 contained [R35M] (in 3A) alone, 7 featured [D51G] (in 3C) alone, and 9 contained the [R35M:D51G] combination. Five clones had additional mutations that were not apparent through sequencing of the bulk PCR product. As with mutant 4, all major and minor mutations in 3A occurred in the amino terminus (Fig. 3). Interestingly, 18 out of 23 clones contained mutations in 3C; of these, 8 had no accompanying mutations in 3A. In addition to the major [D51G] mutation, three minor mutations were identified in 3C through clonal analysis. Except for [F4S], these occurred in close proximity to [D51G] ([M49T] and [V54A]) (Table 3). Curiously, individual clones in adapted mutant 4 and 5 populations did not overlap. The predominant mutation site in the amino terminus of 3A was R35 in both populations; however, seven of eight mutant 4 clones featured [R35T], whereas mutant 5 clones contained exclusively [R35M]. Most clones in mutant 5 populations, but none in mutant 4 viruses, contained mutations in 3C.
Mutations in 3A to 3C compensate UTR-mediated growth defects in neuroblastoma cells.
To investigate the effects of the adaptation mutations on viruses with variable UTRs, we engineered a panel of recombinants (Fig. 4). We tested the mutations by (i) probing their effects on the HRV2 IRES:ΔZ combination by placing them in RICO-ΔZ, (ii) evaluating their effects on growth deficits mediated by the HRV2 IRES alone in CBV-RICO, (iii) assessing the effect on ΔZ alone in CBV-ΔZ, and (iv) excluding general effects on CBV3 growth in Sk-N-Mc cells in a wt background.
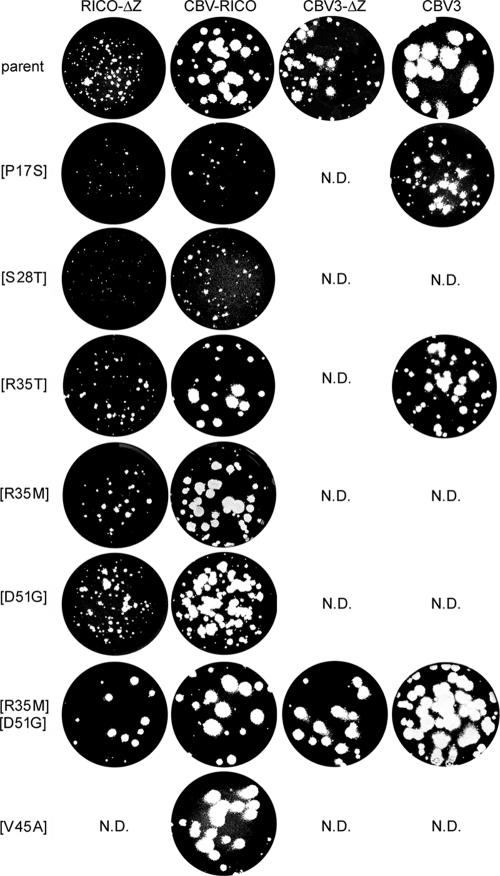
FIG. 4.
Plaque phenotypes of RICO-ΔZ, CBV-RICO, CBV-ΔZ, wt CBV3, and their variants containing the identified amino acid exchanges; all amino acid exchanges affect 3A except [D51G], which occurs in 3C. Individual constructs were derived as described in Materials and Methods, and their plaque phenotype was determined by analyzing cell lysates obtained 48 h after transfection of infectious in vitro transcript genomic RNA. N.D., not done.
A series of RICO-ΔZ and CBV-RICO viruses were generated containing each major amino acid exchange individually or the [R35M:D51G] combination, with the exception of [V45A] (in 3A), which was tested only in its parent, CBV-RICO (Fig. 4). We did not generate viruses containing mutations only present in minute proportions in mixed populations (Tables 2 and 3). All viruses were propagated as described in Materials and Methods to generate viral stock. The genotype of stock virus was confirmed by RT-PCR sequencing of the 3A-3C coding region from total RNA isolated from infected HeLa cells, and the plaque phenotype in HeLa cells was characterized before assessment of one-step growth kinetics in HeLa and Sk-N-Mc cells. With the exception of [P17S] and [S28T], all mutations resulted in unchanged or slightly enlarged plaque phenotypes for both CBV-RICO and RICO-ΔZ (Fig. 4).
We tested CBV-RICO and RICO-ΔZ containing five individual adaptive mutations and one combined adaptive mutation in 3A to 3C. All mutations had comparable effects on propagation in HeLa and Sk-N-Mc cells; for brevity, only the results for [R35M:D51G] are shown (Fig. 5A and B). [R35M:D51G] is present in ∼40% of the adapted mutant 5 population (Table 3). The variants exhibited growth in HeLa cells equivalent to their parents (Fig. 5A), but propagation of both CBV-RICO and RICO-ΔZ was markedly increased in Sk-N-MC cells (Fig. 5B). At 6 h p.i., [R35M:D51G] yielded ∼10- and ∼25-fold-higher titers of CBV-RICO and RICO-ΔZ, respectively (Fig. 5B). The levels of this increase are representative for all other mutants. Although [R35M:D51G] occurred with mutant 5 (containing a truncated SLD Z lacking the proximal stem; Fig. 1A), it partially compensates the growth deficit of RICO-ΔZ, which lacks SLD Z altogether.
FIG. 5.
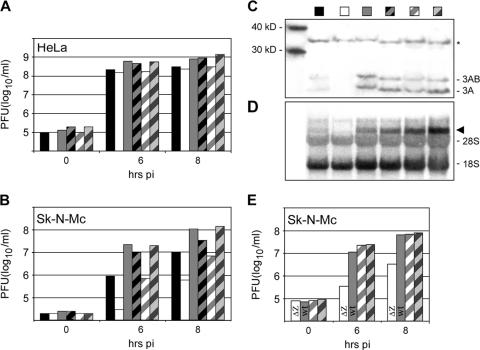
Growth phenotypes of CBV3 variants carrying adaptation mutations [R35M:D51G]. Growth properties of CBV-RICO (solid black bars), RICO-ΔZ (solid white bars), wt CBV3 (solid gray bars), and their [R35M:D51G] variants (hatched bars in identical colors) in (A) HeLa cells and (B) Sk-N-Mc cells. (C) Western blot of CBV3 3A/3AB and (D) Northern blot of plus-strand viral RNA in infected cell lysates 8 h p.i. The identities of the viruses tested are indicated above the lanes; labeling corresponds to that in panels A and B. The asterisk indicates a nonspecific band detected by anti-3A antibody used as loading control, and the black arrowhead indicates viral plus-strand RNA. The bands corresponding to 3AB/3A proteins, as well as 28S and 18S rRNAs are indicated to the right of the gels in panels C and D. The positions of molecular mass markers (in kilodaltons) are shown to the left of the gel in panel C. (E) One-step growth curves in Sk-N-Mc cells of CBV-ΔZ (ΔZ) (solid white bars), wt CBV3 (solid gray bars), and their [R35M:D51G] variants (hatched bars in identical colors).
Next, we assessed translation and RNA replication of CBV-RICO, RICO-ΔZ, and CBV3 with [R35M:D51G] in infected Sk-N-Mc cells (Fig. 5C and D). At 8 h p.i., viral translation was abundant with wt CBV3, barely detectable with CBV-RICO, and not apparent with RICO-ΔZ (Fig. 5C). Translation was enhanced with CBV-RICO and RICO-ΔZ containing [R35M:D51G], but not their wt CBV3 counterpart. We also conducted strand-specific Northern blots of total cellular RNA from Sk-N-Mc cells infected with the constructs listed above at 8 h p.i. (Fig. 5D). We were unable to detect minus-strand RNA for any of the constructs (data not shown). Analyses of plus-strand RNA replication are consistent with the effects observed on viral translation: [R35M:D51G] significantly elevates viral genome replication of CBV-RICO and RICO-ΔZ in Sk-N-Mc cells. CBV3 containing [R35M:D51G] yielded more plus-strand viral RNA than wt CBV3 in Sk-N-Mc cells, but this did not lead to increased viral translation (Fig. 5C) or progeny yield (Fig. 5B).
Thus, adaptation by [R35M:D51G] occurred with RICO-ΔZ as well as with CBV-RICO (Fig. 5B). However, proportionally, the adaptation mutations benefited the HRV2 IRES:ΔZ combination to a greater extent than the HRV2 IRES alone (Fig. 5B). To test whether [R35M:D51G] compensates for ΔZ alone, we examined CBV-ΔZ containing [R35M:D51G]. As with CBV-RICO or RICO-ΔZ, this produced a slightly enlarged plaque phenotype in HeLa cells (Fig. 4), although growth kinetics in HeLa cells were unaffected (data not shown). Growth analysis of wt CBV3, CBV-ΔZ, and their [R35M:D51G] counterparts in Sk-N-Mc cells showed that ΔZ on its own diminishes CBV3 propagation and that this defect is fully compensated by [R35M:D51G] (Fig. 5E). Indeed, CBV-ΔZ containing [R35M:D51G] grows in Sk-N-Mc cells as efficiently as wt CBV3 does (Fig. 5E). Our findings suggest that adaptive mutations in 3A to 3C separately complement growth defects due to the HRV2 IRES and ΔZ and synergistically promote propagation of viruses containing the HRV2 IRES:ΔZ combination.
Adaptation mutations in 3A to 3C do not affect propagation potential of wt CBV3.
Does adaptation compensate for 5′ and 3′UTR cis-acting elements deficient in Sk-N-Mc cells or does it generally improve growth of CBV3? To answer this question, we inserted [P17S], [R35T], and [R35M:D51G] into CBV3 (Fig. 4). All mutations tested had comparable effects on the growth of wt CBV3 in Sk-N-Mc cells, but only the results for [R35M:D51G] are shown (Fig. 5). None of the mutations tested affected CBV3 growth in HeLa or Sk-N-Mc cells (Fig. 5A, B, and E). We observed a slightly reduced plaque phenotype with the mutants compared to that of wt CBV3 (Fig. 4). Plaque size was also reduced for CBV-RICO and RICO-ΔZ carrying [P17S] (although this mutation improves growth in Sk-N-Mc cells), but both [R35T] and [R35M:D51G] substantially increase plaque sizes of CBV-RICO and RICO-ΔZ. Our findings show that the adaptation mutations do not affect propagation of CBV3 containing its cognate UTRs in Sk-N-Mc cells.
DISCUSSION
We analyzed adaptation of CBV3 constructs with recombinant 5′ and 3′UTRs conferring growth defects in Sk-N-Mc neuroblastoma cells. Our work shows the following. (i) The IRES and 3′UTR coordinately control viral translation and replication (10). (ii) The efficiency of viral UTRs in directing viral propagation varies in a cell type-specific manner. (iii) Cell type-specific growth deficits due to incompatible UTRs are complemented by mutations in viral proteins 3A/3AB and 3C/3CD. (iv) Mutants 3A to 3C separately complement a “neuron-incompetent” IRES and 3′UTR and synergistically rescue deficits conferred by their combination in a CBV3 background.
Adaptation of four constructs featuring various SLD Z defects (outright deletion [RICO-ΔZ], the PV 3′UTR naturally devoid of SLD Z [RICO-PV], weakening [mutant 4] or deletion [mutant 5] of the proximal stem) each yielded mutations in 3A and/or 3C. Curiously, adapted mutant 5 viruses predominantly featured mutations in 3C (either alone or combined with mutations in 3A), while adapted mutant 4 viruses exclusively contained mutations in 3A. Moreover, the particular mutations in 3A of both adapted viruses were nonoverlapping. This is surprising, considering the minor differences of mutants 4 and 5 in the proximal stem of SLD Z (Fig. 1B). Our data suggest an intricate relationship of viral nonstructural proteins with both viral UTRs, which is adjusted by adaptation mutations in these proteins in nonpermissive cells.
3A or its precursor, 3AB, has been implicated in diverse functions. A conserved hydrophobic domain mediates homodimerization (25) and membrane insertion (53) and tethers replication complexes to the characteristic saccular membrane network forming in EV-infected cells (reviewed in reference 13). Accordingly, mutations in the hydrophobic domain have been associated with RNA replication deficits (17, 18, 51). In addition, 3AB and 3CD have been demonstrated to bind to both the 3′UTR and cloverleaf of the PV genome (3, 21). As a further sign of involvement in RNA replication, 3AB directly interacts with 3D and stimulates its function (25, 36, 44). 3AB probably is the precursor for VPg, the primer for minus-strand synthesis (38), and its interaction with 3D could position VPg for efficient priming. Relating to its penchant for membrane insertion, 3A mediates vesicular transport and protein secretion defects (11, 12).
All 3A adaptation mutations described here occur in an amino-terminal portion that is highly conserved among rhinoviruses and EVs (Fig. 3) (49, 55). Thus, all adaptation mutations occurred in parts of the protein not implicated in membrane tethering, protein-protein interacting, or vesicular transport rearranging functions (56). Mutations in 3A/3AB affecting these basic roles likely would generally alter viral growth, rather than produce cell type-specific effects in Sk-N-Mc cells. We therefore assume that genetic adaptation involves a separate function of 3A/3AB that is likely based on RNA-protein interactions. Since we did observe dual adaptation mutations in 3A to 3C, this function may involve cooperative effects of 3AB and/or 3CD (interaction of these proteins has been reported previously [32]). Adaptive mutations in 3C occurred in regions distant from the proteolytic active site. Amino acids 49 and 51 in PV 3C are situated in the reverse turn connecting two neighboring β-strands (33) and do not contribute significantly to the secondary structure. The minor [F4S] mutation (Table 3) disrupts predicted intermolecular interactions within the 3C asymmetric dimer. Interestingly, the minor mutation [V54A] was shown previously to affect processing of the PV 3C:3D cleavage (9).
Our sequencing data show that viruses emerging after eight passages in Sk-N-Mc cells are heterogeneous populations, sometimes including wt and adapted variants. Mixed populations of mutant 5 grew more efficiently in Sk-N-Mc cells than homogeneous virus with the dominant adaptation mutations [R35M:D51G], suggesting that heterogeneous virus swarms are more adept at adjusting to the growth conditions within a particular cell type. The importance of genetic adaptability in EV pathogenesis has recently come under scrutiny (40, 41, 54), and our findings may demonstrate possible mechanisms of tissue type-specific adaptation of genetically diverse (quasispecies) viruses.
Genetic adaptation in our study is a cell type-specific phenomenon, evident in cells of neuronal origin. A recurrent problem in studies of neuronal EV growth phenotypes is the choice of cell lines. While there is no optimal solution, a number of tissue culture systems have been devised. These include neuroblastoma cell lines that recapitulate neuron-deficient growth of the live attenuated PV vaccines (1, 26) and laboratory-generated attenuated recombinants (2, 19, 20). However, they are cancerous cells spontaneously interconverting between fibroblast- and neuron-like phenotypes upon prolonged passage (45, 47). We previously investigated HEK-293 cells, shown to be of neuroblastic origin by their originator (48), as an alternative (8). We decided against the use of HEK-293 cells in this study, because of the unfavorable growth properties of CBV3 in these cells (data not shown). CBV-RICO, due to repression of the HRV2 IRES (30, 31), barely propagates in these cells and even after >30 h p.i. produces no cytopathic effect at all. However, even the growth kinetics and cytopathic effect of CBV3 are severely depressed compared to those of Sk-N-Mc cells. The reasons for this are unknown; HEK-293 cells express the CBV/adenovirus receptor CAR (34), excluding attachment and entry defects. Analyses in HEK-293 cells would require drawn-out procedures in a tissue culture system that is not representative for EV growth in a susceptible host. Moreover, we originally described the cooperative effect of the HRV2 IRES:ΔZ on CBV3 growth in Sk-N-Mc neuroblastoma cells (10). To minimize the type-specific interconversion problem of Sk-N-Mc cells, we used cultures at passage numbers below 5 for all growth curve and passaging experiments.
The role of the 3′UTR in viral replication and pathogenesis has been studied before. EVs are surprisingly adept at accepting foreign 3′UTRs (10, 46). However, disruption of the authentic EV/HRV 3′UTR structure yielded viruses with substantial propagation deficits, indicating that the structural arrangement of these elements is important for proper growth (46). Our research demonstrates that the absence of growth deficits in HeLa cells (used in similar investigations [46]) does not necessarily exclude a role for 3′UTR structure in viral propagation. Similarly, a mutant PV lacking the entire 3′UTR exhibited a minor replication defect in HeLa cells but major growth reduction in Sk-H-Sh neuroblastoma cells (6). This is also evident from earlier studies with CBV-ΔZ (29). CBV-ΔZ grew with wt kinetics in African green monkey kidney cells but was significantly less virulent in infected mice, evident by decreased pancreatic and cardiac replication/histological lesions (29). The authors of that study did not attempt to adapt their ΔZ deletion variant.
The cell type-specific performance of viral UTRs and their interaction with viral replication factors are obvious parameters dictating EV virulence and, ultimately, pathogenesis. What then are the mechanisms controlling viral translation and genome replication? Our findings suggest a scenario of puzzling intricacy: cell type-specific UTR competence suggests the involvement of host factors, e.g., in a neuron-specific RNP producing polysome exclusion of viral templates by the DRBP76-NF45 heterodimer (30, 31). Host protein interactions with viral RNA may compete with viral RNA-binding proteins (e.g., 3AB/3CD), may interfere with proper template configuration, or may specifically repress translation initiation (31) or genome replication. Both genome replication and translation may involve long-range interactions of distant UTR elements, possibly resulting in template circularization. Thus, RNA-protein (host or viral) interactions at either terminus may disturb proper function of both UTRs by interfering with long-range contacts. The main viral proteins implicated in interactions with the viral genome, 3AB and 3CD, also interact with each other and mediate association with cellular substructures. They are multifunctional factors giving rise to proteolytic products with redundant or nonoverlapping functions. The functions of all components involved in the viral life cycle are dynamic, since the conditions for translation and genome replication evolve with the progress of viral changes of the intracellular milieu. Considering this thicket of interrelating factors, it is inconceivable that manipulation of any part of this system (e.g., ΔZ in our study) causes solitary defects of distinct viral functions that can be empirically dissected. In other words, translation of a self-replicating template may be controlled by the very fact that it is replicating and vice versa.
Acknowledgments
We used an infectious CBV3(0) clone which was generously provided by N. Chapman (University of Nebraska, Omaha). Anti-CBV 3A antibodies were kindly provided by K. Klingel (University of Tuebingen).
This research was supported by PHS grants CA87537 and NS20023 (M.G.), an Underrepresented Minority Supplement to CA87537 (P.F.D.S.). We gratefully acknowledge a grant from Alex's Lemonade Stand Foundation.
Footnotes
Published ahead of print on 30 May 2007.
REFERENCES
- 1.Agol, V. I., S. G. Drozdov, T. A. Ivannikova, M. S. Kolesnikova, M. B. Korolev, and E. A. Tolskaya. 1989. Restricted growth of attenuated poliovirus strains in cultured cells of a human neuroblastoma. J. Virol. 63:4034-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agol, V. I., E. V. Pilipenko, and O. R. Slobodskaya. 1996. Modification of translational control elements as a new approach to design of attenuated picornavirus strains. J. Biotechnol. 44:119-128. [DOI] [PubMed] [Google Scholar]
- 3.Andino, R., G. E. Rieckhof, P. L. Achacoso, and D. Baltimore. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andino, R., G. E. Rieckhof, and D. Baltimore. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63:369-380. [DOI] [PubMed] [Google Scholar]
- 5.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, D. M., S. E. Kauder, C. T. Cornell, G. M. Jang, V. R. Racaniello, and B. L. Semler. 2004. Cell-dependent role for the poliovirus 3′ noncoding region in positive-strand RNA synthesis. J. Virol. 78:1344-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, S., M. Mulvey, I. Mohr, and M. Gromeier. 2007. Attenuation of herpes simplex virus neurovirulence with picornavirus cis-acting genetic elements. J. Virol. 81:791-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, S. A., J. Lin, E. Y. Dobrikova, and M. Gromeier. 2005. Genetic determinants of cell type-specific poliovirus propagation in HEK 293 cells. J. Virol. 79:6281-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dewalt, P. G., and B. L. Semler. 1987. Site-directed mutagenesis of proteinase 3C results in a poliovirus deficient in synthesis of viral RNA polymerase. J. Virol. 61:2162-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobrikova, E., P. Florez, S. Bradrick, and M. Gromeier. 2003. Activity of a type 1 picornavirus internal ribosomal entry site is determined by sequences within the 3′ nontranslated region. Proc. Natl. Acad. Sci. USA 100:15125-15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doedens, J. R., T. H. Giddings, Jr., and K. Kirkegaard. 1997. Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J. Virol. 71:9054-9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doedens, J. R., and K. Kirkegaard. 1995. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14:894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egger, D., R. Gosert, and K. Bienz. 2004. Role of cellular structures in viral RNA replication, p. 247-254. In B. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, DC.
- 14.Florez, P. M., O. M. Sessions, E. J. Wagner, M. Gromeier, and M. A. Garcia-Blanco. 2005. The polypyrimidine tract binding protein is required for efficient picornavirus gene expression and propagation. J. Virol. 79:6172-6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamarnik, A. V., and R. Andino. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74:2219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giachetti, C., S. S. Hwang, and B. L. Semler. 1992. cis-acting lesions targeted to the hydrophobic domain of a poliovirus membrane protein involved in RNA replication. J. Virol. 66:6045-6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giachetti, C., and B. L. Semler. 1991. Role of a viral membrane polypeptide in strand-specific initiation of poliovirus RNA synthesis. J. Virol. 65:2647-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gromeier, M., L. Alexander, and E. Wimmer. 1996. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc. Natl. Acad. Sci. USA 93:2370-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gromeier, M., B. Bossert, M. Arita, A. Nomoto, and E. Wimmer. 1999. Dual stem loops within the poliovirus internal ribosomal entry site control neurovirulence. J. Virol. 73:958-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris, K. S., W. Xiang, L. Alexander, W. S. Lane, A. V. Paul, and E. Wimmer. 1994. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 269:27004-27014. [PubMed] [Google Scholar]
- 22.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang, S. K., H. G. Krausslich, M. J. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitamura, N., B. L. Semler, P. G. Rothberg, G. R. Larsen, C. J. Adler, A. J. Dorner, E. A. Emini, R. Hanecak, J. J. Lee, S. van der Werf, C. W. Anderson, and E. Wimmer. 1981. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature 291:547-553. [DOI] [PubMed] [Google Scholar]
- 25.Lama, J., A. V. Paul, K. S. Harris, and E. Wimmer. 1994. Properties of purified recombinant poliovirus protein 3AB as substrate for viral proteinases and as co-factor for RNA polymerase 3Dpol. J. Biol. Chem. 269:66-70. [PubMed] [Google Scholar]
- 26.La Monica, N., and V. R. Racaniello. 1989. Differences in replication of attenuated and neurovirulent polioviruses in human neuroblastoma cell line SH-SY5Y. J. Virol. 63:2357-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melchers, W. J., J. M. Bakkers, H. J. Bruins Slot, J. M. Galama, V. I. Agol, and E. V. Pilipenko. 2000. Cross-talk between orientation-dependent recognition determinants of a complex control RNA element, the enterovirus oriR. RNA 6:976-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melchers, W. J., J. G. Hoenderop, H. J. Bruins Slot, C. W. Pleij, E. V. Pilipenko, V. I. Agol, and J. M. Galama. 1997. Kissing of the two predominant hairpin loops in the coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J. Virol. 71:686-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merkle, I., M. J. van Ooij, F. J. van Kuppeveld, D. H. Glaudemans, J. M. Galama, A. Henke, R. Zell, and W. J. Melchers. 2002. Biological significance of a human enterovirus B-specific RNA element in the 3′ nontranslated region. J. Virol. 76:9900-9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merrill, M. K., E. Y. Dobrikova, and M. Gromeier. 2006. Cell-type-specific repression of internal ribosome entry site activity by double-stranded RNA-binding protein 76. J. Virol. 80:3147-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merrill, M. K., and M. Gromeier. 2006. The double-stranded RNA binding protein 76:NF45 heterodimer inhibits translation initiation at the rhinovirus type 2 internal ribosome entry site. J. Virol. 80:6936-6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molla, A., K. S. Harris, A. V. Paul, S. H. Shin, J. Mugavero, and E. Wimmer. 1994. Stimulation of poliovirus proteinase 3Cpro-related proteolysis by the genome-linked protein VPg and its precursor 3AB. J. Biol. Chem. 269:27015-27020. [PubMed] [Google Scholar]
- 33.Mosimann, S. C., M. M. Cherney, S. Sia, S. Plotch, and M. N. James. 1997. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J. Mol. Biol. 273:1032-1047. [DOI] [PubMed] [Google Scholar]
- 34.Nemerow, G. R. 2000. Cell receptors involved in adenovirus entry. Virology 274:1-4. [DOI] [PubMed] [Google Scholar]
- 35.Parsley, T. B., J. S. Towner, L. B. Blyn, E. Ehrenfeld, and B. L. Semler. 1997. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124-1134. [PMC free article] [PubMed] [Google Scholar]
- 36.Paul, A. V., X. Cao, K. S. Harris, J. Lama, and E. Wimmer. 1994. Studies with poliovirus polymerase 3Dpol. Stimulation of poly(U) synthesis in vitro by purified poliovirus protein 3AB. J. Biol. Chem. 269:29173-29181. [PubMed] [Google Scholar]
- 37.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paul, A. V., J. H. van Boom, D. Filippov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 39.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 40.Pfeiffer, J. K., and K. Kirkegaard. 2006. Bottleneck-mediated quasispecies restriction during spread of an RNA virus from inoculation site to brain. Proc. Natl. Acad. Sci. USA 103:5520-5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfeiffer, J. K., and K. Kirkegaard. 2005. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog. 1:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pilipenko, E. V., S. V. Maslova, A. N. Sinyakov, and V. I. Agol. 1992. Towards identification of cis-acting elements involved in the replication of enterovirus and rhinovirus RNAs: a proposal for the existence of tRNA-like terminal structures. Nucleic Acids Res. 20:1739-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pilipenko, E. V., K. V. Poperechny, S. V. Maslova, W. J. Melchers, H. J. Slot, and V. I. Agol. 1996. Cis-element, oriR, involved in the initiation of (−) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary (′kissing') interactions. EMBO J. 15:5428-5436. [PMC free article] [PubMed] [Google Scholar]
- 44.Plotch, S. J., and O. Palant. 1995. Poliovirus protein 3AB forms a complex with and stimulates the activity of the viral RNA polymerase, 3Dpol. J. Virol. 69:7169-7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rettig, W. J., B. A. Spengler, P. G. Chesa, L. J. Old, and J. L. Biedler. 1987. Coordinate changes in neuronal phenotype and surface antigen expression in human neuroblastoma cell variants. Cancer Res. 47:1383-1389. [PubMed] [Google Scholar]
- 46.Rohll, J. B., D. H. Moon, D. J. Evans, and J. W. Almond. 1995. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J. Virol. 69:7835-7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ross, R. A., B. A. Spengler, and J. L. Biedler. 1983. Coordinate morphological and biochemical interconversion of human neuroblastoma cells. J. Natl. Cancer Inst. 71:741-747. [PubMed] [Google Scholar]
- 48.Shaw, G., S. Morse, M. Ararat, and F. L. Graham. 2002. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 16:869-871. [DOI] [PubMed] [Google Scholar]
- 49.Strauss, D. M., L. W. Glustrom, and D. S. Wuttke. 2003. Towards an understanding of the poliovirus replication complex: the solution structure of the soluble domain of the poliovirus 3A protein. J. Mol. Biol. 330:225-234. [DOI] [PubMed] [Google Scholar]
- 50.Teterina, N. L., D. Egger, K. Bienz, D. M. Brown, B. L. Semler, and E. Ehrenfeld. 2001. Requirements for assembly of poliovirus replication complexes and negative-strand RNA synthesis. J. Virol. 75:3841-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teterina, N. L., M. S. Rinaudo, and E. Ehrenfeld. 2003. Strand-specific RNA synthesis defects in a poliovirus with a mutation in protein 3A. J. Virol. 77:12679-12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Todd, S., J. S. Towner, D. M. Brown, and B. L. Semler. 1997. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J. Virol. 71:8868-8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Towner, J. S., T. V. Ho, and B. L. Semler. 1996. Determinants of membrane association for poliovirus protein 3AB. J. Biol. Chem. 271:26810-26818. [DOI] [PubMed] [Google Scholar]
- 54.Vignuzzi, M., J. K. Stone, J. J. Arnold, C. E. Cameron, and R. Andino. 2006. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439:344-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wessels, E., R. A. Notebaart, D. Duijsings, K. Lanke, B. Vergeer, W. J. Melchers, and F. J. van Kuppeveld. 2006. Structure-function analysis of the coxsackievirus protein 3A: identification of residues important for dimerization, viral RNA replication, and transport inhibition. J. Biol. Chem. 281:28232-28243. [DOI] [PubMed] [Google Scholar]
- 56.Xiang, W., A. Cuconati, D. Hope, K. Kirkegaard, and E. Wimmer. 1998. Complete protein linkage map of poliovirus P3 proteins: interaction of polymerase 3Dpol with VPg and with genetic variants of 3AB. J. Virol. 72:6732-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeng, Y., E. J. Wagner, and B. R. Cullen. 2002. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9:1327-1333. [DOI] [PubMed] [Google Scholar]