Abstract
It is not understood how immune inflammation influences the pathogenesis of severe acute respiratory syndrome (SARS). One area of strong controversy is the role of interferon (IFN) responses in the natural history of SARS. The fact that the majority of SARS patients recover after relatively moderate illness suggests that the prevailing notion of deficient type I IFN-mediated immunity, with hypercytokinemia driving a poor clinical course, is oversimplified. We used proteomic and genomic technology to systematically analyze host innate and adaptive immune responses of 40 clinically well-described patients with SARS during discrete phases of illness from the onset of symptoms to discharge or a fatal outcome. A novel signature of high IFN-α, IFN-γ, and IFN-stimulated chemokine levels, plus robust antiviral IFN-stimulated gene (ISG) expression, accompanied early SARS sequelae. As acute illness progressed, SARS patients entered a crisis phase linked to oxygen saturation profiles. The majority of SARS patients resolved IFN responses at crisis and expressed adaptive immune genes. In contrast, patients with poor outcomes showed deviated ISG and immunoglobulin gene expression levels, persistent chemokine levels, and deficient anti-SARS spike antibody production. We contend that unregulated IFN responses during acute-phase SARS may culminate in a malfunction of the switch from innate immunity to adaptive immunity. The potential for the use of the gene signatures we describe in this study to better assess the immunopathology and clinical management of severe viral infections, such as SARS and avian influenza (H5N1), is therefore worth careful examination.
Severe acute respiratory syndrome coronavirus (SARS CoV) causes a spectrum of disease ranging from flu-like symptoms and viral pneumonia to acute respiratory distress syndrome and fatal outcomes (14, 16, 23, 31, 41). The mechanisms by which SARS CoV causes severe illness in humans are largely unknown. SARS CoV takes hold in the airways and other organs via its main putative receptor, angiotensin-converting enzyme 2 (ACE2), expressed on many cell types, including pneumocytes, enterocytes, and endothelial cells (19, 25, 32). SARS CoV appears to evade innate immunity during the first 10 days of infection during a period of widespread inflammation and steadily increasing viral load (39, 52). The consequent immune inflammation and hypercytokinemia, or “cytokine storm,” during the course of SARS has been illustrated (22, 27, 33, 37, 51), but the molecular and cellular basis of how SARS CoV impacts host defense, resulting in a poor prognosis, is not understood. One particular area of controversy is the role of interferon (IFN) responses in human host immune responses against SARS CoV.
Type I IFNs, such as IFN-α and -β, are critical to innate immune responses against viral and other microbial infections and act in concert with IFN-γ in the activation of antiviral IFN-stimulated genes (ISGs) and the immunomodulation of innate and adaptive immunity (3, 36, 42, 48). It has been proposed that deficient type I IFN responses may play a role in SARS pathogenesis (5, 8, 56). This hypothesis, however, has been largely based on in vitro studies. The fact that the majority of SARS patients recover after relatively moderate illness suggests that the notion of deficient type I IFN-mediated immunity and high expression of other cytokines driving a poor clinical course in vivo is oversimplified.
The emergence of new global health threats, such as avian influenza (H5N1), has refocused our attention on acquiring a better understanding of how emerging respiratory viruses can cause severe immunopathology in humans (35). To examine the extent of atypical IFN-mediated immune responses during severe respiratory disease in humans, we have analyzed global gene and protein expression profiles in SARS patients of different clinical evolutions with emphasis on characterizing type I and type II IFN responses and ISG signatures in concert with the development of innate and adaptive host immune responses.
MATERIALS AND METHODS
SARS patients.
Fifty Toronto-area SARS patients were enrolled without bias to age, sex, or previous medical history. SARS CoV infection in each patient was confirmed by positive PCR and/or seroconversion results. Ten healthy volunteers, five males and five females (median age, 28 years), were also enrolled. Informed consent was obtained from all subjects under the approval of the Research Ethics Boards of the University Health Network (UHN) and participating Toronto-area hospitals.
Specimen collection.
Peripheral blood was collected from SARS patients at admission to hospital and every 5 to 7 days thereafter until they were discharged or a fatal outcome occurred. Samples were stabilized and processed for further analysis within 2 to 3 h of collection. RNA was stabilized and purified by using Paxgene blood collection tubes and RNA kits (QIAGEN, Mississauga, ON, Canada). Plasma was obtained by centrifugation. Unless otherwise stated, replicate measurements were not often possible due to sample volume limitations.
Microarray analysis.
Detailed microarray procedures are posted at the UHN Microarray Facility website (http://www.microarrays.ca). Briefly, RNA was amplified by using MessageAMP antisense RNA kits (Ambion, Austin, TX). Sufficient RNA yields were obtained for 60 samples from 40 SARS patients at time points throughout illness (1 sample from 30 patients and 2 to 5 samples from 10 patients) and 10 samples from 10 healthy controls. UHN human single-spotted model 19k cDNA microarray slides containing 15,930 unique and sequence-verified expressed sequence tags were used for hybridizations. Samples were hybridized against amplified external reference RNA (Stratagene, La Jolla, CA) as a means to assess gene expression data derived from different experiments (20, 44). Each data set was filtered for bad spots, normalized with LOWESS (Sub-Grid), and log transformed (log2) using GeneTraffic version 3.1-4 (Iobion Informatics, La Jolla, CA) for further analysis with GeneLinker Platinum version 4.6 (Improved Outcomes, Kingston, Canada). Genes with missing values in over 20% of the data sets were removed. Missing values were estimated by the nearest-neighbors method. Agglomerative hierarchical clustering with Pearson correlation and average linkage distance metrics was used to organize data sets. GO annotation (http://www.geneontology.org), the DAVID Bioinformatics Database (12), the Interferon Stimulated Gene Database (13), and Genatlas (http://www.dsi.univ-paris5.fr/genatlas) were used to classify genes by related functional annotation and cellular expression. Ingenuity Pathway Analysis 5.0 software (Ingenuity Systems Inc., Redwood City, CA) was used to map and organize gene expression data into canonical pathways.
CBA.
Plasma proteins were assayed by human Th1/Th2 cytokine, inflammation, and chemokine cytometric bead array (CBA) kits (BD Biosciences, San Jose, CA) according to the manufacturer's protocols. Detection limits were 2 to 5 pg/ml. Results were generated using BD CBA analysis software.
IFN-α ELISA.
IFN-α was assayed using human IFN-α serum sample enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems Inc., Minneapolis, MN) according to the manufacturer's high-sensitivity protocol. Assay sensitivity was in the range of 5 to 500 pg/ml. Plasma samples were diluted 1:2 in phosphate-buffered saline.
Anti-SARS CoV spike ELISA.
SARS CoV spike-specific antibodies (Abs) were quantified in plasma by direct ELISA per the manufacturer's instructions (Imgenex, San Diego, CA). Assay sensitivity was in the range of 2 to 200 ng/100 μl. Samples were diluted 1:5 in phosphate-buffered saline.
QRT-PCR method.
Quantitative real-time PCR (QRT-PCR) was performed on amplified RNA by using an ABI-PRISM 7900HT sequence detection system and SYBR green PCR master mix (Applied Biosystems, Foster City, CA). One microgram of total RNA was reverse transcribed in a 20-μl reaction mixture. The standard curve and samples were run in triplicate. Each QRT-PCR was performed in a volume of 10 ml with 0.25 ml of cDNA, 1 ml primer, and 5 ml of SYBR green PCR master mix in ABI-PRISM optical 384 well plates. Primers specific for human glyceraldehyde-3-phosphate dehydrogenase mRNA were used to normalize samples. All primers were used at a final concentration of 500 nM. The 5′-3′ sequences of primer pairs were as follows: glyceraldehyde-3-phosphate dehydrogenase mRNA primers, ACC CAG AAG ACT GTG GAT GG (forward) and TTC TAG ACG GCA GGT CAG GT (reverse); CIG5 primers, CTG AGA GGG CCA GAT GAG AC (forward) and AGA AAT GGC TCT CCA CCT GA (reverse); DIABLO primers, GGA GCC AGA GCT GAG ATG AC (forward) and ATC TGT GCT TCT GCC AGC TT (reverse); G1P2 primers, GAA TTC CAG GTG TCC CTG AG (forward) and GCC CTT GTT ATT CCT CAC CA (reverse); IFNAR1 primers, GTG GAA CAG GAG CGA TGA GT (forward) and ATC TGA GCT TTG CGA AAT GG (reverse); MX1 primers, GGG AAG GAA TGG GAA TCA GT (forward) and ATG CTG AGA GCC TCT GTG GT (reverse); IFNGR1 primers, TTG GAT TCC AGT TGT TGC TG (forward) and GGC TCT TCA CAG ACC ACC TC (reverse); MT1G primers, GCA AGT GCA AAG AGT GCA AA (forward) and CAG CTG CAC TTC TCC GAT G (reverse); PSME1 primers, AGA AGA AGG GGG AGG ATG AA (forward) and GCT GGT CAT CAG CTC AAA CA (reverse); superoxide dismutase 2 (SOD2) primers, GAC AAA CCT CAG CCC TAA CG (forward) and CCT TGC AGT GGA TCC TGA TT (reverse); SERPING1 primers, AAC ACT ACC CCG CAT CAA AG (forward) and CTG CAC TTC AAA GAC CAG CA (reverse); CCR1 primers, AGC CTT CAC TTT CCT CAC GA (forward) and AGG GGG TCC AAA AGA GAA AA (reverse); LTBR primers, AAG CCG AGC TCA AAG ATG AA (forward) and TCA GCA TGG TTC CTG ACA TC (reverse); MT2A primers, TCC TGC AAA TGC AAA GAG TG (forward) and CAG CAG CTG CAC TTG TCC (reverse); CCL2 primers, TCT GTG CCT GCT GCT CAT AG (forward) and TGG AAT CCT GAA CCC ACT TC (reverse); CD58 primers, TCA GCT GTT TTT CCC AAC AA (forward) and TTT GGC GAT TCC ATT TCA TAC (reverse); VAV2 primers, TCA AGG TGC ATC ACA GCT TC (forward) and CCA TCC TGG ACC TTC AGT GT (reverse); MT1B primers, TCC TGC AAG TGC AAA GAG TG (forward) and CTG ATG AGC CTT TGC AGA CA (reverse); CXCL10 primers, TCC ACG TGT TGA GAT CAT TGA (forward) and TCT TGA TGG CCT TCG ATT CTG (reverse); CXCR3 primers, GGT GCC CTC TTC AAC ATC AAC (forward) and GGT GGC ATG AAC TAT GTT CAG GTA (reverse).
Statistical methods.
Unless otherwise stated, the Mann-Whitney rank sum test for two independent populations was used for statistical analysis via SPSS for Windows V13.0 software (SPSS Inc., Chicago, IL). The statistical significance of discriminating genes in the microarray experiments was determined by analysis of variance (ANOVA) (F-test, assuming Gaussian distribution). For both tests, a P value of ≤0.05 was considered significant.
Microarray data accession number.
Newly obtained data sets are available at the GEO microarray data repository (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE5972.
RESULTS
Clinical evolution of SARS.
All SARS patients presented to a hospital within a median of 2 days since the onset of symptoms (DSO), with fever (Table 1) and various flu-like symptoms. Symptoms progressed rapidly during acute SARS infection, with fever peaking at a median of 4 DSO, chest radiographic involvement peaking at a median of 6 DSO, and the arterial oxygen saturation (SO2) nadir occurring at a median of 8 DSO in surviving patients (Table 1). Nasopharyngeal viral titers peaked at a median of 7.5 DSO, followed shortly by peak viral titers in stools at a median of 12 DSO (Table 1).
TABLE 1.
SARS patient demographics and treatment and clinical characteristics
| Parametera | Value for patient group
|
||
|---|---|---|---|
| All | Nonsevere SARS | Severe SARS | |
| SARS patient demographics and disease course | |||
| Total no. of patients | 50 | 30 | 20 |
| No. of males/no. of females | 22/28 | 10/20 | 12/8 |
| Median yr of age (range) | 48 (24-83) | 44 (24-80) | 63 (31-83) |
| Median no. of days of illness (range) | 16 (7-136) | 15 (7-27) | 38 (15-136)b |
| No. of patients with: | |||
| Increased O2 supportc (%) | 20 (40) | 0 (0) | 20 (100) |
| Mechanical intubation (%) | 16 (32) | 0 (0) | 16 (80) |
| No. of patients who died (%) | 6 (12) | 0 (0) | 6 (32) |
| SARS patient clinical parameters | |||
| Incidence of fever at presentation (%) | 50 (100) | 30 (100) | 20 (100) |
| Median DSO for peak temp (range) | 4 (3-9) | 3 (3-8) | 4 (3-9) |
| Median no. of days of duration of fever of ≥38°C (range) | 11 (3-36) | 8 (3-15) | 18 (6-36)b |
| No. of patients with CXR positive for infiltrate at any time (%) | 47 (94) | 27 (90) | 20 (100) |
| Median DSO for peak chest radiographic involvement (range) | 6 (1-21) | 6 (1-21) | 7 (2-12) |
| Median no. of days from first positive CXR to improvement (range) | 10 (1-108) | 6 (1-23) | 20 (12-108)b |
| Median DSO for SO2 nadir (range) | 8 (4-24) | 8 (4-18) | 9 (5-24)b |
| Median % SO2 at nadir (range) | 93 (71-96) | 94 (92-96) | 82 (71-90)b |
| No. of patients with severe hypoxemia (i.e., SO2 < 91%) (%) | 19 (38) | 0 (0) | 19 (95) |
| Median duration of hypoxemia (SO2 ≤ 96%) in days (range) | 16 (8-35) | 13 (8-28) | 30 (15-35)b |
| DSO for median peak viral titer (range) in: | |||
| Stool | 12 (5-18) | 12 (5-18) | 13.5 (6-16) |
| Nasopharynx | 7.5 (7-8) | 7.5 (7-8) | 7.5 (7-8) |
| Incidence of lymphopenia (ALC ≤ 1,000/mm3) (%) | 48 (96) | 28 (93) | 20 (100) |
| Median DSO for peak lymphopenia (range) | 8 (3-23) | 5 (3-17) | 14 (7-23) |
| Median no. of days for duration of lymphopenia (range) | 9 (3-60) | 7 (3-18) | 27 (7-60)b |
| SARS patient treatment | |||
| No. of patients given corticosteroid treatment (%) | 37 (74) | 17 (57) | 20 (100) |
| Median DSO of first corticosteroid dose (range) | 6 (2-13) | 7 (2-13) | 6 (2-12) |
| Median no. of days of corticosteroid treatment (range) | 8 (3-114) | 8 (3-18) | 9 (3-114)b |
| Oral prednisone treatment | |||
| No. of patients given treatment (%) | 26 (52) | 17 (57) | 9 (45) |
| Median peak daily dosage in mg (range) | 100 (50-200) | 100 (50-200) | 100 (50-200) |
| Intravenous methylprednisolone treatment | |||
| No. of patients given treatment (%) | 27 (54) | 8 (27) | 19 (95) |
| Median peak daily dosage in mg (range) | 125 (50-500) | 80 (80-500) | 375 (50-500) |
DSO, days since onset of symptoms; CXR, chest X-ray; ALC, absolute lymphocyte count; SO2, arterial O2 saturation.
Data from surviving patients only.
Supplemental O2 rate of ≥4 liters/min or mechanical ventilation.
Given that SARS is primarily a respiratory tract infection hallmarked by progressive respiratory failure, we felt that O2 support and O2 saturation levels would be important patient variables in our model. We considered patients to have severe SARS if they required increased O2 support (mechanical ventilation and/or supplemental oxygen support of ≥4 liters/min via nasal prong) at any time during their clinical course (Table 1). As shown in Table 1, severe-SARS patients were significantly older than nonsevere-SARS patients (medians of 63 years of age and 44 years of age, respectively; P ≤ 0.05) and, among survivors, had significantly longer disease courses (>2-fold; P ≤ 0.05). We noted in our retrospective analysis that all patients with severe SARS had SO2 levels that fell below 91% within 8 DSO. Conversely, all nonsevere-SARS cases experienced abnormal SO2 levels (≤96%) within 8 DSO but never had a drop in SO2 levels below 91% throughout their clinical course (Table 1). Our retrospective analysis of O2 saturation in nonsevere and severe-SARS patients appeared to be in line with hypoxemia thresholds previously proposed in prognostic models of O2 saturation and SARS outcome, i.e., an SO2 of <95% for hypoxemia due to SARS (2, 10) and an SO2 of <90% as criteria for severe SARS (57). Our data further indicated that a crisis phase began in SARS patients by 8 DSO (also corresponding to the median SO2 nadir for surviving patients), at which time patients that maintained their oxygen saturations completely recovered and returned to normal SO2 after a duration of a median 13 days for nonsevere-SARS patients and a median 30 days for severe-SARS patients (Table 1). However, 80% of severe-SARS patients ultimately required mechanical ventilation during crisis, and 32% of severe-SARS patients died (Table 1).
Microarray analysis of SARS disease course.
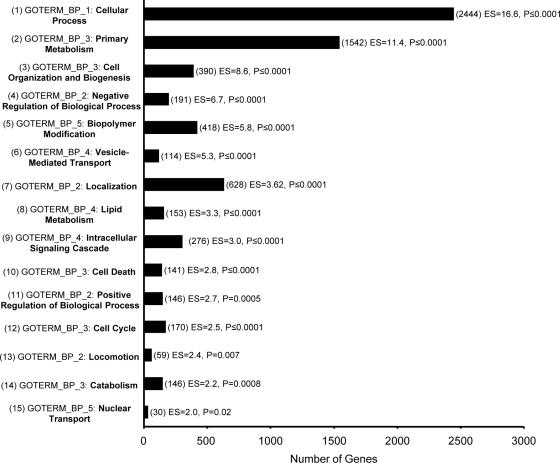
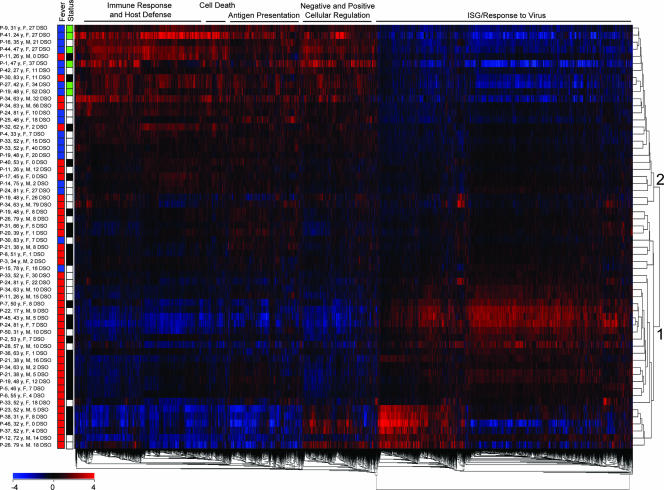
Examining host responses throughout the SARS disease course required a framework derived from objective clinical parameters. We first performed ANOVA (F-test) on all SARS data sets based on the presence or absence of a fever of ≥38°C at the time of patient sampling as an indicator of acute viral illness, which identified 5,555 significantly different genes between the two groups (P ≤ 0.05). Figure 1 shows the top 15 functional annotation clusters, in order of enrichment score, identified in a general analysis using the DAVID Bioinformatics Database (12). More-specific information was obtained following a two-way hierarchical gene clustering analysis as shown in Fig. 2. First, two clusters of SARS patient gene profiles were noted (Fig. 2). The majority of samples in cluster 1 were from SARS patients with fever (32/34) and in the precrisis phase of illness (21/34), while the majority of samples in cluster 2 were from patients who had resolved fever (16/26) and were either in the crisis phase of SARS or being discharged from the hospital (21/26). Second, five gene clusters of related functions corresponding to immune response and host defense, cell death, antigen presentation, cellular regulation, and ISG/response to virus were identified by DAVID analysis (Fig. 2). Most notably, the ISG/response-to-virus gene cluster was found to be strongly upregulated in samples from cluster 1, i.e., those from precrisis SARS patients with fever, and strongly downregulated in samples from cluster 2, i.e., those from SARS patients at crisis or discharge from the hospital and who had resolved fever. On the other hand, the immune response and host defense, cell death, antigen presentation, and cellular regulation gene clusters were strongly upregulated after SARS patients resolved fever (Fig. 2). The uppermost gene expression profiles in cluster 1 and the bottommost profiles in cluster 2 by position, as shown in Fig. 2, most likely represented SARS patients in transition between these two groups. Interestingly, all precrisis SARS patients had fever, while resolution of fever was associated with either discharge from hospital or the SARS crisis phase (Fig. 2). Indeed, six SARS patients who had resolved fever were receiving increased O2 support at the time of sampling (data not shown). To better understand the association of individual genes and gene families with the clinical evolution of SARS, we decided to use SARS phase and patient SO2 profiles as more-specific variables.
FIG. 1.
General functional annotation clusters identified by microarray analysis of SARS patients with or without fever. A total of 5,555 genes were identified by ANOVA (F-test, P < 0.05) based on the presence or absence of a fever of ≥38°C from 60 data sets for 40 SARS patients throughout illness and were uploaded to the DAVID Bioinformatics Database. A total of 4,559 genes were recognized and mapped for functional annotation analysis. The GO term (GOTERM) member and number of genes associated with the largest category of genes within the cluster are shown for each of the top 15 functional annotation clusters in order of enrichment score (ES). Note that functional annotation clustering reduces, but does not eliminate, overlapping GO annotation, so the total number of genes displayed may exceed the number uploaded. P values refer to a one-tailed Fisher's exact probability value used for gene enrichment analysis by the DAVID system.
FIG. 2.
Two-way hierarchical cluster analysis of microarray data sets from SARS patients with or without fever. A total of 5,555 genes were identified by ANOVA (F-test, P < 0.05) based on the presence (red boxes in the “Fever” column) or absence (blue boxes in the “Fever” column) of a fever of ≥38°C from 60 data sets for 40 SARS patients throughout illness. SARS patient data sets were normalized gene by gene against the means for healthy controls (n = 10). Patient data sets were analyzed by two-way hierarchical clustering (red boxes indicate upregulation and blue boxes indicate downregulation of genes of the indicated functions) and labeled by the presence or absence of fever. The general status of a SARS patient (for the “Status” column, black boxes indicate the precrisis phase, white boxes indicate crisis and green boxes indicate patients at discharge) is also noted as defined in the text. The major GO terms according to DAVID for five clusters have been placed above the heat map, and two patient clusters are labeled as described in the text.
We therefore classified SARS patient samples based on the phase of illness (precrisis or crisis) and SO2 at the time of sample collection for additional ANOVA. Also, the variable of corticosteroid treatment during precrisis SARS was considered, since 74% of all SARS patients were ultimately treated with corticosteroids during the crisis phase (Table 1). Group 1 included 14 samples from 14 non-corticosteroid-treated, precrisis SARS patients with a median of 2 DSO and a median age of 51 years. Group 2 consisted of seven samples from seven corticosteroid-treated precrisis SARS patients with a median of 5 DSO and a median age of 52 years. All patients in groups 1 and 2 exhibited fever and an SO2 of ≥91% at the time and were not receiving increased O2 support. Group 3 included 13 samples from 13 SARS patients classified as being at crisis with a median of 10 DSO and a median age of 57 years. There were no significant differences in age between the three groups of SARS patients. Seven patients in group 3 (P-12, P-24, P-26, P-28, P-33, P-34, and P-50) had severe SARS as defined above. Despite increased O2 support, five of these seven SARS patients (P-12, P-24, P-28, P-33, and P-50) had an SO2 of <91% at the time of sampling, and three (P-24, P-28, and P-33) had a fatal outcome.
ANOVA of data sets for SARS patients in groups 1, 2, and 3 compared to healthy controls identified 5,993, 3,136, and 5,329 significantly different (P ≤ 0.05) genes, respectively (data not shown). Many ISGs, including CCL2 chemokine, radical S-adenosyl methionine domain containing 2 (CIG5), colony-stimulating factor 2 receptor alpha (CSF2RA), IFN-induced protein with tetratricopeptide repeats 3 (IFIT3), IFN-induced transmembrane protein 1 (IFITM1), metallothionein 1B (MT1B), metallothionein 1H (MT1H), myxovirus (influenza virus) resistance A (MXA), and syndecan 1 (SDC1) genes, in precrisis SARS patients were significantly upregulated relative to those genes in healthy controls (Table 2). CCL2, CIG5, MT1B, and MXA genes were also significantly upregulated in the corticosteroid-treated group. Overall, the gene expression profiles from the non-corticosteroid-treated and corticosteroid-treated precrisis SARS groups were highly similar, although the gene expression ratios from the corticosteroid-treated group tended to be decreased by approximately 10 to 40% relative to those of non-corticosteroid-treated precrisis SARS patients (Table 2). During the crisis phase, the levels of expression of the CCL2, CIG5, and MXA genes were similar between SARS patients and healthy controls, while MT1B remained upregulated. Other ISGs, such as CCL19 chemokine and IFN-γ receptor 1 (IFNGR1) genes, as well as major histocompatibility complex (MHC) class I and II genes (human leukocyte antigens [HLAs]) and cell death regulators (BCL2-associated athanogene 1 [BAG1], BAG3, BAG4, and BAG5 genes) were generally downregulated throughout SARS (Table 2). The immune response and host defense genes encoding immunoglobulin (Ig) heavy constant gamma 3 (IGHG3), SOD1, signal transducer and activator of transcription 6 (STAT6), and VAV1 oncogene proteins were generally downregulated during the course of SARS, while CXCL14 chemokine, serpin peptidase inhibitor, clade G, member 1 (SERPING1), and VAV2 oncogene proteins were upregulated in non-corticosteroid-treated precrisis SARS patients. Metallothioneins involved in homeostasis (MT1F, MT1G, and MT2A) were also upregulated in non-corticosteroid-treated precrisis SARS patients.
TABLE 2.
Gene expression during SARS disease course
| Gene product designation (by function or type) | Name/description | Unigene designation | Gene expressiona
|
|||||
|---|---|---|---|---|---|---|---|---|
| Precrisis group
|
Precrisis steroid-treated group
|
Crisis-phase group
|
||||||
| Mean ratio | P | Mean ratio | P | Mean ratio | P | |||
| Antigen presentation | ||||||||
| HLA-B | Major histocompatibility complex I-B | Hs.77961 | −0.63 | 0.046 | −1.39 | 0.002 | −0.88 | 0.001 |
| HLA-C | Major histocompatibility complex I-C | Hs.449621 | −0.59 | 0.003 | −0.95 | 0.008 | NS | |
| HLA-DMB | Major histocompatibility complex II-DM beta | Hs.351279 | −1.01 | <0.001 | −1.03 | <0.001 | −0.86 | <0.001 |
| HLA-DPA1 | Major histocompatibility complex II-DP alpha 1 | Hs.347270 | −1.40 | <0.001 | −1.67 | <0.001 | −1.80 | <0.001 |
| HLA-DPB1 | Major histocompatibility complex II-DP beta 1 | Hs.485130 | −1.28 | 0.002 | −1.44 | 0.03 | −1.88 | 0.003 |
| HLA-DQA1 | Major histocompatibility complex II-DQ alpha 1 | Hs.198253 | NS | NS | −1.19 | 0.005 | ||
| HLA-DQB1 | Major histocompatibility complex II-DQ beta 1 | Hs.409934 | −0.92 | 0.030 | −1.04 | 0.001 | −1.39 | 0.011 |
| HLA-DRB3 | Major histocompatibility complex II-DR beta 3 | Hs.308026 | NS | NS | −1.18 | 0.004 | ||
| HLA-E | Major histocompatibility complex I-E | Hs.118354 | −0.84 | <0.001 | −1.18 | <0.001 | −0.76 | <0.001 |
| Cell death | ||||||||
| BAG1 | BCL2-associated athanogene protein | Hs.377484 | −1.11 | <0.001 | −1.61 | <0.001 | −1.10 | <0.001 |
| BAG3 | BCL2-associated athanogene 3 protein | Hs.643507 | −0.86 | 0.001 | −1.18 | 0.006 | −1.03 | <0.001 |
| BAG4 | BCL2-associated athanogene 4 protein | Hs.194726 | −1.07 | 0.02 | −1.30 | <0.001 | −0.92 | 0.017 |
| BAG5 | BCL2-associated athanogene 5 protein | Hs.5443 | −1.11 | <0.001 | −0.70 | <0.001 | −0.76 | <0.001 |
| CD14 | CD14 molecule | Hs.163867 | NS | NS | −0.83 | 0.001 | ||
| Homeostasis | ||||||||
| MT1F | Metallothionein 1F | Hs.513626 | 1.04 | <0.001 | NS | 0.49 | 0.011 | |
| MT1G | Metallothionein 1G | Hs.433391 | 1.18 | <0.001 | NS | 0.26 | 0.045 | |
| MT2A | Metallothionein 2A | Hs.534330 | 1.19 | <0.001 | 0.85 | 0.013 | 0.74 | 0.045 |
| Immune response and host defense | ||||||||
| CXCL14 | CXC chemokine ligand 14 | Hs.483444 | 0.93 | 0.001 | NS | NS | ||
| IGHG3 | Ig heavy constant gamma 3 | Hs.564623 | −0.83 | 0.005 | −1.56 | 0.005 | −1.12 | 0.012 |
| SERPIN G1 | Serpin peptidase inhibitor, clade G, member 1 | Hs.384598 | 0.95 | <0.001 | NS | 0.50 | 0.017 | |
| SOD1 | Superoxide dismutase 1, soluble | Hs.443914 | −1.10 | <0.001 | −1.15 | 0.009 | −1.19 | <0.001 |
| SOD2 | Superoxide dismutase 2, mitochondrial | Hs.487046 | −1.16 | <0.001 | NS | NS | ||
| STAT6 | Signal transducer and activator of transcription 6 | Hs.524518 | −1.77 | <0.001 | −1.66 | <0.001 | −1.30 | <0.001 |
| TAP1 | Ag peptide transporter 1 | Hs.352018 | NS | NS | −0.73 | 0.032 | ||
| VAV1 | Vav 1 oncogene protein | Hs.116237 | −1.03 | <0.001 | −0.95 | 0.02 | −1.09 | <0.001 |
| VAV2 | Vav 2 oncogene protein | Hs.369921 | 0.84 | 0.05 | NS | NS | ||
| Interferon-stimulated gene products | ||||||||
| CCL19 | CC chemokine ligand 19 | Hs.50002 | −0.93 | <0.001 | −1.05 | <0.001 | −0.57 | <0.001 |
| CCL2 | CC chemokine ligand 2 | Hs.303649 | 1.32 | <0.001 | 0.72 | <0.001 | NS | |
| CIG5 | Radical S-adenosyl methionine domain-containing 2 | Hs.17518 | 1.65 | <0.001 | 1.34 | <0.001 | 0.37 | 0.049 |
| CSF2RA | Colony-stimulating factor 2 receptor alpha | Hs.520937 | 0.6 | <0.001 | 0.53 | 0.004 | 0.51 | <0.001 |
| IFIT3 | IFN-induced protein with tetratricopeptide repeats 3 | Hs.47338 | 0.85 | 0.001 | NS | NS | ||
| IFITM1 | IFN-induced transmembrane protein 1 | Hs.458414 | 0.81 | 0.02 | NS | NS | ||
| IFNGR1 | IFN-γ receptor 1 | Hs.520414 | −0.62 | 0.013 | NS | NS | ||
| MT1B | Metallothionein 1B | Hs.36102 | 1.53 | <0.001 | 0.84 | 0.003 | 0.7 | 0.001 |
| MT1H | Metallothionein 1H | Hs.2667 | 1.21 | <0.001 | NS | NS | ||
| MXA | Myxovirus resistance 1 | Hs.517307 | 1.5 | <0.001 | 1.17 | <0.001 | NS | |
| PBEF | Pre-B-cell colony-enhancing factor | Hs.239138 | −1.48 | <0.001 | −1.02 | 0.022 | NS | |
| PSME1 | Proteasome activator subunit 1 | Hs.75348 | −0.94 | <0.001 | −1.14 | <0.001 | −1.02 | <0.001 |
| SDC1 | Syndecan 1 | Hs.82109 | 1.24 | 0.006 | NS | NS | ||
| SDCBP | Syndecan binding protein (syntenin) | Hs.200804 | −1.18 | <0.001 | −1.17 | 0.013 | −1.10 | 0.005 |
Gene expression ratios are F-tested by ANOVA against, and relative to, means for healthy controls (n = 10). The numbers of patients in the precrisis group, the precrisis group treated with steroids, and the crisis-phase SARS patient group were 17, 7, and 13, respectively. Mean gene expression values were considered biologically significant at 0.58 (upregulated 1.5-fold) and −0.58 (downregulated 1.5-fold). Boldface indicates upregulated genes (≥1.5-fold). Boldface with underlining indicates downregulated genes (≥1.5-fold). NS, nonsignificant.
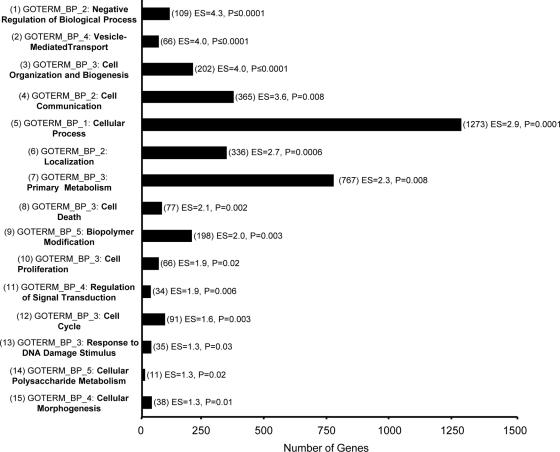
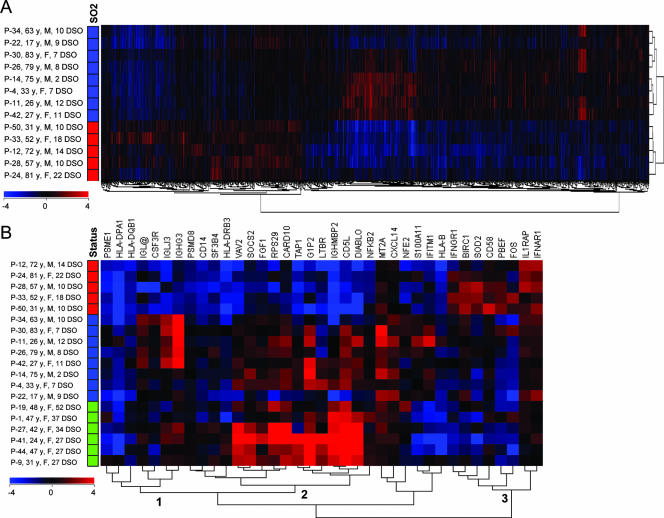
Lastly, an ANOVA between SARS patients in the crisis phase with an SO2 of ≥91% (n = 8) and those who, despite increased O2 support, had an SO2 of <91% (n = 5) at the time of sampling identified 2,487 significantly different genes (P ≤ 0.05). Again, the top 15 functional annotation clusters resulting from DAVID analysis in order of enrichment score are shown in Fig. 3. Data sets from non-corticosteroid-treated, precrisis SARS patients and healthy controls were highly similar (data not shown); however, a two-way hierarchical gene clustering analysis of SARS patients at crisis revealed two clusters of patient gene profiles, corresponding to those with an SO2 of ≥91% and those with an SO2 of <91% at the time of sampling (Fig. 4A). There were no significant differences in age between SARS patients with an SO2 of ≥91% and those with an SO2 of <91% in this group. A closer look at the genes in this data set revealed that the CD58, IFN (alpha, beta, and omega) receptor 1 (IFNAR1), and IFNGR1 ISGs in SARS patients with an SO2 of <91% were significantly upregulated relative to these genes in patients with an SO2 of ≥91% (Table 3). Interestingly, the vast majority of remaining ISGs in SARS patients with an SO2 of <91% were downregulated in comparison to these genes in patients with an SO2 of ≥91%. While the metallothionein MT2A was significantly upregulated throughout the disease course in SARS patients compared to the expression levels in healthy controls (Table 2), it was upregulated only in SARS patients at crisis with an SO2 of ≥91% at the time (Table 3). MHC class I and II genes were downregulated in all SARS patients during crisis; however, most gene expression levels were lower still in patients with an SO2 of <91%. Proapoptotic regulators, including caspase recruitment domain family, member 10 (CARD10), CD14, Diablo homolog (Drosophila sp.) (DIABLO) and lymphotoxin beta receptor (LTBR), were downregulated in patients with an SO2 of <91% and had returned to neutral levels in patients with an SO2 of ≥91% at the time. Conversely, the antiapoptotic baculoviral IAP repeat-containing 1 (BIRC-1) gene was significantly upregulated in SARS patients at crisis with an SO2 of <91%. Most immune response and host defense genes had returned to neutral expression levels in SARS patients with an SO2 of ≥91% during crisis; however, the CXCL14 gene, the Ig lambda joining 3 (IGLJ3) gene, and the VAV2 oncogene were upregulated in SARS patients with an SO2 of ≥91% versus patients with an SO2 of <91% at the time. Proinflammatory mediators, such as interleukin 1 receptor accessory protein (IL1RAP) and SOD2, in SARS patients during crisis with a persistent SO2 of <91% remained upregulated compared to these mediators in patients with an SO2 of ≥91%. Many immune response and host defense genes, including nuclear factor of kappa light polypeptide gene enhancer in B cells 2 (NF-KB2; critical to the acute-phase response), suppressor of cytokine signaling 2 (SOCS2; regulates cytokine signal transduction), and transporter 1 (TAP1; involved in antigen transport) genes, were downregulated in the SARS patients with an SO2 of <91% compared to those with an SO2 of ≥91%. Moreover, Ig genes in SARS patients with an SO2 of <91% were significantly downregulated compared to these genes in patients with an SO2 of ≥91%.
FIG. 3.
General functional annotation clusters identified by microarray analysis of SARS disease course. ANOVA was performed based on SO2 level at the time of SARS patient sampling during crisis (five patients with an SO2 of <91% and eight patients with an SO2 of ≥91%), resulting in the identification of 2,487 genes (P < 0.05). These genes were then uploaded to the DAVID Bioinformatics Database, and 2,123 were recognized and mapped for functional annotation analysis. The GO term (GOTERM) member and number of genes associated with the largest category of genes within the cluster are shown for each of the top 15 functional annotation clusters in order of enrichment score (ES). P values refer to a one-tailed Fisher's exact probability value used for gene enrichment analysis by the DAVID system.
FIG. 4.
Microarray analysis of the SARS disease course. (A) ANOVA was performed based on the SO2 level at the time of SARS patient sampling during crisis (five patients with an SO2 of <91% and eight patients with an SO2 of ≥91%), resulting in the identification of 2,487 genes (P < 0.05). SARS patient data sets, normalized to the means for healthy controls (n = 10), were two-way hierarchically clustered and labeled by SO2 level (red boxes indicate an SO2 of <91% and blue boxes indicate an SO2 of ≥91%). (B) Thirty-seven genes listed in Table 3 were hierarchically clustered alongside patient data sets (n = 19) ordered by the patient's general status at the time (red boxes indicate an SO2 of <91%, blue boxes indicate an SO2 of ≥91%, green boxes indicate patients at discharge). Three clusters discussed in the text are marked.
TABLE 3.
Gene expression in SARS patients at crisis
| Gene product designation (by function or type) | Name/description | Unigene designation | Mean gene expression ratio for patients with an SO2 ofa:
|
P | Cell type(s) expressing protein | Mean adjusted gene expression ratio for patients with an SO2 of <91%b | P | |
|---|---|---|---|---|---|---|---|---|
| ≥91% | <91% | |||||||
| Antigen presentation | ||||||||
| HLA-B | Major histocompatibility complex I-B | Hs.77961 | −0.55 | −1.40 | 0.014 | Leukocyte | −1.75 | 0.001 |
| HLA-DPA1 | Major histocompatibility complex II-DP alpha 1 | Hs.347270 | −1.31 | −2.58 | 0.042 | B cell and monocyte | −2.21 | NS |
| HLA-DQB1 | Major histocompatibility complex II-DQ beta 1 | Hs.409934 | −1.13 | −1.82 | 0.048 | B cell and monocyte | −1.45 | NS |
| HLA-DRB3 | Major histocompatibility complex II-DR beta 3 | Hs.308026 | −0.64 | −2.03 | 0.004 | B cell and monocyte | −1.66 | 0.024 |
| Cell death | ||||||||
| BIRC1 | Baculoviral IAP repeat-containing 1 | Hs.191356 | 0.13 | 0.81 | 0.015 | Unknown (tissue) | ||
| CARD10 | Caspase recruitment domain family, member 10 | Hs.57973 | 0.43 | −0.59 | 0.004 | Unknown (tissue) | ||
| CD14 | CD14 molecule | Hs.163867 | −0.56 | −1.26 | <0.001 | Monocyte | −1.39 | <0.001 |
| DIABLO | Diablo homolog (Drosophila) | Hs.169611 | 0.27 | −1.81 | 0.012 | Unknown (tissue) | ||
| LTBR | Lymphotoxin beta receptor | Hs.1116 | −0.23 | −1.30 | 0.049 | Leukocyte | −1.65 | 0.013 |
| Homeostasis—MT2A | Metallothionein 2A | Hs.534330 | 1.06 | 0.24 | 0.038 | Unknown (tissue) | ||
| Immune response and host defense | ||||||||
| CD5L | CD5 molecule-like | Hs.134035 | 0.46 | −1.58 | 0.001 | Unknown (tissue) | ||
| CSF3R | Colony-stimulating factor 3 receptor | Hs.524517 | −0.10 | −1.28 | 0.001 | Unknown (tissue) | ||
| CXCL14 | CXC chemokine ligand 14 | Hs.483444 | 0.66 | −0.07 | 0.003 | Unknown (tissue) | ||
| FGF1 | Fibroblast growth factor 1 | Hs.483635 | 0.03 | −0.68 | 0.004 | Unknown (tissue) | ||
| FOS | V-fos-FBJ murine osteosarcoma viral oncogene homolog | Hs.25647 | −1.06 | 0.04 | 0.049 | Unknown (tissue) | ||
| IGHG3 | Ig heavy constant gamma 3 | Hs.564623 | −0.44 | −2.20 | 0.003 | B cell | −1.83 | 0.004 |
| IGHM-BP2 | Ig mu binding protein 2 | Hs.503048 | −0.53 | −2.63 | 0.005 | B cell | −2.26 | 0.014 |
| IGL@ | Ig lambda locus | Hs.449585 | 0.4 | −1.01 | 0.013 | B cell | −0.64 | 0.050 |
| IGLJ3 | Ig lambda joining 3 | Hs.449601 | 0.67 | −1.37 | <0.001 | B cell | −1.00 | <0.001 |
| IL1RAP | Interleukin 1 receptor accessory protein | Hs.478673 | 0.33 | 0.83 | 0.042 | Unknown (tissue) | ||
| NF-KB2 | Nuclear factor of kappa light polypeptide gene enhancer in B cells 2 | Hs.73090 | −0.17 | −0.86 | 0.003 | Unknown (tissue) | ||
| SOCS2 | Suppressor of cytokine signaling 2 | Hs.485572 | 0.25 | −0.66 | <0.001 | T cell | −0.47 | 0.003 |
| SOD2 | Superoxide dismutase 2, mitochondrial | Hs.487046 | −0.67 | 0.58 | 0.004 | Unknown (tissue) | ||
| TAP1 | Ag peptide transporter 1 | Hs.352018 | −0.17 | −1.63 | <0.001 | B cell | −1.26 | <0.001 |
| VAV2 | Vav 2 oncogene protein | Hs.369921 | 0.64 | −1.15 | <0.001 | Multiple cell types | ||
| Interferon-stimulated gene products | ||||||||
| CD58 | CD58 molecule | Hs.34341 | −0.05 | 0.61 | 0.014 | Lymphocyte | 0.73 | 0.006 |
| G1P2 | IFN-α-inducible protein (clone IFI-15K) | Hs.458485 | 0.92 | −1.21 | 0.001 | Unknown (tissue) | ||
| IFITM1 | IFN-induced transmembrane protein 1 | Hs.458414 | 0.41 | −0.69 | 0.004 | Leukocyte | −1.04 | <0.001 |
| IFNAR1 | IFN (alpha, beta and omega) receptor 1 | Hs.529400 | 0.39 | 0.93 | 0.022 | Multiple cell types | ||
| IFNGR1 | IFN-γ receptor 1 | Hs.520414 | −0.41 | 0.68 | 0.035 | T cell | 0.87 | <0.001 |
| NF-E2 | Nuclear factor (erythroid-derived 2), 45 kDa | Hs.75643 | −0.06 | −0.91 | 0.015 | Megakaryocyte | ||
| PBEF | Pre-B-cell colony-enhancing factor | Hs.239138 | −0.65 | 0.44 | 0.008 | Unknown (tissue) | ||
| PSMD8 | Proteasome 26S subunit, non-ATPase, 8 | Hs.78466 | −0.26 | −0.93 | <0.001 | Unknown | ||
| PSME1 | Proteasome activator subunit 1 | Hs.75348 | −0.75 | −1.44 | 0.023 | Unknown | ||
| RPS29 | Ribosomal protein S29 | Hs.539 | 0.1 | −0.90 | 0.043 | Unknown (tissue) | ||
| S100A11 | S100 calcium-binding protein A11 | Hs.151973 | 0.25 | −0.69 | 0.008 | Leukocyte | −1.04 | 0.001 |
| SF3B4 | Splicing factor 3b, subunit 4, 49 kDa | Hs.25797 | −0.35 | −1.13 | 0.01 | Unknown (tissue) | ||
Gene expression ratios are F-tested by ANOVA and are relative to means for healthy controls (n = 10). The numbers of SARS patients at crisis with an SO2 of ≥91% and those with an SO2 of <91% at the time were eight and five, respectively. Mean gene expression values were considered biologically significant at 0.58 (upregulated 1.5-fold) and −0.58 (downregulated 1.5-fold). Boldface indicates upregulated genes (≥1.5-fold). Boldface with underlining indicates downregulated genes (≥1.5-fold). NS, nonsignificant.
Gene expression values were adjusted where possible relative to the major expressing cell type identified by Genatlas and reanalyzed by ANOVA as described in Results.
Gene expression data from Table 3, along with additional data from SARS patients being discharged from the hospital, are summarized in Fig. 4B. Cluster 1, containing Ig genes, was found to be expressed in crisis SARS patients with an SO2 of ≥91% and at discharge but downregulated in patients with an SO2 of <91%. Cluster 2, containing immune response and host defense and cell death genes, was expressed in SARS patients with an SO2 of ≥91% and even more so at discharge but again downregulated in patients with a persistent SO2 of <91% during crisis. This is in marked contrast to cluster 3, of which ISGs and genes for several proinflammatory mediators were highly expressed by crisis-phase SARS patients with a persistent SO2 of <91% (Fig. 4B).
A potential caveat existed for the analysis of SARS patients at crisis in that the gene expression may have been influenced by peripheral blood mononuclear cell (PBMC) subpopulations of different origins. While not significant, an ∼3-fold decrease (P = 0.08) in mean absolute lymphocyte count was noted for SARS patients at crisis with an SO2 of <91% compared to those with an SO2 of ≥91% at the time (data not shown). A complete phenotypic analysis was not possible during the SARS outbreak due to sample volume limitations. To examine this issue, however, we used the data set comparing the mean gene expression levels of CD markers in the severely (<91% SO2) and nonseverely (≥91% SO2) hypoxemic SARS patients at crisis to make adjustments where possible to the appropriate data point relative to the major expressing cell type identified by Genatlas (Table 3). Lymphocyte gene ratios were adjusted according to the difference (log2) in CD99 gene expression (+0.12), leukocyte gene ratios according to the difference in CD48 gene expression (−0.35), B-cell gene ratios according to the difference in CD79a/b gene expression (+0.37), T-cell gene ratios according to the difference in CD3D gene expression (+0.19), and monocyte/macrophage gene ratios according to the difference in CD163 gene expression (−0.13); the data were then reanalyzed by ANOVA. Following this correction, the differences in downregulated MHC class I and II gene expression between crisis-phase SARS patients with an SO2 of <91% and those with an SO2 of ≥91% at the time were narrowed; however, Ig genes remained significantly downregulated in the severely hypoxemic patients (Table 3). Indeed, proinflammatory ISGs (CD58 and IFNGR1 genes) remained highly expressed by SARS patients with a persistent SO2 of <91% during crisis. Lastly, we verified changes in microarray gene expression by using QRT-PCR for 17 genes listed in Materials and Methods in four patient groups (three patients per group), i.e., (i) precrisis SARS patients, (ii) crisis-phase SARS patients with an SO2 of ≥91% at the time, (iii) crisis-phase SARS patients with an SO2 of <91% at the time, and (iv) healthy controls. Whether a gene was upregulated or downregulated and the magnitude of the changes (n-fold) for each gene consistently matched the microarray data, with only three minor discrepancies (data not shown).
Cytokine and anti-SARS CoV spike Ab levels in SARS patient plasma.
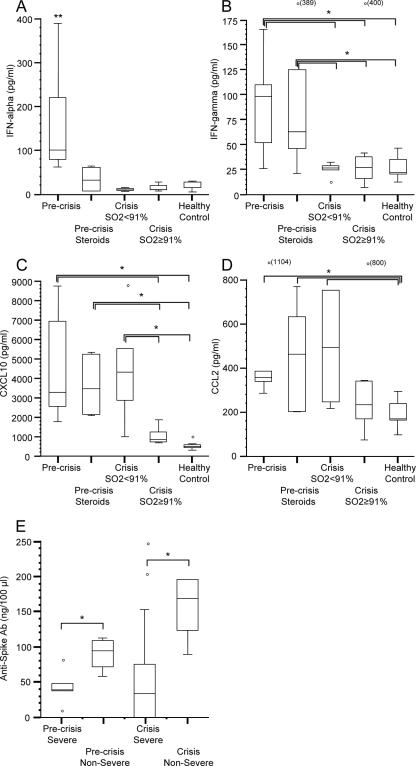
We expanded our analysis of IFN responses during the clinical evolution of SARS by measuring protein expression of cytokines and chemokines in the plasma of SARS patients by using the same groupings as the microarray analysis. IFN-α was significantly increased (∼4-fold; P < 0.05) in non-corticosteroid-treated, precrisis SARS patients compared to those that were treated with corticosteroids, all SARS patients at crisis and healthy controls (Fig. 5A). Plasma levels of IFN-γ were significantly increased (∼4-fold; P < 0.05) in non-corticosteroid-treated and corticosteroid-treated precrisis patients compared to those for crisis-phase SARS patients and healthy controls (Fig. 5B). The amounts of CXCL10, a proinflammatory chemokine induced by IFNs, in precrisis SARS patients and patients with a persistent SO2 of <91% at crisis were increased compared to those in patients with an SO2 of ≥91% at crisis and healthy controls (Fig. 5C). Protein levels of the CCL2 chemokine, another proinflammatory ISG, in precrisis SARS patients were significantly elevated (∼2-fold; P < 0.05) compared to those in healthy controls (Fig. 5D). The amounts of CCL2 protein in corticosteroid-treated precrisis-phase SARS patients and patients with an SO2 of <91% at crisis were also significantly increased relative to that in healthy controls (P < 0.05).
FIG. 5.
Analysis of cytokine and anti-SARS CoV spike Ab levels in the plasma of SARS patients. (A through D) Five patient groups for cytokine analysis were chosen: (i) non-corticosteroid-treated, precrisis SARS (n = 8); (ii) corticosteroid-treated, precrisis SARS (n = 6); (iii) crisis with an SO2 of ≥91% at the time (n = 8); (iv) crisis with an SO2 of <91% at the time (n = 5); and (v) healthy controls (n = 10). The box-and-whisker plots present the 50% interquartile ranges, maximum values excluding outliers (o), and medians. There were no significant differences in protein levels between healthy controls and SARS patients at discharge (data not shown). (E) Four patient groups emerged during analysis of plasma anti-SARS spike Ab levels: (i) precrisis severe-SARS patients (n = 5; median age, 57 years); (ii) precrisis nonsevere-SARS patients (n = 4; median age, 37 years); (iii) severe-SARS patients at crisis (n = 8; median age, 37 years); and (iv) nonsevere SARS patients at crisis (n = 5, median age, 37 years). Anti-SARS spike Abs were first detected in patients at a median of 6 DSO regardless of severity. Anti-SARS spike Abs were not detected in healthy controls (data not shown). *, P < 0.05; **, P value of <0.05 in pairwise comparison to each of the other four groups.
The SARS CoV spike protein is highly immunogenic, eliciting neutralizing Abs in humans in a non-strain-specific manner (38). We profiled anti-SARS CoV spike Abs in plasma samples from 17 SARS patients throughout illness and found that any differences in Ab levels were associated with the overall severity of SARS as defined above. Note that, similar to the larger cohort described in Table 1, severe-SARS patients were significantly older than nonsevere-SARS patients (P ≤ 0.05). Interestingly, the vast majority of SARS patients (6/6 nonsevere-SARS patients and 8/11 severe-SARS patients) mounted detectable anti-SARS CoV spike Ab titers (Fig. 5E). Nonetheless, nonsevere-SARS patients exhibited significantly higher (∼2- to ∼4-fold; P < 0.05) anti-SARS CoV spike Ab levels in the peripheral plasma than severe-SARS patients throughout SARS.
Collectively, our results identify novel IFN responses and ISG signatures associated with the early clinical course of SARS and suggest that immune response deficiencies develop in severe-SARS patients as the illness progresses.
DISCUSSION
As discussed earlier, type I IFNs play a central role in innate immunity to viral infections and other microbial infections and act in concert with type II IFNs in the immunoregulation of adaptive immune responses against infection (3, 36, 42, 48). Likewise, IFN-γ, the sole type II IFN, is an important contributor to adaptive immunity, especially Th1-type immunity (34), and can markedly potentiate type I IFN antiviral activity (48). At the heart of IFN activity lies the JAK/STAT signaling cascade which links the cognate cell surface receptors for type I and type II IFNs with the transcriptional activation of ISGs, a large family of IFN-stimulated immune mediators with pleiotropic downstream functions in innate and adaptive immunity (4, 42, 45, 48). Since certain STATs, such as STAT1, are common between type I and type II IFN signaling cascades, the IFN and ISG networks have multifaceted and interactive antiviral and immunomodulatory effects and play critical roles in the course of a host response to viral infection.
It has been proposed, largely based on in vitro studies, that deficient type I IFN production may undermine innate immune responses during early SARS immunopathogenesis (5, 8, 22, 27, 28, 33, 56). More recently, plasmacytoid dendritic cells have been shown to be capable of type I IFN production upon infection with SARS CoV in vitro (6). Our study, however, is the first to measure plasma levels of IFN-α, a key type I IFN, during the natural history of human SARS. Evasion of innate immunity by SARS CoV may benefit in part by downregulation of IFN-α-mediated innate immunity; however, our results argue that SARS patients mount robust type I and type II IFN responses, exhibit high plasma levels of the IFN-stimulated chemokines CXCL10 and CCL2, and express antiviral ISGs, such as CIG5 and MXA genes, during acute illness (precrisis). While high CCL2 and CXCL10 expression levels and MXA single-nucleotide polymorphisms have been associated with SARS severity and susceptibility, respectively (18, 21, 49), these gene expression signatures again hallmark the precrisis phase of SARS when IFN-mediated innate immune responses and symptoms are progressing. In the crisis period, cytokine and chemokine levels subside, as most SARS patients recover in conjunction with post-peak viral titers (see Fig. 5 and Table 1). However, high circulating levels of the proinflammatory chemokines CXCL10 and CCL2 and robust expression of distinct ISGs, e.g., those encoding IFNAR1, IFNGR1, and the costimulatory molecule CD58, persist in SARS patients with unrelenting severe hypoxemia (e.g., an SO2 of <91%) during crisis. Moreover, severe-SARS patients who died exhibited significantly higher levels of CXCL10 during crisis than those that recovered (median, 3,703 pg/ml versus 1,730 pg/ml, respectively; P = 0.027 [data not shown]). Other ISGs returned to neutral expression levels in SARS patients with an SO2 of ≥91% at crisis relative to healthy controls (Table 3) and were generally absent at discharge from the hospital (Fig. 4B), indicating that resolution of the IFN and ISG activity generated during the innate immune response to SARS CoV is associated with recovery.
Impaired regulation of HLA and Ig genes and the coinciding detrimental effect on antigen presentation and Ab production, i.e., key components of the adaptive immune response, have been implicated in the pathogenesis of several viruses (15, 43, 53). Indeed, severe-SARS patients are deficient in anti-SARS CoV spike Ab titers (Fig. 5E), and the most severely ill SARS patients are deficient in HLA and Ig gene expression during the crisis phase (Table 3). Likewise, interference with apoptosis regulation is a common mechanism of viral evasion (1). The resolution of IFN responses during recovery in conjunction with upregulated proapoptotic genes and MHC, Ig, and anti-spike Ab production further suggests that a critical switch from IFN-driven innate immunity to adaptive immune responses is necessary for clearance of SARS CoV and recovery from infection. Given this suggestion, is interesting to note that the use of corticosteroids in the treatment of SARS remains in debate (46). Corticosteroid treatment was not a significant discriminating factor in our microarray analysis of SARS patients at crisis and we were unable to link corticosteroids to any significant decrease in circulating chemokines during crisis (data not shown). We found, however, that corticosteroid treatment moderately downregulates proinflammatory gene expression during acute-phase SARS (Table 2), albeit alongside a corresponding decrease in potentially beneficial antiviral ISG and IFN-α (Fig. 5A) responses. Since corticosteroid treatment may delay adaptive immunity and viral clearance if administered during acute-phase SARS (29), our results further emphasize the need for clearer recommendations regarding immunosuppressive therapy of severe viral infections in humans (46).
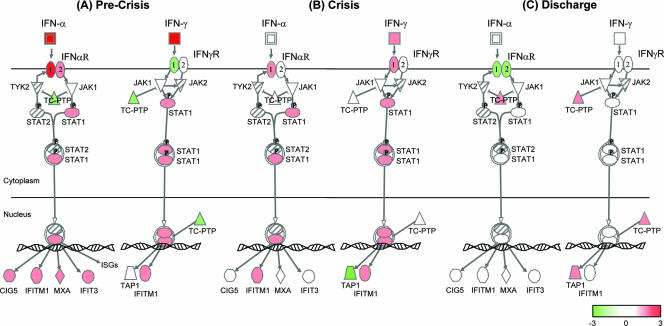
We used the IFN signaling canonical pathway available in Ingenuity Pathway Analysis software to summarize our cytokine and microarray ANOVA data (SARS patients versus healthy controls) during the clinical evolution of SARS (Fig. 6). We propose that hyperinnate immune responses during early (precrisis) SARS infection, hallmarked by robust and concurrent expression of IFN-α and IFN-γ and high levels of IFN-stimulated chemokines and ISG products (Fig. 6; see above), may preclude the development of effective adaptive immunity. The inability to clear SARS CoV from the lungs during crisis in patients with severe SARS may be the event that breaks down homeostatic regulation of developing innate and adaptive immune responses. In agreement, TAP1, a crucial component of the TAP heterodimer responsible for cell surface expression of MHC class I molecules, is downregulated in SARS patients during crisis despite continued upregulation of ISGs (Fig. 6 and Table 2). On the other hand, TAP1 is upregulated in SARS patients at discharge, and ISG expression returned to healthy control levels (Fig. 6).
FIG. 6.
Model of IFN signaling during the clinical evolution of SARS. The IFN signaling canonical pathway from Ingenuity Pathway Analysis software was used to model the microarray ANOVA data (mean ratios) from precrisis SARS patients (n = 14), crisis-phase SARS patients (n = 13), and SARS patients at discharge (n = 6) compared to healthy controls (10 patients), as well as the mean change (n-fold; see scale) of the same groups from the IFN-α and IFN-γ cytokine analysis. Red indicates upregulated molecules, green indicates downregulated molecules, and hatched molecules were not represented on the microarray. (A) The precrisis phase of SARS is hallmarked by robust expression of IFN-α and IFN-γ and high transcriptional activation of ISGs. (B) Despite continued upregulation of ISGs in SARS patients during crisis, TAP1, a component of the TAP heterodimer responsible for cell surface expression of MHC class I molecules, is downregulated. (C) For SARS patients that recover, TAP1 is upregulated at discharge from hospital and ISG expression has returned to healthy control levels.
Previous microarray studies have been unable to discriminate differences in the developing host response to SARS CoV, in particular, developing IFN-mediated innate and adaptive immune responses, due to insufficient patient numbers, time points, clinical information, or microarray platform coverage of IFN and/or immunity-related genes (30, 40, 55). For example, Yu et al. profiled PBMCs from one acute-phase SARS patient, one convalescent-phase SARS patient, and one healthy control and found that proinflammatory cytokine genes, such as interleukin-1, tumor necrosis factor alpha, and interleukin-8 genes, were upregulated during acute-phase SARS but that, surprisingly, IFN-induced genes, such as the CXCL10 gene, were downregulated. Also, in an analysis of PBMCs from 10 SARS-infected patients and 4 healthy controls, Reghunathan et al. found that acute-phase SARS was associated with strong proinflammatory gene responses, albeit in the complete absence of cytokine gene expression (40). Lastly, Lee et al. compared PBMC profiles from 25 recovering SARS patients to those of healthy controls and developed a distinctive gene signature to discriminate SARS patients from non-SARS patients; however, this study's primary goal was to develop a gene signature of severe SARS infection rather than model host immune responses (30). Collectively, our results are significant in identifying IFN-mediated gene and protein expression signatures associated with the clinical course of SARS in a substantial cohort of patients and in identifying atypical innate and adaptive immune responses in patients with severe SARS.
In a companion study of the Toronto SARS cohort, SARS CoV was consistently found in the lungs of deceased patients (16), including the three SARS patients with a fatal outcome (P-24, P-28, and P-33) in our study. While it remains to be determined why some patients experience severe forms of SARS, of the five patients in the severely hypoxemic SARS group (Fig. 4A) who suffered lengthy hospital stays or succumbed to SARS, one had asthma, two were of advanced age, one had borderline diabetes mellitus, and one was a recent lung transplant recipient. As noted earlier, severe-SARS patients in our study were significantly older than nonsevere-SARS patients and immunosenescence may partially explain the reduced anti-SARS CoV spike Ab titers in severe-SARS patients described in Fig. 5E. Advanced age appears to be a strong predictor of a poor outcome due to SARS infection (9, 10, 17, 50, 57). Nonetheless, the presence of comorbidities, particularly diabetes mellitus, hypertension, and immunosuppression, are also important factors (7, 10, 26, 54). Indeed, age was not a significant discriminating factor in our global gene expression analysis of SARS patients at crisis, with the young and elderly alike exhibiting severe SARS (Fig. 4A); however, our SARS patient cohort may not have been large enough to identify a significant association. Interestingly, the 57-year-old lung transplant recipient (patient P-28) who quickly succumbed to SARS at 14 DSO exhibited high levels of CXCL10 and CCL2 throughout SARS in our study despite preexisting immunosuppressive therapy and particularly high viral loads at the time of death (patient A113) (16). Moreover, CXCL10 and its receptor, CXCR3, were highly expressed in the lungs of deceased SARS patients in our study and not in control cadaveric lungs (data not shown). These results suggest that hyperchemokinemia correlates with viral load as has been observed by others in human H5N1 infections (11). While it is generally accepted that cytokines and chemokines can control the transition of innate to adaptive immunity during microbial infection of the lung (47), deciding whether the high expression levels of chemokines in patients with SARS at crisis is a cause or effect of severe illness will require additional study. Similarly, identifying patients at high risk for persistent viral burden and self-sustaining IFN-mediated immunopathogenesis remains an important research question.
In conclusion, our analysis has identified gene signatures and protein profiles associated with nonsevere and severe clinical courses of SARS in humans. While we do not know the exact mechanism that leads to a malfunction in the switch from innate to adaptive immune responses in severe-SARS patients, it is conceivable that factors described above, such as age and comorbidities, heavily influence one's ability to mount properly regulated IFN responses during the course of acute SARS and effective adaptive immune responses against SARS CoV during crisis. In a similar fashion, the severity and outcome of the 1918 influenza virus infection in a macaque model may be determined by dysregulated IFN responses that arise, likewise via an unknown mechanism, during host innate immunity (24). Parallels between the immunopathology of severe viral respiratory illnesses, such as SARS, 1918 influenza, and avian influenza (H5N1), suggest a common relationship in the evolution of these viruses in different human host populations. The potential for the gene signatures we describe in this study to better assess the immunopathology and management of severe viral infections is therefore worth careful examination.
Acknowledgments
Additional contributors from the Canadian SARS Research Network include D. Low, A. Rachlis, A. Simor, J. Butany, T. Mazzulli, J. de Jager, E. Phillips, L. Dresser, M. Gerson, I. Kitai, B. Mederski, M. Loutfy, and D. McRitchie.
This study was supported by the Canadian Institutes of Health Research, Canadian Network for Vaccines and Immunotherapeutics, and Genome Canada/Genome Quebec/Ontario Genomics Institute.
Thanks to G. Boucher for GEO submission assistance. Thanks to members of the Toronto health care community for their valuable assistance, including S. Asa, M. Christian, M. Fearon, F. Jamieson, M. Latchford, A. Musgrave, M. Ofner-Agostini, B. Yaffee, and S. Walmsley. We also express our respect and gratitude to Toronto and area clinicians, nurses, laboratory personnel, and health care workers who cared for SARS patients.
M.J.C. designed and carried out experiments, analyzed data, and wrote the manuscript. L.R. and L.X. carried out microarray data preprocessing. A.D. carried out cytometric bead analysis. J.F.B.-M. and S.M.P. provided clinical design. C.M.C. designed protocols and QRT-PCR primers. M.P.M. carried out chart reviews and curated clinical data. W.L.G., B.M.W., A.H., S.K., A.J.M., and J.B. provided clinical samples and design. S.E.R. provided viral load data. M.E.D., P.W., L.D.G., and R.S. provided genomics analysis support. Y.F. and S.E.B. performed QRT-PCR. C.S. performed the ELISA. D.P. processed samples and raw data. M.L. and M.B.L. helped secure funding and ethics approval. D.J.K. provided laboratory and financial resources, designed experiments, and wrote the manuscript.
Footnotes
Published ahead of print on 30 May 2007.
REFERENCES
- 1.Aubert, M., and K. R. Jerome. 2003. Apoptosis prevention as a mechanism of immune evasion. Int. Rev. Immunol. 22:361-371. [DOI] [PubMed] [Google Scholar]
- 2.Avendano, M., P. Derkach, and S. Swan. 2003. Clinical course and management of SARS in health care workers in Toronto: a case series. Can. Med. Assoc. J. 168:1649-1660. [PMC free article] [PubMed] [Google Scholar]
- 3.Brierley, M. M., and E. N. Fish. 2002. IFN-alpha/beta receptor interactions to biologic outcomes: understanding the circuitry. J. Interferon Cytokine Res. 22:835-845. [DOI] [PubMed] [Google Scholar]
- 4.Brierley, M. M., and E. N. Fish. 2005. Stats: multifaceted regulators of transcription. J. Interferon Cytokine Res. 25:733-744. [DOI] [PubMed] [Google Scholar]
- 5.Castilletti, C., L. Bordi, E. Lalle, G. Rozera, F. Poccia, C. Agrati, I. Abbate, and M. R. Capobianchi. 2005. Coordinate induction of IFN-alpha and -gamma by SARS-CoV also in the absence of virus replication. Virology 341:163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cervantes-Barragan, L., R. Zust, F. Weber, M. Spiegel, K. S. Lang, S. Akira, V. Thiel, and B. Ludewig. 2007. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 109:1131-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, C. Y., C. H. Lee, C. Y. Liu, J. H. Wang, L. M. Wang, and R. P. Perng. 2005. Clinical features and outcomes of severe acute respiratory syndrome and predictive factors for acute respiratory distress syndrome. J. Chin. Med. Assoc. 68:4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung, C. Y., L. L. Poon, I. H. Ng, W. Luk, S. F. Sia, M. H. Wu, K. H. Chan, K. Y. Yuen, S. Gordon, Y. Guan, and J. S. Peiris. 2005. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J. Virol. 79:7819-7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, K. W., T. N. Chau, O. Tsang, E. Tso, M. C. Chiu, W. L. Tong, P. O. Lee, T. K. Ng, W. F. Ng, K. C. Lee, W. Lam, W. C. Yu, J. Y. Lai, and S. T. Lai. 2003. Outcomes and prognostic factors in 267 patients with severe acute respiratory syndrome in Hong Kong. Ann. Intern. Med. 139:715-723. [DOI] [PubMed] [Google Scholar]
- 10.Cowling, B. J., M. P. Muller, I. O. Wong, L. M. Ho, S. V. Lo, T. Tsang, T. H. Lam, M. Louie, and G. M. Leung. 2006. Clinical prognostic rules for severe acute respiratory syndrome in low- and high-resource settings. Arch. Intern. Med. 166:1505-1511. [DOI] [PubMed] [Google Scholar]
- 11.de Jong, M. D., C. P. Simmons, T. T. Thanh, V. M. Hien, G. J. Smith, T. N. Chau, D. M. Hoang, C. N. Van Vinh, T. H. Khanh, V. C. Dong, P. T. Qui, B. Van Cam, D. Q. Ha, Y. Guan, J. S. Peiris, N. T. Chinh, T. T. Hien, and J. Farrar. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 12:1203-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis, G., Jr., B. T. Sherman, D. A. Hosack, J. Yang, W. Gao, H. C. Lane, and R. A. Lempicki. 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4:3. [PubMed] [Google Scholar]
- 13.de Veer, M. J., M. Holko, M. Frevel, E. Walker, S. Der, J. M. Paranjape, R. H. Silverman, and B. R. Williams. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69:912-920. [PubMed] [Google Scholar]
- 14.Ding, Y., L. He, Q. Zhang, Z. Huang, X. Che, J. Hou, H. Wang, H. Shen, L. Qiu, Z. Li, J. Geng, J. Cai, H. Han, X. Li, W. Kang, D. Weng, P. Liang, and S. Jiang. 2004. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J. Pathol. 203:622-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutta, N., A. Gupta, D. N. Mazumder, and S. Banerjee. 2006. Down-regulation of locus-specific human lymphocyte antigen class I expression in Epstein-Barr virus-associated gastric cancer: implication for viral-induced immune evasion. Cancer 106:1685-1693. [DOI] [PubMed] [Google Scholar]
- 16.Farcas, G. A., S. M. Poutanen, T. Mazzulli, B. M. Willey, J. Butany, S. L. Asa, P. Faure, P. Akhavan, D. E. Low, and K. C. Kain. 2005. Fatal severe acute respiratory syndrome is associated with multiorgan involvement by coronavirus. J. Infect. Dis. 191:193-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fowler, R. A., S. E. Lapinsky, D. Hallett, A. S. Detsky, W. J. Sibbald, A. S. Slutsky, and T. E. Stewart. 2003. Critically ill patients with severe acute respiratory syndrome. JAMA 290:367-373. [DOI] [PubMed] [Google Scholar]
- 18.Hamano, E., M. Hijikata, S. Itoyama, T. Quy, N. C. Phi, H. T. Long, L. D. Ha, V. V. Ban, I. Matsushita, H. Yanai, F. Kirikae, T. Kirikae, T. Kuratsuji, T. Sasazuki, and N. Keicho. 2005. Polymorphisms of interferon-inducible genes OAS-1 and MxA associated with SARS in the Vietnamese population. Biochem. Biophys. Res. Commun. 329:1234-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamming, I., W. Timens, M. L. Bulthuis, A. T. Lely, G. J. Navis, and H. van Goor. 2004. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 203:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han, T., C. D. Melvin, L. Shi, W. S. Branham, C. L. Moland, P. S. Pine, K. L. Thompson, and J. C. Fuscoe. 2006. Improvement in the reproducibility and accuracy of DNA microarray quantification by optimizing hybridization conditions. BMC Bioinformatics 7(Suppl. 2):S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, J., D. Feng, S. J. de Vlas, H. Wang, A. Fontanet, P. Zhang, S. Plancoulaine, F. Tang, L. Zhan, H. Yang, T. Wang, J. H. Richardus, J. D. Habbema, and W. Cao. 2006. Association of SARS susceptibility with single nucleic acid polymorphisms of OAS1 and MxA genes: a case-control study. BMC Infect. Dis. 6:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, K. J., I. J. Su, M. Theron, Y. C. Wu, S. K. Lai, C. C. Liu, and H. Y. Lei. 2005. An interferon-gamma-related cytokine storm in SARS patients. J. Med. Virol. 75:185-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang, D. M., D. W. Chamberlain, S. M. Poutanen, D. E. Low, S. L. Asa, and J. Butany. 2005. Pulmonary pathology of severe acute respiratory syndrome in Toronto. Mod. Pathol. 18:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobasa, D., S. M. Jones, K. Shinya, J. C. Kash, J. Copps, H. Ebihara, Y. Hatta, J. H. Kim, P. Halfmann, M. Hatta, F. Feldmann, J. B. Alimonti, L. Fernando, Y. Li, M. G. Katze, H. Feldmann, and Y. Kawaoka. 2007. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445:319-323. [DOI] [PubMed] [Google Scholar]
- 25.Kuba, K., Y. Imai, S. Rao, H. Gao, F. Guo, B. Guan, Y. Huan, P. Yang, Y. Zhang, W. Deng, L. Bao, B. Zhang, G. Liu, Z. Wang, M. Chappell, Y. Liu, D. Zheng, A. Leibbrandt, T. Wada, A. S. Slutsky, D. Liu, C. Qin, C. Jiang, and J. M. Penninger. 2005. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 11:875-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar, D., R. Tellier, R. Draker, G. Levy, and A. Humar. 2003. Severe acute respiratory syndrome (SARS) in a liver transplant recipient and guidelines for donor SARS screening. Am. J. Transplant. 3:977-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lau, Y. L., and J. S. Peiris. 2005. Pathogenesis of severe acute respiratory syndrome. Curr. Opin. Immunol. 17:404-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Law, H. K., C. Y. Cheung, H. Y. Ng, S. F. Sia, Y. O. Chan, W. Luk, J. M. Nicholls, J. S. Peiris, and Y. L. Lau. 2005. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived dendritic cells. Blood 106:2366-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, N., K. C. Allen Chan, D. S. Hui, E. K. Ng, A. Wu, R. W. Chiu, V. W. Wong, P. K. Chan, K. T. Wong, E. Wong, C. S. Cockram, J. S. Tam, J. J. Sung, and Y. M. Lo. 2004. Effects of early corticosteroid treatment on plasma SARS-associated coronavirus RNA concentrations in adult patients. J. Clin. Virol. 31:304-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, Y. S., C. H. Chen, A. Chao, E. S. Chen, M. L. Wei, L. K. Chen, K. D. Yang, M. C. Lin, Y. H. Wang, J. W. Liu, H. L. Eng, P. C. Chiang, T. S. Wu, K. C. Tsao, C. G. Huang, Y. J. Tien, T. H. Wang, H. S. Wang, and Y. S. Lee. 2005. Molecular signature of clinical severity in recovering patients with severe acute respiratory syndrome coronavirus (SARS-CoV). BMC Genomics 6:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lew, T. W., T. K. Kwek, D. Tai, A. Earnest, S. Loo, K. Singh, K. M. Kwan, Y. Chan, C. F. Yim, S. L. Bek, A. C. Kor, W. S. Yap, Y. R. Chelliah, Y. C. Lai, and S. K. Goh. 2003. Acute respiratory distress syndrome in critically ill patients with severe acute respiratory syndrome. JAMA 290:374-380. [DOI] [PubMed] [Google Scholar]
- 32.Li, W., M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, T. C. Greenough, H. Choe, and M. Farzan. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lo, A. W., N. L. Tang, and K. F. To. 2006. How the SARS coronavirus causes disease: host or organism? J. Pathol. 208:142-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malmgaard, L. 2004. Induction and regulation of IFNs during viral infections. J. Interferon Cytokine Res. 24:439-454. [DOI] [PubMed] [Google Scholar]
- 35.Mandavilli, A. 2006. China: open season. Nature 439:382-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen, K. B., W. T. Watford, R. Salomon, S. R. Hofmann, G. C. Pien, A. Morinobu, M. Gadina, J. J. O'Shea, and C. A. Biron. 2002. Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection. Science 297:2063-2066. [DOI] [PubMed] [Google Scholar]
- 37.Nicholls, J. M., L. L. Poon, K. C. Lee, W. F. Ng, S. T. Lai, C. Y. Leung, C. M. Chu, P. K. Hui, K. L. Mak, W. Lim, K. W. Yan, K. H. Chan, N. C. Tsang, Y. Guan, K. Y. Yuen, and J. S. Peiris. 2003. Lung pathology of fatal severe acute respiratory syndrome. Lancet 361:1773-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nie, Y., G. Wang, X. Shi, H. Zhang, Y. Qiu, Z. He, W. Wang, G. Lian, X. Yin, L. Du, L. Ren, J. Wang, X. He, T. Li, H. Deng, and M. Ding. 2004. Neutralizing antibodies in patients with severe acute respiratory syndrome-associated coronavirus infection. J. Infect. Dis. 190:1119-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peiris, J. S., C. M. Chu, V. C. Cheng, K. S. Chan, I. F. Hung, L. L. Poon, K. I. Law, B. S. Tang, T. Y. Hon, C. S. Chan, K. H. Chan, J. S. Ng, B. J. Zheng, W. L. Ng, R. W. Lai, Y. Guan, and K. Y. Yuen. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reghunathan, R., M. Jayapal, L. Y. Hsu, H. H. Chng, D. Tai, B. P. Leung, and A. J. Melendez. 2005. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol. 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salto-Tellez, M., E. Tan, and B. Lim. 2005. ARDS in SARS: cytokine mediators and treatment implications. Cytokine 29:92-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schroder, K., P. J. Hertzog, T. Ravasi, and D. A. Hume. 2004. Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 75:163-189. [DOI] [PubMed] [Google Scholar]
- 43.Shao, L., and K. Sperber. 2002. Impaired regulation of HLA-DR expression in human immunodeficiency virus-infected monocytes. Clin. Diagn. Lab. Immunol. 9:739-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi, L., L. H. Reid, W. D. Jones, R. Shippy, J. A. Warrington, S. C. Baker, P. J. Collins, F. de Longueville, E. S. Kawasaki, K. Y. Lee, Y. Luo, Y. A. Sun, J. M. Willey, R. A. Setterquist, G. M. Fischer, W. Tong, Y. P. Dragan, D. J. Dix, F. W. Frueh, F. M. Goodsaid, D. Herman, R. V. Jensen, C. D. Johnson, E. K. Lobenhofer, R. K. Puri, U. Schrf, J. Thierry-Mieg, C. Wang, M. Wilson, P. K. Wolber, L. Zhang, W. Slikker, Jr., L. Shi, and L. H. Reid. 2006. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 24:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, P. L., G. Lombardi, and G. R. Foster. 2005. Type I interferons and the innate immune response—more than just antiviral cytokines. Mol. Immunol. 42:869-877. [DOI] [PubMed] [Google Scholar]
- 46.Stockman, L. J., R. Bellamy, and P. Garner. 2006. SARS: systematic review of treatment effects. PLoS Med. 3:e343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strieter, R. M., J. A. Belperio, and M. P. Keane. 2003. Host innate defenses in the lung: the role of cytokines. Curr. Opin. Infect. Dis. 16:193-198. [DOI] [PubMed] [Google Scholar]
- 48.Takaoka, A., and H. Yanai. 2006. Interferon signalling network in innate defence. Cell Microbiol. 8:907-922. [DOI] [PubMed] [Google Scholar]
- 49.Tang, N. L., P. K. Chan, C. K. Wong, K. F. To, A. K. Wu, Y. M. Sung, D. S. Hui, J. J. Sung, and C. W. Lam. 2005. Early enhanced expression of interferon-inducible protein-10 (CXCL-10) and other chemokines predicts adverse outcome in severe acute respiratory syndrome. Clin. Chem. 51:2333-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsui, P. T., M. L. Kwok, H. Yuen, and S. T. Lai. 2003. Severe acute respiratory syndrome: clinical outcome and prognostic correlates. Emerg. Infect. Dis. 9:1064-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong, C. K., C. W. Lam, A. K. Wu, W. K. Ip, N. L. Lee, I. H. Chan, L. C. Lit, D. S. Hui, M. H. Chan, S. S. Chung, and J. J. Sung. 2004. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 136:95-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong, R. S., A. Wu, K. F. To, N. Lee, C. W. Lam, C. K. Wong, P. K. Chan, M. H. Ng, L. M. Yu, D. S. Hui, J. S. Tam, G. Cheng, and J. J. Sung. 2003. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ 326:1358-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, C. G., A. Budhu, S. Chen, X. Zhou, N. C. Popescu, K. Valerie, and X. W. Wang. 2006. Effect of hepatitis C virus core protein on the molecular profiling of human B lymphocytes. Mol. Med. 12:47-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, J. K., Y. Feng, M. Y. Yuan, S. Y. Yuan, H. J. Fu, B. Y. Wu, G. Z. Sun, G. R. Yang, X. L. Zhang, L. Wang, X. Xu, X. P. Xu, and J. C. Chan. 2006. Plasma glucose levels and diabetes are independent predictors for mortality and morbidity in patients with SARS. Diabet. Med. 23:623-628. [DOI] [PubMed] [Google Scholar]
- 55.Yu, S. Y., Y. W. Hu, X. Y. Liu, W. Xiong, Z. T. Zhou, and Z. H. Yuan. 2005. Gene expression profiles in peripheral blood mononuclear cells of SARS patients. World J. Gastroenterol. 11:5037-5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ziegler, T., S. Matikainen, E. Ronkko, P. Osterlund, M. Sillanpaa, J. Siren, R. Fagerlund, M. Immonen, K. Melen, and I. Julkunen. 2005. Severe acute respiratory syndrome coronavirus fails to activate cytokine-mediated innate immune responses in cultured human monocyte-derived dendritic cells. J. Virol. 79:13800-13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zou, Z., Y. Yang, J. Chen, S. Xin, W. Zhang, X. Zhou, Y. Mao, L. Hu, D. Liu, B. Chang, W. Chang, Y. Liu, X. Ma, Y. Wang, and X. Liu. 2004. Prognostic factors for severe acute respiratory syndrome: a clinical analysis of 165 cases. Clin. Infect. Dis. 38:483-489. [DOI] [PMC free article] [PubMed] [Google Scholar]