Abstract
Infectious bursal disease virus (IBDV) causes a highly contagious disease in young chicks and leads to significant economic losses in the poultry industry. The capsid protein VP2 of IBDV plays an important role in virus binding and cell recognition. VP2 forms a subviral particle (SVP) with immunogenicity similar to that of the IBDV capsid. In the present study, we first showed that SVP could inhibit IBDV infection to an IBDV-susceptible cell line, DF-1 cells, in a dose-dependent manner. Second, the localizations of the SVP on the surface of DF-1 cells were confirmed by fluorescence microscopy, and the specific binding of the SVP to DF-1 cells occurred in a dose-dependent manner. Furthermore, the attachment of SVP to DF-1 cells was inhibited by an SVP-induced neutralizing monoclonal antibody against IBDV but not by denatured-VP2-induced polyclonal antibodies. Third, the cellular factors in DF-1 cells involved in the attachment of SVP were purified by affinity chromatography using SVP bound on the immobilized Ni2+ ions. A dominant factor was identified as being chicken heat shock protein 90 (Hsp90) (cHsp90) by mass spectrometry. Results of biotinylation experiments and indirect fluorescence assays indicated that cHsp90 is located on the surface of DF-1 cells. Virus overlay protein binding assays and far-Western assays also concluded that cHsp90 interacts with IBDV and SVP, respectively. Finally, both Hsp90 and anti-Hsp90 can inhibit the infection of DF-1 cells by IBDV. Taken together, for the first time, our results suggest that cHsp90 is part of the putative cellular receptor complex essential for IBDV entry into DF-1 cells.
Infectious bursal disease virus (IBDV), a member of genus Avibirnavirus of the family Birnaviridae, causes a highly contagious disease in young chicks (27). Two serotypes (serotypes 1 and 2) of IBDV have been documented. Serotype 1 showed different degrees of pathogenicity and mortality in chicks, and serotype 2 was avirulent. As with all viruses, IBDV needs to penetrate target cells to cause infection. Chicken B lymphocytes are the primary target for virulent serotype 1 strains of IBDV, and the infection causes a functional loss of the bursa of Fabricius and severe immunodepression. However, other susceptible cells have also been reported (17). Viral attachment is the first step in virus infection (41). The distribution of a virus receptor mainly determines the cell and tissue tropism of the virus (1, 11) and the site of pathology associated with infection (9, 26). Thus, study of virus infection at the levels of virus binding, receptor identification, and even uncoating is critical for an understanding of the virus-host cell interactions and pathogenesis of viral disease (24, 32). Additionally, viral entry and uncoating can serve as targets for the development of antiviral drugs (39, 42). Recent advances in the understanding of the viral infection process have made it possible to develop new approaches to block the entry of viruses (31) and to prevent diseases (41). However, the identification of a specific receptor on the surface of a susceptible host cell for the attachment of IBDV has been elusive.
Although pathogenic serotype 1 strains of IBDV replicate efficiently in lymphoid cells of the bursa of Fabricius of chickens, strains of serotypes 1 and 2 are widely propagated in chicken embryo fibroblast (CEF) cells. Nieper and Müller (28) first reported that CEF cells had receptors common to both serotypes and specific ones for each serotype. Specifically, two proteins with molecular masses of 40 and 46 kDa expressed on CEF cells were responsible for the binding of both serotypes. Later, for another very virulent IBDV-permissive chicken B-lymphoblastoid cell line, LSCC-BK3, three proteins with molecular masses of 70, 82, and 110 kDa on the plasma membranes (PM) of LSCC-BK3 were also found to interact with IBDV (38). These studies employed the whole IBDV to study the interaction of IBDV with susceptible cells.
Recently, the crystal structures of IBDV and a subviral particle (SVP) formed by the IBDV external capsid VP2 protein reveal their similar VP2 structures (7, 33). As a single capsid protein (7, 37), VP2 is the primary host-protective immunogen of IBDV and contains the antigenic regions responsible for the elicitation of neutralizing antibodies (13). In addition to epitope recognition by antibodies, the hypervariable region, i.e., amino acids 206 to 350 of VP2, is also presumably responsible for the interaction with the cellular receptors and restriction in infectivity (45). Amino acid substitutions in the hypervariable region of VP2 in different strains appeared to be associated with altered antigenicity and virulence. Thus, SVP was used to replace the whole IBDV virion for the attachment of IBDV to an IBDV-susceptible cell line and for the identification of putative cellular receptors of IBDV.
In the present study, large amounts of homologous SVP formed by VP2-441 (a 441-amino-acid-residue VP2 protein) were prepared. Either uniform VP2-formed SVP or fluorescein isothiocyanate (FITC)-labeled SVP was used to search for possible receptors on the surface of DF-1 cells (14), a spontaneously immortalized cell line derived from primary CEF, which are commonly used for the propagation of IBDV. Our results showed for the first time that chicken heat shock protein 90 (Hsp90) (cHsp90) of DF-1 cells plays a very significant role in the entry into and infection of this cell line by IBDV.
MATERIALS AND METHODS
Viruses and cells.
Recombinant baculovirus stocks were propagated in Spodoptera frugiperda cells (Sf9) (ATCC 1171; American Type Culture Collection) using TNM-FH medium (Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum (FBS) (Biological Industries, Kibbutz Bet Haemek, Israel) in tissue culture flasks (Corning, NY) at 28°C. High-five (Hi-5) cells (purchased from Invitrogen) were routinely cultured and passaged in ESF-921 medium (Expression System LLC, Woodland, CA). DF-1 (chicken fibroblast) cells, kindly provided by Ching-Ho Wang at the Department of Veterinary Medicine, National Taiwan University, were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% inactivated fetal calf serum. CEF cells were derived from 10-day-old embryonated eggs and grown in medium 199 (Gibco) for the propagation of IBDV P3009, a local isolate, by standard procedures (47).
Infection of DF-1 cells with IBDV P3009.
DF-1 cells were grown in monolayers to subconfluency in 75T flasks and then infected with 100 50% tissue culture infective doses (TCID50) of IBDV P3009. At 3 to 4 days postinfection, cells were observed for cytopathic effects (CPEs) using an inverted microscope. After being detached with 5 mM EDTA, cells were collected for immunoblotting assays using polyclonal anti-VP2 or anti-VP3 antibodies (21), and the supernatant, in general with a titer of 6 × 106 PFU/ml, was saved as a viral stock.
Generation of a recombinant baculovirus-expressing IBDV VP2-441-formed SVP.
The generation of the recombinant baculovirus-expressing VP2-441-formed SVP was done according to procedures described previously (47). Briefly, a VP2 gene fragment encoding VP2 with 441 amino acid residues (VP2-441) was generated by PCR using pBluescript-VP2 as a template (6) and a pair of primers: forward primer P4F (CGATCGCTAGCGATGACAAAC) (5′ to 3′) and reverse primer 1323NH (CGAATTCCTATGCTCCTGCAATCTTCAG) (5′ to 3′), respectively. Following the standard procedures, the recombinant transfer plasmid pBB4ΔC11 was obtained by inserting the PCR-generated VP2-441 gene fragment into a transfer vector, pBlueBac4 (47), and a recombinant baculovirus was obtained by cotransfecting plasmid pBB4ΔC11 with linear Autographa californica multiple nucleopolyhedrovirus DNA into Sf9 cells as described in the manual provided by the supplier (Invitrogen). Putative recombinant baculoviruses were then plaque purified. The expression of the VP2-441 monomer protein in Sf9 or Hi-5 cells was characterized by Western blotting using a polyclonal antibody, anti-VP2 (21).
Production and purification of SVP and preparation of FITC-conjugated SVP.
SVP formed by VP2-441 was produced and purified by using a previously established protocol (20). Purified SVP was characterized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) staining with silver nitrate, Western blotting, and negative-stain electron microscopy (21). For conjugation of SVP with FITC (Sigma), 4 mg of SVP was resuspended in 500 mM carbonate-bicarbonate buffer (pH 9.5), mixed with 400 μg of FITC, and incubated at room temperature for 1 h (18). The salt was then removed by use of a dialysis method. Finally, the FITC-conjugated SVP was stored in 10 mM TRIZAM 7∼9 buffer (pH 8.2) (Sigma) and was quantified by SDS-PAGE (4).
Infection inhibition by SVP.
DF-1 cells were seeded into 96-well-plates with DMEM supplemented with 10% FBS for 2 h. Next, for each 96-well plate, the medium was removed, and DF-1 cells in each well were incubated with the medium containing a concentration of SVP increasing from 0.01 to 20 μg/100 μl at 4°C for 60 min. After the SVP-containing media were removed, the cells were infected with 10 TCID50 of IBDV P3009 per well at 4°C for 1 h and then washed three times with fresh culture medium. Fresh medium was then added to the infected cells, and the infection was allowed to proceed for 96 h at 39°C. After infection, cells were observed under an inverted microscope for CPEs. For each plate, the number of CPE-positive wells was determined by staining the cells with 1.5% crystal violet in 50% ethanol for 20 min. The infection efficiency of each plate was calculated by counts of the number of wells that were CPE positive in that plate divided by 96 (the total number of the wells in a 96-well plate). The experiment was repeated once, with a similar result. For bovine serum albumin (BSA), the procedures were similar, except that SVP was replaced with 1 μg of BSA.
Preparation of anti-SVP monoclonal antibodies.
Monoclonal antibodies (MAbs) against SVP were obtained from BALB/c mice immunized with SVP four times at 2-week intervals. The hybridoma cell lines were prepared and subcloned as described previously (10), with the following modifications. After spleen cells were fused with the NS1 myeloma cell line, the fused cells were cultured in hypoxanthine-aminopterin-thymidine-free medium for 24 h, and 2× hypoxanthine-aminopterin-thymidine medium was then added to each well. The ascites fluids containing high concentrations of MAbs were produced in BALB/c mice primed with pristane (Sigma). Several MAbs (MAbSVP-1, MAbSVP-4, MAbSVP-10, MAbSVP-12, MAbSVP-13 MAbSVP-16, MAbSVP-17, and MAbSVP-29) against VP2-441-formed SVP were generated. These MAbs were confirmed by Western blotting and also screened for their SVP binding activity by enzyme-linked immunosorbent assay (ELISA) (22). MAbs were then further characterized by virus neutralization (47) and immunoprecipitation (IP). The binding of MAbs to IBDV was examined by IP according to procedures provided by the manufacturer (Amersham Biosciences). The detailed characterization of MAbs by ELISA, virus neutralization, immunoprecipitation, and Western blotting will be published elsewhere.
SDS-PAGE and Western blotting analysis.
SDS-PAGE was performed as described previously (5), with slight modification. Briefly, samples were mixed with reducing sample buffer, boiled, centrifuged, and loaded onto a 12.5% slab gel. Following electrophoresis, the gel was stained by silver nitrate or used for Western blotting analysis by electroblotting onto a polyvinylidene difluoride (PVDF) membrane. To detect IBDV antigens (pVP2/VP2 and VP3), the polyclonal anti-VP2 and anti-VP3 antibodies (21), respectively, were employed as the first antibody. To detect cHsp90, either anti-Hsp90 mouse MAb (anti-Hsp90 MAb from Novus Biologicals, Inc., CO) or rat anti-Hsp90α MAb (Stressgen Biotechnologies, Victoria, British Columbia, Canada) was used as the primary antibody. Although they are not derived from cHsp90, both MAbs can react with cHsp90. Following washing, the membrane was incubated with the appropriate Affinipure secondary antibody conjugated with alkaline phosphatase (Jackson ImmunoResearch, West Grove, PA), and the color was developed as described previously (5).
Fluorescent microscopy and confocal microscopy.
The cells cultivated in monolayers were washed three times with cold Dulbecco's phosphate-buffered saline (PBS) (D-PBS) and chilled to 4°C. Purified FITC-conjugated SVP (40 μg/ml) was added to wells in D-PBS at final volumes of 200 μl per well in both 24-well plates and chamber slides. The plates were incubated for 1 h with gentle agitation on a rocker platform at 4°C. The binding reaction was terminated by gently washing the cells three times with cold D-PBS to remove the unbound SVP. After fixation and mounting, cells were examined using a fluorescent microscope (Nikon TS-2000) or a confocal microscope (Zeiss LSM510). Indirect immunofluorescence assays on subconfluent (75%) DF-1 cells in 24-well plates or chamber slides (Nunc) were performed as described previously (23, 35), with modifications. Briefly, DF-1 cells were prepared and fixed with 3% paraformaldehyde and 2% sucrose in D-PBS at room temperature for 5 min. To check the permeability of the fixed cells, they were reacted with the first antibody, a mouse-raised anti-β-actin MAb (diluted 1:2,500; Sigma), washed three times with D-PBS, and then reacted with the secondary antibody, a rhodamine-coupled anti-mouse immunoglobulin G (IgG) (diluted 1:500; Sigma). To locate the position of cHsp90 on the PM of DF-1, the fixed cells were reacted with a rat-raised anti-Hsp90α MAb (diluted 1:500 in PBS with BSA; Stressgen Biotechnologies) for 30 min and then detected with Alexa Fluor 555 goat anti-rat IgG (diluted 1:1,000; Molecular Probes, Inc.). Alternatively, the fixed cells were reacted with the purified FITC-conjugated SVP (40 μg/ml) for 10 min to check the binding of SVP to the PM. After mounting, cells were examined using a fluorescent microscope (Nikon TS-2000) or a confocal microscope (FluoView FV1000; Olympus).
Flow cytometry analysis.
Flow cytometry was used for SVP binding assays and inhibition of SVP binding with a neutralizing MAb. For the SVP binding assay, cells were harvested using 5 mM EDTA, washed, and resuspended at 106 cells per ml in D-PBS at 4°C. One million cells were incubated with 0, 1, 2, 4, 8, 16, 32, and 64 μg of FITC-conjugated SVP in 100 μl of D-PBS at 4°C for 1 h. After centrifugation, supernatants were completely discarded; cells were washed once with ice-cold D-PBS and resuspended in 1 ml D-PBS. Fluorescent spectra were analyzed by flow cytometry (Cytomics FC500; Beckman), counting 10,000 cells per sample. For inhibition of SVP binding to DF-1 cells with a neutralizing MAb (MAbSVP-4) or a polyclonal antibody (anti-VP2) without neutralizing activity, 16 μg of FITC-conjugated SVP was mixed well with various titers (10−1, 10−2, and 10−3) of MAbSVP-4 or anti-VP2 (as a negative control) in 100 μl D-PBS and then incubated for 1 h at 4°C for the neutralization reaction. Cells were then added and incubated for 1 h. After centrifugation, supernatants were completely discarded, and cells were washed three times with ice-cold D-PBS and resuspended in 1 ml D-PBS. Fluorescent spectra were analyzed by counting 105 cells per sample using flow cytometry.
Preparation of DF-1 total cell proteins.
DF-1 cells were pelleted at 800 × g for 5 min (KUBOTA 5922) and washed twice with D-PBS to remove EDTA. The cell pellet was then resuspended in RSB-NP-40 (1.5 mM MgCl2, 10 mM Tris-HCl, 10 mM NaCl, and 1% Nonidet P-40) in the presence of protease inhibitor cocktail (2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 2 mM benzonidine, 5 μg of aprotinin per ml, 5 μg pepstatin per ml, 5 μg leupeptin per ml, and 5 μg chymostatin per ml [MiniComplete; Roche]) for 30 min. The supernatant was collected by centrifugation at 12,000 × g for 15 min at 4°C to remove nuclei and debris. The amount of total cellular proteins was measured using a BCA protein assay kit (Pierce).
To obtain biotinylated membrane proteins, 2.5 mg of biotin sulfo-succinimidyl ester (Sulfo NHS-biotin; Sigma) in 10 ml PBS was added to approximately 1 × 107 DF-1 cells and incubated in the dark for 1 h at room temperature. Cells were stained with trypan blue before and after cell biotinylation to check the membrane integrity. The biotinylated cells were then collected and lysed by sonication at 4°C for 20 s. The membrane pellet was collected by centrifugation and resuspended in RSB-NP-40. Soluble membrane protein extracts were obtained by centrifugation at 12,000 × g for 15 min at 4°C, and the amount of membrane proteins was determined by BCA protein assay.
Affinity isolation of putative virus receptor proteins.
SVP was purified by immobilized metal-ion affinity chromatography (IMAC) as described above. Ni-Sepharose High Performance resin (GE Healthcare) was saturated with SVP by incubating 1 ml of 2 mg SVP in native binding buffer (20 mM NaH2PO4, 0.5 M NaCl [pH 7.8]) with 400 μl of preequilibrated resin in a column (1.5-cm diameter, 7-cm height) for 30 min at room temperature. The unbound fraction was removed by washing the resin with native binding buffer, and the SVP-bound resin was equilibrated with the native binding buffer. Afterward, 4 mg of DF-1 total protein extract was diluted in 5 ml of native binding buffer, added into the column, and mixed by inverting the column for 30 min at room temperature. Nonspecific binding to the resin was avoided by washing the column with 10 volumes of the native binding buffer and was followed by 10 volumes of native wash buffer 1 (20 mM NaH2PO4, 0.5 M NaCl [pH 6.3]). To remove more nonspecific binding, the column was washed with 10 volumes of native wash buffer 2 (20 mM NaH2PO4, 0.5 M NaCl [pH 6.0]). Subsequently, protein-bound and free SVPs were eluted in four 1,000-μl fractions with elution IMAC buffer (20 mM NaH2PO4, 0.5 M NaCl [pH 4.0]). Finally, the resin was washed with 5 mM EDTA to remove the immobilized ions. The molecules eluted by the eluation IMAC buffer were concentrated in a Centricon 10 centrifuge microconcentrator (Amicon, Beverly, MA) and analyzed by SDS-PAGE gels stained with Coomassie blue. The 90-kDa band was excised; after in-gel digestion with trypsin, the peptides were analyzed by liquid chromatography tandem mass spectrometry, performed on an electrospray ionization quadrupole quadrupole time-of-flight mass spectrometer. The protein identification was performed at the Proteomic Mass Spectrometry Core Laboratory of the Biotechnology Center at National Chung-Hsing University, Taichung, Taiwan. The interaction of biotinylated membrane proteins with immobilized SVP was done essentially as described above. Transferred biotinylated proteins obtained in the elution fraction were detected using streptavidin-alkaline phosphatase, and color was developed as described previously (5).
Pull-down assay by immunoprecipitation.
One hundred micrograms of DF-1 total proteins extracted using a lysis buffer (150 mM NaCl, 50 mM Tris-HCl [pH 8.0], 1 mM phenylmethylsulfonyl fluoride, and 1% Nonidet P-40) was well reacted with 100 μg of SVP for 60 min at 4°C. Purified MAbSVP-1 (5 μg) was then added and gently mixed for 60 min at 4°C. Thirty microliters of protein G-Sepharose 4 Fast Flow (GE Healthcare) was then added. The mixture was gently mixed for 1 h at 4°C and centrifuged at 12,000 × g for 20 s to collect the pellet. The pellet was washed three times with 1 ml lysis buffer and once with wash buffer (50 mM Tris-HCl [pH 8.0]) and then centrifuged at 12,000 × g for 20 s to save the beads. The final pellet was suspended in 30 to 40 μl of sample buffer, heated to 95°C for 3 min, and then centrifuged at 12,000 × g for 20 s to remove the beads. The supernatant was analyzed by SDS-PAGE and Western blotting.
VOPBA and far-Western assay.
A virus overlay protein binding assay (VOPBA) was performed to identify cellular proteins involved in IBDV entry. Briefly, total proteins from the elution fraction containing the 90-kDa protein were separated by SDS-12.5% PAGE and transferred onto PVDF membranes as stated above. After overnight incubation of transferred proteins with 4% BSA (Sigma) in PBS at 4°C, the membranes were blocked overnight at 4°C with 5% low-fat milk and washed three times with PBS. Afterwards, membranes were incubated with 2 × 104 PFU of IBDV P3009 in PBS with 5% low-fat milk for 5 h at 39°C. After being washed three times with PBS, the membrane was incubated with anti-VP2 polyclonal antibody diluted 1:4,000 in PBS for 2 h at 4°C and with a goat polyclonal anti-rabbit IgG antibody conjugated to alkaline phosphatase diluted 1:4,000 (Jackson ImmunoResearch) in PBS for 2 h at 4°C. For the far-Western assay, all procedures were the same except that a mixture of 10 μg of SVP in PBS was used instead of IBDV. As a negative control, membranes with total DF-1 cell proteins were incubated with anti-VP2 polyclonal antibody, and the same anti-rabbit polyclonal antibody conjugated with alkaline phosphatase as described above was then used as the secondary antibody. Color was developed in an alkaline phosphatase buffer containing nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolylphosphate (BCIP) (5).
Infection inhibition by anti-Hsp90 and Hsp-90.
DF-1 cells were seeded in 24-well plates with DMEM supplemented with 10% FBS for 5 h. The medium was then removed, and DF-1 cells were incubated with an increasing concentration of rat anti-Hsp90α monoclonal antibody (from 0.5 to 15 μg/250 μl) at 4°C for 60 min with one repeat. Similarly, the cells were treated with different concentrations of mouse monoclonal MAbSVP-1 as a control. After the media were removed, the cells were infected with 100 TCID50 of IBDV P3009 per well at 4°C for 1 h and then washed three times with fresh culture medium. Fresh medium was then added to the infected cells, and the infection was allowed to proceed for 72 to 96 h at 39°C. After infection, the supernatant was collected, and the viral infectivity titer was measured by a standard end-point method using DF-1 cells as a host.
Additionally, 100 TCID50 of IBDV P3009 were incubated with increasing concentrations (from 250 to 4,000 ng) of recombinant Hsp90α protein (Stressgen Biotechnologies) prior to infection. These Hsp90α-containing viral mixtures were added to infect DF-1 cells seeded in 24-well plates for 1 h. Similarly, cells were treated with 100 TCID50 of IBDV P3009 preincubated with different concentrations of BSA as a control. Later, infected cells were washed three times with culture medium. Fresh medium was then added to infected cells, and infection was allowed to proceed for 72 to 96 h at 39°C. After infection, the supernatant was collected, and the viral infectivity titer was measured as described above.
RESULTS
DF-1 cells are susceptible to IBDV.
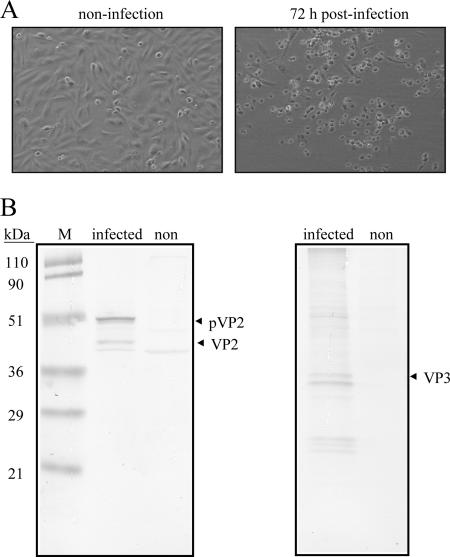
Several cell lines, including DF-1, Vero, Hi-5, and Sf9 cells, were infected with a local isolate of IBDV P3009 to evaluate their susceptibility to infection with IBDV. Among the cell lines tested, DF-1 cells were found to be the most susceptible, because a titer of 6 × 106 PFU/ml of IBDV could be obtained only from the infected DF-1 or CEF cells. CPE of DF-1 cells was observed at 3 days (72 h) postinfection (Fig. 1A, right). By Western blot analysis (Fig. 1B), two major structural proteins, pVP2/VP2 and VP3, with molecular masses of 50/43 kDa and 33 kDa, respectively, were generated in the infected DF-1 cells but not in the control (uninfected) group. This result confirms that DF-1 cells allow the entry of IBDV, which results in the successful replication of the virus. These data also imply that DF-1 cells may bear receptors for the binding and entry of IBDV.
FIG. 1.
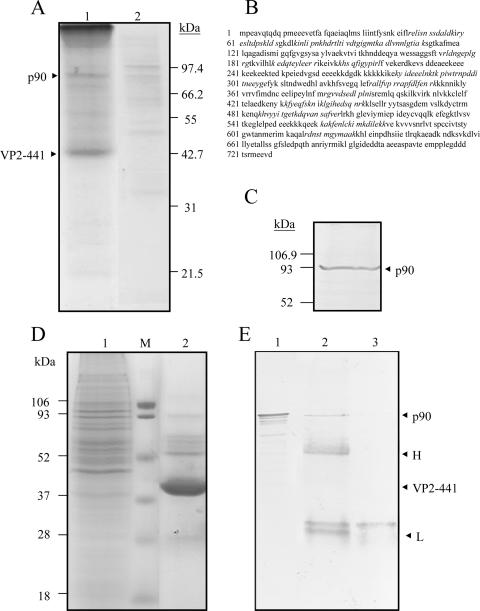
Infection of DF-1 cells with a local IBDV strain, strain P3009. (A) CPE of DF-1 cells caused by IBDV infection. More cells with a round shape (CPE) (right) were seen, and some infected cells died 72 h postinfection; cells in the noninfected group were still healthy (left). (B) Western blot of IBDV-infected DF-1 cells. Two major structural proteins, pVP2/VP2 and VP3, with molecular masses of 50/43 kDa and 33 kDa, respectively, generated in the IBDV-infected DF-1 cells were recognized by anti-VP2 (left) and anti-VP3 (right) polyclonal antibodies, respectively, and neither pVP2/VP2 nor VP3 was observed in the noninfected group. Molecular mass standards are shown on the left in kDa.
Expression and purification of SVP formed by VP2-441.
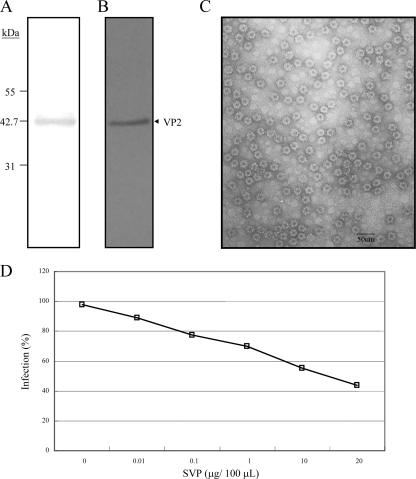
To study the attachment and entry of SVP to the DF-1 cells, SVP was produced by expressing VP2 with 441 amino acid residues, i.e., VP2-441 (∼43 kDa) (Fig. 2A), in Hi-5 cells. The expressed VP2-441 could be recognized by a polyclonal anti-VP2 antibody (Fig. 2B) and self-assembled into the form of SVP (Fig. 2C). The VP2-441-formed SVP was purified by IMAC and then concentrated by membranes with a 100-kDa-molecular-mass cutoff. Under the electron microscope, SVP is homogenous in morphology, and its size is about 25 nm in diameter (Fig. 2C). In our previous study of X-ray structures (19), the strong negative charge on the outer surface of the SVP accounts for the SVP affinity for a Ni-nitrilotriacetic acid (NTA) column at pH 7.8 and the SVP elution from the column at pH 4.0 despite the lack of a His tag in the protein.
FIG. 2.
Purity analysis of VP2-441 SVP prepared by IMAC and inhibition of IBDV infection of DF-1 cells by SVP. IMAC-purified SVP was concentrated through a centrifugal filter (100 kDa) and analyzed by SDS-PAGE with silver staining (A) and Western blotting with anti-VP2 polyclonal antibody (B). Molecular mass standards are shown on the left in kDa. (C) Electron micrograph with ×100,000 magnification (scale bar, 50 nm) and negatively stained with 2% uranyl acetate. (D) Dose-dependent inhibition of IBDV infection by SVP. DF-1 cells were preincubated with different concentrations of SVP (0, 0.01, 0.1, 1, 10, and 20 μg/100 μl) for 1 h at 4°C and then infected with 10 TCID50 of IBDV P3009 for 1 h. CPE was determined using crystal violet staining 96 h after infection. Two separate experiments showed similar results.
Inhibition of the IBDV infection of DF-1 cells by SVP.
To verify whether SVP could interact with the cellular receptors for IBDV, an inhibitory experiment was performed to demonstrate that VP2-441-formed SVP could be a competitor for the infection of DF-1 cells by IBDV. DF-1 cells were preincubated with increasing concentrations of the SVP and then infected with IBDV for 72 to 96 h. The CPE of DF-1 was observed under a microscope. Results showed that SVP was able to block IBDV infection in a dose-dependent manner. At a concentration of 20 μg/ml, SVP inhibited the infectivity of the viruses by ∼60% compared to the control group (Fig. 2D). In contrast, BSA had no effect on the infectivity of IBDV (data not shown), suggesting that the binding of SVP is specific. We surmise that SVP, which has a three-dimensional VP2 conformation similar to that of IBDV (7, 37), may compete with IBDV by occupying the same IBDV cellular receptor(s) and reduce the IBDV infection of DF-1 cells.
DF-1 cells have a limited capacity to bind to SVP.
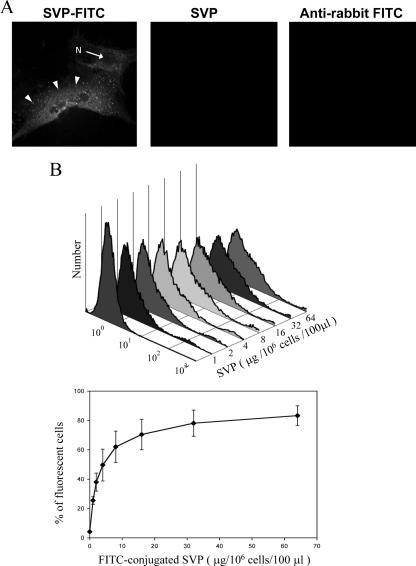
The binding of FITC-labeled SVP to the PM of DF-1 cells was demonstrated by laser scanning confocal microscope. Preliminary results from fluorescence microscopy showed that DF-1 cells display strong fluorescence following the incubation of FITC-labeled SVP (40 μg/50 μl). The confocal images further showed that FITC-labeled SVP could be visualized on the surface of DF-1 cells, as indicated in Fig. 3A. However, no stained cell was detected with the incubation of either unlabeled SVP or FITC-labeled anti-rabbit IgG with this cell line. This result supports that the binding of SVP to DF-1 cells is specific. To determine the particle binding efficiency of DF-1 cells, different amounts of FITC-labeled SVP were incubated with a constant number of cells (106 cells), and the cells bound with FITC-labeled particles were counted by flow cytometry (Fig. 3B, top). The result showed that the binding of SVP to DF-1 cells is dose dependent (Fig. 3B). At low concentrations of SVP, binding increased with increasing particle concentrations. However, the binding was saturated when approximately 32 μg of SVP/100 μl was incubated with 106 cells (Fig. 3B, bottom).
FIG. 3.
Characterization of SVP binding to DF-1 cells by confocal microscopy (A) and flow cytometry (B). DF-1 cells grown on microscope slides were incubated with FITC-labeled SVP (left), unlabeled SVP (middle), and FITC-labeled goat anti-rabbit IgG (right) for 60 min at 4°C. Cells were fixed in 4% paraformaldehyde for 60 min after the removal of unbound SVP and antibody. Cells were examined using a confocal microscope (Zeiss LSM510) at ×630 magnification or a fluorescent microscope (Nikon TS-2000) at ×400 magnification. The cell nucleus (N) and the represented endosome (arrows) are indicated. (B) Saturation of SVP binding to DF-1 cells. Aliquots of 106 DF-1 cells incubated with increasing amounts of the FITC-conjugated SVPs were submitted to one-color fluorescence flow cytometric analysis. A typical flow cytometry histogram is shown on the top. The percentage of cell-bound SVP (FITC-positive cells) was determined in three separate experiments. Average values are shown with standard deviations (bottom).
Binding of SVP to DF-1 cells was inhibited by a neutralizing MAb but not by a polyclonal antibody.
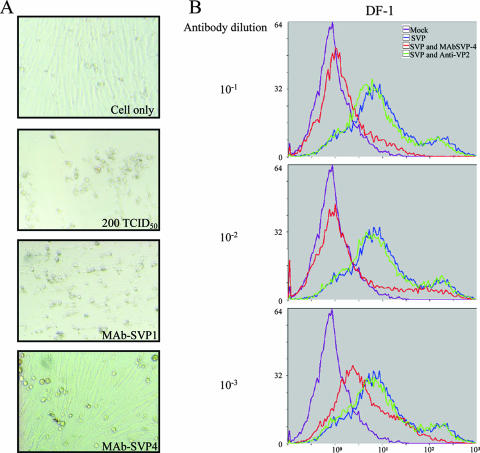
To further characterize the binding of SVP to the DF-1 cell line, purified anti-VP2 monoclonal and polyclonal antibodies were used in the SVP binding inhibition test. As described in Materials and Methods, several MAbs specific for VP2-441-formed SVP were generated to characterize their antigenic regions. Among them, MAbSVP-1 and MAbSVP-4 are the only two MAbs that have a higher ELISA titer (approximately 103 and 104, respectively). In addition, both MAbs can precipitate IBDV (data not shown). These two MAbs were further tested for their virus-neutralizing activities. As shown in Fig. 4A, MAbSVP-4 is able to inhibit the infection of CEF (and DF-1) cells by IBDV (data not shown) and has a neutralizing titer of at least 103. However, MAbSVP-1 as well as a polyclonal antibody (anti-VP2) that was induced by a denatured VP2 fragment (22) have almost no neutralizing activity (Fig. 4A). Therefore, MAbSVP-4 and polyclonal anti-VP2 antibody were used to compare their ability to inhibit SVP binding to DF-1 cells. MAbSVP-4 blocked the binding of the FITC-labeled SVP to DF-1 cells in a dose-dependent manner at a dilution of 10−1 to 10−3 of the MAbSVP-4 antibody (Fig. 4B). In contrast, the anti-VP2 polyclonal antibody did not significantly affect the binding of the labeled SVP (Fig. 4B). Since MAbSVP-4 may block the infection of IBDV through the interaction with the cellular receptors of IBDV, it is very possible that SVP interacts with the same cellular receptors on DF-1 cells.
FIG. 4.
Dose-dependent inhibition of SVP binding to DF-1 cells by MAbSVP-4, a neutralizing MAb. (A) MAbSVP-4 protects CEF cells from IBDV infection. The IBDV neutralization capabilities of MAbSVP-4, MAbSVP-1, and anti-VP2 were measured by virus neutralization assays. Serum samples were diluted twofold, starting at a 1:16 dilution, in Medium 199 and mixed with 200 TCID50 of IBDV P3009 per well (final serum dilution, 1:10,240) at 37°C for 1 h. The antibody-virus mixture was then added to 2 × 104 CEF cells in Medium 199 containing 10% fetal calf serum per well in a 96-well culture plate. After 4 days at 37°C, cell monolayers were observed under a microscope for CPE and were then washed with PBS and stained for 20 min with 1.5% crystal violet in 50% ethanol. The titer of neutralizing activity was evaluated by visual screening of the infected monolayers, and the end point was calculated as the reciprocal value of the highest serum dilution that causes a 50% reduction of the cell monolayer. Results for MAbSVP-4 with a dilution factor of 1,024 and for MAbSVP-1 with a dilution factor of 16 (same as anti-VP2) are shown. CPE of cells incubated with a mixture of MAbSVP-1 (or anti-VP2) and virus could be observed as clearly as that in the negative control, where CEF cells were infected with only 200 TCID50 of the virus. A similar result was not shown for the cells treated with a mixture of anti-VP2 and virus. In contrast, with MAbSVP-4 neutralizing IBDV and protecting the cells from the infection, cells incubated with MAbSVP-4 and virus complex were as healthy as cells of the control group (top). (B) Flow cytometry histograms. Shown are staining DF-1 cells with FITC-conjugated SVP preincubated with either different concentrations of MAbSVP-4 or anti-VP2. Violet, control cells; blue, cells incubated with FITC-labeled SVP; red, cells incubated with FITC-labeled SVP and MAbSVP-4; green, cells incubated with FITC-labeled SVP and anti-VP2 polyclonal antibody.
Purification of cellular proteins associated with SVP and identification of cHsp90α in elution fractions from DF-1 cell extracts.
Since SVP partially inhibits the infection of DF-1 cells by IBDV and induces a MAb with neutralizing activity against IBDV, we suspect that SVP can interact with the cellular receptors of IBDV. An affinity chromatography approach, recently employed for the isolation of dengue virus receptor (36), was set up using SVP coupled on Ni-NTA resins as bait to isolate the receptors. Four milligrams of total protein extract from DF-1 cells diluted in an IMAC native binding buffer was passed through a column packaged with SVP-coupled resins. The column was then washed with two buffers (native wash buffers 1 and 2) to eliminate more nonspecific binding (data not shown). Finally, proteins that strongly interact with SVP were eluted in one step by a native elution buffer (20 mM H2PO4 and 0.5 M NaCl [pH 4.0]) in which most of the SVP were eluted. Several cellular proteins were coeluted, and a 90-kDa protein (p90) was the dominant one among them (Fig. 5A, lane 1). As expected, this 90-kDa protein was only slightly detected when the extracts were passed through the column in the absence of SVP (Fig. 5A, lane 2).
FIG. 5.
Isolation of cHsp90 with affinity for SVP. (A) Affinity isolation of a p90 protein with SVP as the IMAC ligand. A procedure described in the text was set up for affinity isolation of putative IBDV receptors using SVP as the ligand bound to the immobilized Ni2+ ions. In brief, total proteins from DF-1 cells were passed through the affinity chromatographic column. After washing with two wash buffers, the elution of SVP and its associated factors was accomplished by using an IMAC elution buffer. The fractions collected were concentrated, and 40 μl of the concentrate (lane1) and, as a control, 50 μl of the concentrated elution fraction obtained in the absence of SVP (lane 2) were separated by SDS-12.5% PAGE and stained with Coomassie blue (left). Positions of a p90 protein and the monomer VP2-441 that forms SVP are indicated on the left. (B) Identification of p90 by mass spectrometry. The protein sequence of cHsp90 was derived from Protein Data Bank accession number P11501. Peptide sequences of p90 identified by mass spectrometry are shown in italics. (C) Identification of p90 as cHsp90 by Western blotting. An aliquot of 40 μl of the concentrated elution from the affinity column chromatography with SVP was separated by SDS-12.5% PAGE, transferred onto a PVDF membrane, and incubated with an anti-Hsp90 MAb. The membranes were then incubated with a second antibody coupled to alkaline phosphatase and developed in a buffer containing NBT and BCIP. Molecular mass markers are indicated on the left. An arrow indicates the position of p90 (cHsp90). (D) Isolation of p90 from DF-1 cells by immunoprecipitation. Total proteins from DF-1 cells (lane 1) and the precipitated immune complexes (lane 2) were separated by SDS-12.5% PAGE and stained with Coomassie blue (left). The precipitated immune complexes in the presence of SVP (lane 2) and without SVP (lane 3) were separated by SDS-12.5% PAGE, transferred onto a PVDF membrane, and incubated with anti-Hsp90. (E) Western blots were developed as described above. As expected, cHsp90 (p90) was identified in the total DF-1 lysate (lane 1). Molecular mass markers are indicated on the left. Arrows indicate positions of proteins of the immune complexes.
To identify the p90 protein, the protein band in the gel was excised, and after alkylation and in-gel digestion with trypsin, the peptide mass map was analyzed by mass spectrometry. This mass spectrum was compared with protein databases, and the p90 protein was identified as being cHsp90α. Peptide sequences identified by mass spectrometry are shown in Fig. 5B. To further corroborate this identification, transferred proteins obtained in elution fractions from DF-1 cell lysates were probed with a commercially available anti-Hsp90 MAb followed with the corresponding alkaline phosphatase-conjugated secondary antibody. The monoclonal anti-Hsp90 antibody specifically recognized the p90 protein in the elution fraction obtained from DF-1 cells (Fig. 5C).
To alternatively verify the interaction between SVP and cHsp90 in DF-1 cell lysates, the SVP was used as bait in the pull-down IP assays. The protein band pattern pulled down in SVP complexes in which p90 was noticed was found (Fig. 5D, lane 2). Notably, other protein bands with molecular masses of 72, 68, and 62 kDa were also observed. The anti-Hsp90 MAb recognized the 90-kDa protein in pulled-down SVP elutes from DF-1 cells (Fig. 5E, lane 2), while no protein bands were recognized by anti-Hsp90 when the resin was incubated with the lysates in the absence of SVP (Fig. 5E, lane 3). Thus, this p90 protein was identified as being cHsp90, which could interact with SVP as demonstrated by the pull-down IP assay. As expected, the presence of cHsp90 in total extracts from DF-1 cells was also observed (Fig. 5E, lane 1).
cHsp90 is present on the surface of DF-1 cells.
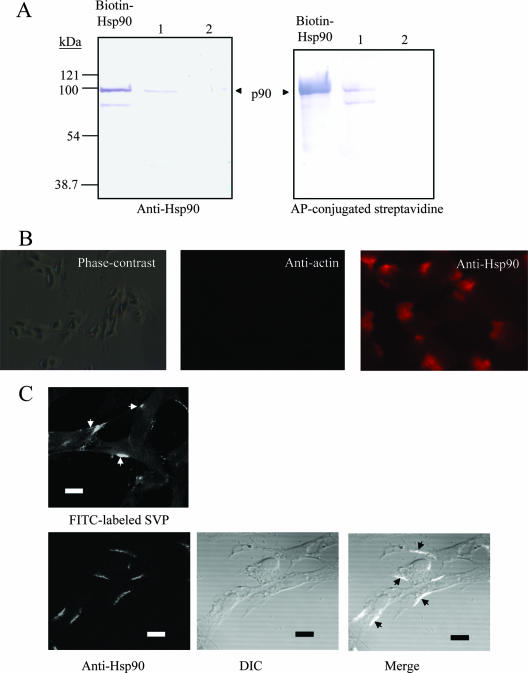
To demonstrate that p90 (cHsp90) is a surface protein of DF-1 cells, a biotinylation of membrane proteins from DF-1 cells was performed. After biotin labeling, cell membrane proteins were prepared and passed through the Ni-NTA-SVP column. The 90-kDa protein obtained in the elution fraction was revealed with anti-Hsp90 (Fig. 6A, left, lane 1) and alkaline phosphatase-conjugated streptavidin (Fig. 6A, right, lane 1), indicating that it is located on the surface of DF-1 cells because it could be labeled by biotin under a nonpermeable condition and because it could be specifically purified by the immobilized SVP. In contrast, the 90-kDa protein was barely detectable using Ni-NTA resin in the absence of SVP (Fig. 6A, lane 2). As a control, no band with a similar molecular mass was detected by alkaline phosphatase-conjugated streptavidin in total nonbiotinylated extract (data not shown). Alternatively, the presence of cHsp90 was corroborated by an indirect immunofluorescence assay on the surface of nonpermeable DF-1 cells. Cells treated with anti-actin MAb have no detectable fluorescence (Fig. 6B, middle), suggesting that the integrity of the cells is preserved. Under the same conditions, anti-Hsp90 MAb was able to detect cHsp90 protein (Fig. 6B, right), indicating that the identification of cHsp90 by anti-Hsp90 MAb is specific and that cHsp90 is located on the surface of DF-1 cells. The characteristic distributions of cHsp90 among the PM of DF-1 cells are indicated in Fig. 6C (bottom right), suggesting that cHsp90 locates only in certain areas of the PM. As a comparison, the binding of SVP to the PM was also demonstrated in Fig. 6C (top left). The similar binding pattern of SVP or anti-Hsp90 to the PM suggests that SVP may colocalize with cHsp90 on the surface of DF-1 cells.
FIG. 6.
Surface localization of the 90-kDa protein from DF-1 cells. (A) Purification of biotinylated membrane-bound cHsp90 of DF-1 cells by SVP. Surface-biotinylated proteins from DF-1 were passed through a column with SVPs coupled to Ni-NTA-Sepharose. Aliquots of 40 μl of the elution fraction (lane 1), the elution fractions obtained in the absence of SVP (as controls) (lane 2), and a biotinylated recombinant Hsp90 (lane Biotin-Hsp90) were separated by SDS-12.5% PAGE, transferred onto a PVDF membrane, and stained with anti-Hsp90 MAb and a proper secondary antibody (left) or streptavidin-alkaline phosphatase (AP) (right), NBT, and BCIP. Molecular mass markers are indicated on the left. The arrow indicates the migration of eluted p90 present in DF-1 cells. An unidentified product has also been visualized. (B) Localization of cHsp90 on the surface of DF-1 by indirect immunofluorescence. Nonpermeabilized cells were incubated with anti-β-actin MAb (middle) or anti-Hsp90 (right), followed by staining with a proper secondary antibody, and examined by fluorescence microscopy at ×600 magnification. A phase-contrast image of cells is shown on the left. (C) Characteristic distribution of cHsp90 among the PM of DF-1 cells. DF-1 cells grown on microscope slides were prefixed and then incubated with FITC-labeled SVP or treated with anti-Hsp90 as described above. After mounting, cells were examined using a confocal microscope (FluoView FV1000; Olympus). Characteristic distributions of SVP (top) and cHsp90 (bottom) among the PM are indicated by arrows. Differential interference contrast (DIC) was used to yield a better image when viewed in bright-field illumination. Bar, 10 μm.
Identification of the virus binding protein by VOPBAs.
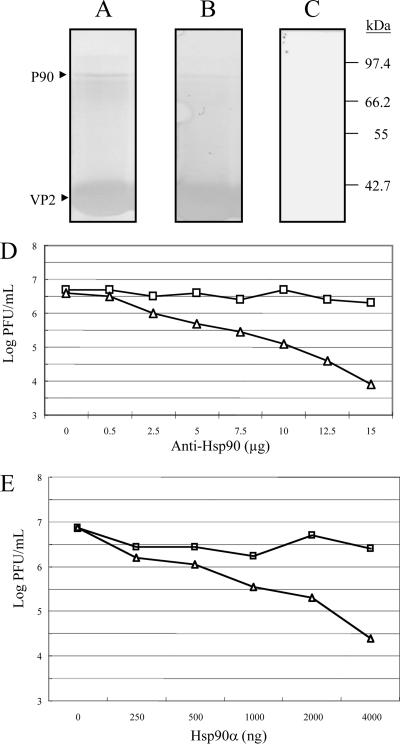
A VOPBA was performed to confirm that the 90-kDa protein could be recognized by IBDV. The elution fraction containing this 90-kDa protein was separated by SDS-12.5% PAGE and transferred onto a nitrocellulose membrane. The membrane was incubated with either IBDV P3009 or SVP and afterwards with a polyclonal anti-VP2 polyclonal antibody. Either IBDV or SVP was able to bind to the 90-kDa protein (Fig. 7A and B, respectively), while in the absence of IBDV, the antibody was unable to react with any band (Fig. 7C).
FIG. 7.
VOPBA and far-Western assay of p90 and infection inhibition assays of recombinant Hsp90 protein and anti-Hsp90 in DF-1 cells. Elution fractions containing proteins from DF-1 showing affinity to SVP bound to the immobilized Ni2+ ions were separated by SDS-12.5% PAGE and transferred onto PVDF membranes. Membranes were incubated with (A) or without (C) 2 × 104 PFU of IBDV and later with a rabbit polyclonal anti-VP2 antibody and goat anti-rabbit IgG coupled to alkaline phosphatase; color was developed with BCIP and NBT. A similar procedure was performed for the far-Western assay (B), except that IBDV was replaced with SVP. Molecular mass markers are indicated on the right. p90 (marked with an arrow) from DF-1 cells was recognized by IBDV (A) and SVP (B). Notably, some other proteins were slightly recognized. (D) Infection inhibition assays with anti-Hsp90 antibody. DF-1 cells were preincubated with different concentrations of SVPMAb-1 (square, mouse monoclonal antibody as a control) or anti-Hsp90 antibody (triangle) for 1 h at 4°C. Subsequently, cells were infected with 100 TCID50 of IBDV. Culture supernatants were collected 96 h after infection, and the resulting infectious virus titer was determined. Each point represents the average of two separate experiments. (E) Infection inhibition assays of recombinant Hsp90. Recombinant Hsp90 protein (Hsp90α) of different concentrations or BSA (as a negative control) was preincubated with 100 TCID50 of IBDV for 1 h at 39°C prior to its incubation with DF-1 cells. Culture supernatants were collected 96 h after infection, and the resulting infectious virus titer was determined. Each point represents the average of two separate experiments.
IBDV infection is inhibited by a recombinant Hsp90α protein and an anti-Hsp90α monoclonal antibody.
To further confirm that cHsp90α is a functional part of the virus receptor complex, anti-Hsp90α MAb was used to evaluate the role of cHsp90α in the infection of DF-1 cells by IBDV. The anti-Hsp90α MAb reduced the infectivity of IBDV P3009 to DF-1 cells in a dose-dependent manner, up to 2.5 logs in infectious virus yields evaluated by the end-point dilution method (Fig. 7D). This reduction in virus yields was not observed for the other control MAb, MAbSVP-1 (Fig. 7D), suggesting that cHsp90α is specifically involved in the entry of IBDV into DF-1 cells. Additionally, a recombinant Hsp90α was used as a competitor in an infection assay. This commercial recombinant human Hsp90α was used because it has a high level of sequence homology to cHsp90α. As shown in Fig. 7E, this recombinant human Hsp90α was able to inhibit the infection of DF-1 cells by IBDV in a dose-dependent manner. Compared to BSA, a reduction of infectious virus yields of approximately 2 logs was observed at a concentration of 4 μg for both proteins.
DISCUSSION
Virus receptors on host cells are essential for the determination of virus host range and tissue tropism. For IBDV, little is known about its receptor. In this study, we established a system to identify the putative IBDV receptors from DF-1 cells, a cell line derived from primary CEF cells (14). CEF cells are generally used for the propagation of IBDV; however, the preparation is time-consuming. Here, we first demonstrated that DF-1 cells are IBDV susceptible. Our unpublished data also indicated that DF-1 cells are more favorable for the propagation of IBDV P3009 than Vero cells; therefore, DF-1 cells are used to purify the IBDV receptor and are routinely employed in our laboratory for the propagation of IBDV.
Virus-like particles derived from Norwalk virus (43, 48), JC virus (18), and human papillomavirus (46) have been utilized for the study of viral receptors; however, those virus-like particles are morphologically similar to their parent virions. In our study, SVP is unique because the size of SVP (∼25 nm) is over two times smaller than that of IBDV (∼65 nm). Despite the smaller size, the preparation of SVP is much simpler than that of IBDV capsid or IBDV-like particles (virus-like particles) (21). There are two more reasons why SVP was used. First, SVP can inhibit the infection of DF-1 cells by IBDV (Fig. 2D) and freshly prepared CEF cells (data not shown) in a dose-dependent manner. However, the differences between T=1 SVP and T=13 IBDV capsid may account for the incomplete inhibition of IBDV infection of DF-1 cells by SVP (Fig. 2D). Second, SVP, similarly to IBDV, is able to induce a high titer of neutralizing antibody (47). This is first time that a neutralizing MAb, i.e., MAbSVP-4, was generated using SVP as an antigen. This result further suggests that SVP carries the neutralizing epitopes that could be viral receptor-binding domains, as indicated by studies of other viruses (12, 34). Taken together, we suspect that SVP, as IBDV, is able to interact with cellular receptors.
Interaction between SVP and DF-1 cells: implication for the entry of IBDV.
Many viruses gain entry into the cell through the endosomal pathway (40). For a prototype virus of the family Birnaviridae, infectious pancreatic necrosis virus penetration occurs through receptor-mediated endocytosis in CHSE-214 cells (8). Similar to infectious pancreatic necrosis virus, the early stages of the infection of CHSE-214 cells by marine birnavirus, which also belongs to the genus Aquabirnavirus, showed virus penetration by vesicular peripheral compartments by electron microscopy (15). While adsorption/penetration mechanisms of IBDV are not fully characterized so far, the specificity of IBDV binding to an N-glycosylated receptor of LSCC-BK3 cells has been observed using biotinylated IBDV, an IBDV-specific chicken serum, MAb GI-27, and flow cytometry (29). As indicated by our binding experiments, the binding of SVP to DF-1 cells is saturated and specific. These results indicate that the binding of SVP is also receptor mediated. To demonstrate our theory, SVP was utilized as a bait to isolate cHsp90α as one of the binding molecules on the surface of DF-1 cells. Because cHsp90α can also be recognized by IBDV, cHsp90α may thus mediate the entrance of IBDV besides SVP. Furthermore, the infection of IBDV was also inhibited by an anti-Hsp90 MAb and a recombinant Hsp90α protein, an analogue of cHsp90α, in a dose-dependent manner. Thus, a receptor-mediated endocytosis pathway is proposed for the entry of IBDV or SVP in which cHsp90α is the putative receptor (41).
cHsp90α as a receptor component.
Hsp90 is one of the abundant and evolutionarily conserved proteins in most species. Even though cHsp90 functions mainly as a cytoplasmic chaperone (30), several studies suggest that human Hsp90 exists on the cell surface (3, 35, 44). To demonstrate that is also true for DF-1 cells, indirect immunofluorescence assays using anti-Hsp90α MAb and biotinylation of membrane proteins were performed, and the presence of cHsp90α on the surface of DF-1 was detected. Human Hsp90 and Hsp70 have recently been identified as being components of the dengue virus receptor complex in human cells (35). Additionally, they are also the CD14-independent cell surface functional receptors for lipopolysaccharide in human monocytes/macrophages (44). Previous results also indicated that both Hsp's are associated with membrane microdomains (lipid rafts) in response to dengue virus infection (35). Lipid rafts are specific membrane domains enriched in certain lipids, cholesterol, and proteins, and they have been identified as being the gateway for the entry and exit of filoviruses (2). Both cHsp90α and cHsp90β have been identified in chicken cells (16, 25); however, the chance of cHsp90β being an IBDV receptor is low because the peptide sequences identified by mass spectrometry did not match cHsp90β. Although we identify the cHsp90α as being a putative IBDV receptor, the presence of other coreceptors is possible because some minor bands were also identified by affinity chromatography (Fig. 5A) and pull-down assays (Fig. 5D) or the overlay assay (Fig. 7A). Interestingly, it has been reported previously that proteins in LSCC-BK3 plasma membranes with molecular masses of 70, 82, and 110 kDa can specifically bind to virulent IBDV (38). We speculate that two proteins with molecular masses of 70 and 82 kDa are cHsp70 (or cHsc70) and cHsp90α, respectively. In this context, the putative receptor complex formed by cHsp90α and other components may also serve as a gateway for the entry of IBDV into DF-1 cells.
Although our data appear to be sound for DF-1 cells, other avian cells as well as additional negative control cells will be tested to identify coreceptors and to eliminate any nonspecific interactions. As Hsp90 is a chaperone and hence binds many proteins quite tightly, the involvement of other factors is likely. The finding that commercial human Hsp90 can inhibit infection (Fig. 7E) also suggests that the interaction between IBDV P3009 and Hsp90 is not chicken specific and that another receptor is controlling IBDV infection in the host. The detailed characterization of a cell that is nonpermissive to IBDV P3009, e.g., Vero cells, the introduction of chicken Hsp90 into that cell, and the subsequent rescue of infection will give critical functional relevance to the studies.
In summary, several lines of evidence have been provided for cHsp90 as a putative receptor component. First of all, SVP can inhibit IBDV infection of an IBDV-susceptible cell line, DF-1 cells. Also, the attachment of SVP to the surface of DF-1 cells was confirmed by immunofluorescence microscopy, and the binding of SVP to DF-1 cell surfaces was demonstrated by Western blotting (data not shown) and flow cytometry. Furthermore, the attachment of SVP to DF-1 cells was inhibited by a neutralizing MAb against IBDV but not a denatured VP2-induced polyclonal antibody. These results as well as data from an IBDV infection-inhibitory experiment suggest that the attachment of IBDV to susceptible cells is similar to that of VP2-formed SVP. Moreover, cHsp90 was purified by an affinity chromatography approach (utilizing SVP bound on immobilized Ni2+ ions) and identified by mass spectrometry. Both biotinylation experiment and indirect fluorescence assays indicated that cHsp90 is located on the surface of DF-1 cells. VOPBA assays also support the conclusion that cHsp90 interacts with IBDV. In addition to virus overlay blots, the inhibitory effect of anti-Hsp90 or a recombinant Hsp90 has confirmed that cHsp90 is a functional part of the receptor complex for IBDV. Further studies will be necessary to demonstrate whether other putative coreceptors are involved in IBDV infection of DF-1 cells or other IBDV-susceptible cell lines. This will facilitate the design of new antiviral agents and vaccines that could interfere with viral entry into target cells once the receptor complex is defined.
Acknowledgments
This research was supported by grants from the National Science Council (grants NSC 93-2313-B-005-070 to M.-Y.W. and NSC 92-2751-B-001-017-Y to A.H.-J.W. and M.-Y.W.).
We gratefully acknowledge critical review of the manuscript by Bing-Hui Liu (Department of Life Sciences, Chung Shan Medical University) and technical assistance by Pei-Chi Chao (Laboratory of Electron Microscopy, National Science Council, National Chung Hsing University).
Footnotes
Published ahead of print on 23 May 2007.
REFERENCES
- 1.Bass, D. M., and H. B. Greenberg. 1992. Strategies for the identification of icosahedral virus receptors. J. Clin. Investig. 89:3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bavari, S., C. M. Bosio, E. Wiegand, G. Ruthel, A. B. Will, T. W. Geisbert, M. Hevey, C. Schmaljohn, A. Schmaljohn, and M. J. Aman. 2002. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 195:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrd, C. A., W. Bornmann, H. Erdjument-Bromage, P. Tempst, N. Pavletich, N. Rosen, C. F. Nathan, and A. Ding. 1999. Heat shock protein 90 mediates macrophage activation by Taxol and bacterial lipopolysaccharide. Proc. Natl. Acad. Sci. USA 96:5645-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, C. S., S. Y. Suen, S. Y. Lai, G. R. Chang, T. C. Lu, M. S. Lee, and M. Y. Wang. 2005. Purification of capsid-like particles of infectious bursal disease virus (IBDV) VP2 expressed in E. coli with a metal-ion affinity membrane system. J. Virol. Methods 130:51-58. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, Y. S., M. S. Lee, S. Y. Lai, S. R. Doong, and M. Y. Wang. 2001. Separation of pure and immunoreactive virus-like particles using gel filtration chromatography following immobilized metal ion affinity chromatography. Biotechnol. Prog. 17:318-325. [DOI] [PubMed] [Google Scholar]
- 6.Chung, Y. T., S. L. Yu, H. K. Shieh, and L. H. Lee. 1995. Characterization of the nucleotide sequences of the VP2 and a part of VP4 gene of infectious bursal disease virus strain P3009 isolated in Taiwan. Taiwan J. Vet. Med. Anim. Husb. 65:205-213. [Google Scholar]
- 7.Coulibaly, F., C. Chevalier, I. Gutsche, J. Pous, J. Navaza, S. Bressanelli, B. Delmas, and F. A. Rey. 2005. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell 120:761-772. [DOI] [PubMed] [Google Scholar]
- 8.Couve, E., J. Kiss, and J. Kuznar. 1992. Infectious pancreatic necrosis virus internalization and endocytic organelles in CHSE-214 cells. Cell Biol. Int. Rep. 16:899-906. [DOI] [PubMed] [Google Scholar]
- 9.Garzino-Demo, A., and R. C. Gallo. 2003. HIV receptors on lymphocytes. Curr. Opin. Hematol. 10:279-283. [DOI] [PubMed] [Google Scholar]
- 10.Hadas, E., and G. Theilen. 1987. Production of monoclonal antibodies. The effect of hybridoma concentration on the yield of antibody-producing clones. J. Immunol. Methods 96:3-6. [DOI] [PubMed] [Google Scholar]
- 11.Haywood, A. M. 1994. Virus receptors: binding, adhesion strengthening, and changes in viral structure. J. Virol. 68:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He, Y., H. Lu, P. Siddiqui, Y. Zhou, and S. Jiang. 2005. Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies. J. Immunol. 174:4908-4915. [DOI] [PubMed] [Google Scholar]
- 13.Heine, H. G., M. Haritou, P. Failla, K. Fahey, and A. Azad. 1991. Sequence analysis and expression of the host-protective immunogen VP2 of a variant strain of infectious bursal disease virus which can circumvent vaccination with standard type I strains. J. Gen. Virol. 72:1835-1843. [DOI] [PubMed] [Google Scholar]
- 14.Himly, M., D. N. Foster, I. Bottoli, J. S. Iacovoni, and P. K. Vogt. 1998. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248:295-304. [DOI] [PubMed] [Google Scholar]
- 15.Imajoh, M., K. Yagyu, and S. Oshima. 2003. Early interactions of marine birnavirus infection in several fish cell lines. J. Gen. Virol. 84:1809-1816. [DOI] [PubMed] [Google Scholar]
- 16.Jerome, V., C. Vourc'h, E. E. Baulieu, and M. G. Catelli. 1993. Cell cycle regulation of the chicken hsp90 alpha expression. Exp. Cell Res. 205:44-51. [DOI] [PubMed] [Google Scholar]
- 17.Kibenge, F. S., and P. K. McKenna. 1992. Isolation and propagation of infectious bursal disease virus using the ovine kidney continuous cell line. Avian Dis. 36:256-261. [PubMed] [Google Scholar]
- 18.Komagome, R., H. Sawa, T. Suzuki, Y. Suzuki, S. Tanaka, W. J. Atwood, and K. Nagashima. 2002. Oligosaccharides as receptors for JC virus. J. Virol. 76:12992-13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, C. C., T. P. Ko, C. C. Chou, M. Yoshimura, S. R. Doong, M. Y. Wang, and A. H. Wang. 2006. Crystal structure of infectious bursal disease virus VP2 subviral particle at 2.6Å resolution: implications in virion assembly and immunogenicity. J. Struct. Biol. 155:74-86. [DOI] [PubMed] [Google Scholar]
- 20.Lee, C. C., T. P. Ko, M. S. Lee, C. C. Chou, S. Y. Lai, A. H. Wang, and M. Y. Wang. 2003. Purification, crystallization and preliminary X-ray analysis of immunogenic virus-like particles formed by infectious bursal disease virus (IBDV) structural protein VP2. Acta Crystallogr. D Biol. Crystallogr. 59:1234-1237. [DOI] [PubMed] [Google Scholar]
- 21.Lee, M. S., S. R. Doong, S. Y. Lai, J. Y. Ho, and M. Y. Wang. 2006. Processing of infectious bursal disease virus (IBDV) polyprotein and self-assembly of IBDV-like particles in Hi-5 cells. Biotechnol. Prog. 22:763-769. [DOI] [PubMed] [Google Scholar]
- 22.Lee, M. S., M. Y. Wang, Y. J. Tai, and S. Y. Lai. 2004. Characterization of particles formed by the precursor protein VPX of infectious bursal disease virus in insect Hi-5 cells: implication on its proteolytic processing. J. Virol. Methods 121:191-199. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Barragan, J. J., and R. M. del Angel. 2001. Identification of a putative coreceptor on Vero cells that participates in dengue 4 virus infection. J. Virol. 75:7818-7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathis, J. M., M. A. Stoff-Khalili, and D. T. Curiel. 2005. Oncolytic adenoviruses—selective retargeting to tumor cells. Oncogene 24:7775-7791. [DOI] [PubMed] [Google Scholar]
- 25.Meng, X., V. Jerome, J. Devin, E. E. Baulieu, and M. G. Catelli. 1993. Cloning of chicken hsp90 beta: the only vertebrate hsp90 insensitive to heat shock. Biochem. Biophys. Res. Commun. 190:630-636. [DOI] [PubMed] [Google Scholar]
- 26.Miller, A. D. 2003. Identification of Hyal2 as the cell-surface receptor for jaagsiekte sheep retrovirus and ovine nasal adenocarcinoma virus. Curr. Top. Microbiol. Immunol. 275:179-199. [DOI] [PubMed] [Google Scholar]
- 27.Müller, H., M. R. Islam, and R. Raue. 2003. Research on infectious bursal disease—the past, the present and the future. Vet. Microbiol. 97:153-165. [DOI] [PubMed] [Google Scholar]
- 28.Nieper, H., and H. Müller. 1996. Susceptibility of chicken lymphoid cells to infectious bursal disease virus does not correlate with the presence of specific binding sites. J. Gen. Virol. 77:1229-1237. [DOI] [PubMed] [Google Scholar]
- 29.Ogawa, M., T. Yamaguchi, A. Setiyono, T. Ho, H. Matsuda, S. Furusawa, H. Fukushi, and K. Hirai. 1998. Some characteristics of a cellular receptor for virulent infectious bursal disease virus by using flow cytometry. Arch. Virol. 143:2327-2341. [DOI] [PubMed] [Google Scholar]
- 30.Passinen, S., J. Valkila, T. Manninen, H. Syvala, and T. Ylikomi. 2001. The C-terminal half of Hsp90 is responsible for its cytoplasmic localization. Eur. J. Biochem. 268:5337-5342. [DOI] [PubMed] [Google Scholar]
- 31.Pierson, T. C., and R. W. Doms. 2003. HIV-1 entry inhibitors: new targets, novel therapies. Immunol. Lett. 85:113-118. [DOI] [PubMed] [Google Scholar]
- 32.Pomeranz, L. E., A. E. Reynolds, and C. J. Hengartner. 2005. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. 69:462-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pous, J., C. Chevalier, M. Ouldali, J. Navaza, B. Delmas, and J. Lepault. 2005. Structure of birnavirus-like particles determined by combined electron cryomicroscopy and X-ray crystallography. J. Gen. Virol. 86:2339-2346. [DOI] [PubMed] [Google Scholar]
- 34.Prabakaran, P., J. Gan, Y. Feng, Z. Zhu, V. Choudhry, X. Xiao, X. Ji, and D. S. Dimitrov. 2006. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J. Biol. Chem. 281:15829-15836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reyes-del Valle, J., S. Chavez-Salinas, F. Medina, and R. M. del Angel. 2005. Heat shock protein 90 and heat shock protein 70 are components of dengue virus receptor complex in human cells. J. Virol. 79:4557-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reyes-del Valle, J., and R. M. del Angel. 2004. Isolation of putative dengue virus receptor molecules by affinity chromatography using a recombinant E protein ligand. J. Virol. Methods 116:95-102. [DOI] [PubMed] [Google Scholar]
- 37.Saugar, I., D. Luque, A. Ona, J. F. Rodriguez, J. L. Carrascosa, B. L. Trus, and J. R. Caston. 2005. Structural polymorphism of the major capsid protein of a double-stranded RNA virus: an amphipathic alpha helix as a molecular switch. Structure (Cambridge) 13:1007-1017. [DOI] [PubMed] [Google Scholar]
- 38.Setiyono, A., T. Hayashi, T. Yamaguchi, H. Fukushi, and K. Hirai. 2001. Detection of cell membrane proteins that interact with virulent infectious bursal disease virus. J. Vet. Med. Sci. 63:219-221. [DOI] [PubMed] [Google Scholar]
- 39.Sia, S. K., P. A. Carr, A. G. Cochran, V. N. Malashkevich, and P. S. Kim. 2002. Short constrained peptides that inhibit HIV-1 entry. Proc. Natl. Acad. Sci. USA 99:14664-14669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sieczkarski, S. B., and G. R. Whittaker. 2002. Influenza virus can enter and infect cells in the absence of clathrin-mediated endocytosis. J. Virol. 76:10455-10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, A. E., and A. Helenius. 2004. How viruses enter animal cells. Science 304:237-242. [DOI] [PubMed] [Google Scholar]
- 42.Smith, B. J., J. L. McKimm-Breshkin, M. McDonald, R. T. Fernley, J. N. Varghese, and P. M. Colman. 2002. Structural studies of the resistance of influenza virus neuramindase to inhibitors. J. Med. Chem. 45:2207-2212. [DOI] [PubMed] [Google Scholar]
- 43.Tamura, M., K. Natori, M. Kobayashi, T. Miyamura, and N. Takeda. 2000. Interaction of recombinant Norwalk virus particles with the 105-kilodalton cellular binding protein, a candidate receptor molecule for virus attachment. J. Virol. 74:11589-11597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Triantafilou, K., M. Triantafilou, and R. L. Dedrick. 2001. A CD14-independent LPS receptor cluster. Nat. Immunol. 2:338-345. [DOI] [PubMed] [Google Scholar]
- 45.van Loon, A. A., N. de Haas, I. Zeyda, and E. Mundt. 2002. Alteration of amino acids in VP2 of very virulent infectious bursal disease virus results in tissue culture adaptation and attenuation in chickens. J. Gen. Virol. 83:121-129. [DOI] [PubMed] [Google Scholar]
- 46.Volpers, C., F. Unckell, P. Schirmacher, R. E. Streeck, and M. Sapp. 1995. Binding and internalization of human papillomavirus type 33 virus-like particles by eukaryotic cells. J. Virol. 69:3258-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, M. Y., Y. Y. Kuo, M. S. Lee, S. R. Doong, J. Y. Ho, and L. H. Lee. 2000. Self-assembly of the infectious bursal disease virus capsid protein, rVP2, expressed in insect cells and purification of immunogenic chimeric rVP2H particles by immobilized metal-ion affinity chromatography. Biotechnol. Bioeng. 67:104-111. [DOI] [PubMed] [Google Scholar]
- 48.White, L. J., J. M. Ball, M. E. Hardy, T. N. Tanaka, N. Kitamoto, and M. K. Estes. 1996. Attachment and entry of recombinant Norwalk virus capsids to cultured human and animal cell lines. J. Virol. 70:6589-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]