Abstract
The hepatitis C virus glycoprotein E2 receptor-binding domain is encompassed by amino acids 384 to 661 (E2661) and contains two hypervariable sequences, HVR1 and HVR2. E2 sequence comparisons revealed a third variable region, located between residues 570 and 580, that varies widely between genotypes, designated here as igVR, the intergenotypic variable region. A secreted E2661 glycoprotein with simultaneous deletions of the three variable sequences retained its ability to bind CD81 and conformation-dependent monoclonal antibodies (MAbs) and displayed enhanced binding to a neutralizing MAb directed to E2 immunogenic domain B. Our data provide insights into the E2 structure by suggesting that the three variable regions reside outside a conserved E2 core.
Hepatitis C virus (HCV) is a member of the Flaviviridae family and is classified into six major genotypes and numerous subtypes that differ in nucleotide sequence by up to 35% and 25%, respectively (37). The virus encodes two envelope glycoproteins, E1 (polyprotein residues 191 to 383 [H77c numbering is used throughout this study]) and E2 (residues 384 to 746) that function in viral entry as noncovalently associated heterodimers (1) (Fig. 1A). Glycoprotein E2 attaches the virus to host cell receptors that include the tetraspanin CD81 (33), claudin-1 (14), and the high-density lipoprotein receptor scavenger receptor, class B type I (SR-B1) (35), while E1 contains an internal fusion peptide-like sequence and membrane-proximal heptad repeat, both containing residues essential for viral entry function (9, 16).
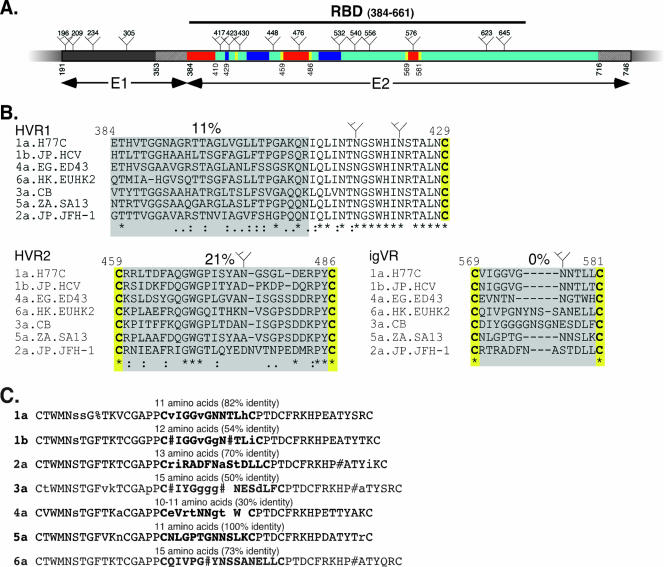
FIG. 1.
(A) Schematic representation of the N-terminal portion of the HCV polyprotein showing E1 in dark gray and E2 in cyan. The locations of putative N-linked glycosylation sites in H77c are shown ( ). Variable regions present within E2 are shown in red and proximal conserved cysteine residues in yellow. The locations of three discontinuous CD81 binding regions are shown in blue, and the hatched regions represent transmembrane domains of E1 and E2. The E2 RBD, residues 384 to 661, is indicated by a line. The numbering is according to the prototype 1a strain H77c. (B) Intergenotypic alignment of the HCV E2 variable regions HVR1, HVR2, and igVR. Representative isolates from each genotype of HCV were aligned using ClustalX. Symbols show conserved (*), semiconserved (:), and weakly conserved (.) residues. The percentage of identity is shown above each alignment, and conserved glycosylation sites are indicated (
). Variable regions present within E2 are shown in red and proximal conserved cysteine residues in yellow. The locations of three discontinuous CD81 binding regions are shown in blue, and the hatched regions represent transmembrane domains of E1 and E2. The E2 RBD, residues 384 to 661, is indicated by a line. The numbering is according to the prototype 1a strain H77c. (B) Intergenotypic alignment of the HCV E2 variable regions HVR1, HVR2, and igVR. Representative isolates from each genotype of HCV were aligned using ClustalX. Symbols show conserved (*), semiconserved (:), and weakly conserved (.) residues. The percentage of identity is shown above each alignment, and conserved glycosylation sites are indicated ( ). (C) Intragenotypic alignment of the igVR region. Sequences were aligned using MULTALIGN (4a), and the consensus sequence is shown. The length of the igVR and the percentage of identity are shown above each alignment. Symbols show N, D, Q, or E at conserved positions (#) and F/Y conserved positions (%). Uppercase letters indicate >90% identity, and lowercase letters 50% to 90% identity. A space shows a <50%-conserved residue or an insertion/deletion.
). (C) Intragenotypic alignment of the igVR region. Sequences were aligned using MULTALIGN (4a), and the consensus sequence is shown. The length of the igVR and the percentage of identity are shown above each alignment. Symbols show N, D, Q, or E at conserved positions (#) and F/Y conserved positions (%). Uppercase letters indicate >90% identity, and lowercase letters 50% to 90% identity. A space shows a <50%-conserved residue or an insertion/deletion.
The receptor-binding domain (RBD) of E2 is encompassed by polyprotein residues 384 to 661 (E2661) (Fig. 1A). Recombinant forms of E2661 RBD are efficiently secreted from transfected cells and are able to interact with CD81, SR-B1, and other cell surface molecules (4, 33, 35). The E2 RBD contains two hypervariable regions, HVR1 (residues 384 to 410) and HVR2 (residues 474 to 482) (21, 42). Hypervariable region 1, located at the N terminus of E2, is the most variable region in the HCV genome, is highly immunogenic, and rapidly accumulates neutralization escape mutations (15). Despite the high level of amino acid variability in HVR1, there is an overall conservation of basic residues that are important for viral entry (3, 32). HVR1 also appears to play a role in the enhancement of viral entry via high-density lipoproteins present in human serum, which upregulate the SR-B1-mediated endocytosis of virions (2, 7, 26, 29, 40).
Hypervariable region 2 is located within the region flanked by Cys-459 to Cys-486 (21). Although originally described as a 7-residue sequence, comparison of E2 sequences from different HCV genotypes suggests it may extend from residue 461 to 481 (Fig. 1B and data not shown). The degree of sequence identity across the Cys-459 to Cys-486 region ranges from 39% (genotypes 1a and b) to 93% (genotype 5a), and the region is 28 to 30 residues in length (data not shown). In comparison to the HVR1 sequence, the sequence of HVR2 is relatively stable within HCV-infected people (30), although an accumulation of mutations at this location has been shown to correlate with responsiveness to alpha interferon treatment (19). An N-linked glycosylation site is conserved in genotypes 1a, 4a, 6a, 3a, and 2a, while the G468WG motif is conserved in all isolates, suggesting that structural features of HVR2 are necessary for E1E2 function (Fig. 1B and data not shown). A third hypervariable region (residues 431 to 466) has recently been reported based on the analysis of 391 sequences from 17 subjects. Although this region contained a high rate of nonsynonymous versus synonymous base changes, the corresponding amino acid substitutions were conservative, and overall hydropathy was conserved (38).
The alignment of E2 sequences representing the six major genotypes of HCV reveals a previously undescribed variable region between polyprotein residues 570 and 580 that is relatively conserved within a genotype but varies across genotypes due to amino acid insertions and deletions (Fig. 1B). We have designated amino acids 570 to 580 the intergenotypic variable region (igVR). Examination of the corresponding region from all six genotypes of HCV (Fig. 1B) and divergent isolates therein (Fig. 1C) shows that the igVR length ranges from 10 amino acids (genotype 4a) to 15 amino acids (genotypes 3a and 6a). All isolates contain a single conserved N-linked glycosylation site at position 576. Similar to HVR2, igVR is also flanked by conserved cysteine residues (Cys-569 and Cys-581), suggesting that these sequences form disulfide-constrained loops.
The three-dimensional structure of E2 is currently not available; however, it is fundamental to understanding HCV attachment and entry and thus, the rational design of antiviral agents that block these critical first stages of infection. Although E2661 is a lead candidate for structural studies, as it contains the functional RBD, the presence of mobile solvent-exposed variable loops and a high degree of glycosylation are likely to confound crystallization attempts. Essential to obtaining crystals suitable for three-dimensional structural determination of the human immunodeficiency virus type 1 (HIV-1) receptor-binding glycoprotein, gp120, was the replacement of three of the five variable loops with short linker motifs to reduce the glycoprotein's flexibility for crystallization while maintaining correct disulfide bonding (25). In spite of these deletions, the gp120 core attained its global fold and revealed the neutralization epitopes and RBDs.
We therefore examined whether the conserved E2 core domain retained its ability to form complex structures, such as conformational monoclonal antibody (MAb) epitopes and the CD81 receptor-binding site, following deletion of the three variable regions. As a starting point to a crystallization strategy, HVR1 (polyprotein residues 387 to 409), HVR2 (residues 460 to 485), and igVR (residues 570 to 580) were replaced—individually and in combination—with a short, flexible Gly-Ser-Ser-Gly linker that theoretically provides up to 14.4 Å of flexible spacer length to facilitate correct folding and disulfide pairing. Deletions were performed in the context of the pcE2661myc expression vector that directs the expression of soluble E2661 of the H77c isolate (8). The E2661 protein is translocated into the lumen of the endoplasmic reticulum via an N-terminal tissue plasminogen activator leader sequence, and a C-terminal myc epitope tag (13) facilitates detection and purification (E2661myc). The first three amino acids of E2 (E384TH) were included in all constructs to ensure efficient signal peptide cleavage and glycoprotein secretion.
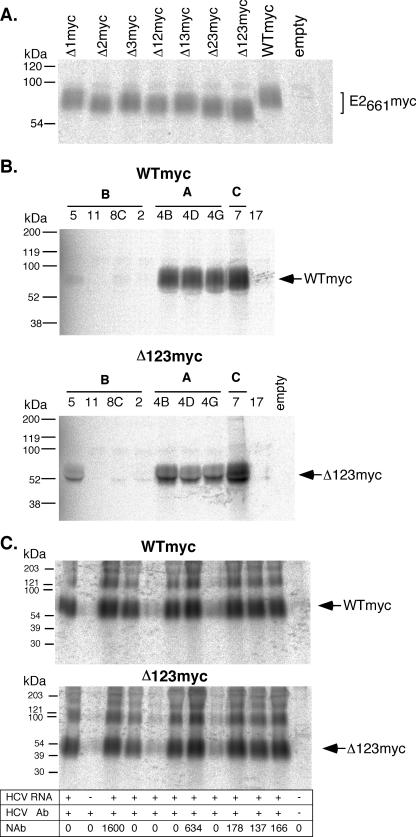
The levels of expression and secretion of the E2661myc deletion constructs were analyzed by radioimmunoprecipitation and nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as previously described (10). Briefly, HEK 293T cells were labeled at 24 h posttransfection with Tran-35S-label (ICN, Costa Mesa, CA). The labeled E2661myc was then immunoprecipitated from the tissue culture fluid, using the conformation-dependent MAb H53, obtained from Jean Dubuisson (6), revealing that all single and multiple variable region E2661myc deletion constructs were expressed and secreted at similar levels (Fig. 2A).
FIG. 2.
Recognition of E2661myc constructs with conformation-dependent MAbs and human serum. (A) Metabolically labeled E2661myc was immunoprecipitated from the tissue culture fluid of transfected cells with the conformation-dependent E2-specific MAb H53 and analyzed by SDS-PAGE under nonreducing conditions in 10 to 15% polyacrylamide gradient gels, followed by scanning in a phosphorimager. (B) Wild-type (WTmyc) and Δ123-deleted E2661myc (Δ123 myc) were metabolically labeled and immunoprecipitated with the “CBH” panel of conformation-dependent human MAbs specific to three immunogenic domains (A, B, and C) of E2 (23). Specific CBH MAbs are indicated above the lanes. Immunoprecipitated proteins were analyzed by SDS-PAGE under nonreducing conditions in 10 to 15% polyacrylamide gradient gels, followed by scanning in a phosphorimager. (C) Immunoprecipitation of WTmyc and Δ123myc proteins by a panel of human sera obtained from HCV-infected individuals. Immunoprecipitated proteins were analyzed by SDS-PAGE under nonreducing conditions in 10 to 15% polyacrylamide gradient gels, followed by scanning in a phosphorimager. Indicated below each lane are the HCV RNA status, presence of HCV-specific antibody detected by using a Bio-Rad Monolisa, Abbot Murex, or Chiron RIBA assay, and the 50% neutralizing antibody titer (NAb) for that serum sample. Molecular mass markers are indicated to the left.
To further examine the effects of these deletions on the conformation of E2, a panel of conformation-dependent human MAbs, obtained from Steven Foung (18), were used to immunoprecipitate biosynthetically labeled WTmyc and Δ123myc (lacking all three variable regions) E2661myc glycoproteins, and analysis by nonreducing SDS-PAGE was performed. This panel of conformation-dependent MAbs are specific to epitopes representing three distinct immunogenic domains of E2—A, B, and C (22, 23). The WTmyc protein was efficiently immunoprecipitated by MAbs to domains A (CBH-4B, -4D, and -4G) and C (CBH-7) but reacted weakly with one domain B MAb, CBH-5. Three other domain B MAbs (CBH-2, -8C, and -11) did not recognize WTmyc, consistent with the observations of Keck et al. with the genotype 1a E2661 (23). The Δ123myc protein exhibited an antigenic profile similar to that of the WTmyc protein (Fig. 2B). Of particular interest was the finding that simultaneous deletion of the three variable sequences led to better recognition by the immunogenic domain B-neutralizing MAb CBH-5 (24), suggesting that this neutralization epitope of E2 may be occluded by the surface-exposed variable regions. A panel of sera obtained from HCV-infected individuals was used to compare the global antigenic profiles of biosynthetically labeled WTmyc and Δ123myc by immunoprecipitation. The sera were screened for the presence of neutralizing antibodies towards H77c E1E2 pseudotyped retoviral particles (Fig. 2C) as described previously (17), with 50% neutralization titers ranging from 0 to 1,600 observed. The reactivity pattern of WTmyc was almost identical to that of Δ123myc, indicating that the antigenic structure of the two proteins is similar (Fig. 2C).
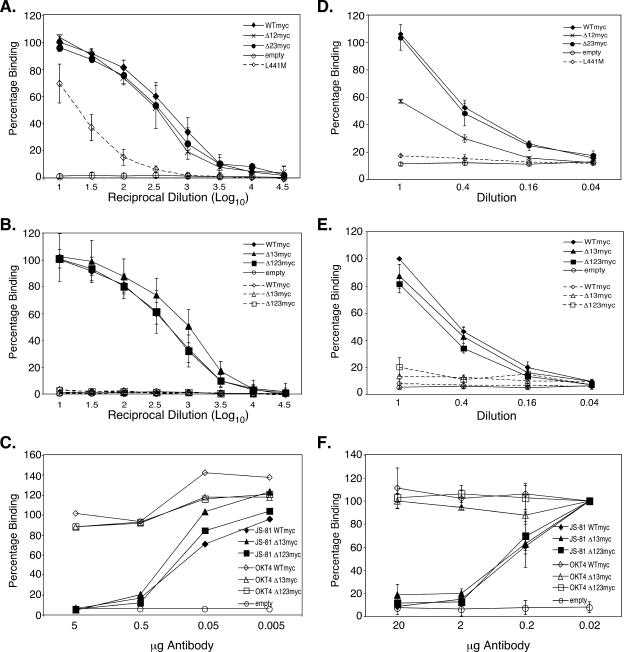
E2-CD81 interactions are critical for HCV infection and have also been implicated in immunopathogenic responses to and immune evasion by HCV in vitro (5, 27, 39, 41). The discontinuous CD81 binding sequences G436WLAGLFY (8), W420 (31), and Y527SWGANDTD (31) form a composite receptor-binding site and are located in conserved regions that alternate with HVR1, HVR2, and igVR (Fig. 1A). We first examined whether the variable region deletions affected the CD81 receptor-binding function by using a chimera composed of the CD81 large extracellular loop (LEL), residues 113 to 201, encoding the E2 binding site and linked to the maltose-binding protein (MBP-LEL113-201) (12). E2661myc proteins from the transfected 293T cell culture supernatants were concentrated approximately 10-fold at 3 days posttransfection. The input E2661myc proteins were normalized following SDS-PAGE under nonreducing conditions, transfer to nitrocellulose, and immunoblotting with 9E10 or H52. The monomeric E2661myc band intensity was quantitated by using a Licor Odyssey system. Equivalent amounts of serially diluted monomeric wild-type and variable region-deleted E2661myc proteins were applied to solid-phase dimeric MBP-LEL113-201, and binding was detected using MAb H53 and horseradish peroxidase-conjugated rabbit anti-mouse immunoglobulin (12). The E2661myc proteins containing single (data not shown) or multiple deletions of the three variable regions, including Δ123myc, showed wild-type levels of binding to MBP-LEL113-201 (Fig. 3A and B). WTmyc, Δ13myc, and Δ123myc did not interact with MBP-LEL113-201 containing the F186S mutation in the E2 binding site (12) (Fig. 3B). Furthermore, E2661myc containing the L441M mutation in the CD81 binding site had a 2-log reduction in the level of binding to wild-type MBP-LEL113-201 (Fig. 3A) (8). MAb JS-81 (BD Biosciences), but not anti-CD4 MAb OKT4, blocked the binding of WTmyc, Δ123myc, and Δ13myc to CD81 in a dose-dependent manner, confirming the specificity of the assay (Fig. 3C).
FIG. 3.
Ability of E2661myc with HVR deletions to interact with CD81. (A) Abilities of WTmyc, Δ12myc, Δ23myc, and E2661myc proteins containing the mutation L441M that disrupts interaction with CD81 to interact with wild-type recombinant MBP-LEL113-201. E2661myc was serially diluted in MBP-LEL113-201-coated enzyme immunoassay plates. The data are the averages of the results of two to seven independent experiments and are shown as the mean percentages of the levels of binding relative to the level of binding of WTmyc, with error bars showing the standard deviations. (B) Abilities of WTmyc, Δ13myc, and Δ123myc to interact with wild-type recombinant MBP-LEL113-201 (solid lines) or recombinant MBP-LEL113-201 containing the F186S mutation that disrupts E2 binding (dashed lines). E2661myc was serially diluted in MBP-LEL113-201-coated enzyme immunoassay plates. The data are the averages of the results of two to seven independent experiments and are shown as the mean percentages of the levels of binding relative to the level of binding of WTmyc, with error bars showing the standard deviations. (C) Inhibition of binding of WTmyc, Δ13myc, and Δ123myc to wild-type recombinant MBP-LEL113-201 by anti-CD81 MAb JS-81. Various amounts of JS-81 or anti-CD4 MAb OKT4 were added to MBP-LEL113-201-coated immunoassay plates, followed by a constant amount of E2661myc protein. Bound E2661myc was detected with biotinylated 9E10 followed by horseradish peroxidase-conjugated avidin. Results shown are representative of two independent experiments. (D and E) Dilutions of E2661myc were applied to ice cold, human CD81-transfected CHO-K1 cells and incubated on ice for 4 h. After plates were washed, 1 × 106 cpm 125I-9E10 was added and plates were incubated for 1 h at room temperature, washed, and counted in a Packard Auto GammaCounter. (D) Abilities of WTmyc, Δ12myc, Δ23myc, and E2661myc protein containing the mutation L441M to interact with full-length CD81 transfected into CHO-K1 cells. The data shown are the mean percentages of the levels of binding relative to the level of binding of the wild type, with error bars showing the standard errors, from two to five independent experiments. (E) Abilities of WTmyc, Δ123myc, and Δ13myc to interact with full-length human CD81 (solid line) or F186S-CD81 (dashed line) transfected into CHO-K1 cells. The data shown are the mean percentages of the levels of binding relative to the level of binding of the wild type, with error bars showing the standard errors, from two to five independent experiments. (F) Inhibition of binding of WTmyc, Δ13myc, and Δ123myc to full-length human CD81 transfected into CHO-K1 cells by anti-CD81 MAb JS-81. Various amounts of JS-81 or OKT4 were applied to transfected cells, followed by a constant amount of E2661myc proteins for 4 h on ice. Bound E2661myc was detected by using iodinated 9E10 as described above. The data shown are the mean percentages of the levels of binding relative to the level of binding with no antibody present, with error bars showing the standard deviations, from two independent experiments.
To confirm these results in the context of the full-length CD81 receptor, we performed a cell surface binding assay using CHO-K1 cells transfected with a full-length CD81 expression vector as previously described (8). Equivalent amounts of monomeric, secreted E2661myc were serially diluted and incubated with CD81-transfected CHO-K1 cells on ice for 4 h. Bound E2661myc was detected by using 125I-labeled MAb 9E10. The assay's specificity was confirmed by the lack of binding by WTmyc, Δ13myc, and Δ123myc to CHO-K1 cells transfected with CD81 containing the mutation F186S in the E2 binding site (Fig. 3E) (12). Furthermore, E2661myc containing the L441M mutation in the CD81 binding site lacked binding activity for wild-type-CD81-transfected CHO-K1 cells (8) (Fig. 3D). Finally, the binding of WTmyc, Δ13myc, and Δ123myc to wild-type-CD81-transfected CHO-K1 cells was inhibited in a dose-dependent manner by anti-CD81 MAb JS-81 (BD Biosciences) but not anti-CD4 MAb OKT4 (Fig. 3F).
Although we did not detect variations in binding to recombinant dimeric MBP-LEL113-201 for singly and multiply deleted E2661myc constructs, differences in binding to surface-expressed CD81 were observed, perhaps reflecting subtle differences in LEL structure when it is expressed in isolation versus in native tetraspanin (11). The results shown in Fig. 3D and E indicate that Δ23myc, Δ13myc, Δ123myc, and WTmyc displayed similar CD81 binding profiles, whereas the deletion of HVR1 plus HVR2 from E2661myc (Δ12myc) caused an approximately 50% reduction in CD81 binding compared to binding by Δ23myc (P = 0.035). By contrast, the CD81 binding abilities of Δ13myc (which exhibited a binding curve identical to that of WTmyc) and Δ123myc were not significantly different (P = 0.62), indicating that the presence of igVR compromises the CD81 binding function when HVR1 and HVR2 are absent in Δ12myc (Fig. 3E). Although HVR1, HVR2, and igVR are not required for the core folding properties of E2661, these data point to a functional interaction between igVR and one or both of HVR1 and HVR2 such that the CD81 binding site is properly formed or becomes fully accessible to the receptor.
The abilities of WTmyc and Δ123myc to bind full-length CD81 and MBP-LEL113-201 at similar levels suggests that the CD81 binding site is significantly exposed in the presence of the three variable sequences in the H77c sequence used here. However, previous studies have revealed differences in the CD81 binding capacity of E2661 and in the infectivities of E1E2-pseudotyped retroviruses prepared from different genotypes of HCV (28, 34, 36). For example, Roccasecca et al. found that the binding of genotype 1b E2661 to CD81 can be improved by the deletion of HVR1 residues 384 to 410, while the replacement of HVR1 and HVR2 of the genotype 1b strain with those from genotype 1a results in increased binding to CD81 (34). A possible function of HVR1, HVR2, and the igVR may be to shield the underlying CD81 binding site from neutralizing antibodies with movement of the variable regions required for attachment to cellular receptors. Precedence for this is found with HIV-1 gp120, where the first, second, and third variable regions occlude the chemokine receptor binding sites and conserved cryptic neutralization epitopes present on the underlying core domain (20, 25, 43, 44). Such genotype- or strain-specific differences in the ability of E2 to directly interact with CD81 may be related to variations in the flexibility, length, and glycosylation of the E2 variable regions and their capacity to modulate exposure of the conserved CD81 binding site. The ∼35-amino-acid V3 loop of HIV-1 gp120 protrudes some 30Å from the core (20). The 11- to 16-residue igVR and the 26-residue region containing HVR2 may also project a considerable distance from the E2 core, sterically occluding the underlying CD81 binding site. It is now important to examine how the three variable regions modulate CD81 binding in various genotypes of HCV.
In this study, we have identified an E2 RBD core structure, lacking the three variable regions, which retained the conformational characteristics of the wild-type E2661 RBD. We anticipate that further refinement of this E2 RBD construct will lead to the crystallization and structural determination of E2.
Acknowledgments
We sincerely thank Jean Dubuisson and Steven Foung for the provision of antibodies and Patricia Vietheer for technical assistance.
This work was supported by National Health and Medical Research Council project grants 296200 and 433913 and by the Australian Centre for HIV and Hepatitis Virology Research.
Footnotes
Published ahead of print on 20 June 2007.
REFERENCES
- 1.Bartosch, B., and F. L. Cosset. 2006. Cell entry of hepatitis C virus. Virology 348:1-12. [DOI] [PubMed] [Google Scholar]
- 2.Bartosch, B., G. Verney, M. Dreux, P. Donot, Y. Morice, F. Penin, J. M. Pawlotsky, D. Lavillette, and F. L. Cosset. 2005. An interplay between hypervariable region 1 of the hepatitis C virus E2 glycoprotein, the scavenger receptor BI, and high-density lipoprotein promotes both enhancement of infection and protection against neutralizing antibodies. J. Virol. 79:8217-8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callens, N., Y. Ciczora, B. Bartosch, N. Vu-Dac, F.-L. Cosset, J.-M. Pawlotsky, F. Penin, and J. Dubuisson. 2005. Basic residues in hypervariable region 1 of hepatitis C virus envelope glycoprotein E2 contribute to virus entry. J. Virol. 79:15331-15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cocquerel, L., C. Voisset, and J. Dubuisson. 2006. Hepatitis C virus entry: potential receptors and their biological functions. J. Gen. Virol. 87:1075-1084. [DOI] [PubMed] [Google Scholar]
- 4a.Combet, C., C. Blanchet, C. Geourjon, and G. Deléage. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25:147-150. [DOI] [PubMed] [Google Scholar]
- 5.Crotta, S., A. Stilla, A. Wack, A. D'Andrea, S. Nuti, U. D'Oro, M. Mosca, F. Filliponi, R. M. Brunetto, F. Bonino, S. Abrignani, and N. M. Valiante. 2002. Inhibition of natural killer cells through engagement of CD81 by the major hepatitis C virus envelope protein. J. Exp. Med. 195:35-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deleersnyder, V., A. Pillez, C. Wychowski, K. Blight, J. Xu, Y. S. Hahn, C. M. Rice, and J. Dubuisson. 1997. Formation of native hepatitis C virus glycoprotein complexes. J. Virol. 71:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dreux, M., T. Pietschmann, C. Granier, C. Voisset, S. Ricard-Blum, P. E. Mangeot, Z. Keck, S. Foung, N. Vu-Dac, J. Dubuisson, R. Bartenschlager, D. Lavillette, and F. L. Cosset. 2006. High density lipoprotein inhibits hepatitis C virus-neutralizing antibodies by stimulating cell entry via activation of the scavenger receptor BI. J. Biol. Chem. 281:18285-18295. [DOI] [PubMed] [Google Scholar]
- 8.Drummer, H. E., I. Boo, A. L. Maerz, and P. Poumbourios. 2006. A conserved Gly436-Trp-Leu-Ala-Gly-Leu-Phe-Tyr motif in hepatitis C virus glycoprotein E2 is a determinant of CD81 binding and viral entry. J. Virol. 80:7844-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drummer, H. E., I. Boo, and P. Poumbourios. 2007. Mutagenesis of a conserved fusion peptide like motif and membrane proximal heptad repeat region of hepatitis C virus glycoprotein E1. J. Gen. Virol. 88:1144-1148. [DOI] [PubMed] [Google Scholar]
- 10.Drummer, H. E., A. Maerz, and P. Poumbourios. 2003. Cell surface expression of functional hepatitis C virus E1 and E2 glycoproteins. FEBS Lett. 546:385-390. [DOI] [PubMed] [Google Scholar]
- 11.Drummer, H. E., K. A. Wilson, and P. Poumbourios. 2005. Determinants of CD81 dimerization and interaction with hepatitis C virus glycoprotein E2. Biochem. Biophys. Res. Commun. 328:251-257. [DOI] [PubMed] [Google Scholar]
- 12.Drummer, H. E., K. A. Wilson, and P. Poumbourios. 2002. Identification of the hepatitis C virus E2 glycoprotein binding site on the large extracellular loop of CD81. J. Virol. 76:11143-11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evan, G. I., G. K. Lewis, G. Ramsay, and J. M. Bishop. 1985. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 5:3610-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans, M. J., T. von Hahn, D. M. Tscherne, A. J. Syder, M. Panis, B. Wolk, T. Hatziioannou, J. A. McKeating, P. D. Bieniasz, and C. M. Rice. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801-805. [DOI] [PubMed] [Google Scholar]
- 15.Farci, P., A. Shimoda, A. Coiana, G. Diaz, G. Peddis, J. C. Melpolder, A. Strazzera, D. Y. Chien, S. J. Munoz, A. Balestrieri, R. H. Purcell, and H. J. Alter. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339-344. [DOI] [PubMed] [Google Scholar]
- 16.Garry, R. F., and S. Dash. 2003. Proteomics computational analyses suggest that hepatitis C virus E1 and pestivirus E2 envelope glycoproteins are truncated class II fusion proteins. Virology 307:255-265. [DOI] [PubMed] [Google Scholar]
- 17.Grollo, L., J. Torresi, H. Drummer, W. Zeng, N. Williamson, and D. C. Jackson. 2006. Exploiting information inherent in binding sites of virus-specific antibodies: design of an HCV vaccine candidate cross-reactive with multiple genotypes. Antivir. Ther. 11:1005-1014. [PubMed] [Google Scholar]
- 18.Hadlock, K. G., R. E. Lanford, S. Perkins, J. Rowe, Q. Yang, S. Levy, P. Pileri, S. Abrignani, and S. K. Foung. 2000. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J. Virol. 74:10407-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofmann, W. P., C. Sarrazin, B. Kronenberger, B. Schonberger, K. Bruch, and S. Zeuzem. 2003. Mutations within the CD81-binding sites and hypervariable region 2 of the envelope 2 protein: correlation with treatment response in hepatitis C virus-infected patients. J. Infect. Dis. 187:982-987. [DOI] [PubMed] [Google Scholar]
- 20.Huang, C. C., M. Tang, M. Y. Zhang, S. Majeed, E. Montabana, R. L. Stanfield, D. S. Dimitrov, B. Korber, J. Sodroski, I. A. Wilson, R. Wyatt, and P. D. Kwong. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato, N., Y. Ootsuyama, S. Ohkoshi, T. Nakazawa, H. Sekiya, M. Hijikata, and K. Shimotohno. 1992. Characterization of hypervariable regions in the putative envelope protein of hepatitis C virus. Biochem. Biophys. Res. Commun. 189:119-127. [DOI] [PubMed] [Google Scholar]
- 22.Keck, Z.-Y., T.-K. Li, J. Xia, B. Bartosch, F.-L. Cosset, J. Dubuisson, and S. K. H. Foung. 2005. Analysis of a highly flexible conformational immunogenic domain A in hepatitis C virus E2. J. Virol. 79:13199-13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keck, Z. Y., A. Op De Beeck, K. G. Hadlock, J. Xia, T. K. Li, J. Dubuisson, and S. K. Foung. 2004. Hepatitis C virus E2 has three immunogenic domains containing conformational epitopes with distinct properties and biological functions. J. Virol. 78:9224-9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keck, Z. Y., J. Xia, Z. Cai, T. K. Li, A. M. Owsianka, A. H. Patel, G. Luo, and S. K. Foung. 2007. Immunogenic and functional organization of hepatitis C virus (HCV) glycoprotein E2 on infectious HCV virions. J. Virol. 81:1043-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lavillette, D., Y. Morice, G. Germanidis, P. Donot, A. Soulier, E. Pagkalos, G. Sakellariou, L. Intrator, B. Bartosch, J. M. Pawlotsky, and F. L. Cosset. 2005. Human serum facilitates hepatitis C virus infection, and neutralizing responses inversely correlate with viral replication kinetics at the acute phase of hepatitis C virus infection. J. Virol. 79:6023-6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazzocca, A., S. C. Sciammetta, V. Carloni, L. Cosmi, F. Annunziato, T. Harada, S. Abrignani, and M. Pinzani. 2005. Binding of hepatitis C virus envelope protein E2 to CD81 up-regulates matrix metalloproteinase-2 in human hepatic stellate cells. J. Biol. Chem. 280:11329-11339. [DOI] [PubMed] [Google Scholar]
- 28.McKeating, J. A., L. Q. Zhang, C. Logvinoff, M. Flint, J. Zhang, J. Yu, D. Butera, D. D. Ho, L. B. Dustin, C. M. Rice, and P. Balfe. 2004. Diverse hepatitis C virus glycoproteins mediate viral infection in a CD81-dependent manner. J. Virol. 78:8496-8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meunier, J. C., R. E. Engle, K. Faulk, M. Zhao, B. Bartosch, H. Alter, S. U. Emerson, F. L. Cosset, R. H. Purcell, and J. Bukh. 2005. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc. Natl. Acad. Sci. USA 102:4560-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakazawa, T., N. Kato, Y. Ootsuyama, H. Sekiya, T. Fujioka, A. Shibuya, and K. Shimotohno. 1994. Genetic alteration of the hepatitis C virus hypervariable region obtained from an asymptomatic carrier. Int. J. Cancer 56:204-207. [DOI] [PubMed] [Google Scholar]
- 31.Owsianka, A. M., J. M. Timms, A. W. Tarr, R. J. Brown, T. P. Hickling, A. Szwejk, K. Bienkowska-Szewczyk, B. J. Thomson, A. H. Patel, and J. K. Ball. 2006. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J. Virol. 80:8695-8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Penin, F., C. Combet, G. Germanidis, P. O. Frainais, G. Deleage, and J. M. Pawlotsky. 2001. Conservation of the conformation and positive charges of hepatitis C virus E2 envelope glycoprotein hypervariable region 1 points to a role in cell attachment. J. Virol. 75:5703-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 34.Roccasecca, R., H. Ansuini, A. Vitelli, A. Meola, E. Scarselli, S. Acali, M. Pezzanera, B. B. Ercole, J. McKeating, A. Yagnik, A. Lahm, A. Tramontano, R. Cortese, and A. Nicosia. 2003. Binding of the hepatitis C virus E2 glycoprotein to CD81 is strain specific and is modulated by a complex interplay between hypervariable regions 1 and 2. J. Virol. 77:1856-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scarselli, E., H. Ansuini, R. Cerino, R. M. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw, M. L., J. McLauchlan, P. R. Mills, A. H. Patel, and E. A. McCruden. 2003. Characterisation of the differences between hepatitis C virus genotype 3 and 1 glycoproteins. J. Med. Virol. 70:361-372. [DOI] [PubMed] [Google Scholar]
- 37.Simmonds, P. 2004. Genetic diversity and evolution of hepatitis C virus—15 years on. J. Gen. Virol. 85:3173-3188. [DOI] [PubMed] [Google Scholar]
- 38.Troesch, M., I. Meunier, P. Lapierre, N. Lapointe, F. Alvarez, M. Boucher, and H. Soudeyns. 2006. Study of a novel hypervariable region in hepatitis C virus (HCV) E2 envelope glycoprotein. Virology 352:357-367. [DOI] [PubMed] [Google Scholar]
- 39.Tseng, C. T., and G. R. Klimpel. 2002. Binding of the hepatitis C virus envelope protein E2 to CD81 inhibits natural killer cell functions. J. Exp. Med. 195:43-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voisset, C., N. Callens, E. Blanchard, A. Op De Beeck, J. Dubuisson, and N. Vu-Dac. 2005. High density lipoproteins facilitate hepatitis C virus entry through the scavenger receptor class B type I. J. Biol. Chem. 280:7793-7799. [DOI] [PubMed] [Google Scholar]
- 41.Wack, A., E. Soldaini, C. Tseng, S. Nuti, G. Klimpel, S. Abrignani, C. Agrati, C. Nisii, A. Oliva, G. D'Offizi, C. Montesano, L. P. Pucillo, and F. Poccia. 2001. Binding of the hepatitis C virus envelope protein E2 to CD81 provides a co-stimulatory signal for human T cells. Eur. J. Immunol. 31:166-175. [DOI] [PubMed] [Google Scholar]
- 42.Weiner, A. J., M. J. Brauer, J. Rosenblatt, K. H. Richman, J. Tung, K. Crawford, F. Bonino, G. Saracco, Q. L. Choo, M. Houghton, et al. 1991. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology 180:842-848. [DOI] [PubMed] [Google Scholar]
- 43.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 44.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]