Abstract
The Ebola virus (EBOV) genome encodes for several proteins that are necessary and sufficient for replication and transcription of the viral RNAs in vitro; NP, VP30, VP35, and L. VP30 acts in trans with an RNA secondary structure upstream of the first transcriptional start site to modulate transcription. Using a bioinformatics approach, we identified a region within the N terminus of VP30 with sequence features that typify intrinsically disordered regions and a putative RNA binding site. To experimentally assess the ability of VP30 to directly interact with the viral RNA, we purified recombinant EBOV VP30 to >90% homogeneity and assessed RNA binding by UV cross-linking and filter-binding assays. VP30 is a strongly acidophilic protein; RNA binding became stronger as pH was decreased. Zn2+, but not Mg2+, enhanced activity. Enhancement of transcription by VP30 requires a RNA stem-loop located within nucleotides 54 to 80 of the leader region. VP30 showed low binding affinity to the predicted stem-loop alone or to double-stranded RNA but showed a good binding affinity for the stem-loop when placed in the context of upstream and downstream sequences. To map the region responsible for interacting with RNA, we constructed, purified, and assayed a series of N-terminal deletion mutations of VP30 for RNA binding. The key amino acids supporting RNA binding activity map to residues 26 to 40, a region rich in arginine. Thus, we show for the first time the direct interaction of EBOV VP30 with RNA and the importance of the N-terminal region for binding RNA.
Ebola viruses (EBOV) are nonsegmented, negative-stranded RNA viruses, which together with Marburg virus, constitute the family Filoviridae (18). Filoviruses cause severe and lethal hemorrhagic fevers in humans and nonhuman primates and, as such, are classified as biosafety level 4 (BSL-4) agents (11). Of the eight proteins encoded by the EBOV genome, the L protein, VP35, the N protein (NP), and VP30 are proposed to form a ribonucleocapsid complex, along with the genomic RNA (10, 28). The L protein bears the enzymatic activities required for transcription and replication, whereas VP30 and VP35 are implicated in regulation of transcription and replication (28, 41). Although the NP is necessary for transcription, its role is still unknown (28). Phosphorylation of VP30 positively regulates its binding to NP and negatively regulates its transcriptional activity (26). In addition, enhancement of transcription by VP30 requires a putative RNA secondary structure located within nucleotides (nt) 54 to 80 of the leader region (44). Deletion of the predicted RNA secondary structure permits VP30-independent transcription of viral messengers (44). These reported activities of VP30 suggest the possibility of a direct interaction of VP30 with EBOV RNA in its role in transcription. In addition, the N terminus of VP30 contains a Zn2+ binding Cys3-His motif (25) and is rich in basic amino acids (34), which can interact with nucleic acid, and provided impetus for our studies here. By analogy, a homologue of VP30, the M2-1 protein of human respiratory syncytial virus (hRSV), has a Cys3-His motif that binds hRSV leader RNA with an apparent dissociation constant of 90 nM (8, 14). Mutation of three cysteine residues of the Cys3-His motif led to a significant reduction of hRSV recovery (39).
Recent publications with the minigenome system for EBOV suggest at least two possible mechanisms that VP30 may use in its transcriptional regulatory role. One possible mechanism could be that VP30 interacts with one or more of the other nucleocapsid proteins, polymerase, NP or VP35, and promotes increased stability of the transcriptional complex. An increase in the number of active transcription initiation complexes or the increased stability of the transcriptional elongation complex would yield higher levels of mRNA, which is the reported role of VP30 (28). Muhlberger and coworkers looked for truncated transcripts to determine whether VP30 had a role in promoting transcriptional elongation but did not observe any differences in the presence or absence of VP30 (44). Alternatively, VP30 may interact directly with viral RNA(s) to regulate transcription. In this model, VP30 could operate in one or more ways, such as protection of synthesized transcripts from degradation and turnover, stabilization of the genomic template to enhance transcript synthesis by the polymerase, or binding to RNA to recruit the other nucleocapsid proteins. We test here the hypothesis that VP30 may directly interact with RNA and thereby promote transcription. We therefore searched the VP30 sequence for signatures and features typical of RNA-binding proteins. After having identified a putative RNA binding motif within an intrinsically disordered region of VP30, we expressed EBOV VP30, along with a series of deletion constructs, in a bacterial expression system and purified them to homogeneity. We used UV cross-linking and filter-binding assays to examine their ability to bind RNA and demonstrate for the first time the RNA binding activity of EBOV VP30. We also mapped this activity to the N-terminal region of VP30.
MATERIALS AND METHODS
Bioinformatics analysis of EBOV VP30.
The VP30 sequences used in the present study were retrieved from the VaZYMoLo (12) and RefSeq (30) databases. The accession numbers are as follows: VAZy178 (strain Zaire), VAZy399 (strain Reston), and NC_006432.1 (strain Sudan). Hydrophobic cluster analysis was carried out with the program DRAWHCA (5). Secondary structure predictions were performed with PSI-PRED (24) and the Predict Protein Server (33). Prediction of NORS (for nonregular secondary structure) regions was done with the NORSp server (http://cubic.bioc.columbia.edu/services/NORSp) (23).
Disorder predictions based on the net charge hydrophobicity method (40) and amino acid composition analysis were carried out as previously described (17). The protein sequence was submitted to the Disembl (http://www.dis.embl.de) (22), Disopred2 (http://bioinf.cs.ucl.ac.uk/disopred) (43), Globplot (http://globplot.embl.de) (21), FoldIndex (http://bip.weizmann.ac.il/fldbin/findex) (46), IUPred (http://iupred.enzim.hu) (9), and RONN (http://www.strubi.ox.ac.uk/RONN) (45) servers using the default settings. The PONDR server (http://www.pondr.com) (19, 31, 32) was used to run both VL-XT and VS-L1 predictions using the default parameters. Access to PONDR was provided by Molecular Kinetics (IUETC, Indianapolis, IN).
Cloning of the EBOV VP30 gene.
The gene coding for VP30 of EBOV Zaire strain Mayinga (EBOV-Z) was amplified by using total RNA isolated from Vero E6 cells infected with EBOV-Z (a gift from Captain Timothy Nelle) under BSL-4 conditions. EBOV-Z-infected cells were treated with TRIzol reagent to isolate the total RNA as described by the manufacturer (Invitrogen, Carlsbad, CA). The EBOV-Z VP30 gene was amplified from total RNA by using reverse transcription-PCR and Pfx polymerase (Invitrogen). The primers used (5′-CCG GCC ATG GAA GCT TCA TAT GAG AG AGGA CG-3′ and 5′-CCT CGA GAG GGG TAC CCT CAT CAG ACC ATG AGC-3′) were complementary to the 5′ and 3′ ends of the coding sequence (in italics) for EBOV-Z VP30. All primers contained the recognition site for either NcoI or XhoI restriction endonucleases (underlined). The blunt-ended VP30 amplicon obtained from PCR was ligated directly into pCR-Blunt II-TOPO as described by the manufacturer (Invitrogen). Plasmid DNA containing the desired insert was isolated, and the insert DNA was excised by NcoI/XhoI digestion. The insert was ligated into pET19b, which allowed for the desired insert to be excised by XbaI/XhoI digestion and subcloned into pET21b. The resulting final construct pET-VP30 (EBOV-Z) was confirmed by restriction analysis and DNA sequencing using a 3100 genetic analyzer (Applied Biosystems, Foster City, CA) and found to be the gene coding for EBOV-Z VP30 fused with a C-terminal His6 tag.
Construction of deletion mutants.
Deletions of the VP30 gene were constructed with the BD In-Fusion PCR cloning kit (BD Biosciences, San Jose, CA). Primer design was done according to the BD In-Fusion Dry-Down PCR cloning kit user manual from BD Biosciences. Primers were synthesized and purified at IDT (Coralville, IA). The following forward primers were used for creating each mutant, VP30Δ1-11, 5′-CCG GCG ATG GCC ATG GGC GCT GCC AGA CAG CAT TCA A-3′; VP30Δ1-26, 5′-CCG GCG ATG GCC ATG GGC GCA CGA TCA TCA TCC AGA A-3′; VP30Δ1-40, 5′-CCG GCG ATG GCC ATG GGC CCA TCA AGG AGC GCC TCA-3′; VP30Δ1-59, 5′-CCG GCG ATG GCC ATG GGC GTT GAA CCA TTA ACA GTT CCT CCA-3′; VP30Δ1-72, 5′-CCG GCG ATG GCC ATG GGC CCG ACC TTG AAA AAA GGA TTT TTG T-3′; VP30Δ1-90, 5′-CCG GCG ATG GCC ATG GGC CAG TTG GAG AGT TTA ACT GAT AG-3′; and VP30Δ1-106, 5′-CCG GCG ATG GCC ATG GGC ACT TGT GGA TCA GTA GAA CAA CA-3′. The reverse primer used in conjunction with each forward primer was 5′-GGT GGT GCT CGA GAG GGG TAC CCT CAT CAG ACC A-3′. In brief, the 5′ end of the forward and reverse primers for cloning of the truncated VP30 gene fragments into pET22b vector share 15 bases of sequence homology with the sequences on either side of the site of amplification and one extra base that recreated NcoI and XhoI sites. Two additional nucleotides were added to the primer design that introduced an additional Gly after the first amino acid in each mutated protein. PCR was performed with HiFi Taq DNA polymerase (Invitrogen) for 25 cycles to amplify the VP30 gene fragments. PCR products were purified with a PCR purification kit (QIAGEN, Valencia, CA). The fragments were cloned into pET22b at the NcoI and XhoI sites. Recombination between pET22b and PCR products was carried out according to the BD In-Fusion cloning procedure in the BD In-Fusion Dry-Down PCR cloning kit user manual. The reaction mixtures were digested with DpnI and transformed into DH5α-competent cells. Resultant colonies were screened by PCR, and their coding sequences were confirmed by DNA sequencing. Plasmids containing VP30 fragments were transformed into BL21(DE3) (Novagen, San Diego, CA), selected for ampicillin resistance, and examined for their ability to express proteins of the correct size.
Construction of site-directed mutations in VP30.
Primers were designed to introduce the site-directed mutations into pET-VP30 according to the QuikChange multi-site-directed mutagenesis kit from Stratagene (La Jolla, CA). The primer (IDT) sequences (nucleic acid substitutions underlined), named by the corresponding amino acid mutations, were: R28A, 5′-GAC CAC CAT GTT CGA GCA GCA TCA TCA TCC AGA GAG AAT TAT C-3′; R32A, 5′-GAG CAC GAT CAT CAT CCG CAG AGA ATT ATC GAG GTG-3′; R36A, 5′-CAT CCA GAG AGA ATT ATG CAG GTG AGT ACC GTC AAT CAA G-3′; R40A, 5′-GAA TTA TCG AGG TGA GTA CGC TCA ATC AAG GAG CGC CTC AC-3′; R28A/R32A, 5′-GAG CAG CAT CAT CAT CCG CAG AGA ATT ATC GAG GTG-3′; and R32A/R36A/R40A, 5′-CAT CCG CAG AGA ATT ATG CAG GTG AGT ACG CTC AAT CAA G-3′. Colonies were screened by DNA sequencing for the required mutations. The quadruple mutant, R28A/R32A/R36A/R40A, was constructed by sequential cloning of each mutation until all four were obtained in a single clone.
Expression and purification of wild-type and mutant VP30.
Wild-type and mutant VP30 proteins were expressed in BL21(DE3) cells by the autoinduction method as described by Studier (38), with the following modifications. Tryptone was used instead of NZ amine in making ZYM 5052 media. For the optimization of autoinduction, cells were cultured in a 500-ml volume. Cells were harvested by centrifugation, lysed in 100 μl of 1× sample buffer (100 mM Tris [pH 6.8], 2% sodium dodecyl sulfate [SDS], 5% β-mercaptoethanol, 15% glycerol, 0.01 mg of bromophenol blue/ml, and 4 M urea), and subjected to SDS-polyacrylamide gel electrophoresis (PAGE). For the purification, bacterial cells were grown in 500 ml of medium at 37°C and harvested after 24 h by centrifugation at 7,000 rpm for 10 min. The pellet was resuspended in 15 ml of phosphate-buffered saline on ice. A total of 0.2 mg of lysozyme (Sigma, St. Louis, MO)/ml was added, and cells were lysed at 4°C for 45 min. Cells were further lysed by douncing for 10 min and sonication for 1 min, three times, with a 2-min interval on ice. Insoluble material was collected by centrifugation at 30,000 × g for 30 min. The pellet was resuspended in 15 ml of buffer A (20 mM Tris [pH 8.0], 1 M NaCl, 8 M urea, 0.1 mM EDTA, 1.0 mM dithiothreitol [DTT], 1 ml of protease inhibitor cocktail [Sigma], and 5 mM imidazole) and mixed on a platform shaker for about 1 h at room temperature. The mixture was sonicated again as described above and centrifuged at 30,000 × g for 30 min at 4°C. The supernatant was collected and loaded onto 1 ml of Ni-NTA resin (QIAGEN) which was pre-equilibrated with buffer A. The column was washed with 10 ml of buffer A and then 10 ml of buffer A containing 50 mM imidazole. The protein was eluted from the column with 10 ml of buffer A containing 1 M imidazole. One-milliliter fractions were collected, examined by SDS-PAGE, and quantified by the Bradford method (Bio-Rad, Hercules, CA). The eluted fractions with approximately more than 90% purity were taken and diluted to a uniform concentration of 0.02 mg/ml and dialyzed consecutively against 2 liters of buffer B (40 mM HEPES [pH 7.4], 1 M NaCl, 20 mM BME, protease inhibitor cocktail, 50 μM ZnCl2, 10% glycerol, and 0.1% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}) with 3, 2, 1, and 0 M urea each for 24 h at 4°C. The protein fractions were dialyzed twice at 4°C in 2 liters of buffer C (40 mM HEPES [pH 7.4], 200 mM NaCl, 1.0 mM DTT, protease inhibitor cocktail, 50 μM ZnCl2, and 10% glycerol) for a total of 24 h. Then, 0.76 μg of protein in 1× sample buffer was loaded for SDS-PAGE and Western blot analysis. For Western blot analysis of VP30, we used a mouse monoclonal anti-His tag antibody for the primary (Sigma) and an anti-mouse immunoglobulin G coupled with alkaline phosphatase for the secondary antibody. The alkaline phosphatase was developed in situ on nitrocellulose membrane with BCIP/NBT substrate (Sigma).
For additional purification of the VP30 by gel filtration, 3 ml of VP30 eluted from the Ni-NTA resin and dialyzed as described above was concentrated to 300 μl with a Centricon-10 (Millipore, Billerica, MA) and passed through a Sephacryl S400HR (Sigma) column pre-equilibrated with buffer A. Then, 1-ml fractions were collected while eluting the column with buffer A.
RNA substrates and 32P end labeling.
RNAs were synthesized by IDT (Coralville, IA) with the EBOV Zaire strain leader sequence (5′-UUUUGUGUGCGAAUAACUAUGAGGAAGAUUAAUAAUUUCCUCUC; NCBI accession no. AY354458). A total of 0.67 pmol of EBOV leader (L) (+)36-80 RNA was end labeled with polynucleotide kinase (New England Labs, Ipswich, MA) in a 150-μl reaction for 1 h in nuclease-free buffers with 0.2234 μCi of [32P]ATP (Perkin-Elmer, Wellesley, MA) at 37°C. 32P-unincorporated nucleotides were removed by passing the reaction through a G-25 spin column (Perkin-Elmer). The RNA was heated for 15 min at 85°C and cooled to 4°C slowly (0.4°C/min) in a thermocycler in 150 mM NaCl. Typically, the probes had a specific activity of 0.1425 μCi/pmol RNA.
UV cross-linking.
In a standard reaction, a 49 nM concentration of 32P end-labeled RNA was preincubated with 142 nM VP30 in nuclease-free reaction buffer (40 mM HEPES [pH 7.4], 150 mM NaCl, 25 mM KCl, 1 mM DTT, and 5% glycerol) at 37°C for 15 min. The RNA-VP30 complexes were covalently cross-linked by exposing binding reactions to 1.8 kJ of UV light (Hoeffer UVC500, GE Health Care Biosciences Corp., Piscataway, NJ). Unbound RNA was digested with 50 U of RNase A (Ambion, Austin, TX) at 37°C for 30 min. Reactions were stopped by adding 5× sample buffer (1× sample buffer is 100 mM Tris [pH 6.8], 2% SDS, 5% β-mercaptoethanol, 15% glycerol, 0.01 mg of bromophenol blue/ml, and 4 M urea). Reactions were heated to 95°C and separated by SDS-12% PAGE. Gels were dried, and the results were visualized by exposure to autoradiographic film or by using a Storm 840 phosphorimager (GE Healthcare Bio-Sciences Corp.). Signals were quantified by using ImageQuant TL (GE Healthcare Bio-Sciences Corp.). Then, 0.075 μCi of 14C protein marker (Amersham Biosciences, Pittsburgh, PA) was loaded, along with samples, for calculating the apparent molecular mass of the complex.
To study the ability of VP30 to bind to EBOV leader RNA under different pH conditions, reaction buffers were made with 40 mM PIPES (pH 5.8 and 6.8), 40 mM morpholinepropanesulfonic acid (pH 7.0 and 7.4), and 40 mM Tris, (pH 7.8, 8.0, and 8.4). For the Zn2+ chelator treatments, 15 μM 1,7-phenanthroline and 1,10-phenanthroline in dimethyl sulfoxide (DMSO; 10% total) were preincubated with VP30 at 37°C in the reaction buffer for 15 min before the addition of the end-labeled RNA. A reaction with 10% DMSO without chelators was included as a positive control.
Filter binding.
VP30 protein was serially diluted in buffer C and incubated with RNA at 37°C for 15 min in reaction buffer (40 mM HEPES [pH 7.4], 150 mM NaCl, 25 mM KCl, 1 mM DTT, and 5% glycerol). The reactions were slot blotted onto nitrocellulose membrane (0.2-μm pore size; Bio-Rad), which was prewet with reaction buffer at room temperature. Membranes were washed with 250 μl of reaction buffer and air dried. Background contribution of nonspecifically bound RNA was measured by filtering the whole reaction in the absence of protein. The signals were quantified by using a Storm phosphorimager as described above. The apparent dissociation constant (Kd) was calculated by fitting a sigmoid binding curve to the empirical data using the Origin program version 7 (Origin Lab Corp., Northampton, MA). The apparent Kd corresponds to the concentration of VP30 protein required to obtain half-saturation, assuming the complex formation obeys a simple bimolecular equilibrium. Another assumption made was that the complex did not dissociate during filtration.
RESULTS
Bioinformatic analysis of VP30.
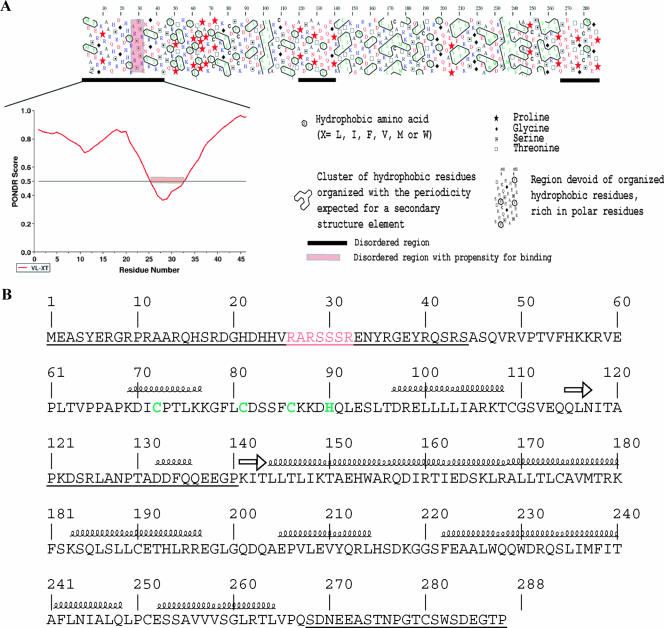
Examination of the EBOV (strain Zaire) VP30 sequence using hydrophobic cluster analysis, revealed the presence of two structured domains (amino acids [aa] 45 to 119 and 141 to 267), typically enriched in hydrophobic clusters (Fig. 1A). Three disordered regions depleted in such clusters were observed: they span residues 1 to 44, 120 to 140, and 268 to 288 (Fig. 1A). Hydrophobic cluster analysis carried out on two other EBOV VP30 sequences (strains Reston and Sudan) revealed the same overall modular organization as for the Zaire strain (data not shown). The three intrinsically disordered regions were also predicted by using ab initio methods as detailed in Materials and Methods. Analysis of the 1-50 region by the net charge/hydrophobicity method (40) confirmed that this region is intrinsically unstructured. Analysis of the deviation in amino acid composition of the disordered N-terminal region indicated that it is depleted in order-promoting residues (W, C, Y, V, I, L and F) and enriched in disorder-promoting residues (R, G, Q, P, K, E, D, and S), with arginine being highly over-represented (four times more represented than in globular proteins) (data not shown). Further, there was no predicted secondary structure elements in the 1-50 region (Fig. 1B). A region with no regular secondary structure and exposed to the solvent is predicted within aa 1 to 97. However, the presence of a Cys3-His type zinc-finger motif in the 72-90 region (Fig. 1B) prevents meaningful predictions of disorder within this latter region. Indeed, metal-binding regions may have features of disordered regions but can be stabilized upon binding to ions, thus leading to gain of structure (40).
FIG. 1.
(A) The top panel shows a hydrophobic cluster analysis plot of Zaire EBOV VP30 (accession no. VAZy178). Conventions are explained (or detailed) in the caption. Globular regions are characterized by a thick distribution of hydrophobic clusters, while unstructured regions are poor or devoid of hydrophobic clusters. Consistently predicted disordered regions are indicated by a black bar under the plot. The bottom panel shows a PONDR VL-XT prediction of the region spanning residues 1 to 45. The threshold of significance for disorder prediction is set to 0.5. The sharp drop in the prediction, encompassing residues 26 to 32, is indicated by a pink bar. These latter residues are also highlighted in the hydrophobic cluster analysis plot using the same color code. (B) Primary structure of Zaire EBOV VP30 (accession no. VAZy178). Consistently predicted disordered regions are underlined. Residues belonging to the potential RNA binding site are indicated in pink, while residues building up the Cys3-His zinc finger are indicated in green. Predicted secondary structure elements are also shown.
It has long been noticed that PONDR can highlight potential molecular recognition elements; these elements are regions within an intrinsically disordered protein that have a propensity to bind to a partner and thereby to undergo induced folding (i.e., a disorder-to-order transition) (3, 4, 13, 29). Potential binding sites appear as sharp drops in the middle of long disordered regions (for examples, see reference 29). Notably, analysis of the PONDR VL-XT prediction for the 1-45 region indicates the presence of such a drop within residues 26 to 32 (Fig. 1A), supporting a predicted role for this short stretch in binding to a partner/ligand. The occurrence of a zinc-finger motif—an evolutionary conserved motif in proteins with RNA processing functions—downstream of the N-terminal intrinsically disordered region, together with the enrichment in arginine residues, supports the hypothesis that the ligand could be RNA. Notably, the hydrophobic cluster analysis indicates that the 26-32 region does not exhibit clear structural propensities, suggesting that this region could be involved in a disorder-to-order transition upon binding to RNA without, however, gaining any regular secondary structure elements.
Altogether, these results suggest that the N-terminal region of VP30 has the sequence properties that typify intrinsically disordered regions and possesses a putative binding site within residues 26 to 32. This site may act in synergy with the zinc-finger motif located in the proximity.
Expression and purification of VP30.
EBOV VP30 was expressed in BL21(DE3) cells, and expression was controlled by the autoinduction method as previously described (38). Optimum induction occurred after a 24-h incubation at 37°C (data not shown). Additional incubation to 48 h did not yield higher levels of recombinant protein. Most of the VP30 protein was expressed as inclusion bodies, and we were unable to identify conditions that resulted in soluble recombinant protein (data not shown). Thus, we used denaturing conditions to purify VP30, since native conditions failed to purify it (data not shown). Nickel affinity chromatography was used in the purification procedure (Fig. 2A). Yields typically ranged from 200 to 280 μg from a 500-ml culture. The protein fractions were refolded by using a slow renaturation procedure that we have used successfully for several retroviral integrases (1, 2, 15, 42) and hantaviral N proteins (16, 36). Human immunodeficiency virus type 1 (HIV-1) integrase was included as a refolding control in each purification, as well as the vector alone, which provided a negative control for subsequent studies. Since VP30 has been shown to be a Zn2+ binding protein (25), we included Zn2+ ion in all of the buffers used for refolding.
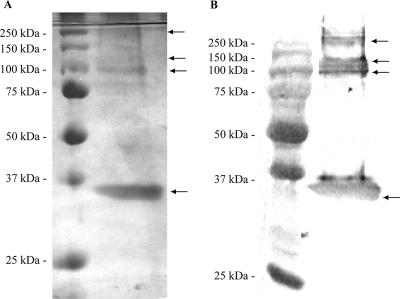
FIG. 2.
(A) Silver-stained SDS-PAGE analysis of purified VP30. Protein was purified by nickel affinity chromatography from BL21(DE3) cells expressing VP30 in lane 2 and from cells with vector alone (V) in lane 3. (B) Western blot analysis of purified VP30. Purified VP30 was separated by SDS-12% PAGE and transferred to nitrocellulose membrane. The second lane shows VP30 detected with anti-His6 monoclonal antibody coupled with alkaline phosphate enzyme. Prestained molecular mass marker is shown in lane 1.
High-molecular-mass bands, higher than that expected for VP30, were observed in all preparations of VP30 at approximately 100, 135, and 250 kDa (Fig. 2A). In addition, these higher molecular mass bands were detected by Western blot analysis of the VP30 protein with an anti-His6 antibody (Fig. 2B). The fractions from the Ni-NTA chromatography were pooled, dialyzed, concentrated to 300 μl, and gel filtrated through a 50-ml Sephacryl 200 column in buffer A (data not shown). Analysis of the eluted fractions showed the presence of the higher-molecular-mass complexes, which suggested that they were oligomers of VP30. To confirm the identity of the bands, the high-molecular-mass bands visualized in SDS-PAGE analyses were isolated, and their identity was confirmed by matrix-assisted laser desorption ionization-time of flight spectroscopy. The protein match searches for each of the four major bands visualized by SDS-PAGE (Fig. 2A) matched the EBOV VP30 with a protein score of 101 to 105 (data not shown). The presence of the higher-molecular-mass bands in the SDS-PAGE suggests that VP30 forms stable oligomers, since strong denaturing conditions of 4 M urea in the 1× sample buffer and boiling were not able to dissociate the oligomers completely. Overall, these results reflect a purity of ca. 90%, as determined by SDS-PAGE analysis.
Analysis of RNA-VP30 interaction.
VP30-dependent transcriptional activity depends on an RNA secondary structure formation at the position encompassing nt 54 to 80 in the antigenome (44). We hypothesized that VP30 may interact directly with RNA within this leader region. A synthetic RNA probe of the EBOV leader (nt 36 to 80), EBOV L(+)36-80, was used to probe the ability of VP30 to interact with RNA. We used the positive-sense viral RNA since we hypothesized a transcriptional control through a posttranscriptional mechanism. EBOV L(+)36-80 contains the predicted secondary structure (54-80) and has an additional 18-nt extension at the 5′ end (Fig. 3A). The RNA and VP30 were assembled in nuclease-free reaction mixtures at 37°C for 15 min, which was followed by UV irradiation, and then subjected to autoradiography. Autoradiography revealed a major band of 35 kDa, together with some additional higher-molecular-mass bands (Fig. 3B, lane 3), which correspond to the VP30-RNA complex monomer and oligomer conformations of VP30. A bacterial culture with vector alone was included as a mock purification for possible background contaminants. These protein preparations did not show any detectable bands in the UV cross-linking assay (Fig. 3B, lane 1). In addition, bovine serum albumin, included as a negative control, was not able to form a complex with the RNA probe (Fig. 3B, lane 2). We also used a nonspecific RNA (NS RNA) in these studies. The NS RNA was obtained through a reiterative process of submitting sequences to MFOLD that might minimize secondary structures and to contain a higher concentration of C rather than G. This NS RNA showed lower binding levels in comparison to an EBOV-specific RNA for competing with VP30-RNA complex in a filter-binding assay (Fig. 3Aiii and C). This shows that VP30-RNA interaction is more specific to EBOV RNAs.
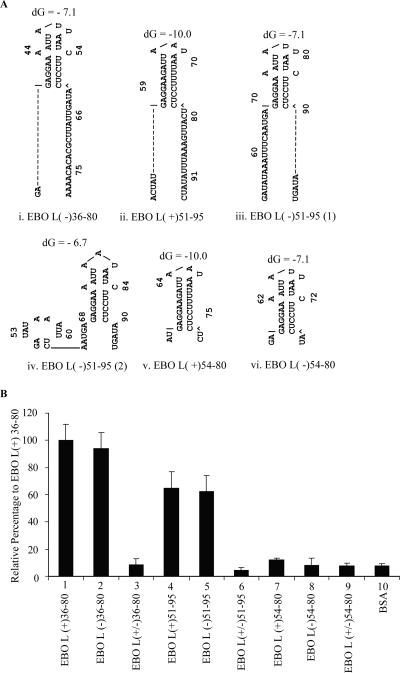
FIG. 3.
(A) Predicted MFOLD structure of EBOV L(+)36-80 and NS RNA. Two different secondary structures were predicted for this RNA probe, with an additional small stem-loop at the 5′ end (1 and 2) differing in the base pairing of the 5′ stem-loop. The free energy (dG) for each structure in kilojoules is given above each structure. (B) Analysis of RNA binding activity of purified VP30. Proteins purified from BL21(DE3) with vector alone (V, lane 1) or vector with VP30 (lane 3) were incubated for 10 min with 49 nM EBOV L(+)36-80 RNA at 37°C and cross-linked with 1.8 kJ of UV light. Complexes were separated on SDS-12% PAGE and subjected to phosphorimaging as described in Materials and Methods. (C) Competition of VP30-RNA interaction with specific and nonspecific RNA. Complexes formed from VP30 and radiolabeled RNA [EBOV L(+)36-80] were challenged by addition of 5 × 10−6 M to 1.6 × 10−9 M nonlabeled EBOV L(+)36-80 RNA and a nonspecific RNA. RNA-protein complex was then captured onto nitrocellulose membrane by filtration and phosphorimaged. The error bar represents the standard deviation for each reaction.
Optimization of ionic strength, metal ion, and pH in VP30-RNA binding assays.
The reaction conditions for the interaction of VP30 with RNA were titrated with NaCl, pH, and magnesium to optimize RNA binding levels. The NaCl concentration was titrated from 50 to 400 mM, and the potassium chloride concentration was titrated from 100 to 0 mM. The highest level of VP30-RNA interaction was observed at 150 mM NaCl and 25 mM KCl (data not shown). The RNA binding activity was reduced to the nondetectable levels when NaCl concentrations reached 400 mM.
Mg2+ ion concentrations were also examined from 0 to 12.5 mM, with salt concentrations at 150 mM NaCl and 25 mM KCl. The reaction without Mg2+ ion showed the highest activity, with threefold-higher activity than the reaction performed at 1.25 mM MgCl2 (data not shown). The increase in Mg2+ ion concentration to 12.5 mM reduced the activity significantly (data not shown).
The optimal pH was assessed from pH 5.8 to 9.3 in reactions of VP30 and EBOV L(+)36-80 RNA. No Mg2+ was used in these reactions, and the salt concentrations were held at 150 mM NaCl and 25 mM KCl. The lowest pH tested had the highest activity, suggesting that low pH values favor the VP30-RNA interaction (data not shown).
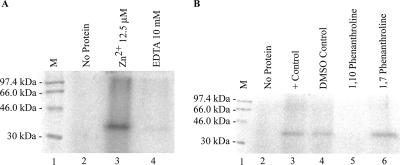
Requirement of Zn2+ ion.
VP30 has been shown to be a Zn2+ binding protein by colorimetric assays (25). To test whether Zn2+ was required for the RNA binding activity of VP30, we refolded denatured Ni-affinity-purified VP30 in the presence or absence of ZnCl2. VP30 was refolded in two sets, with one set having 50 μM ZnCl2 in the buffer B and buffer C and the other set without ZnCl2 but with 10 mM EDTA. Both sets had the same concentrations of wild-type VP30 protein and were dialyzed at 4°C with stepwise lowering of urea concentration as described in Materials and Methods. The protein refolded in the presence of EDTA had only negligible binding activity (Fig. 4A, lane 4). However, the protein refolded in the presence of ZnCl2 bound RNA (Fig. 4A, lane 3). In addition, we tested the Zn2+ chelators, 1,10-phenanthroline, and as a control, the structural analog 1,7-phenanthroline. Hence, 1,10-phenanthroline chelates Zn2+ very effectively and should ablate RNA binding ability if the Zn2+ binding Cys3-His motif is involved. However, its structural analog 1,7-phenanthroline has no significant chelating abilities and should not interfere with Zn2+ chelation and this motif (20). Only reactions with the nonchelating 1,7-phenanthroline showed VP30-RNA binding (Fig. 4B). The positive control reaction with 10% DMSO had no significant difference in activity compared to the reaction without (Fig. 4B, lanes 3 and 4).
FIG. 4.
Influence of Zn2+ on VP30-EBOV leader RNA binding activity. VP30 was incubated with 32 nM EBOV L(+)36-80 RNA. (A) Effect of EDTA in the refolding buffer. VP30 was dialyzed in the presence of 50 μM ZnCl2 (lane 3) or 10 mM EDTA (lane 4). (B) Influence of Zn2+ chelator on EBOV L(+)36-80 RNA interactions. VP30 was treated with a Zn2+ chelator, 1,10-phenanthroline (lane 5), and nonchelator 1,7-phenanthroline (lane 6). 14C molecular mass markers (M) are shown in lanes 1 in both panels A and B.
Specificity of VP30's interaction with RNA.
To further examine the substrate preferences of VP30, substrates were designed within the region of nt 34 through 95 so as to create substrates with the stem-loop, double-stranded RNAs, as well as substrates with 3′ or 5′ extensions of the stem-loop (Fig. 3A and 5A). In addition to the secondary structure predicted at the position from nt 56 to 79, an additional stem-loop is predicted with MFOLD for EBOV L(+)36-80 at the 5′ end at nt 37 to 48 (Fig. 3A). MFOLD secondary structure predictions for the EBOV L(−)36-80 sequence showed a secondary structure from nt 38 to 60, a single-strand region from 60 to 80, and two nucleotide extensions at the 5′ end (Fig. 5Ai). EBOV L(+)51-95 has a 3′ single-strand extension of 17 nt in addition to 5 nt at the 5′ end, prior to the single secondary structure from 56 to 79 (Fig. 5Aii). EBOV L(−)51-95 has a 5-nt extension at the 3′ end and a 17-nt extension at the 5′ end in addition to the secondary structure. Two alternate MFOLD structures were predicted for EBOV L(−)51-95, one with a second stem-loop structure at the 5′ end (Fig. 5Aiii and iv). EBOV L(+)54-80 had minimal 5′ or 3′ extensions predicted within its secondary structure (Fig.5Av). EBO L(−)54-80 was predicted to have two nucleotide extensions at both ends (Fig.5Avi). Double-stranded RNAs were prepared by mixing together these complementary RNAs.
FIG. 5.
(A) Secondary structure prediction of RNA substrates used in VP30 interaction studies. The structure was predicted by MFOLD program version 3.2. Two different conformations were predicted for probes EBOV L(+)51-95 (subpanels iii and iv). The free energy (dG) for each structure in kilojoules is given above each structure. (B) RNA binding activity of VP30 with single-stranded and double-stranded RNAs. RNA-protein complexes were captured on nitrocellulose membranes and exposed to a phosphor screen. The average pixel value obtained for VP30 with probe EBOV L(+)36-80 was taken as 100%, and the relative percentage was calculated for other reactions. The error bar represents the standard deviation for each reaction.
Filter-binding assays were used to explore the substrate preference of VP30. For comparison purposes, the filter-binding signal obtained for the probe EBOV L(+)36-80 was taken as 100%, and the relative signal was calculated for the other RNAs. Surprisingly, only the substrates that were predicted to have either 5′ or 3′ extensions from the secondary stem-loop structure showed VP30 binding activity (Fig. 5B, lanes 1, 2, 4, and 5). The RNA substrate with the shortest 5′ or 3′ extension had only negligible activity (Fig. 5B, lanes 7 and 8). Finally, we tested the interaction of VP30 with double-stranded RNA. Double-stranded RNAs showed only negligible activity compared to the single-stranded RNA probes (Fig. 5B, lanes 3, 6, and 9). Taken together, these data suggest that VP30 had very low affinity to the predicted stem-loop or to double-stranded RNA.
The RNA interacting domain of VP30 maps to N terminus.
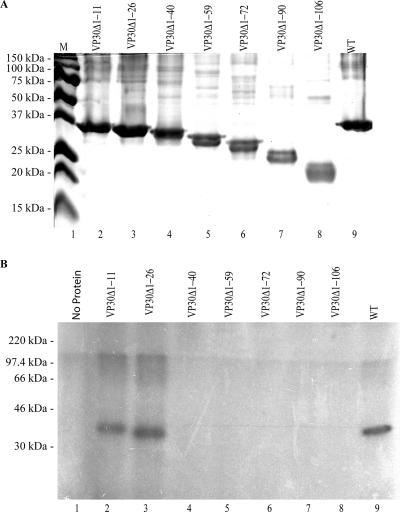
VP30 has a highly basic N terminus, which also contains a Zn2+ binding motif as mentioned previously (25, 34). Indications provided by the bioinformatics analysis for EBOV VP30, together with the observed higher affinity for RNA at low pH values, suggested a role for the N terminus in this interaction. To test this hypothesis, we constructed a series of deletion mutations in the N terminus. Each mutant was purified under identical conditions and refolded at the same concentration as that of the wild-type protein (Fig. 6A). Three independent experiments yielded 90% or more homogeneous protein. As with the wild-type VP30, higher oligomeric complexes were detected by SDS-PAGE analysis of mutant proteins (Fig. 6A).
FIG. 6.
(A) VP30 and N-terminal deletions of VP30 purified by nickel affinity chromatography. Molecular mass markers (M) are shown in lane 1. (B) UV cross-linking assay of VP30 wild type (WT) and VP30 N-terminal deletions with VP30-EBOV L(+) RNA. The RNA binding affinity of each VP30 mutated protein was measured with EBOV L(+)36-80 RNA. Reactions were assembled and UV cross-linked as described in Materials and Methods.
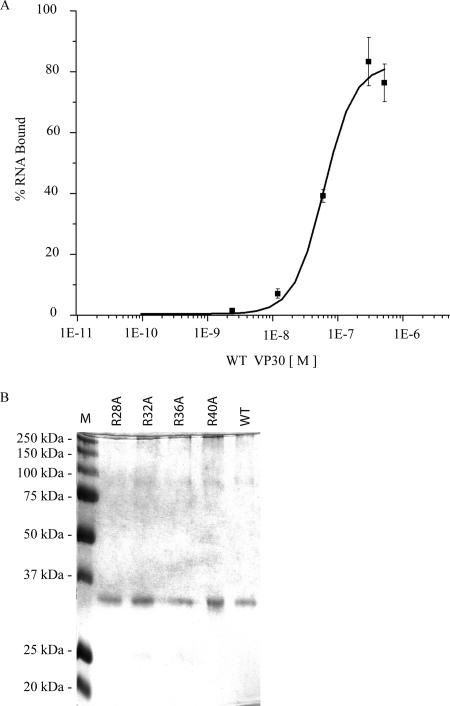
The purified VP30 mutant proteins were examined for their ability to interact with RNA at a concentration of 0.2 μg each in a 40-μl total volume by using UV cross-linking assay. Only the mutant proteins, VP30Δ1-11 and VP30Δ1-26, were active for RNA binding by UV cross-linking. The proteins with deletions from positions 27 through 106 had no detectable RNA binding activity (Fig. 6B). This suggests that the amino acids at the positions from 27 to 40 are important for the RNA interaction. To explore the potential of the arginine residues within this region to be involved in the observed RNA binding, we made the following VP30 mutants; R28A, R32A, R36A and R40A. Each mutant was purified and refolded under identical conditions and concentrations as that of the wild-type protein (Fig. 7B). In addition to the single mutants, we made a double mutant (R28A/R32A), a triple mutant (R28A/R32A/R40A), and a quadruple mutant (R28A/R32A/R36A/R40A). We used UV cross-linking and/or filter binding to probe the activity of each mutant (data not shown). Three of the single amino acid mutants—R28A, R32A, and R36A—showed an RNA binding affinity similar to the wild-type VP30; however, the binding activity of R40A was reduced fourfold (Table 1). The apparent Kd values were measured by filter binding since UV cross-linking does not provide this level of sensitivity.
FIG. 7.
(A) Saturation curve of VP30. VP30 was titrated from 9.52 × 10−11 M to 5.2 × 10−7 M with 7.9 nM 5′ end-labeled EBOV L(+)36-80 RNA. The percentage of total RNA bound is plotted against the concentration of VP30 in a logarithmic scale. The y axis shows the percentage of RNA bound, and the x axis shows the molar concentration of VP30. (B) VP30 and VP30 mutants purified by nickel affinity chromatography. A prestained molecular mass marker is shown in lane 1.
TABLE 1.
Apparent dissociation constants for wild-type VP30 and substitution mutants of VP30
| Protein | Kd (nM) |
|---|---|
| Wild-type VP30 | 61 |
| R28A | 73 |
| R32A | 92 |
| R36A | 65 |
| R40A | 255 |
Binding affinity of the wild type and substitution mutants of VP30.
The apparent dissociation constants of the wild-type VP30 protein was calculated from the filter-binding assay data. EBOV L(+)36-80 RNA (7.9 nM) was titrated with increasing concentrations of WT VP30 (9.52 × 10−11 M to 5.2 × 10−7 M). The apparent dissociation constants of VP30 mutants R28A, R32A, R36A, and R40A were calculated by titrating EBO L(+)36-80 RNA (7.9 nM) with purified mutant proteins (6.0 × 10−11M to 8.6 × 10−11 M) using a filter-binding assay. The apparent Kd, determined as the concentration at which half-saturation is obtained, was averaged and calculated from three replicate values by using the Origin program (Fig. 7A and Table 1).
DISCUSSION
Several lines of evidence suggested a direct interaction of VP30 with RNA in its role in transcriptional regulation of EBOV. As a first step toward studying the function of VP30, we carried out a bioinformatics analysis of the VP30 sequence in order to unveil possible RNA-binding motifs. This led us to identify three intrinsically disordered regions. Further analysis of the N-terminal disordered region pointed out the presence of a short amino acid stretch (residues 26 to 32) possibly involved in binding to a ligand. The presence of a zinc-finger motif downstream of this binding motif, as well as the relative enrichment of this region in arginine residues, suggests that the ligand could be RNA. After purifying a series of EBOV VP30 deletion proteins, we experimentally confirmed this prediction. The results presented here represent the first report of the biochemical purification of the EBOV VP30 protein and of its RNA binding activity.
Denaturation followed by renaturation has been adopted previously for the elucidation of RNA binding activity of the hRSV M2-1 protein (8). A similar method was adopted for the purification of highly insoluble VP30 protein. The VP30 protein was shown to have a strong propensity to form oligomers, as judged by both SDS-PAGE and gel filtration. The purity of the protein was estimated to be 90%. Two assays, UV cross-linking and filter binding, were developed to assess VP30's ability to interact with RNA.
The RNA interaction observed in vitro was independent of the presence of Mg2+ ions. However, the binding activity of VP30 was dependent on the presence of Zn2+ ion that was added during refolding of the protein. Moreover, preincubation of the protein refolded in the presence of Zn2+ with Zn2+ chelators abolished the RNA binding activity. Interaction of VP30 with RNA was strongly dependent on pH, and higher RNA binding activity was observed at a lower pH. It is possible that at the lower pH, the highly basic amino acids at the N terminus become more protonated and thus provide more affinity for the VP30 for interaction with the negatively charged RNA. The RNA binding was observed at the low ionic concentration range tested, with the higher ionic strength abrogating the RNA interaction completely.
One of the important characteristic features of filoviral transcription start signals is that each has a predicted stable RNA secondary structure (27, 34). Even though the function of most of these secondary structures is not known, the transcriptional activity of EBOV requires VP30 and the presence of sequences surrounding the transcription initiation site of NP gene in the leader region (44). We hypothesized that VP30 binds to the stem-loop of the leader sequence. However, we determined that VP30 had a lower binding activity for the stem-loop region or double-stranded RNA, suggesting that VP30 does not bind to the stem or loop region in the leader RNA. Our data suggest that the 3′ or 5′ extensions were required for VP30 binding, which implies that VP30 interacts outside the stem-loop or that these regions may require the formation of complex structures with the step-loop. It may also simply require a free 5′-end. With the exception of EBOV L(+)36-80 and EBOV L(−)51-95, all substrates had only linear 3′ or 5′ extensions based on MFOLD predictions. However, both EBOV L(+)36-80 and EBOV L(−)51-95 probes had single-strand regions predicted in the 5′ extensions. This suggests that VP30 may require the regions within the predicted single-strand regions of the RNA substrates. Weik et al. have previously shown that the deletion of the stem-loop destroys the VP30-dependent transcription (44). Recently, it has been shown that VP35, another key component required for the transcription and replication of EBOV, binds double-stranded RNA (6). It is possible that the VP30 acts as a regulator for VP35 and prevent VP35 stalling at the double-strand regions of the RNA, by virtue of its lower affinity for such regions, and thus allowing transcription to continue. Further studies are needed to completely elucidate the role of VP30 in EBOV transcription.
The presence of the Cys3-His motif and the enrichment in basic amino acids in the N terminus led us to postulate that the RNA interacting domain of VP30 is in the N terminus. Experiments showed that VP30 proteins with N-terminal deletions from 26 to 40 were inactive for RNA binding. This strongly suggests that residues 26 to 40 are important for the RNA interaction of VP30. VP30 has tyrosine (35 and 39) as well as arginine (28, 32, 36, and 40) residues therein. Moreover, the RXRSXXS motif spanning residues 27 to 33 of EBOV VP30 is highly conserved in filoviruses (Fig. 8). Notably, R40A, but none of the other arginine residues, showed an ∼4-fold increase in its Kd, which suggests its involvement in RNA binding. Furthermore, our Zn2+ chelation studies suggest that Cys3-His motif also plays a role in RNA binding. The absence of RNA binding activity of the proteins, VP30Δ1-40 and VP30Δ1-59, which still possess the Cys3-His motif, suggests that a Cys3-His motif alone does not have RNA binding activity. Hence, the RNA binding domain may result from three-dimensional arrangement of these two regions. The fact that the activity of VP30 was strongly dependent on the presence of Zn2+ ions suggests that the intact Zn2+-bound Cys3-His motif may be required for the interaction of the protein with RNA, albeit indirectly. As with the EBOV VP30 protein, the RNA binding domain of M2-1 protein of hRSV was mapped to aa 59 to 85 within the N terminus (8). The Cys3-His domain of M2-1 protein present in the first 30 amino acids was not required for the RNA binding activity of M2-1 (8). Interestingly, residues 29 to 31 and serine residues at positions 42, 44, and 46 have been shown to be phosphorylated and involved in modulating the transcriptional activity of EBOV VP30 (26). Hence, phosphorylation of VP30 might lead to a block in RNA binding activity and thus of transcription.
FIG. 8.
Amino acid alignment of the N terminus of VP30 from published filovirus sequences. The sequences of VP30 for the Zaire (aa 21 to 61), Marburg (aa 33 to 78), Reston (aa 21 to 61), and Sudan (aa 26 to 58) viruses were aligned by using Vector NTI (version 9.1) software.
The dissociation constant of VP30-RNA interaction was comparable to the dissociation constants calculated for the M2-1 protein of hRSV-RNA interaction (8) and for other viral RNA binding proteins of RNA viruses. For example, the dissociation constant of Hantaan virus nucleocapsid protein has been reported to be 14 nM when the sodium chloride concentration was kept at 100 mM (35). Similarly the purified HIV-1 Tat protein interacts with TAR RNA with an apparent dissociation constant of 10 nM (37). The NS1 protein of influenza A virus on the other hand, interacts with double-stranded RNA with an apparent dissociation constant of 1 μM (7). Comparison of these dissociation constants shows that VP30 binds RNA with a rather high affinity, which is comparable to that of other RNA binding proteins of RNA viruses.
In summary, we show for the first time the direct interaction of VP30 with RNA. In addition, we show the importance of the N-terminal region of VP30 for this activity and the lower affinity of VP30 for stem-loop and double-stranded RNA regions. Current efforts are under way to build upon these biochemical studies in order to further elucidate the role of the RNA binding domain using the minigenome reporter systems. The information obtained here will prove valuable in mapping these complex interactions within the context of the cell.
A structural study either by nuclear magnetic resonance or X-ray crystallography of VP30 will be important in elucidating role of the two regions shown here to be involved in RNA binding.
Acknowledgments
This study was performed and funded under a collaborative agreement between USAMRIID and SRI project no. 1119.
This study was performed while S.S. held a National Research Council postdoctoral fellowship at USAMRIID.
Footnotes
Published ahead of print on 13 June 2007.
REFERENCES
- 1.Balakrishnan, M., and C. B. Jonsson. 1997. Functional identification of nucleotides conferring substrate specificity to retroviral integrase reactions. J. Virol. 71:1025-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balakrishnan, M., D. Zastrow, and C. B. Jonsson. 1996. Catalytic activities of the human T-cell leukemia virus type II integrase. Virology 219:77-86. [DOI] [PubMed] [Google Scholar]
- 3.Bourhis, J., K. Johansson, V. Receveur-Bréchot, C. J. Oldfield, A. K. Dunker, B. Canard, and S. Longhi. 2004. The C-terminal domain of measles virus nucleoprotein belongs to the class of intrinsically disordered proteins that fold upon binding to their physiological partner. Virus Res. 99:157-167. [DOI] [PubMed] [Google Scholar]
- 4.Callaghan, A. J., J. P. Aurikko, L. L. Ilag, J. Gunter Grossmann, V. Chandran, K. Kuhnel, L. Poljak, A. J. Carpousis, C. V. Robinson, M. F. Symmons, and B. F. Luisi. 2004. Studies of the RNA degradosome-organizing domain of the Escherichia coli ribonuclease RNase E. J. Mol. Biol. 340:965-979. [DOI] [PubMed] [Google Scholar]
- 5.Callebaut, I., G. Labesse, P. Durand, A. Poupon, L. Canard, J. Chomilier, B. Henrissat, and J. P. Mornon. 1997. Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell Mol. Life Sci. 53:621-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardenas, W. B., Y. M. Loo, M. Gale, Jr., A. L. Hartman, C. R. Kimberlin, L. Martinez-Sobrido, E. O. Saphire, and C. F. Basler. 2006. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J. Virol. 80:5168-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chien, C. Y., Y. Xu, R. Xiao, J. M. Aramini, P. V. Sahasrabudhe, R. M. Krug, and G. T. Montelione. 2004. Biophysical characterization of the complex between double-stranded RNA and the N-terminal domain of the NS1 protein from influenza A virus: evidence for a novel RNA-binding mode. Biochemistry 43:1950-1962. [DOI] [PubMed] [Google Scholar]
- 8.Cuesta, I., X. Geng, A. Asenjo, and N. Villanueva. 2000. Structural phosphoprotein M2-1 of the human respiratory syncytial virus is an RNA binding protein. J. Virol. 74:9858-9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dosztanyi, Z., V. Csizmok, P. Tompa, and I. Simon. 2005. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21:3433-3434. [DOI] [PubMed] [Google Scholar]
- 10.Elliott, L. H., M. P. Kiley, and J. B. McCormick. 1985. Descriptive analysis of Ebola virus proteins. Virology 147:169-176. [DOI] [PubMed] [Google Scholar]
- 11.Feldmann, H., H. Bugany, F. Mahner, H. D. Klenk, D. Drenckhahn, and H. J. Schnittler. 1996. Filovirus-induced endothelial leakage triggered by infected monocytes/macrophages. J. Virol. 70:2208-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferron, F., C. Rancurel, S. Longhi, C. Cambillau, B. Henrissat, and B. Canard. 2005. VaZyMolO: a tool to define and classify modularity in viral proteins. J. Gen. Virol. 86:743-749. [DOI] [PubMed] [Google Scholar]
- 13.Garner, E., P. Romero, A. K. Dunker, C. Brown, and Z. Obradovic. 1999. Predicting binding regions within disordered proteins. Genome Inform. Ser. Workshop Genome Inform. 10:41-50. [PubMed] [Google Scholar]
- 14.Hardy, R. W., and G. W. Wertz. 2000. The Cys(3)-His(1) motif of the respiratory syncytial virus M2-1 protein is essential for protein function. J. Virol. 74:5880-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonsson, C. B., G. A. Donzella, E. Gaucan, C. M. Smith, and M. J. Roth. 1996. Functional domains of Moloney murine leukemia virus integrase defined by mutation and complementation analysis. J. Virol. 70:4585-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jonsson, C. B., J. Gallegos, P. Ferro, W. Severson, X. Xu, and C. S. Schmaljohn. 2001. Purification and characterization of the Sin Nombre virus nucleocapsid protein expressed in Escherichia coli. Protein Expr. Purif. 23:134-141. [DOI] [PubMed] [Google Scholar]
- 17.Karlin, D., F. Ferron, B. Canard, and S. Longhi. 2003. Structural disorder and modular organization in Paramyxovirinae N and P. J. Gen. Virol. 84:3239-3252. [DOI] [PubMed] [Google Scholar]
- 18.Kiley, M. P., E. T. Bowen, G. A. Eddy, M. Isaacson, K. M. Johnson, J. B. McCormick, F. A. Murphy, S. R. Pattyn, D. Peters, O. W. Prozesky, R. L. Regnery, D. I. Simpson, W. Slenczka, P. Sureau, G. van der Groen, P. A. Webb, and H. Wulff. 1982. Filoviridae: a taxonomic home for Marburg and Ebola viruses? Intervirology 18:24-32. [DOI] [PubMed] [Google Scholar]
- 19.Li, X., P. Romero, M. Rani, A. K. Dunker, and Z. Obradovic. 1999. Predicting protein disorder for N, C, and internal regions. Genome Inform. Ser. Workshop Genome Inform. 10:30-40. [PubMed] [Google Scholar]
- 20.Lin, R. S., C. Rodriguez, A. Veillette, and H. F. Lodish. 1998. Zinc is essential for binding of p56lck to CD4 and CD8α. J. Biol. Chem. 273:32878-32882. [DOI] [PubMed] [Google Scholar]
- 21.Linding, R., L. J. Jensen, F. Diella, P. Bork, T. J. Gibson, and R. B. Russell. 2003. Protein disorder prediction: implications for structural proteomics. Structure 11:1453-1459. [DOI] [PubMed] [Google Scholar]
- 22.Linding, R., R. B. Russell, V. Neduva, and T. J. Gibson. 2003. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 31:3701-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, J., H. Tan, and B. Rost. 2002. Loopy proteins appear conserved in evolution. J. Mol. Biol. 322:53-64. [DOI] [PubMed] [Google Scholar]
- 24.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 25.Modrof, J., S. Becker, and E. Muhlberger. 2003. Ebola virus transcription activator VP30 is a zinc-binding protein. J. Virol. 77:3334-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Modrof, J., E. Muhlberger, H. D. Klenk, and S. Becker. 2002. Phosphorylation of VP30 impairs Ebola virus transcription. J. Biol. Chem. 277:33099-33104. [DOI] [PubMed] [Google Scholar]
- 27.Muhlberger, E., S. Trommer, C. Funke, V. Volchkov, H. D. Klenk, and S. Becker. 1996. Termini of all mRNA species of Marburg virus: sequence and secondary structure. Virology 223:376-380. [DOI] [PubMed] [Google Scholar]
- 28.Muhlberger, E., M. Weik, V. E. Volchkov, H. D. Klenk, and S. Becker. 1999. Comparison of the transcription and replication strategies of Marburg virus and Ebola virus by using artificial replication systems. J. Virol. 73:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oldfield, C. J., Y. Cheng, M. S. Cortese, P. Romero, V. N. Uversky, and A. K. Dunker. 2005. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry 44:12454-12470. [DOI] [PubMed] [Google Scholar]
- 30.Pruitt, K. D., T. Tatusova, and D. R. Maglott. 2005. NCBI reference sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33:D501-D504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romero, P., Z. Obradovic, and K. A. Dunker. 1997. Sequence data analysis for long disordered regions prediction in the calcineurin family. Genome Inform. Ser. Workshop Genome Inform. 8:110-124. [PubMed] [Google Scholar]
- 32.Romero, P., Z. Obradovic, X. Li, E. C. Garner, C. J. Brown, and A. K. Dunker. 2001. Sequence complexity of disordered proteins. Proteins 42:38-48. [DOI] [PubMed] [Google Scholar]
- 33.Rost, B. 1996. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 266:525-539. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez, A., M. P. Kiley, B. P. Holloway, and D. D. Auperin. 1993. Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res. 29:215-240. [DOI] [PubMed] [Google Scholar]
- 35.Severson, W., L. Partin, C. S. Schmaljohn, and C. B. Jonsson. 1999. Characterization of the Hantaan nucleocapsid protein-ribonucleic acid interaction. J. Biol. Chem. 274:33732-33739. [DOI] [PubMed] [Google Scholar]
- 36.Severson, W., X. Xu, M. Kuhn, N. Senutovitch, M. Thokala, F. Ferron, S. Longhi, B. Canard, and C. B. Jonsson. 2005. Essential amino acids of the Hantaan virus N protein in its interaction with RNA. J. Virol. 79:10032-10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slice, L. W., E. Codner, D. Antelman, M. Holly, B. Wegrzynski, J. Wang, V. Toome, M. C. Hsu, and C. M. Nalin. 1992. Characterization of recombinant HIV-1 Tat and its interaction with TAR RNA. Biochemistry 31:12062-12068. [DOI] [PubMed] [Google Scholar]
- 38.Studier, F. W. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41:207-234. [DOI] [PubMed] [Google Scholar]
- 39.Tang, R. S., N. Nguyen, X. Cheng, and H. Jin. 2001. Requirement of cysteines and length of the human respiratory syncytial virus M2-1 protein for protein function and virus viability. J. Virol. 75:11328-11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uversky, V. N., J. R. Gillespie, and A. L. Fink. 2000. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 41:415-427. [DOI] [PubMed] [Google Scholar]
- 41.Volchkov, V. E., V. A. Volchkova, E. Muhlberger, L. V. Kolesnikova, M. Weik, O. Dolnik, and H. D. Klenk. 2001. Recovery of infectious Ebola virus from complementary DNA: RNA editing of the GP gene and viral cytotoxicity. Science 291:1965-1969. [DOI] [PubMed] [Google Scholar]
- 42.Wang, T., S. John, S. Archuleta, and C. B. Jonsson. 2004. Rapid, high-throughput purification of HIV-1 integrase using microtiter plate technology. Protein Expr. Purif. 33:232-237. [DOI] [PubMed] [Google Scholar]
- 43.Ward, J. J., L. J. McGuffin, K. Bryson, B. F. Buxton, and D. T. Jones. 2004. The DISOPRED server for the prediction of protein disorder. Bioinformatics 20:2138-2139. [DOI] [PubMed] [Google Scholar]
- 44.Weik, M., J. Modrof, H. D. Klenk, S. Becker, and E. Muhlberger. 2002. Ebola virus VP30-mediated transcription is regulated by RNA secondary structure formation. J. Virol. 76:8532-8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, Z. R., R. Thomson, P. McNeil, and R. M. Esnouf. 2005. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics 21:3369-3376. [DOI] [PubMed] [Google Scholar]
- 46.Zeev-Ben-Mordehai, T., E. H. Rydberg, A. Solomon, L. Toker, V. J. Auld, I. Silman, S. Botti, and J. L. Sussman. 2003. The intracellular domain of the Drosophila cholinesterase-like neural adhesion protein, gliotactin, is natively unfolded. Proteins 53:758-767. [DOI] [PubMed] [Google Scholar]