Abstract
Mason-Pfizer monkey virus (M-PMV) capsids that have assembled in the cytoplasm must be transported to and associate with the plasma membrane prior to being enveloped by a lipid bilayer during viral release. Structural studies have identified a positive-charge density on the membrane-proximal surface of the matrix (MA) protein component of the Gag polyprotein. To investigate if basic amino acids in MA play a role in intracellular transport and capsid-membrane interactions, mutants were constructed in which lysine and arginine residues (R10, K16, K20, R22, K25, K27, K33, and K39) potentially exposed on the capsid surface were replaced singly and in pairs by alanine. A majority of the charge substitution mutants were released less efficiently than the wild type. Electron microscopy of mutant Gag-expressing cells revealed four distinct phenotypes: K16A and K20A immature capsids accumulated on and budded into intracellular vesicles; R10A, K27A, and R22A capsid transport was arrested at the cellular cortical actin network, while K25A immature capsids were dispersed throughout the cytoplasm and appeared to be defective at an earlier stage of intracellular transport; and the remaining mutant (K33A and K39A) capsids accumulated at the inner surface of the plasma membrane. All mutants that released virions exhibited near-wild-type infectivity in a single-round assay. Thus, basic amino acids in the M-PMV MA define both cellular location and efficiency of virus release.
Mason-Pfizer monkey virus (M-PMV), an immunosuppressive betaretrovirus, catalyzes the membrane envelopment of a preassembled spherical protein shell (capsid) to release infectious virions. In contrast, viruses such as human immunodeficiency virus type 1 (HIV-1) simultaneously assemble an immature capsid and extrude the plasma membrane (20). The presence of myristic acid and a basic domain on many retroviral matrix (MA) proteins has led to the hypothesis that a bipartite signal initiates the molecular interactions necessary for a myristylated Gag polyprotein to associate with the plasma membrane and instigate the processes of capsid assembly and budding (C-type morphology) or membrane extrusion (B/D-type morphology) (5, 65, 66).
M-PMV assembles capsids from its Gag polyproteins in a pericentriolar region of the cytoplasm prior to transport to the plasma membrane and viral budding (49). Transport of capsids from the assembly site to the plasma membrane is dependent on a functional endosomal pathway, and release of capsids is seven times more efficient in the presence of the viral envelope glycoprotein (48). The envelope glycoprotein must enter the endosomal pathway, following cleavage into the surface (gp70) and transmembrane (gp22) subunits in the Golgi body, in order to be incorporated into capsids (50, 52).
The M-PMV MA is comprised of four helical domains arranged in two perpendicularly aligned pairs, with two distinct positively charged regions located on opposite sides of the molecule (7). The positively charged region which contains basic side chains of amino acids from helices A and B is analogous to the N-terminal positive-charge region seen in the MA structures of HIV-1 (19, 59) and simian immunodeficiency virus (35). Unlike the latter retroviruses, which simultaneously assemble their protein shell and extrude the plasma membrane, M-PMV must wrap the lipid bilayer around a 90-nm preformed protein shell. We have hypothesized previously (40, 56) that a driving force for this wrapping process could be the exposure and insertion of the 14-carbon saturated fatty acid (myristate) moiety, which is covalently attached to a glycine residue at position 2 of the Gag polyprotein, into the plasma membrane. Myristylation of MA is not required for immature capsid assembly but is needed for transport and release of capsids; a glycine-to-valine mutant that is defective for myristic acid attachment assembles capsids, but these remain at a perinuclear region of the cell (41). MA mutants that have a more hydrophobic core are myristylated and transported to the plasma membrane but are defective at an early stage of budding. The phenotype of these mutants is consistent with the hypothesis that myristic acid is sequestered within the MA domain upon capsid assembly to yield a protein conformation that is conducive for both capsid transport and initiation of envelopment at the plasma membrane (40, 56).
Protein sequestration of an attached myristic acid moiety was first observed in nuclear magnetic resonance analyses of recoverin, a myristylated cellular protein found in retinal rod cells (37). These studies showed that the N-terminal myristate moiety was buried within the protein in the absence of calcium and extruded from the protein when calcium was bound (1, 58, 67). A similar but distinct myristyl switch mechanism has been described for the myristylated alanine-rich protein kinase C substrate (MARCKS) protein (22, 47). In this case, myristate is sequestered within the protein core prior to an electrostatic interaction of the positively charged, basic effector domain (residues 151 to 175) of MARCKS with acidic phospholipid head groups on the inner leaflet of the plasma membrane (13, 36, 57, 60, 62).
The presence of myristic acid and a basic domain on many retroviral MA proteins favors an electrostatic switch mechanism similar to that of the MARCKS protein during Gag-membrane interactions (7, 30, 34, 35, 55, 59). Covalent attachment of myristate to the matrix domain of HIV-1 Gag is required for the Gag polyprotein to multimerize and assemble capsids at the plasma membrane (5, 14). A deletion mutation that disrupts the structure of the HIV-1 matrix domain without altering myristylation redirects capsid assembly and membrane extrusion to the endoplasmic reticulum (10). Moreover, HIV-1 capsid assembly is redirected to multivesicular bodies when basic residues on the outer surface of the matrix domain are replaced with acidic residues (29) or when hydrophobic residues that face the core of the MA domain are replaced with less-hydrophobic residues (11, 27). These biochemical data suggest that myristylation and positive-charge residues in the MA domain of HIV-1 Gag direct protein association with the plasma membrane.
In a manner similar to that of the MARCKS protein, which is triggered to associate with the plasma membrane following interaction with acidic phospholipids, the cellular location at which HIV-1 assembles and buds also appears to be dependent on a phospholipid—phosphoinositol-4,5-bisphosphate [PI(4,5)P2]. Depletion of this molecule from the plasma membrane or enrichment at intracellular membranes alters HIV-1 Gag accumulation and budding to an intracellular site (26). Moreover, recent in vitro studies have shown that PI(4,5)P2 interacts directly with HIV-1 MA, inducing a conformational change in the protein which results in myristate exposure (43). Thus, it is possible that, in the case of retroviral budding, the basic residues on the membrane-proximal surface of the MA protein interact with PI(4,5)P2 to initiate the process of myristate extrusion.
Although the structural studies above are consistent with a myristyl-switch mechanism that is triggered by phospholipid binding, M-PMV provides a unique system to investigate the role of basic residues in the MA domain of Gag in both intracellular trafficking and membrane envelopment since capsid assembly, transport, and budding are discrete events. To further investigate the hypothesis that positively charged amino acids of the M-PMV MA interact electrostatically with the inner leaflet of the plasma membrane to facilitate membrane extrusion, we have replaced arginine and lysine residues in helices A and B, singly and in pairs, with the neutral amino acid alanine. These mutants fall into four major phenotypes: those defective for intracellular transport, those that associate with and bud into intracellular vesicles, those that are arrested at the cortical actin layer adjacent to the plasma membrane, and those that are arrested at an early stage of plasma membrane association. These results demonstrate that basic amino acid residues in M-PMV MA influence multiple steps in intracellular targeting and transport as well as facilitate electrostatic capsid-membrane interactions required to initiate membrane envelopment.
MATERIALS AND METHODS
Cells and antibodies.
COS-1 and 293T cells were obtained from the American Type Culture Collection (Manassas, VA) and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (HyClone Laboratories, Logan, UT), 100 U/ml penicillin G sodium, and 100 μg/ml streptomycin sulfate (Gibco BRL, Rockville, MD). Anti-M-PMV mouse monoclonal antibody 10.10, which binds to the p12 domain of M-PMV Gag, was used at a concentration of 20 μg/ml (44). Polyclonal rabbit anti-Pr78 (M-PMV Gag) serum 3493 (44) was diluted 1:500. The mouse monoclonal anti-CD63 antibody (clone H5C6) was used at a concentration of 1 μg/ml (BD Biosciences, San Jose, CA). Fluorescein isothiocyanate-goat anti-rabbit immunoglobulin G was purchased from Zymed Laboratories (San Francisco, CA). Alexa Fluor 594-goat anti-mouse immunoglobulin G was purchased from Molecular Probes Inc. (Eugene, OR).
Construction of plasmids and mutant proviruses.
Mutant derivatives of the M-PMV proviral vector were constructed using pNCS (56), a plasmid containing a gag fragment corresponding to nucleotides 351 to 1167 of pSARM4 (51). The desired codon(s) was generated in pNCS by PCR-directed mutagenesis. For each single mutant, a pair of complementary and reverse-oriented mutagenic primers with a 2-bp change compared to the wild-type sequence was designed. The sequences of the mutagenic oligonucleotides used were as follows: R10A, 5′-GCAGGCTTTAGCGACAGGGGGAG-3′; K16A, 5′-GAACAATTGGCGCAGGCTTTAAAG-3′; K20A, 5′-GCAGGCTTTAGCGA-CACGGGGAG-3′; R22A, 5′-GGCTTTAAAGACAGCGGGAGTAAAGG-3′; K25A, 5′-GACACGGGGAGTAGCGGTTAAATATGC-3′; K27A, 5′-GGGAGTAAAGGTTCATAT-GCTGATC-3′, K33A, 5′-GCTGATCTTTTGGCATTTTTTTGATTTTGTGAAGG-3′, and K39A, 5′-GATTTTGTGGCGGATACTTGTC-3′. Double mutants, with the exception of K20A/R22A (5′-GGCTTTAGCGACAGCGGGAGTAAAGG-3′), R22A/K25A (5′-GAC-AGCGGGAGTAGCGGTTAAATATGC-3′), and K25A/K27A (5′-GACACGGGGAGTAG-CGGTTGCATATGCTG-3′), were generated by including two single mutagenic primer pairs in the PCR. Multiple-round PCR using Pfu-Turbo DNA polymerase (Stratagene, La Jolla, CA) incorporated the mutation from the primers into the pNCS plasmid. Following amplification, the PCR product was digested with DpnI to remove the methylated wild-type template, leaving the mutated pNCS vector intact. The nucleotide sequence of the mutated region of each gag construct was determined to confirm that the desired base pair changes were introduced. Subsequently, the EagI-to-PacI mutated M-PMV gag fragment corresponding to nucleotides 407 to 750 was reengineered into the M-PMV proviral expression vector pSARM4.
For the single-round infectivity assay, the M-PMV mutants constructed above were cloned into the pSARM-EGFP (enhanced green fluorescent protein) vector (25) via the EcoRI (nucleotide 8217)-to-SacI (nucleotide 1163) fragment of the pSARM4 expression vector.
Metabolic labeling and immunoprecipitation of Gag.
293T cells were transfected with wild-type or mutant proviral constructs by the Fugene 6 method (Roche Molecular Biochemicals, Indianapolis, IN). Approximately 24 h posttransfection, 293T cells expressing the wild-type or mutant M-PMV viral proteins were starved for 10 min with methionine- and cysteine-deficient DMEM (Gibco) and then pulse-labeled for 15 min at 37°C with 100 μCi of [35S]methionine-[35S]cysteine protein-labeling mix (Perkin-Elmer NEN, Boston, MA). The radioactive medium was removed at the end of the pulse period; cells were then chased in complete DMEM for 1, 2, 4, or 24 h. Pulse cells were washed with cold Tris-buffered saline and lysed in 200 μl of 1% sodium dodecyl sulfate (SDS) in phosphate-buffered saline (PBS) for 5 min at room temperature. Cell lysates were then scraped from the well, transferred to Eppendorf tubes, and boiled for 5 min. The volume of each cell lysate was then adjusted to 1 ml by adding 800 μl of lysis buffer A (1% Triton X-100, 1% sodium deoxycholate, 0.05 M NaCl, 0.025 M Tris, pH 8.0). RNase-free DNase (10 U; Stratagene) was added to each lysate prior to a 1-h incubation at room temperature. Chase cells were processed in the same manner as the pulse cells. The culture medium of the chase cells was filtered through a 0.45-μm-pore-size filter and then adjusted to contain 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS. Viral proteins were immunoprecipitated from the pulse, chase, and chase cell media with polyclonal rabbit anti-Pr78 (M-PMV Gag) serum 3493 (44) and separated by SDS-polyacrylamide gel electrophoresis (PAGE).
Quantitation of Gag polyprotein processing and release.
The SDS-polyacrylamide gels were dried, and the radiolabeled protein bands were quantitated on a Packard Cyclone system using OptiQuant software (Perkin-Elmer, Shelton, CT). For each time point, band intensities for Pr78 (Gag), Pr95 (Gag-Pro), Pr180 (Gag-Pro-Pol), and p27 (CA) were acquired for pulse-labeled cells, pulse-chase cells, and the chase culture medium. The quantitated results of individual band intensities were adjusted to reflect the number of methionine residues present in each protein; the number was divided by the sum of the adjusted band intensities and then multiplied by 100 to calculate the percentage of each individual protein. The percent total Gag precursor is the summation of the percent Pr78 (Gag), Pr95 (Gag-Pro), and Pr180 (Gag-Pro-Pol). The percent total CA is the summation of the percent p27 associated with the pulse-chase cells and released into the culture medium. The calculations assume that all labeled Gag is incorporated into mature virions and does not take into consideration labeled Gag proteins that undergo degradation. These calculations are sufficient to compare changes in the rate of Gag processing for mutant M-PMV Gag to changes in that for wild-type M-PMV Gag.
For analysis of virus release, the intensity of the p27 band present in culture supernatants was quantitated for each time point and normalized to the intensity of the Pr78 band in the pulse-labeled cells, based on the number of methionine residues in each.
Electron microscopy.
COS-1 cells transiently expressing the wild-type or mutant M-PMV proviral constructs were grown as a monolayer on 13-mm Thermonox plastic coverslips (Nunc, Rochester, NY) and fixed for 4 h in freshly prepared 2.5% glutaraldehyde in 0.1 M cacodylate buffer. The cells were postfixed in 1% osmium tetroxide, dehydrated in an ethanol series (30, 50, 70, 80, 90, and 100%), and embedded in fresh EMBED 812 resin in a labeled Beem capsule. Ultrathin sections (90 to 100 nm) of COS-1 cells expressing wild-type and mutant M-PMV were cut with a diamond knife on an RMC MT 7000 Ultramicrotome and picked up on 200-mesh copper grids. Several sections were collected onto multiple grids for each sample. A JEOL JEM-1210 analytical transmission electron microscope operated at 90 kV was used to view the sections. For each sample three to seven transfected cells were identified and images of M-PMV capsids with the cell were captured on film. The developed negatives were then scanned with an AGFA Duoscan T2500 scanner to generate high-resolution image files. Adobe Photoshop CS was then used to adjust the black, white, and gray tonal ranges of the images for better visualization of M-PMV capsids within the cell.
Immunofluorescence microscopy.
Indirect immunofluorescent staining was carried out as described previously (56) with the following modifications. To look at the distribution of wild-type and mutant Gag proteins in conjunction with cellular Rab-GFP fusion proteins, COS-1 cells were transfected with wild-type or mutant M-PMV constructs and Rab11-GFP, Rab4-GFP, Rab5-GFP, or Rab7-GFP (54). The cells were grown on no. 1.5 12-mm glass coverslips (Fisher, Pittsburgh, PA) for 24 h and then fixed at room temperature for 15 min in freshly prepared 4% formaldehyde (Tousimis, Rockville, MD) in PBS. For primary antibody staining, coverslips were incubated in 100 μl of 20-μg/ml (anti-M-PMV Gag) monoclonal antibody 10.10 diluted in PBS containing 2.5% goat serum (Abcam, Cambridge, MA) and 0.2% Tween 20 (Sigma) for 45 min at 37°C. Since the GFP fluorescence from the Rab-GFP fusion proteins was not quenched in these experiments, a primary anti-GFP antibody was not used. The coverslips were mounted in Prolong antifade reagent (Invitrogen, Carlsbad, CA) to prevent quenching of the fluorescence signal. The distribution of wild-type and mutant Gag and CD63, a member of the tetraspanin transmembrane 4 superfamily found on multivesicular bodies (9, 12), was examined as follows. COS-1 cells expressing wild-type or mutant M-PMV proviral constructs were fixed as described above and then permeabilized and blocked with 5% goat serum in PBS containing 0.2% Tween 20. Coverslips were incubated in a primary antibody mixture (100 μl) containing a 1:500 dilution of a rabbit polyclonal anti-M-PMV Gag antibody and a 1-μg/ml human anti-CD63 monoclonal antibody diluted in PBS containing 5% goat serum and 0.2% Tween 20. For secondary antibody staining, coverslips were incubated in 100 μl of a 20-μg/ml Alexa 594-goat anti-mouse immunoglobulin G antibody and a 6-μg/ml fluorescein isothiocyanate-labeled goat anti-rabbit immunoglobulin G antibody diluted in PBS containing 5% goat serum in 0.2% Tween 20. Coverslips were mounted in Prolong antifade reagent. Images were visualized with a multiple-wavelength, wide-field, three-dimensional microscopy system (Intelligent Imaging Innovations, Denver, CO), which was based on a Zeiss 200 M inverted microscope (Carl Zeiss, Thornwood, NY). Immunofluorescent samples were visualized using a 63× oil objective with a numerical aperture of 1.4. Images of successive 0.2-μm optical sections were captured with a cooled Coolsnap HQ charge-coupled device camera which contained an Orca-ER chip. A standard Sedat filter set (Chroma Technology, Rockingham, UT) was used to eliminate cross talk of fluorescent emissions. The images were deconvoluted using the constrained iterative algorithm (46) and analyzed using Slidebook software (Intelligent Imaging Innovations).
Single-round infectivity assay.
293T cells were plated in vented T75 flasks at a density of 1.3 × 106 cells. Twenty-four hours later, they were cotransfected by the Fugene 6 method with both the pSARM-EGFP expression vector, containing either the wild-type or the mutant gag gene, and the glycoprotein expression vector pTM0 (4). At 72 h after cotransfection, culture supernatants were collected and filtered through a 0.45-μm filter. Capsid protein (p27) for each sample was normalized by Western blotting, and the volume of culture supernatant used to infect COS-1 cells was adjusted such that equivalent amounts of virus were added for wild type and each mutant. After 48 h, the cells were washed twice with PBS, and 500 μl of PBS-1 mM EDTA was added to resuspend the cells. A final concentration of 4% paraformaldehyde in PBS was used to fix the cells prior to analysis of GFP expression on a BD FACSCalibur flow cytometer.
Release of wild-type and K16A capsids from cells depleted of plasma membrane pools of PI(4,5)P2.
Pulse-chase experiments were executed as described under “Metabolic labeling and immunoprecipitation of Gag” with the following modification. Cells were transfected with a 5:1 ratio of vectors containing the gene for either the wild-type phosphoinositide (PI)-5-phosphatase IV enzyme or the inactive delta-1 derivative (26) and proviral vectors of either wild-type or K16A M-PMV. Release of virus was quantitated as described previously.
RESULTS
Construction of mutants for arginine and lysine residues in M-PMV Gag matrix domain.
To investigate the role of positively charged amino acids in the matrix domain of M-PMV Gag in intracellular transport and capsid-membrane interactions, two arginine residues (R10 and R22) and six lysine residues (K16, K20, K25, K27, K33, and K39) were replaced individually and in pairwise combinations with the smaller, nonpolar alanine residue (Fig. 1A). The van der Waals electrostatic potential map generated in Swiss-PDB viewer (http://www.expasy.org/spdbv/) (16) predicts a large density of positive charge on helices A and B of the matrix protein (7) (Fig. 1B), in which the side chains of R10 and R22 are oriented to the outermost surface of helix A, side chains of K16 and K20 are on the opposing side of helix A from R10 and R22, lysine residues at positions 25 and 27 are in the loop between helices A and B, and K33 and K39 are located in helix B (Fig. 1C). The confirmed M-PMV MA mutants R10A, K16A, K20A, R22A, K25A, K27A, K33A, K39A, R10A/K16A, K16A/K20A, K20A/R22A, R22A/K25A, K25A/K27A, K27A/K33A, and K33A/K39A as well as the previously described matrix mutant R55W (39) and the wild type were studied in the context of an infectious provirus.
FIG. 1.
Location of arginine and lysine residues in the M-PMV matrix domain. (A) Schematic diagram of the M-PMV Gag polyprotein. Myristate is shown attached to the N-terminal matrix domain (MA) of Gag. The primary sequence of MA is shown with arginine and lysine residues in bold and numbered. The lines show the four helices of M-PMV MA. (B) Electrostatic potential map of M-PMV MA protein. A large positive potential (blue) is depicted surrounding the Arg and Lys residues in helices 1 and 2. The negative-charge potential is shown in red. (C) Positions of arginine and lysine residues in the context of the M-PMV matrix protein nuclear magnetic resonance structure.
Synthesis, expression, and release of wild-type and mutant M-PMV Gag.
The synthesis and stability of wild-type and mutant Gag polyproteins with a decreased positive charge in the MA domain were assayed in a pulse-chase experiment. 293T cells transfected with wild-type and mutant proviral constructs were pulse-labeled with [35S]methionine and then chased for 1 and 4 h in complete medium. Cell lysates from pulse-labeled cells and pulse-chase cells, as well as culture medium from the latter, were collected. M-PMV gag gene products were immunoprecipitated as described in Materials and Methods and then subjected to SDS-PAGE and imaged using a Cyclone phosphorimager. During the pulse, M-PMV Gag polyprotein (Pr78) was synthesized at similar levels for the wild type and each of the mutants (Fig. 2A). After a 4-h chase, the mutant Gag polyprotein precursors varied significantly in the extent to which they were processed into mature products. Mutants R10A, R22A, K25A, and K27A and six of the seven double mutants exhibited much higher levels of residual Pr78 in the pulse-chase cell lysates than did the wild type or the other single mutants (Fig. 2B). As shown below, these processing-deficient mutants are defective in virion release, consistent with our previous observation that the Gag polyprotein is proteolytically cleaved only after release of virions (3, 31, 53) and that Gag is not cleaved in mutants blocked in a late stage of budding (40, 64).
FIG. 2.
Synthesis, processing, and release of wild-type (WT) and mutant M-PMV Gag. 293T cells were transfected with wild-type and mutant M-PMV proviral genomes. Viral proteins were metabolically labeled with [35S]methionine and then immunoprecipitated from cell lysates and analyzed by SDS-PAGE. (A) Viral proteins immunoprecipitated from lysates of pulse-labeled cells. Positions of the wild-type and mutant viral precursor proteins Pr180 (Gag-Pro-Pol), Pr95 (Gag-Pro), and Pr78 (Gag) are shown. (B) Viral proteins immunoprecipitated from cells pulse-labeled and then chased for 4 h. Viral precursor proteins and capsid (p27CA), the major cleavage product, are shown. (C) M-PMV p27CA immunoprecipitated from the culture medium collected after the 4-h chase. (D) Band intensities were quantitated for the pulse-labeled Gag (Pr78) and for the p27 (CA) released into the culture medium at 1 and 4 h after pulse-label. The percent capsid protein released into culture supernatants after a 1-h chase (gray bars) and a 4-h chase (black bars) for a representative experiment is shown for the wild type and each of the positive-charge mutants.
Release of virus was assessed by calculating the percentage of the Gag precursor proteins immunoprecipitated from the pulse-labeled cells that were shed as capsid protein (p27) into the culture medium during the chase. Similar amounts of virion-associated p27 were released for the wild type and MA mutant K39A after a 4-h chase (Fig. 2C and D), while mutants K16A, K20A, R10A/K16A, and K16A/K20A were released with efficiencies of 50 to 70% of that of the wild type. Mutant K33A exhibited the lowest release of this intermediate group—33% of that of the wild type. In contrast, the release of mutants R10A, K25A, K27A, and K33A/K39A was reduced by more than 90% from that of the wild type, and that of the remaining double mutants, as well as R22A, was abrogated completely. Essentially the same results were observed after a 1-h chase (Fig. 2D), with the exception that the K16A and R10A/K16A mutants showed release equivalent to that of the wild type at this time point. In addition, the R55W mutant was released 2.5 times faster than the wild type, consistent with our previous observations for this C-type morphogenesis mutant (39).
Kinetics of Gag precursor processing.
The kinetics of precursor processing for wild-type and mutant Gag precursor proteins were investigated by pulse-chase experiments in which cell-associated and virion-associated proteins were analyzed after a 1-, 2-, or 4-h chase. Cell lysates from pulse-labeled cells and pulse-chase cells, as well as culture medium from the latter, were collected for each time point. M-PMV gag gene products were immunoprecipitated, subjected to SDS-PAGE, and imaged using a phosphor screen. OptiQuant software was used to obtain the band intensities for Gag (Pr78), Gag-Pro (Pr95), Gag-Pro-Pol (Pr180), and capsid (p27CA). The percentage of Gag precursor molecules and the percent CA molecules relative to total Gag were calculated for each chase time point as described in Materials and Methods.
The half-life of Gag precursor processing for wild-type M-PMV was 1.5 h (Fig. 3), and similar kinetics were observed for K20A, K33A, and K39A and the double mutants R10A/K16A and K16A/K20A. In contrast, accelerated kinetics (half-life of less than 1 h) were observed for MA mutants K16A and R55W (Fig. 3). Reduced processing kinetics were observed for K25A, K27A, and K33A/K39A, with only 20 to 30% of Pr78 processed at 4 h (Fig. 3). An even greater defect was observed for R10A, whereas for mutants K25A/K27A and K27A/K33A and those that contain the R22A substitution, no processing of Gag was detected 4 h after the pulse-label (Fig. 3).
FIG. 3.
Kinetics of Gag precursor processing for M-PMV basic mutants. 293T cells transfected with an M-PMV proviral genome were metabolically labeled with [35S]methionine and then chased for 0, 1, 2, and 4 h. Viral proteins were immunoprecipitated from cell lysates and culture medium at each time point and following SDS-PAGE were quantitated. The processing curves from a representative experiment are shown for the wild type and the positive-charge mutants. Black solid squares represent percentage of the total Gag precursor at each time point. Gray open circles represent percent total capsid protein.
MA positive-charge mutants accumulate at multiple intracellular locations.
The diverse kinetics of precursor processing and the extent of virion release suggested that intracellular transport of Gag might be altered by the positive-charge substitutions. To determine the intracellular location of mutant capsids at high resolution, thin sections of transfected cells expressing the wild-type or mutant M-PMV provirus were analyzed by transmission electron microscopy. Cells expressing wild-type M-PMV exhibited few immature capsids in the cytoplasm and a large number of capsids in the process of budding or already released into the intercellular space (Fig. 4). As reported previously (39), cells expressing the R55W matrix mutant provirus exhibited a C-type assembly pattern with dense patches of Gag assembling at the plasma membrane (Fig. 4). This contrasts with the budding of fully assembled capsids in wild-type-infected cells.
FIG. 4.
Transmission electron microscopy of COS-1 cells expressing wild-type and mutant M-PMV proviruses. COS-1 cells expressing wild-type or mutant M-PMV proviral genomes were fixed 24 h posttransfection in 2.5% glutaraldehyde and postfixed in 1% osmium tetroxide to preserve proteins and membranes, respectively, prior to embedding and thin sectioning. Bars, 200 nm. WT, wild type; N, nucleus; PM, plasma membrane. The white arrowhead depicts a C-type assembly (K16A). Black arrowheads denote particles that have budded intracellularly (K16A and K16A/K20A).
Four distinct positive-charge mutant phenotypes were observed. Capsids that contained the R10A and K27A substitution were found adjacent to or traversing through what appeared to be cortical actin, the dense filamentous actin (F-actin) network at the cell periphery (17, 42, 63) (Fig. 4). This phenotype was more pronounced for R22A, K20/R22A, and R22A/K25A, with capsids accumulating in the cytoplasm under the cortical actin (Fig. 4).
A transport-defective phenotype with capsids scattered throughout the cytoplasm or in small clusters in the cytoplasm near the plasma membrane was observed for the K25A mutant (Fig. 4). This phenotype was also observed for the K25A/K27A mutant; however, in general, fewer capsids were scattered in the cytoplasm and larger accumulations close to the plasma membrane were observed (data not shown).
Mutants K33A, K39A, and K33A/K39A had a phenotype of immature capsid accumulation at the plasma membrane (Fig. 4).
A dramatically different phenotype in which immature capsids were found accumulating around intracellular vesicles was observed when lysine at position 16 or 20 was replaced with alanine (Fig. 4). Both D-type assembly and C-type assembly were detected at these intracellular membranes in cells expressing K16A. Mutant K20A capsids that accumulated intracellularly were all of the D-type morphology. In cells expressing each of these single mutants, capsids were seen budding both into and within intracellular vesicles. This aberrant intracellular budding was augmented for capsids that contained the double substitution K16A/K20A (Fig. 4), and C-type assembly on intracellular vesicles was also observed for this double mutant.
K16A and K20A associate with Rab11, Rab5, and CD63-positive vesicles.
Our laboratory has shown previously that wild-type M-PMV Gag colocalizes with Rab11 at the pericentriolar recycling endosome and that capsid transport from the pericentriolar region to the plasma membrane is dependent on a functional endosomal pathway (48). To investigate if the intracellular membranes around which K16A, K20A, and K16A/K20A mutant capsids cluster are derived from the endosomal pathway, the distribution of immunostained wild-type and mutant M-PMV Gag was compared to that of Rab11-GFP (recycling endosome), Rab4-GFP (recycling endosome), Rab5-GFP (sorting endosome), Rab7-GFP (lysosome) (54), and immunostained CD63 protein (multivesicular bodies) (9, 12).
COS-1 cells expressing wild-type M-PMV and Rab11-GFP had a distribution of anti-p12 Gag staining that colocalized with the Rab11-GFP signal primarily at a perinuclear location (Fig. 5A). A section of the cell was deconvoluted to remove out-of-focus light, and intensities from Rab11-GFP (green channel) and anti-p12 staining of M-PMV Gag (red channel) were associated in a maximum-intensity projection image (yellow pixels in merged channel denoted by arrows). Similar associations of K16A, K20A, and K16A/K20A mutant Gag and Rab11-GFP were seen. In contrast, neither wild-type nor mutant Gag proteins appeared to be associated with either the Rab4-GFP or the Rab7-GFP signal. While we did not observe colocalization of Rab5-GFP on sorting endosomes with wild-type Gag (Fig. 5B), this was observed with K16A (Fig. 5C), K20A, and K16A/K20A.
FIG. 5.
Immunofluorescent staining of M-PMV Gag. COS-1 cells were either cotransfected with wild-type (WT) or mutant M-PMV proviral genome and a Rab-GFP construct or transfected with only wild-type or mutant provirus. In both cases, cells were fixed 24 h later and cotransfected cells were immunostained for Gag with an anti-p12 monoclonal antibody. Cells expressing only wild-type or mutant provirus were immunostained for both Gag (anti-Pr78 polyclonal antibody) and CD63 (anti-CD63 monoclonal antibody). A single optical section of a cell is shown in the leftmost panel, from which the area within the square was deconvoluted using a constrained iterative algorithm with the Slidebook software. A maximum-intensity projection image of the deconvoluted sections was obtained, and each channel is shown separately and then merged. The white arrowheads depict staining that contributes to a signal of M-PMV associated with a cellular marker in the merged image (yellow pixels). Bars, 10 μm. (A) WT-Rab11; (B) WT-Rab5; (C) K16A-Rab5; (D) WT-CD63; (E) K16A/K20A-CD63.
For studies of M-PMV Gag association with CD63, we utilized an anti-Pr78 polyclonal antibody to detect M-PMV Gag and a monoclonal antibody to detect CD63.
The wild-type Gag does not associate with CD63 (Fig. 5D). In contrast, staining of the K16A, K20A, and K16A/K20A mutant Gags did overlap with that of CD63; this distribution is seen in a deconvoluted section of the cell, strongly supporting the association of these mutant capsids with CD63-positive vesicles (Fig. 5E).
Infectivity of mutant virions.
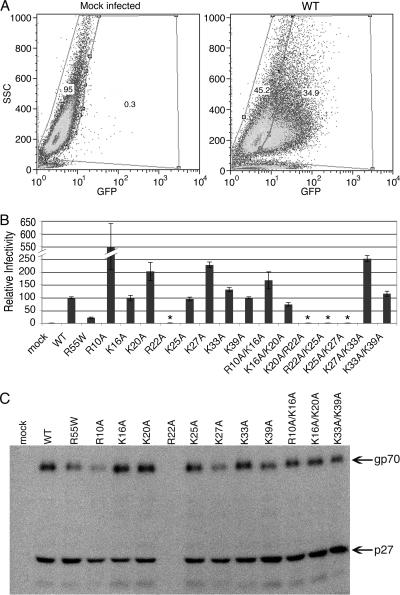
Because several of the basic amino acid mutants appeared to exhibit altered trafficking within the cell, it was of interest to determine the infectivity and glycoprotein incorporation of the released virions. For this purpose the mutations were engineered into the pSARM-EGFP vector, which expresses EGFP in place of Env. Wild-type and mutant EGFP-proviruses were cotransfected into 293T cells with an Env expression vector, and virus-containing supernatants were collected after 72 h and, after normalization for p27 content, used to infect COS-1 cells. At 24 h postinfection, cells were removed from the plate and analyzed by flow cytometry. A representative gating is shown in Fig. 6A for wild-type virus, where approximately 35% of the cells are infected. Figure 6B shows the normalized results from four independent experiments for all of the mutants and wild type. Interestingly, with the exception of K16A/K20A, which exhibits approximately 75% of wild-type infectivity, none of the mutants that release virions show any defect in this single-cycle infection assay. In contrast, mutants R10A and K27A and the double mutants R10A/K16A and K27A/K33A, which are for the most part inefficiently released and appear to inefficiently traverse the cortical actin, exhibit enhanced infectivity (two- to fivefold that of the wild type). The basis for this increased infectivity is not apparent but does not seem to reflect Env incorporation since both R10A and K27A appear to incorporate less gp70 than the wild type does (Fig. 6C). Even though both K16A and K20A associate with intracellular vesicles, these mutants exhibit the same (K16A) or higher (K20A) infectivity than does the wild type. Virions released from cells transfected with these mutant proviruses incorporate Env at levels similar to that observed for the wild type (Fig. 6C).
FIG. 6.
Relative infectivity of positive-charge M-PMV mutants determined by a single-round assay. 293T cells were cotransfected with mutant and wild-type (WT) pSARM-EGFP and pTMO vectors. At 72 h posttransfection supernatants were filtered and normalized by Western blotting for p27. Equivalent amounts of p27 were used to infect COS-1 cells. Twenty-four hours later, cells were harvested and the percentage of GFP-expressing cells was quantitated by fluorescence-activated cell sorter analysis. The mean percentage (± standard deviation) of infectivity for each mutant relative to the wild type from four independent experiments is shown. (A) Representative gating for mock- and wild-type-infected cells. (B) Percent infectivity relative to the wild type for each of the positive-charge mutants. Asterisks denote mutants that were not released into the medium and therefore are not infectious. (C) Glycoprotein incorporation for the wild type and positive-charge mutants. Briefly, 293T cells were transfected with either the wild-type or the mutant proviral expression vector. Twenty-four hours posttransfection, the cells were labeled with [3H]leucine, and 6 h later, the supernatant was collected and virus was pelleted through a 25% (wt/vol) sucrose cushion. M-PMV viral proteins that were pelleted were immunoprecipitated with a goat anti-M-PMV antiserum and analyzed by 12% SDS-PAGE and fluorography. The SU (gp70) of Env and the capsid (p27) proteins are shown.
Release of wild-type and K16A mutant M-PMV Gag in cells depleted in plasma membrane pools of PI(4,5)P2.
The negatively charged phospholipid PI(4,5)P2 is mainly localized to the plasma membrane of the cell and has been shown to target HIV-1 Gag to the plasma membrane (26). To ascertain if PI(4,5)P2 is involved in release of capsids for wild-type M-PMV and the positive-charge mutant K16A, which buds into internal vesicles, we coexpressed PI-5-phosphatase IV, which when overexpressed depletes plasma membrane pools of PI(4,5)P2 (26). 293T cells were cotransfected with an expression vector containing the coding region for the active phosphatase and the proviral expression vectors for either the wild type or the M-PMV positive-charge mutant K16A. In conjunction, cells were cotransfected with a derivative of the PI-5-phosphatase IV vector, which lacks the coding region for the active site of the enzyme (ΔPI-5-phosphatase IV or delta-1) together with either the proviral vector for wild-type M-PMV or K16A. Sixteen hours later, transfected cells were pulse-labeled with [35S]methionine and then chased for 1, 2, and 4 h in complete medium. Cell lysates from pulse-labeled cells and pulse-chase cells, as well as culture medium from the latter, were collected and analyzed as described previously. Release of virus was assessed by calculating the percentage of the Gag precursor proteins immunoprecipitated from the pulse-labeled cells that were shed as capsid protein (p27) into the culture medium during the chase.
Depletion of the cellular pools of PI(4,5)P2 by overexpression of the active form of PI-5-phosphatase IV in cells expressing either the wild-type M-PMV or the K16A mutant resulted in an overall decrease in release of pulse-labeled Gag (Fig. 7). For the wild type, the release of pulse-labeled Gag was reduced by 90% during the 4-h chase. In contrast, the release of pulse-labeled Gag was reduced by only 30% during the 4-h chase for the K16A mutant (Fig. 7).
FIG. 7.
Release of wild-type (WT) and mutant M-PMV Gag in cells depleted in plasma membrane pools of PI(4,5)P2. 293T cells were cotransfected with the wild-type or K16A mutant proviral expression vector in combination with either the delta-1 or phosphatase IV vectors (26). Sixteen hours later, viral proteins were metabolically labeled with [35S]methionine-cysteine and then immunoprecipitated from cell lysates as well as supernatant culture medium at 1, 2, and 4 h postlabeling. The percent pulse-labeled capsid protein released in a representative experiment at 1 (light gray bars), 2 (dark gray bars), and 4 (black bars) h is shown for WT-delta-1, WT-phosphatase IV, K16A-delta-1, and K16A-phosphatase IV.
DISCUSSION
In this study, the positive charge of the N-terminal M-PMV Gag MA domain was modified to determine the role of basic amino acid residues in intracellular trafficking to and release of capsids from the plasma membrane. Previous studies of HIV-1 have suggested that a combination of positive charges and the N-terminal myristic acid on the MA of Gag forms a bipartite signal for transport and attachment of Gag to the plasma membrane (11, 19, 66). Since HIV-1 assembles at the plasma membrane, attachment to the plasma membrane through the bipartite signal of MA may effectively increase the concentration of Gag at a specific site in the membrane to facilitate capsid assembly. In contrast, the myristate and positive charge on M-PMV MA are incorporated into assembled capsids at the pericentriolar region of the cell. These capsids are transported through the cytoplasm and are specifically released at the plasma membrane, and so it is likely that myristate is sequestered in M-PMV MA with the positive charge on the outer surface of the assembled capsid. At the plasma membrane, M-PMV must associate with the membrane and wrap the lipid bilayer around a spherical capsid. The studies here probe the functions of the positive surface charge on the assembled capsid and test the hypothesis that these positive charges facilitate the intimate association of Gag with the membrane and likely trigger the exposure of myristate that is necessary for membrane envelopment.
The MA mutant with a substitution of alanine for lysine at position 25, located in the loop between helices A and B of M-PMV MA (7), delayed intracellular transport of immature capsids so that less than 20% of the pulse-labeled Gag precursors were cleaved after a 4-h chase and an equivalent amount of Gag was released as p27 into the culture medium. This is in contrast to wild-type kinetics, in which 50% of Gag was cleaved at 1.5 h and 60% was released as p27 in the 4-h chase. Within the cell, K25A immature capsids were primarily dispersed throughout the cytoplasm; this phenotype is similar to the previously characterized Y82F transport-defective MA mutant (56). In contrast to Y82F, however, the K25A mutant capsids did not accumulate in the pericentriolar region of the cell, suggesting that capsids can initiate intracellular transport but are inefficiently transferred to the plasma membrane. The transport defect appeared to be enhanced when K25A was combined with other mutations (R22A or K27A). In cells expressing these double mutants, capsids accumulated at a perinuclear region and in groups within the cytoplasm with no release of capsids during the 4-h chase.
Immature capsids accumulating in the initial stages of budding were characteristic of MA mutants with replacements of lysine residues at positions 33 and 39 which are located in helix B. For these mutants, the Gag precursor was cleaved with kinetics similar to those of the wild type; however, 30% of the K33A precursor and 15% of the K39A precursor (versus 10% for the wild type) remained uncleaved after 4 h. This residual uncleaved Gag may, for K33A in particular, represent the capsid population accumulated at the plasma membrane. Since few capsids were observed in the cytoplasm of transfected cells, it is likely that the efficiency of intracellular transport to the plasma membrane for these mutants was similar to that of the wild type.
The accumulation of capsids at the plasma membrane in an early stage of membrane extrusion suggests that these mutants may be defective in an initial electrostatic interaction with the negatively charged inner leaflet of the plasma membrane. Positively charged amino acids in HIV-1 MA have been shown to facilitate Gag-plasma membrane interactions (65, 66), and the basic cluster from the HIV-1 MA can be introduced into the Rous sarcoma virus MA domain of Gag and restore budding of membrane-binding-domain mutants (2). Similarly, loss of positive-charge residues in Rous sarcoma virus MA resulted in defects in Gag association with the plasma membrane, while addition of basic residues to the membrane-binding domain of MA resulted in a dramatic increase of particles released (6). Moreover, for HIV-1, budding from the plasma membrane can be restored for MA mutants that interact with intracellular membranes by addition of a lysine residue in the basic domain (28). Thus, it is possible that the positive charges at positions 33 and 39 of M-PMV MA are critical for capsids to interact with negatively charged phospholipids at the plasma membrane, which in turn could be required to trigger release of myristate from MA into the plasma membrane. A defect in release of myristate for these basic MA mutants would be much like the hydrophobic MA mutants that we have described previously which also exhibited capsid accumulation at the plasma membrane (40, 56). This would also be consistent with the PI(4,5)P2-stimulated release of myristic acid observed in vitro with HIV-1 MA (43).
Immature capsids for the MA mutants R10A, R22A, and K27A were released very inefficiently. Only small amounts of R10A and K27A (approximately 14% of that observed for the wild type) were released at 4 h, although for R10A this increased to 60% of that of the wild type after 24 h. No detectable levels of p27 were released for R22A at either time point. In contrast to the positive-charge mutants that are delayed in the initial stages of membrane envelopment, capsids from this group of mutants can be observed by electron microscopy to be accumulated in the cytoplasm under (R22A) or traversing (R10A and K27A) the peripheral filamentous actin layer located beneath the plasma membrane (17, 42, 63). This suggests that there is a rate-limiting delay in capsid transport through the dense cortical actin patches for these mutants. With R22A, large accumulations of capsids can be observed and few if any reach the plasma membrane. This unusual phenotype is very similar to that of an endogenous virus variant (enJS56A1) of Jaagseike sheep retrovirus in which a similarly located arginine (R21) is replaced by tryptophan (24). Interestingly, this arginine residue is conserved in all betaretroviruses (24).
A block in transport at the cortical actin argues that capsid transport must switch from a primarily microtubule-based system to a primarily actin-based process in order to reach the plasma membrane. While capsid transport from the pericentriolar region to the plasma membrane can be blocked by a combination of microtubule and actin inhibitors (R. A. LaCasse and E. Hunter, unpublished data), these mutants argue that the positive charges within MA directly facilitate capsid transport through the cortical actin patches. It is possible that the arginine residues at positions 10 and 22 as well as the lysine at position 27 in M-PMV MA mediate the association of capsids with actin in a way similar to that of the basic effector domain of the MARCKS protein, which has been shown to bind actin (18).
A striking phenotype was observed for mutants K16A and K20A and the K16A/K20A double mutant, with capsids surrounding and budding into intracellular vesicles. Few if any capsids were released from the plasma membrane. All of these mutants exhibited a slightly less efficient release (60 to 70% of that of the wild type) of capsids during a 4-h chase, but the capsids released had similar levels of gp70 incorporated and were as infectious as the wild type. Processing of K16A Pr78 occurred with kinetics similar to those of R55W, but in contrast to this C-type morphogenesis mutant, we did not observe a more-rapid or more-efficient release of virus for K16A. Thus, it is likely that processing of K16A Gag is initiated following budding into intracellular vesicles but that some of these mature virions are unable to exit the cell.
The K16A, K20A, and K16A/K20A mutant capsids appear to associate with a variety of intracellular membranes and colocalize with sorting endosomes and multivesicular bodies in addition to the recycling endosomes with which wild-type capsids associate (48). This phenotype is similar to one described for an HIV-1 mutant in which lysine residues in MA were replaced with glutamic acid, resulting in Gag colocalization with CD63 (27). MA mutants of HIV-1, which alter or abolish the globular head of MA and in which myristate may be constitutively exposed, could also be seen assembling at and budding into intracellular vesicles (10, 11, 30, 38). Thermodynamic calculations show that a single myristate moiety has insufficient binding energy to facilitate the binding of a protein to a membrane (33), so there is likely to be a requirement for Gag to undergo some oligomerization prior to a high-affinity association with membranes. This is consistent with observations that the I domain of HIV-1 Gag facilitates membrane association through oligomerization of Gag (8, 45). In contrast, assembled M-PMV capsids contain upward of 2,000 myristic acid moieties (32); thus, it is possible that loss of the basic amino acids in helix A results in mutants which are unable to sequester myristic acid and therefore associate with the first available membrane through hydrophobic interactions—yielding the phenotype observed with K16A and K20A. Although this hypothesis is one that we favor, it is also possible that the K16A and K20A M-PMV mutants initiate myristate exposure and intracellular budding through an altered electrostatic interaction between MA and lipids on the endosomal compartments. PI(4,5)P2 has been shown to be important for targeting HIV-1 Gag to the plasma membrane (26). It also appears to be important for budding and release of M-PMV from the plasma membrane. Depletion of PI(4,5)P2 by overexpression of PI-5-phosphatase IV in M-PMV-expressing cells resulted in a 90% decrease in the release of wild-type p27 during a 4-h chase of pulse-labeled Pr78. In contrast, K16A release is inhibited by only 30% under the same conditions, arguing that this mutant is not as dependent on PI(4,5)P2 for its release. It seems likely, because the K16A mutant bypasses the block imposed on wild-type M-PMV by PI-5-phosphatase IV, that it utilizes a distinct pathway for release from the cell, perhaps within endosomal vesicles that are recycled back to the plasma membrane. It remains to be determined whether the K16A and K20A mutants are able to initiate the budding process intracellularly through spontaneous release of myristate or through interaction with an alternate phospholipid molecule such as PI(3,5)P2.
The retroviral budding process has been shown to involve a complex cellular machinery, endosomal sorting complexes required for transport (ESCRT), which is normally involved in multivesicular body formation (21, 61). Retroviral late domains have been shown to interact with a series of ESCRT protein complexes (I to III) that function in the pinching-off stages of capsid release (23), and M-PMV encodes two late-domain motifs (PSAP and PPPY) in the pp24 region, which is C terminal to MA (3, 15, 53, 64). These motifs are necessary for M-PMV to complete the budding process (15, 64). Since K16A and K20A mutants colocalized with vesicles that appeared to be derived from the late endosomal pathway (CD63 multivesicular bodies), it is tempting to speculate that cellular ESCRT components associated with these vesicles might be recruited to facilitate efficient intracellular budding. It has not been established when the ESCRT machinery associates with wild-type M-PMV capsids, since the latter do not normally associate with the multivesicular bodies; however, it is possible that at least some of the ESCRT components associate with M-PMV during capsid assembly in the pericentriolar region of the cell. These initial interactions may then recruit additional components of the ESCRT complex once the myristyl-switch mechanism has initiated membrane envelopment and release of capsids from the plasma membrane.
The results presented here demonstrate that basic amino acids on the outer surface of the MA domain of Gag play critical roles in intracellular targeting, trafficking, and release. They reveal for the first time the critical role that the viral MA protein plays in facilitating transport of capsids across the cortical actin component of the cell's cytoskeleton and support both microtubule- and actin-mediated steps in intracellular transport. Mutation of basic residues in helix A of the M-PMV MA appears to alter the membrane specificity of viral budding, consistent with altered myristate exposure or triggering. In contrast, mutation of positive charges in helix B seems to modulate the efficiency of viral budding at the plasma membrane, without affecting targeting or intracellular trafficking. This could reflect inefficient interactions of capsid with negative phospholipids in the inner leaflet of the plasma membrane and/or the subsequent triggering of myristate exposure necessary for the budding process. Structural studies of the interaction of PI(4,5)P2 with myristylated M-PMV MA should provide further insights into these capsid-membrane interactions.
Acknowledgments
We thank Daniel Kalman, Emory University, for generous access to his fluorescence microscope as well as for his assistance with the fluorescence microscopy experiments. We thank Michael Sakalian and Chisu Song for advice in mutation and vector design; for critical comments on the manuscript, we thank Melissa Alexander, Grace Rong, Benyue Zhang, and Thomas Postler. We also thank Jeannette Taylor at the Integrated Microscopy and Microanalytical Facility as well as Carla Belk and Shaharah Hardy for their technical assistance.
This work was supported by grant R01 CA-27834 from the National Institutes of Health.
Footnotes
Published ahead of print on 27 June 2007.
REFERENCES
- 1.Ames, J. B., R. Ishima, T. Tanaka, J. I. Gordon, L. Stryer, and M. Ikura. 1997. Molecular mechanics of calcium-myristoyl switches. Nature 389:198-202. [DOI] [PubMed] [Google Scholar]
- 2.Bennett, R. P., T. D. Nelle, and J. W. Wills. 1993. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 67:6487-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradac, J., and E. Hunter. 1984. Polypeptides of Mason-Pfizer monkey virus. I. Synthesis and processing of the gag-gene products. Virology 138:260-275. [DOI] [PubMed] [Google Scholar]
- 4.Brody, B. A., and E. Hunter. 1992. Mutations within the env gene of Mason-Pfizer monkey virus: effects on protein transport and SU-TM association. J. Virol. 66:3466-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant, M., and L. Ratner. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 87:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callahan, E. M., and J. W. Wills. 2000. Repositioning basic residues in the M domain of the Rous sarcoma virus Gag protein. J. Virol. 74:11222-11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conte, M. R., M. Klikova, E. Hunter, T. Ruml, and S. Matthews. 1997. The three-dimensional solution structure of the matrix protein from the type D retrovirus, the Mason-Pfizer monkey virus, and implications for the morphology of retroviral assembly. EMBO J. 16:5819-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derdowski, A., L. Ding, and P. Spearman. 2004. A novel fluorescence resonance energy transfer assay demonstrates that the human immunodeficiency virus type 1 Pr55Gag I domain mediates Gag-Gag interactions. J. Virol. 78:1230-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Escola, J. M., M. J. Kleijmeer, W. Stoorvogel, J. M. Griffith, O. Yoshie, and H. J. Geuze. 1998. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 273:20121-20127. [DOI] [PubMed] [Google Scholar]
- 10.Facke, M., A. Janetzko, R. L. Shoeman, and H. G. Krausslich. 1993. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J. Virol. 67:4972-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freed, E. O., J. M. Orenstein, A. J. Buckler-White, and M. A. Martin. 1994. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J. Virol. 68:5311-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukuda, M. 1991. Lysosomal membrane glycoproteins. Structure, biosynthesis, and intracellular trafficking. J. Biol. Chem. 266:21327-21330. [PubMed] [Google Scholar]
- 13.George, D. J., and P. J. Blackshear. 1992. Membrane association of the myristoylated alanine-rich C kinase substrate (MARCKS) protein appears to involve myristate-dependent binding in the absence of a myristoyl protein receptor. J. Biol. Chem. 267:24879-24885. [PubMed] [Google Scholar]
- 14.Gottlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottwein, E., J. Bodem, B. Muller, A. Schmechel, H. Zentgraf, and H. G. Krausslich. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 17.Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science 279:509-514. [DOI] [PubMed] [Google Scholar]
- 18.Hartwig, J. H., M. Thelen, A. Rosen, P. A. Janmey, A. C. Nairn, and A. Aderem. 1992. MARCKS is an actin filament crosslinking protein regulated by protein kinase C and calcium-calmodulin. Nature 356:618-622. [DOI] [PubMed] [Google Scholar]
- 19.Hill, C. P., D. Worthylake, D. P. Bancroft, A. M. Christensen, and W. I. Sundquist. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 93:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter, E., J. Casey, B. Hahn, M. Hayami, B. Korber, R. Kurth, J. Neil, A. Rethwilm, P. Sonigo, and J. Stoye. 2000. Retroviridae, p. 369-387. In M. H. V. van Regenmortel, C. M. Fauquet, D. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, London, United Kingdom.
- 21.Katzmann, D. J., G. Odorizzi, and S. D. Emr. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3:893-905. [DOI] [PubMed] [Google Scholar]
- 22.McLaughlin, S., and A. Aderem. 1995. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem. Sci. 20:272-276. [DOI] [PubMed] [Google Scholar]
- 23.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395-425. [DOI] [PubMed] [Google Scholar]
- 24.Mura, M., P. Murcia, M. Caporale, T. E. Spencer, K. Nagashima, A. Rein, and M. Palmarini. 2004. Late viral interference induced by transdominant Gag of an endogenous retrovirus. Proc. Natl. Acad. Sci. USA 101:11117-11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman, R. M., L. Hall, M. Connole, G. L. Chen, S. Sato, E. Yuste, W. Diehl, E. Hunter, A. Kaur, G. M. Miller, and W. E. Johnson. 2006. Balancing selection and the evolution of functional polymorphism in Old World monkey TRIM5alpha. Proc. Natl. Acad. Sci. USA 103:19134-19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ono, A., S. D. Ablan, S. J. Lockett, K. Nagashima, and E. O. Freed. 2004. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. USA 101:14889-14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ono, A., and E. O. Freed. 2004. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J. Virol. 78:1552-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ono, A., M. Huang, and E. O. Freed. 1997. Characterization of human immunodeficiency virus type 1 matrix revertants: effects on virus assembly, Gag processing, and Env incorporation into virions. J. Virol. 71:4409-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ono, A., J. M. Orenstein, and E. O. Freed. 2000. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J. Virol. 74:2855-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paillart, J. C., and H. G. Gottlinger. 1999. Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of Gag membrane targeting. J. Virol. 73:2604-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker, S. D., and E. Hunter. 2001. Activation of the Mason-Pfizer monkey virus protease within immature capsids in vitro. Proc. Natl. Acad. Sci. USA 98:14631-14636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker, S. D., J. S. Wall, and E. Hunter. 2001. Analysis of Mason-Pfizer monkey virus Gag particles by scanning transmission electron microscopy. J. Virol. 75:9543-9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peitzsch, R. M., and S. McLaughlin. 1993. Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristoylated proteins. Biochemistry 32:10436-10443. [DOI] [PubMed] [Google Scholar]
- 34.Perez-Caballero, D., T. Hatziioannou, J. Martin-Serrano, and P. D. Bieniasz. 2004. Human immunodeficiency virus type 1 matrix inhibits and confers cooperativity on Gag precursor-membrane interactions. J. Virol. 78:9560-9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rao, Z., A. S. Belyaev, E. Fry, P. Roy, I. M. Jones, and D. I. Stuart. 1995. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature 378:743-747. [DOI] [PubMed] [Google Scholar]
- 36.Rauch, M. E., C. G. Ferguson, G. D. Prestwich, and D. S. Cafiso. 2002. Myristoylated alanine-rich C kinase substrate (MARCKS) sequesters spin-labeled phosphatidylinositol 4,5-bisphosphate in lipid bilayers. J. Biol. Chem. 277:14068-14076. [DOI] [PubMed] [Google Scholar]
- 37.Ray, S., S. Zozulya, G. A. Niemi, K. M. Flaherty, D. Brolley, A. M. Dizhoor, D. B. McKay, J. Hurley, and L. Stryer. 1992. Cloning, expression, and crystallization of recoverin, a calcium sensor in vision. Proc. Natl. Acad. Sci. USA 89:5705-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Gottlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhee, S. S., and E. Hunter. 1990. A single amino acid substitution within the matrix protein of a type D retrovirus converts its morphogenesis to that of a type C retrovirus. Cell 63:77-86. [DOI] [PubMed] [Google Scholar]
- 40.Rhee, S. S., and E. Hunter. 1991. Amino acid substitutions within the matrix protein of type D retroviruses affect assembly, transport and membrane association of a capsid. EMBO J. 10:535-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rhee, S. S., and E. Hunter. 1987. Myristylation is required for intracellular transport but not for assembly of D-type retrovirus capsids. J. Virol. 61:1045-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rinnerthaler, G., M. Herzog, M. Klappacher, H. Kunka, and J. V. Small. 1991. Leading edge movement and ultrastructure in mouse macrophages. J. Struct. Biol 106:1-16. [DOI] [PubMed] [Google Scholar]
- 43.Saad, J. S., J. Miller, J. Tai, A. Kim, R. H. Ghanam, and M. F. Summers. 2006. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. USA 103:11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakalian, M., S. D. Parker, R. A. Weldon, Jr., and E. Hunter. 1996. Synthesis and assembly of retrovirus Gag precursors into immature capsids in vitro. J. Virol. 70:3706-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandefur, S., V. Varthakavi, and P. Spearman. 1998. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 72:2723-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scalettar, B. A., J. R. Swedlow, J. W. Sedat, and D. A. Agard. 1996. Dispersion, aberration and deconvolution in multi-wavelength fluorescence images. J. Microsc. 182:50-60. [DOI] [PubMed] [Google Scholar]
- 47.Seykora, J. T., M. M. Myat, L. A. Allen, J. V. Ravetch, and A. Aderem. 1996. Molecular determinants of the myristoyl-electrostatic switch of MARCKS. J. Biol. Chem. 271:18797-18802. [DOI] [PubMed] [Google Scholar]
- 48.Sfakianos, J. N., and E. Hunter. 2003. M-PMV capsid transport is mediated by Env/Gag interactions at the pericentriolar recycling endosome. Traffic 4:671-680. [DOI] [PubMed] [Google Scholar]
- 49.Sfakianos, J. N., R. A. LaCasse, and E. Hunter. 2003. The M-PMV cytoplasmic targeting-retention signal directs nascent Gag polypeptides to a pericentriolar region of the cell. Traffic 4:660-670. [DOI] [PubMed] [Google Scholar]
- 50.Song, C., S. R. Dubay, and E. Hunter. 2003. A tyrosine motif in the cytoplasmic domain of Mason-Pfizer monkey virus is essential for the incorporation of glycoprotein into virions. J. Virol. 77:5192-5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song, C., and E. Hunter. 2003. Variable sensitivity to substitutions in the N-terminal heptad repeat of Mason-Pfizer monkey virus transmembrane protein. J. Virol. 77:7779-7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song, C., K. Micoli, H. Bauerova, I. Pichova, and E. Hunter. 2005. Amino acid residues in the cytoplasmic domain of the Mason-Pfizer monkey virus glycoprotein critical for its incorporation into virions. J. Virol. 79:11559-11568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sonigo, P., C. Barker, E. Hunter, and S. Wain-Hobson. 1986. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell 45:375-385. [DOI] [PubMed] [Google Scholar]
- 54.Sonnichsen, B., S. De Renzis, E. Nielsen, J. Rietdorf, and M. Zerial. 2000. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J. Cell Biol. 149:901-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spearman, P., R. Horton, L. Ratner, and I. Kuli-Zade. 1997. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J. Virol. 71:6582-6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stansell, E., E. Tytler, M. R. Walter, and E. Hunter. 2004. An early stage of Mason-Pfizer monkey virus budding is regulated by the hydrophobicity of the Gag matrix domain core. J. Virol. 78:5023-5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stumpo, D. J., J. M. Graff, K. A. Albert, P. Greengard, and P. J. Blackshear. 1989. Molecular cloning, characterization, and expression of a cDNA encoding the “80- to 87-kDa” myristoylated alanine-rich C kinase substrate: a major cellular substrate for protein kinase C. Proc. Natl. Acad. Sci. USA 86:4012-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanaka, T., J. B. Ames, T. S. Harvey, L. Stryer, and M. Ikura. 1995. Sequestration of the membrane-targeting myristoyl group of recoverin in the calcium-free state. Nature 376:444-447. [DOI] [PubMed] [Google Scholar]
- 59.Tang, C., E. Loeliger, P. Luncsford, I. Kinde, D. Beckett, and M. F. Summers. 2004. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. USA 101:517-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taniguchi, H., and S. Manenti. 1993. Interaction of myristoylated alanine-rich protein kinase C substrate (MARCKS) with membrane phospholipids. J. Biol. Chem. 268:9960-9963. [PubMed] [Google Scholar]
- 61.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 62.Wang, J., A. Arbuzova, G. Hangyas-Mihalyne, and S. McLaughlin. 2001. The effector domain of myristoylated alanine-rich C kinase substrate binds strongly to phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 276:5012-5019. [DOI] [PubMed] [Google Scholar]
- 63.Weed, S. A., and J. T. Parsons. 2001. Cortactin: coupling membrane dynamics to cortical actin assembly. Oncogene 20:6418-6434. [DOI] [PubMed] [Google Scholar]
- 64.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yuan, X., X. Yu, T. H. Lee, and M. Essex. 1993. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J. Virol. 67:6387-6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou, W., L. J. Parent, J. W. Wills, and M. D. Resh. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 68:2556-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zozulya, S., and L. Stryer. 1992. Calcium-myristoyl protein switch. Proc. Natl. Acad. Sci. USA 89:11569-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]