Abstract
Hepatitis C virus (HCV) infection induces a wide range of chronic liver injuries; however, the mechanism through which HCV evades the immune surveillance system remains obscure. Blood dendritic cells (DCs) play a pivotal role in the recognition of viral infection and the induction of innate and adaptive immune responses. Several reports suggest that HCV infection induces the dysfunction of DCs in patients with chronic hepatitis C. Toll-like receptor (TLR) has been shown to play various roles in many viral infections; however, the involvement of HCV proteins in the TLR signaling pathway has not yet been precisely elucidated. In this study, we established mouse macrophage cell lines stably expressing HCV proteins and determined the effect of HCV proteins on the TLR signaling pathways. Immune cells expressing NS3, NS3/4A, NS4B, or NS5A were found to inhibit the activation of the TLR2, TLR4, TLR7, and TLR9 signaling pathways. Various genotypes of NS5A bound to MyD88, a major adaptor molecule in TLR, inhibited the recruitment of interleukin-1 receptor-associated kinase 1 to MyD88, and impaired cytokine production in response to TLR ligands. Amino acid residues 240 to 280, previously identified as the interferon sensitivity-determining region (ISDR) in NS5A, interacted with the death domain of MyD88, and the expression of a mutant NS5A lacking the ISDR partially restored cytokine production. These results suggest that the expression of HCV proteins modulates the TLR signaling pathway in immune cells.
Hepatitis C virus (HCV) belongs to the family Flaviviridae and possesses a positive, single-stranded RNA genome that encodes a single polyprotein composed of approximately 3,000 amino acids. HCV polyprotein is processed by host and viral proteases, resulting in 10 viral proteins. Viral structural proteins, including the capsid protein and two envelope proteins, are located in the N-terminal one-third of the polyprotein, followed by nonstructural proteins. HCV infects 170 million people worldwide and frequently leads to cirrhosis and hepatocellular carcinoma (36). In over one-half of patients, acute infection evolves into a persistent carrier state, presumably due to the ability of HCV to incapacitate the activation of the host immune mechanisms. Dendritic cells (DCs) are one type of potent antigen-presenting cell in vivo and play a crucial role in the enhancement and regulation of cell-mediated immune reactions. Since DCs express various costimulatory and/or adhesion molecules, they can activate even naïve T cells in a primary response. The role of the response of HCV antigen-specific T cells in viral clearance or persistence has been investigated extensively in both humans and chimpanzees (6, 27, 48, 51). These studies suggest that acute HCV infections followed by viral clearance are associated with a high frequency of HCV-specific CD4+ and CD8+ T-cell responses that can persist (27, 51), while chronic HCV infections are characterized by weak and restricted CD4+ and CD8+ T-cell responses that are not sustained (51).
Toll-like receptors (TLRs) are membrane-bound receptors that can be activated by the binding of molecular structures conserved among families of microbes. More than 10 different TLRs have been identified to date (2). They are highly conserved among mammals and are expressed in a variety of cell types. TLR binding and stimulation by pathogen-associated molecules is followed by a cascade of intracellular events that culminate in the expression of multiple genes (2). TLR signaling is mediated primarily by the adaptor protein myeloid differentiation factor 88 (MyD88), which triggers the activation of transcription factors, such as NF-κB, that are essential for the expression of proinflammatory cytokine genes (2). This pathway also leads to the potent production of type I interferon (IFN) through the activation of IFN regulatory factor 7 (IRF7) upon stimulation of TLR7 or TLR9 (22). In contrast, Toll/interleukin-1 (IL-1) receptor homology domain-containing adaptor-inducing IFN-β (TRIF/TICAM-1) mediates the production of type I IFNs primarily through the activation of IRF3 in response to TLR3 or TLR4 stimulation (2). Type I IFN induces the maturation of DCs by increasing both the expression of costimulatory molecules such as CD80, CD86, and CD40 and antigen presentation via major histocompatibility complex class I in addition to classical endogenous antigen presentation; it also facilitates the cross-presentation of viral antigens. A cumulative report has shown that DC activation via TLR signaling is a prerequisite for the subsequent induction of vigorous T-cell responses (42). Some viral proteins have been shown to inhibit the TLR-dependent signaling pathway through interactions with the downstream adaptor molecules, suggesting that the alteration of TLR-mediated signals is one of the mechanisms of virus-induced immune modulation (49). Dysfunction of DCs in patients with chronic HCV infection due to immaturation caused by the direct infection of DCs by HCV or by interactions with HCV proteins has been reported previously (4, 21). On the other hand, there have also been contrasting reports suggesting a lack of impairment of DC function in both chimpanzees and humans chronically infected with HCV (26, 32). Thus, at present, alterations in the TLR signaling pathway in the immune cells of patients with chronic hepatitis C virus infection are not well understood.
In the present study, we examined the effect of HCV proteins on TLR function in murine macrophage cell lines stably expressing HCV proteins. The expression of NS3, NS3/4A, NS4B, or NS5A was found to impair the activation of the TLR signaling pathways, and NS5A interacted with MyD88 through the IFN sensitivity-determining region (ISDR) and impaired cytokine production. To the best of our knowledge, this is the first demonstration of NS5A as an immunomodulator of TLR signaling pathways through the direct interaction with an adaptor molecule in immune cells.
MATERIALS AND METHODS
Cell culture.
Human embryonic kidney 293T cells and mouse macrophage RAW264.7 cells were maintained in Dulbecco's modified Eagle's medium (Sigma, St. Louis, MO) containing 10% fetal calf serum. All cells were cultured at 37°C in a humidified atmosphere with 5% CO2.
Plasmids and viruses.
DNA fragments encoding each of the HCV structural and nonstructural proteins were generated from a full-length cDNA clone of genotype 1b strain J1 (1) by PCR using Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA). The fragments were cloned into pCAGGs-puro/N-Flag, in which the sequence encoding a Flag tag is inserted at the 5′ terminus of the cloning site of pCAGGs-puro (37). A protease-deficient NS3/4A mutant with Ser139 replaced with Ala (S139A) was generated by the method of splicing by overlap extension and cloned into pCAGGs-puro. NS5A genes were amplified by PCR from HCV clones of strains of J1 (genotype 1b), H77c (genotype 1a, kindly provided by J. Bukh), and JFH1 (genotype 2a, kindly provided by T. Wakita) and cloned into pcDNA3.1Flag/HA (38). The NS5A deletion mutants were prepared as described previously (16). DNA fragments encoding a human MyD88, human Toll-IL-1 receptor domain-containing adapter protein (TIRAP), and human TRIF-related adapter molecule (TRAM) were amplified by reverse transcription-PCR from total RNA of THP-1 cells and cloned into pcDNA3.1-C-Myc-His (Invitrogen, Carlsbad, CA) and pcDNA3.1Flag/HA. Murine IPS-1 (mIPS-1) was amplified from total RNA of RAW264.7 cells by reverse transcription-PCR and cloned into pcDNA3.1Flag/HA. Human MyD88 deletion mutants and a mIPS-1 mutant with Cys508 replaced by Ala (C508A) were generated by the method of splicing by overlap extension and cloned into pcDNA3.1Flag/HA. pCMVIRAK1-myc and pCMVIRAK4-myc, encoding IL-1 receptor-associated kinase 1 (IRAK-1) and IRAK-4, respectively, were prepared as described previously (53). pEFBossTICAM-I-HA was kindly provided by T. Seya (44). All PCR products were confirmed by sequencing by using an ABI PRSM 310 genetic analyzer (Applied Biosystems, Tokyo, Japan). Vesicular stomatitis virus (VSV) (Indiana strain, NCP12.1) (19) was kindly provided by M. A. Whitt.
Establishment of stable cell lines expressing HCV proteins.
pCAGGs-puro/N-Flag plasmids encoding HCV proteins were transfected into RAW264.7 cells by liposome-mediated transfection using Lipofectamine 2000 (Invitrogen) and selected with 10 μg/ml of puromycin (InvivoGen, San Diego, CA). After about 2 to 3 weeks of selection, several clones were isolated, and cell lysates of each clone were immunoblotted with each of specific mouse anti-HCV antibody (1) or anti-Flag M2 mouse monoclonal antibody (Sigma). Macrophage cell lines stably expressing HCV proteins and a control cell line obtained by transfection with an empty pCAGGs-puro vector were maintained in the presence of puromycin (10 μg/ml) throughout the experiments.
Immunoprecipitation and immunoblotting.
Cells were seeded onto a six-well tissue culture plate 24 h before transfection. The plasmids were transfected by the lipofection method, and the cells were harvested at 48 h posttransfection, washed three times with 1 ml of ice-cold phosphate-buffered saline (PBS), and suspended in 0.4 ml lysis buffer containing 20 mM Tris-HCl (pH 7.4), 135 mM NaCl, 1% Triton X-100, 10% glycerol, and protease inhibitor cocktail tablets (Roche Molecular Biochemicals, Mannheim, Germany). Cell lysates were incubated for 30 min at 4°C and centrifuged at 14,000 × g for 15 min at 4°C. The supernatant was immunoprecipitated with 1 μg of mouse monoclonal anti-Flag M2, anti-hemagglutinin (HA) 16B12 (HA.11; BabCO, Richmond, CA), or anti-hexahistidine (Santa Cruz Biotechnology, Santa Cruz, CA) antibody and 10 μl of protein G-Sepharose 4B Fast Flow beads (Amersham Pharmacia Biotech, Franklin Lakes, NJ) at 4°C for 90 min. The immunocomplex was precipitated with the beads by centrifugation at 5,000 × g for 1 min and then washed five times with 0.4 ml of 20 mM Tris-HCl (pH 7.4) containing 135 mM NaCl and 0.05% Tween 20 (TBST buffer) by centrifugation. The proteins binding to the beads were boiled in 20 μl of sample buffer and then subjected to sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Millipore, Tokyo, Japan). The membranes were blocked with TBST containing 5% skim milk at room temperature for 1 h; incubated with mouse monoclonal anti-Flag M2, anti-HA 16B12, or anti-hexahistidine monoclonal antibody at room temperature for 1 h; and then incubated with horseradish peroxidase-conjugated anti-mouse immunoglobulin G (IgG) antibody at room temperature for 1 h. The cell lines (2 × 106 cells/well) were stimulated with various doses of lipopolysaccharide (LPS) derived from Salmonella enterica serovar Minnesota (Re-595) (Sigma), peptidoglycans (PGN) derived from Staphylococcus aureus (Sigma), R-837 (InvivoGen), or phosphorothioate-stabilized mouse CpG (mCpG) oligodeoxynucleotides (ODN1668) (TCC-ATG-ACG-TTC-CTG-ATG-CT) (Invitrogen) for the times indicated, and the phosphorylation status of extracellular signal-regulated kinase (ERK) was determined by immunoblotting using antibodies specific to ERK1/2 or phosphorylated ERK1/2 (T202/Y204) (Cell Signaling Technology, Inc., Beverly, MA). Cells (1 × 106 cells/well) were treated with various doses of mouse IFN-α (PBL Biomedical Laboratories, New Brunswick, NJ) or VSV for 24 h, and the phosphorylation status of double-stranded RNA-dependent protein kinase (PKR) and signal transducer and activator of transcription 1 (STAT1) was determined by immunoblotting using antibodies specific to STAT1 (Cell Signaling), phosphorylated STAT1 (Cell Signaling), or phosphorylated PKR (BioSource International, Inc., Camarillo, CA). The immune complexes were visualized with Super Signal West Femto substrate (Pierce, Rockford, IL) and detected by using an LAS-3000 image analyzer system (Fujifilm, Tokyo, Japan).
Cytokine production and enzyme-linked immunosorbent assay (ELISA).
To evaluate cytokine production in macrophage cell lines expressing HCV proteins, cells were seeded onto 96-well plates at a concentration of 1 × 105 cells/well and stimulated with various doses of LPS, PGN, R-837, or mCpG. After 24 h of incubation, culture supernatants were collected, and IL-6 production was determined by using an OptEIA mouse IL-6 set purchased from BD Pharmingen (San Diego, CA).
Real-time PCR.
The cell lines (3 × 106 cells/well) were stimulated with R-837, LPS, PGN, mCpG, VSV, and polyinosine-poly(C) [poly(I:C)] (Invivogen) for the times indicated, and the expression of mRNA of cytokines, chemokines, and TLR genes was determined by real-time PCR. Total RNA was prepared from the macrophage cell lines using an RNeasy Mini kit (QIAGEN). First-strand cDNA was synthesized using a ReverTra Ace (TOYOBO, Japan) and oligo(dT)20 primer. Each cDNA was estimated by Platinum SYBR Green qPCR SuperMix UDG (Invitrogen) according to the manufacturer's protocol. Fluorescent signals were analyzed by using an ABI PRISM 7000 apparatus (Applied Biosystems). Mouse Ccl2, IFN-β, IFN-α1, IFN-α4, and IL-1-α genes were amplified with primer pairs 5′-GCATCCACGTGTTGGCTCA-3′ and 5′-CTCCAGCCTACTCATTGGGATCA-3′, 5′-ACACCAGCCTGGCTTCCATC-3′ and 5′-TTGGAGCTGGAGCTGCTTATAGTTG-3′, 5′-AGCCTTGACACTCCTGGTACAAATG-3′ and 5′-TGGGTCAGCTCACTCAGGACA-3′, 5′-GCTCAAGCCATCCTTGTGCTAA-3′ and 5′-CATTGAGCTGATGGAGGTC-3′, and 5′-TTGGTTAAATGACCTGCAACAGGA-3′ and 5′-AGGTCGGTCTCACTACCTGTGATG-3′, respectively. The mouse TLR2, TLR3, TLR4, TLR7, TLR9, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) genes were amplified using primer pairs 5′-AGCTCTTTGGCTCTTCTG-3′ and 5′-AGAACTGGGGGATATGC-3′, 5′-AAATCCTTGCGTTGCGAAGTG-3′ and 5′-TCAGTTGGGCGTTGTTCAAGAG-3′, 5′-GCCTCGAATCCTGAGCAAACA-3′ and 5′-CTTCTGCCCGGTAAGGTCCA-3′, 5′-TCTGCAGGAGCTCTGTCCTTGA-3′ and 5′-CAAGGCATGTCCTAGGTGGTGA-3′, 5′-ACCAATGGCACCCTGCCTAA-3′ and 5′-CGTCTTGAGAATGTTGTGGCTGA-3′, and 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′, respectively. The expression of mRNAs of each of the chemokines, cytokines, and TLR was normalized with that of GAPDH mRNA.
Immunofluorescence microscopy and subcellular localization of HCV proteins in stable macrophage cell lines.
Cells were seeded onto an eight-well chamber slide at 1.5 × 104 cells per well, washed twice with PBS, fixed with PBS containing 4% paraformaldehyde at 18 h of cultivation, and permeabilized with PBS containing 0.5% Triton X-100 at 15 min. The cells were then incubated at 4°C for 1 h with 1 μg of mouse anti-NS5A antibody (Austral Biologicals, San Ramon, CA) or rabbit polyclonal antibody against calregulin (Santa Cruz Biotechnology) in PBS containing 10% fetal calf serum (PBSF) and then incubated at room temperature for 1 h with 0.5 μg of Alexa Fluor 488-conjugated anti-mouse IgG (Molecular Probes) or Alexa Fluor 594-conjugated anti-rabbit IgG (Molecular Probes) after three washes with PBSF. After extensive washing with PBSF, the samples were examined with a FluoView FV1000 laser scanning confocal microscope (Olympus, Japan). To confirm the subcellular localization of the HCV proteins in the macrophage cell lines, each stable cell line was fractionated with a Subcellular Proteome Extraction kit (Calbiochem, Darmstadt, Germany). Stepwise extraction resulted in four distinct fractions, which contained primarily cytosolic, membrane-organelle, nuclear, and cytoskeleton proteins, respectively. Each fraction was concentrated by Microcon (Millipore) and subjected to immunoblotting. PA28α (Biomol International, Plymouth Meeting, PA), calregulin, and histone H1 (Santa Cruz Biotechnology) were used as cytoplasmic, membrane, and nuclear markers, respectively.
RESULTS
Establishment of macrophage cell lines stably expressing HCV proteins.
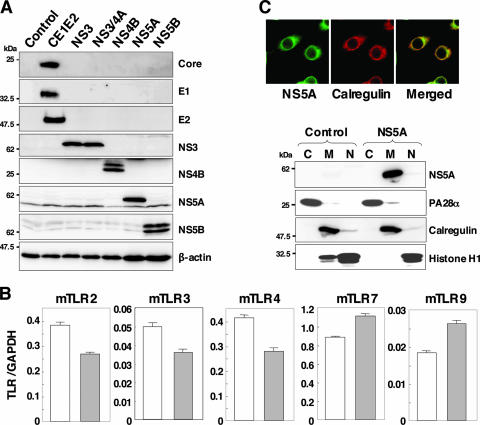
To examine the effect of HCV proteins on the TLR function of immune cells, we established murine macrophage cell lines stably expressing HCV structural or nonstructural proteins. We selected mouse macrophage RAW264.7 cells due to their high level of expression of various TLRs (3) and their high sensitivity to stimulation with TLR ligands. Processed HCV structural and nonstructural proteins were detected in each of the cell lines by immunoblot analyses using specific monoclonal antibodies (Fig. 1A). To examine the effect of HCV proteins on TLR expression in macrophage cell lines, the mRNA of TLRs in cells expressing NS5A was determined by real-time PCR (Fig. 1B). Although slight reductions in TLR2, TLR3, and TLR4 or enhancement of TLR7 and TLR9 was observed, a substantial amount of mRNA of the examined TLRs was detected in the cell lines expressing NS5A and other HCV proteins (data not shown). To determine the subcellular localization of HCV proteins in macrophage cell lines, the expression of HCV proteins was examined by confocal microscopy and cell fractionation (Fig. 1C). HCV NS5A was colocalized with the endoplasmic reticulum marker calregulin in the macrophage cell line as reported previously for human hepatoma cell lines (47). Other HCV proteins exhibited similar localization with NS5A (data not shown). To further confirm the subcellular localization of NS5A proteins, cytoplasmic, membrane-organelle, and nuclear fractions of the cell line expressing NS5A were analyzed by Western blotting. NS5A was detected mainly in the membrane-organelle fraction.
FIG. 1.
Establishment of stable macrophage cell lines expressing HCV proteins. (A) Cell lysates were prepared from macrophage cell lines expressing each of the HCV proteins (4 × 106 cells) and immunoblotted with antibodies against HCV proteins or β-actin. (B) Total RNA was extracted from macrophage cell lines expressing NS5A (gray bars) or control (white bars), and the expression of mRNA of TLRs was determined by real-time PCR. (C) The subcellular localization of NS5A was examined by confocal microscopy. Cells were fixed with 4% paraformaldehyde-PBS, permeabilized with 0.5% Triton X-100, and stained with specific antibodies. Cells expressing NS5A or control cells were extracted into cytosol (C), membrane-organelle (M), and nuclear (N) fractions. Each fraction was concentrated and subjected to immunoblotting with specific antibodies. PA28α, calregulin, and histone H1 were used as markers for cytosol, membrane-organelle, and nuclear fractions, respectively.
Expression of HCV NS3, NS3/4A, NS4B, or NS5A modulates the TLR-dependent signaling pathway in macrophage cell lines.
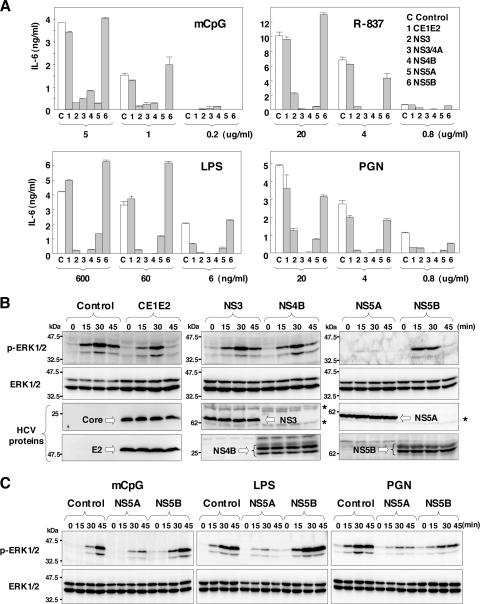
In order to determine the effect of the expression of HCV proteins on the TLR signaling pathway in macrophage cell lines, we examined the ability of HCV proteins to inhibit NF-κB activation via stimulation with various TLR ligands. The macrophage cell lines were stimulated with the TLR ligands, and the production of the proinflammatory cytokine IL-6 in the culture supernatants was determined by ELISA (Fig. 2A). The expression of HCV structural proteins or NS5B had no effect on IL-6 production after stimulation with mCpG, R-837, LPS, or PGN, which are ligands for TLR9, TLR7, TLR4, and TLR2, respectively. On the other hand, the expression of NS3, NS3/4A, NS4B, or NS5A inhibited the production of IL-6 induced by treatment with the ligands. These results indicate that the expression of NS3, NS3/4A, NS4B, and NS5A inhibits the production of IL-6 through the TLR-dependent signaling pathway in macrophage cell lines.
FIG. 2.
Expression of HCV nonstructural proteins modulates IL-6 production and MAPK cascades through the TLR-dependent signaling pathway in macrophage cell lines. (A) Cells were seeded onto 96-well plates (1 × 105 cells/well) and stimulated with the indicated amounts of mCpG, R-837, LPS, or PGN. After 24 h of stimulation, IL-6 production in the culture supernatants was determined by sandwich ELISA. Data are shown as means ± standard deviations (SD). (B) Cells (2 × 106 cells/well) were stimulated with 10 μg/ml of R-837 for the times indicated, and ERK1/2 phosphorylation was determined by immunoblotting with antibodies to ERK and phosphorylated ERK (p-ERK). Asterisks indicate nonspecific bands. (C) Cells (2 × 106 cells/well) were stimulated with 10 μg/ml of mCpG, 25 ng/ml of LPS, or 10 μg/ml of PGN for the times indicated, and ERK1/2 phosphorylation was determined by immunoblotting.
In addition to proinflammatory cytokine production via NF-κB activation, stimulation of TLR also activates mitogen-activated protein kinases (MAPKs). We then examined the activation of ERK, a MAPK signaling pathway, in response to the TLR ligands in the macrophage cells expressing HCV proteins (Fig. 2B). Although the expression of the HCV structural proteins NS3, NS4B, and NS5B did not alter the phosphorylation status of ERK1/2 in response to stimulation with the TLR7 ligand R-837, the expression of NS5A exhibited a clear inhibition of the phosphorylation of ERK1/2. To further examine the effect of NS5A expression on the MAPK cascade in response to the TLR ligands, the cells were treated with mCpG, LPS, and PGN. NS5A expression was found to inhibit the phosphorylation of ERK1/2 in response to stimulation with the ligands for TLR9, TLR4, and TLR2 (Fig. 2C). In contrast, the phosphorylation of c-Jun NH2-terminal kinase in response to stimulation with R-837 was less impaired in the macrophage cell line expressing NS5A (data not shown). These results indicate that the expression of NS3, NS3/4A, NS4B, or NS5A inhibits the production of proinflammatory cytokines and that the expression of NS5A alone induces the inhibition of the MAPK cascade in response to stimulation by various TLR ligands in macrophage cells.
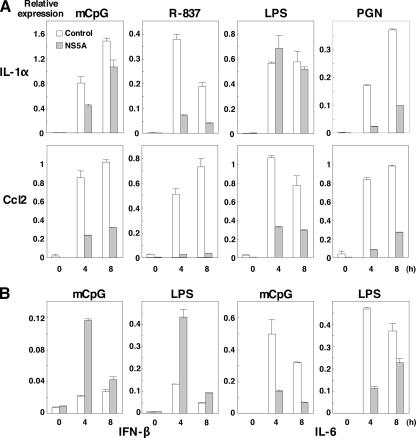
To further examine the effect of NS5A expression on the production of the other proinflammatory cytokines and chemokines in response to TLR ligands, the expression of mRNA of IL-1α and Ccl2 in cells expressing NS5A after stimulation with TLR ligands was determined by real-time PCR (Fig. 3A). Expression of IL-1α and Ccl2 was reduced in cells expressing NS5A by stimulation with mCpG, R-837, LPS, or PGN except for the IL-1α expression by treatment with LPS, probably due to the TRIF-dependent activation of NF-κB. To further confirm the specific inhibition of the MyD88-dependent signaling pathway by NS5A, we examined the effects of NS5A expression in macrophage cells on the MyD88-independent/TRIF-dependent production of IFN-β (Fig. 3B). Although the expression of IL-6 mRNA in cells expressing NS5A was impaired after stimulation with mCpG or LPS, the expression of IFN-β was enhanced. These results suggest that the expression of NS5A specifically inhibits the MyD88-dependent signaling pathway.
FIG. 3.
Effect of NS5A expression on the production of cytokines and chemokines in response to TLR ligands in macrophage cell lines. Cells (3 × 106 cells/well) were stimulated with 10 μg/ml of mCpG, 10 μg/ml of R-837, 25 ng/ml of LPS, and 10 μg/ml of PGN for the times indicated. Total RNA was extracted from macrophage cell lines expressing NS5A (gray bars) or control (white bars), and the expression of mRNA of IL-1α and Ccl2 (A) and IFN-β and IL-6 (B) was determined by real-time PCR.
TLR-dependent and -independent immune activation of macrophage cells expressing NS3/4A or NS5A protein by RNA virus and dsRNA.
TLR3 has been shown to sensitize cells in response to double-stranded RNA (dsRNA) generated by viral infection and a synthetic dsRNA analog, poly(I:C), through an adaptor molecule, TRIF/TICAM-1, but not MyD88. Furthermore, RIG-I and MDA5 have been identified as being cytoplasmic dsRNA detectors responding to poly(I:C) and viral RNAs (57, 58), sensitizing cells through an adaptor molecule, IPS-1/MAVS/VISA/CARDIF, in a TLR-independent manner (24, 35, 46, 55). Recently, HCV NS3/4A protease was shown not only to cleave HCV nonstructural proteins but also to inhibit viral RNA- and dsRNA-induced IFN production through the cleavage of the adaptor molecules TRIF (28) and IPS-1 (29, 30, 33, 35). Moreover, it has been shown that NS3/4A protease inhibits dsRNA-induced immune activation in a protease-dependent manner in human hepatoma cell lines (11).
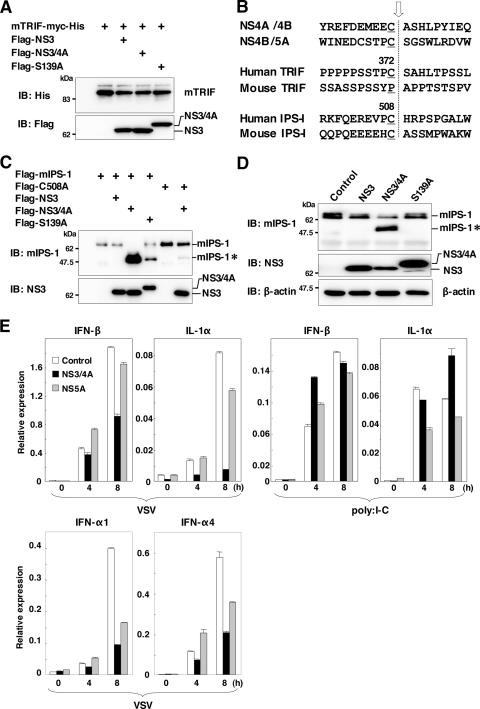
To determine whether murine TRIF (mTRIF) is cleaved by HCV NS3/4A protease, C-terminally His-tagged mTRIF was coexpressed with N-terminally Flag-tagged NS3, NS3/4A, or NS3/4A(S139A) in 293T cells. Immunoblot analyses revealed that mTRIF was not processed by HCV NS3/4A protease, probably due to differences in the amino acid sequences at the cleavage site in mTRIF (Fig. 4A). Amino acid sequences at the cleavage site of human TRIF are Cys372 and Ser373, and those at the cleavage sites of mTRIF are Pro372 and Ala373 (Fig. 4B). These results suggest that HCV NS3/4A protease could not inhibit immune activation through the TLR3-mTRIF-dependent signaling pathway in murine cells. We next determined the processing of IPS-1 by HCV NS3/4A protease. N-terminally Flag-tagged mIPS-1 or its C508A mutant, with Cys508 replaced with Ala to prevent cleavage by HCV NS3/4A protease, was coexpressed with N-terminally Flag-tagged NS3, NS3/4A, or NS3/4A(S139A) in 293T cells. Immunoblot analyses revealed that wild-type mIPS-1 was cleaved in cells coexpressing the active NS3/4A protease but not in those with NS3 (Fig. 4C). mIPS-1 processing was reduced in cells coexpressing NS3/4A(S139A) as well as in those coexpressing mIPS-1(C508A) and NS3/4A (Fig. 4C). Furthermore, we were able to detect cleavage of endogenous mIPS-1 in macrophage cell lines expressing NS3/4A but not in those expressing NS3 or NS3/4A(S139A) (Fig. 4D), indicating that mIPS-1 in murine macrophage cell lines is cleaved by HCV NS3/4A protease, as reported previously for a human hepatoma cell line.
FIG. 4.
TLR-dependent and -independent immune activation of macrophage cells expressing theNS3/4A or NS5A protein by RNA virus and dsRNA. (A) Myc-His-mTRIF was coexpressed with Flag-NS3, -NS3/4A, or -NS3/4A(S139A) in 293T cells and immunoblotted (IB) with antibodies against His and Flag. (B) Alignment of the flanking sequence of NS3 protease cleavage sites of NS4A/4B, NS4B/5A, TRIF, and IPS-1 of human and murine origins. The cleavage site is indicated by an arrow. (C) Flag-mIPS-1 and a mutant with Cys508 replaced with Ala (C508A) were coexpressed with Flag-NS3, -NS3/4A, or -NS3/4A(S139A) in 293T cells and immunoblotted with antibodies against mIPS-1 and NS3. (D) Processing of endogenous mIPS-1. Cell lysates of the macrophage cell lines expressing NS3, NS3/4A, and NS3/4A(S139A) were immunoblotted with antibodies against mIPS-1, NS3, and β-actin. The cleavage product of mIPS-1 is indicated as mIPS-I*. (E) Cells (3 × 106 cells/well) were stimulated with 2 × 105 PFU/ml of VSV or 50 μg/ml of poly(I:C) for the times indicated. Total RNA was extracted from the macrophage cell lines expressing NS3/4A (black bars), NS5A (gray bars), or control (white bars), and the expression of mRNA of IFN-β, IL-1α, IFN-α1, and IFN-α4 was determined by real-time PCR.
We then examined the effect of expression of NS3/4A and NS5A on TLR-dependent and -independent immune activation induced by dsRNA. VSV and poly(I:C) were inoculated into macrophage cell lines, and the expression of mRNA of IFN-β and IL-1α was determined by real-time PCR (Fig. 4E). The macrophage cell lines expressing NS3/4A exhibited inhibition of IL-1α and IFN-β expression upon infection with VSV but not in response to poly(I:C), whereas no inhibition was observed in those expressing NS5A. These results suggest that the invasion of VSV and poly(I:C) is preferentially recognized in RAW cell lines by RIG-1-IPS-1- and TLR3-TRIF-dependent signaling pathways, respectively. Inhibition of IL-1α and IFN-β expression upon infection with VSV but not in response to poly(I:C) is probably due to the selective cleavage of IPS-1 but not TRIF by NS3/4A protease in the macrophage cell lines. In contrast, the expression of NS5A has no effect on both TLR3-TRIF and RIG-I-IPS-1-dependent signaling pathways in macrophage cells.
Although MyD88/IRF7-dependent production of IFN-α upon activation was reported in plasmacytoid DCs (pDCs) (17, 23), it is unclear whether murine macrophage cells are capable of producing IFN-α in a TLR/MyD88/IRF7-dependent manner. To examine the effect of NS5A expression on IFN-α production, the expression of IFN-α1 and IFN-α4 in the macrophage cell line upon infection with VSV was determined (Fig. 4E, bottom). In contrast to the effect on IFN-β production, the expression of NS5A in the macrophage cells reduced the production of IFN-α1 and IFN-α4 upon infection with VSV, although the inhibitory effect was weaker than that of NS3/4A. These results suggest that RAW264.7 cells are capable of producing IFN-α in a TLR/MyD88/IRF7-dependent manner upon infection with VSV as reported for pDCs, and the expression of NS5A partially counteracts this signaling pathway. However, the production of type I IFNs by the treatment with ligands for TLR7 (R-837) and TLR9 (mouse CpG) was weaker than that induced by infection with VSV in macrophage cells (data not shown). Further study is needed to clarify the precise mechanisms of the inhibition of TLR/MyD88/IRF7-dependent IFN-α production by the expression of HCV NS5A in human immune cells.
NS5A interacts with MyD88 in mammalian cells.
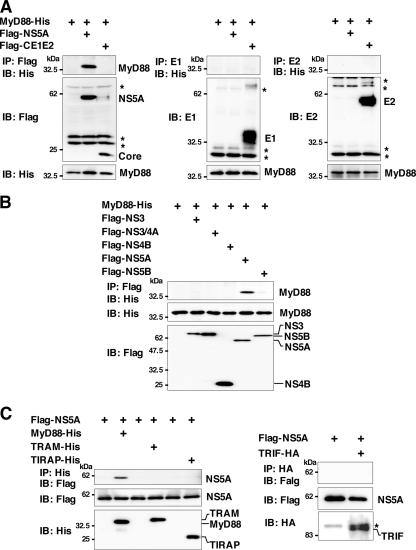
The inhibition of the production of proinflammatory cytokines and chemokines and the MAPK cascade by NS5A expression in response to stimulation by various TLR ligands without participation of TRIF- and IPS-1-dependent signaling pathways suggests that NS5A specifically inhibits the TLR-MyD88-dependent signaling pathway in macrophage cell lines. MyD88 is a critical component of the signaling pathway and leads to the production of proinflammatory cytokines, chemokines, and MAPKs. To determine the effect of the expression of HCV proteins on the TLR signaling pathway in macrophage cell lines, the interaction of the HCV proteins with the adaptor molecules in the signaling pathway of the TLR family was examined by immunoprecipitation analysis. His-tagged MyD88 was coexpressed with Flag-tagged HCV proteins in 293T cells and immunoprecipitated with the indicated antibodies. As shown in Fig. 5A and B, MyD88 was coimmunoprecipitated with NS5A but not with structural and other nonstructural proteins in 293T cells.
FIG. 5.
NS5A interacts with MyD88. MyD88-His was coexpressed with Flag-core/E1/E2 or -NS5A (A) or Flag-NS3, -NS3/4A, -NS4B, -NS5A, or -NS5B (B) in 293T cells; immunoprecipitated (IP) with anti-Flag, E1, or E2 antibody; and immunoblotted (IB) with anti-His antibody. (C) Flag-NS5A was coexpressed with MyD88-His, TRAM-His, TIRAP-His, or TRIF-HA in 293T cells and immunoprecipitated with anti-His or -HA antibody. The immunoprecipitates were immunoblotted with anti-Flag antibody. Asterisks indicate nonspecific bands.
To further confirm the specificity of the interaction of NS5A with MyD88, NS5A was coexpressed with other adaptor molecules in the TLR signaling pathway, TRAM, TIRAP, or TRIF, in 293T cells (Fig. 5C). NS5A interacted with MyD88 but not other adaptor molecules, suggesting that NS5A may inhibit the production of proinflammatory cytokines and chemokines and the phosphorylation of MAPKs through the counteraction of the MyD88-dependent TLR signaling pathway.
NS5A interacts with the death domain of MyD88 through the ISDR and inhibits recruitment of IRAK to MyD88.
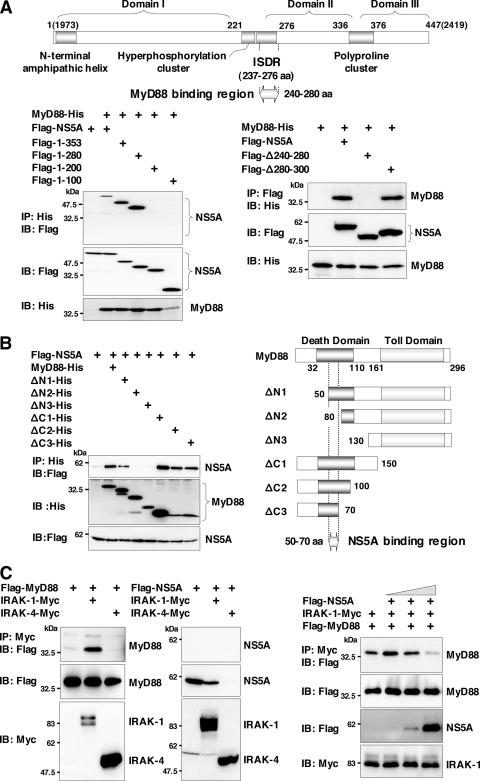
To determine the region of NS5A responsible for the interaction with MyD88, a series of deletion mutants of N-terminal Flag-tagged NS5A was constructed, and its interaction with His-tagged MyD88 was examined (Fig. 6A). The NS5A mutant covering amino acids 1 to 280 but not that covering amino acids 1 to 200 exhibited binding to MyD88, suggesting that amino acid residues 200 to 280 of NS5A are required for the interaction with MyD88. Further mutational analyses of NS5A revealed that amino acid residues 240 to 280, which overlap the ISDR (amino acid residues 237 to 276), which was previously suggested to be involved in IFN resistance (10, 41), are required for the interaction with MyD88 (Fig. 6A). To determine the region of MyD88 responsible for the interaction with NS5A, His-tagged MyD88 mutants were coexpressed with Flag-tagged NS5A in 293T cells and immunoprecipitated with anti-His antibody. A MyD88 deletion mutant lacking amino acids 1 to 50, but not one lacking amino acids 1 to 80, and a mutant possessing amino acids 1 to 70 exhibited binding to NS5A, suggesting that amino acid residues 50 to 70 in the death domain of MyD88 are required for the interaction with NS5A (Fig. 6B).
FIG. 6.
NS5A interacts with the death domain of MyD88 through the ISDR and inhibits recruitment of IRAK to MyD88. (A) The structure of NS5A and the MyD88 binding region are indicated at the top. MyD88-His was coexpressed with C-terminal deletion mutants of Flag-NS5A in 293T cells, immunoprecipitated (IP) with anti-His antibody, and immunoblotted (IB) with anti-Flag antibody (left). MyD88-His was coexpressed with Flag-NS5A deletion mutants (Δ240-280 or Δ280-300) in 293T cells, immunoprecipitated with anti-Flag antibody, and then immunoblotted with anti-His antibody (right). (B) Flag-NS5A was coexpressed with N-terminal or C-terminal deletion mutants of MyD88-His (ΔN1, ΔN2, ΔN3, ΔC1, ΔC2, or ΔC3) in 293T cells, immunoprecipitated with anti-His antibody, and immunoblotted with anti-Flag antibody. The structures of MyD88 and the deletion mutants and the NS5A binding region are indicated on the left. (C) Flag-MyD88 (left) or Flag-NS5A (middle) was coexpressed with IRAK-1-Myc or IRAK-4-Myc in 293T cells, immunoprecipitated with anti-Myc antibody, and immunoblotted with anti-Flag antibody. Flag-MyD88 and IRAK-1-Myc were coexpressed with Flag-NS5A in 293T cells, immunoprecipitated with anti-Myc antibody, and immunoblotted with anti-Flag antibody. The effect of the increase in Flag-NS5A expression on the interaction of MyD88 with IRAK-1 was examined by transfection with 0.1, 0.5, or 2 μg of Flag-NS5A expression plasmid (right).
MyD88 associates with TLRs and acts as an adapter that recruits IRAK, which is known as a key regulator for TLR7- and TLR9-mediated IFN-α production in pDCs (53). To determine the role of NS5A binding to MyD88 in the TLR-MyD88-dependent signaling pathway, we examined the association of IRAK with MyD88 in the presence of NS5A. Flag-tagged MyD88 was coexpressed with Myc-tagged IRAK-1 or IRAK-4 and immunoprecipitated with anti-Myc antibody (Fig. 6C, left). IRAK-1, but not IRAK-4, was coimmunoprecipitated with MyD88. Although NS5A did not bind to IRAK-1, it was not possible to assess the interaction of NS5A with IRAK-4 due to the degradation of NS5A in cells coexpressing IRAK-4 for unknown reasons (Fig. 6C, middle). To examine the interplay between IRAK-1 and MyD88 in the presence of NS5A, Flag-tagged MyD88 and Myc-tagged IRAK-1 were coexpressed with Flag-tagged NS5A in 293T cells. The interaction of IRAK-1 and MyD88 decreased in accord with the increasing NS5A expression complex (Fig. 6C, right), suggesting that the expression of NS5A may interfere with the TLR-MyD88-dependent signaling pathway through the inhibition of the recruitment of IRAK-1 to MyD88.
NS5A of other genotypes also interacts with MyD88 and inhibits the TLR signaling pathway.
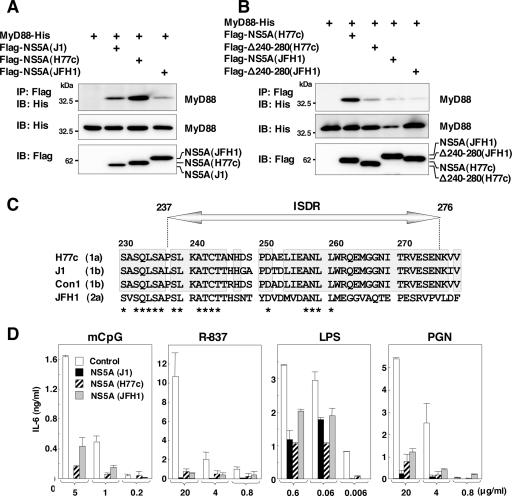
To determine the interaction of MyD88 with NS5A of other genotypes, Flag-tagged NS5A of genotype 1a (H77c) or 2a (JFH1) was coexpressed with His-tagged MyD88 in 293T cells. MyD88 was coprecipitated with the NS5As of genotypes 1a and 2a, although it should be noted that the interaction between the MyD88 and NS5A of genotype 2a was weaker than that of the other genotypes (Fig. 7A). To determine the region of the NS5As of genotype 1a or 2a responsible for the interaction with MyD88, N-terminal Flag-tagged NS5As of genotype 1a or 2a deletion mutants lacking amino acids 240 to 280 (Δ240-280) were constructed, and their interaction with MyD88 was examined. Mutational analyses revealed that amino acid residues 240 to 280 of the NS5As of genotypes 1a and 2a were also required for the interaction with MyD88 (Fig. 7B). Amino acid alignment of the ISDRs of genotypes 1a, 1b, and 2a revealed that the region of genotype 2a was less conserved than those of the other genotypes (Fig. 7C).
FIG. 7.
NS5A of other genotypes also interacts with MyD88 and inhibits the TLR signaling pathway. (A) Flag-NS5As of other genotypes were coexpressed with MyD88-His in 293T cells, immunoprecipitated (IP) with anti-Flag antibody, and immunoblotted (IB) with anti-His antibody. (B) The wild type or a deletion mutant lacking amino acids 240 to 280 of Flag-NS5A of genotype 1a or 2a was coexpressed with MyD88-His in 293T cells, immunoprecipitated with anti-Flag antibody, and immunoblotted with anti-His antibody. (C) Amino acid sequences of ISDR and its adjacent region of strains H77c (genotype 1a), J1 (genotype 1b), Con1 (genotype 1b), and JFH1 (genotype 2a). The conserved amino acids among genotypes 1a and 1b are indicated by boxes. Conserved amino acids among all strains are indicated by asterisks. (D) Macrophage cell lines expressing NS5A of genotypes 1a (H77c), 1b (J1), and 2a (JFH1) were established. Cells were stimulated with the indicated amounts of mCpG, R-837, LPS, or PGN, and the production of IL-6 in the culture supernatants was determined by ELISA 24 h after stimulation. Data are shown as the means ± SD.
To determine the effect of NS5A expression of other genotypes on the TLR signaling pathway, we established macrophage cell lines expressing NS5A of each genotype. NS5A expression for all genotypes was found to inhibit IL-6 production after stimulation with mCpG, R-837, LPS, or PGN (Fig. 7D). Although the association of NS5A of genotype 2a to MyD88 was weaker than that of other genotypes, the expression of genotype 2a NS5A in macrophage cells exhibited comparable inhibition of IL-6 production in response to stimulation by various TLR ligands with those of genotypes 1b and 1a. These results suggest that NS5As of genotypes 1a, 1b, and 2a interact with MyD88 and inhibit the TLR signaling pathway in macrophage cell lines.
ISDR participates in the inhibition of the MyD88-dependent signaling pathway by NS5A.
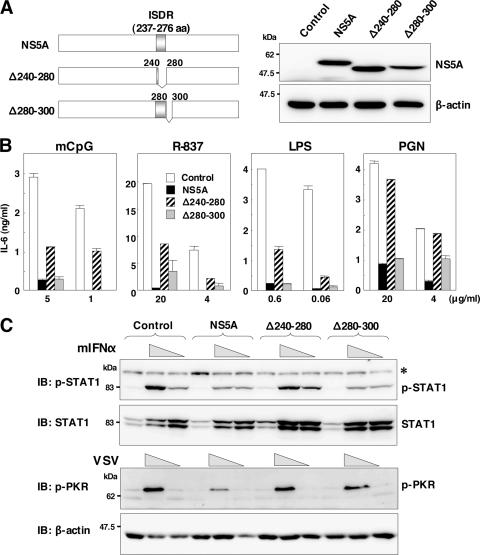
To further confirm the inhibitory effect of NS5A on the TLR signaling pathway, we established macrophage cell lines stably expressing an NS5A mutant lacking the ISDR/MyD88 binding region (Δ240-280) or lacking a region dispensable for the interaction with MyD88 (Δ280-300) (Fig. 8A). The inhibitory effect of TLR signaling in response to stimulation with mCpG, R-837, LPS, or PGN by NS5A was partially restored in the cell line expressing the NS5A lacking the ISDR (Δ240-280), and comparable inhibition was observed in the cell line expressing the NS5A deletion mutant retaining the ISDR (Δ280-300) (Fig. 8B). These results suggest that the interaction of NS5A with MyD88 through the ISDR is responsible for the disruption of the TLR-MyD88-dependent signaling pathway due to the expression of NS5A in macrophage cells. Partial recovery of the TLR signaling pathway by the expression of the NS5A mutant lacking the ISDR suggests the involvement of other inhibitory mechanisms by NS5A.
FIG. 8.
ISDR in NS5A participates in the inhibition of the MyD88-dependent signaling pathway. (A) Structures of NS5A mutants lacking amino acid residues 240 to 280, in which the ISDR/MyD88-interacting region is located (Δ240-280), and lacking amino acid residues 280 to 300 (Δ280-300) (left). Immunoblot analyses of cells expressing wild-type or mutant NS5A (right) are shown. (B) Cells expressing wild-type or mutant NS5A were stimulated with the indicated amounts of mCpG, R-837, LPS, or PGN, and the production of IL-6 in the culture supernatants was determined by ELISA 24 h after stimulation. Data are shown as the means ± SD. (C) Phosphorylation of STAT1 or PKR in response to treatment with murine IFN-α or infection with VSV. The cell lines were stimulated with two doses of murine IFN-α (2 × 103 and 2 × 102 units/ml) or VSV (2 × 107 and 2 × 106 PFU/ml). After 24 h of stimulation, cell extracts were immunoblotted (IB) with specific antibodies. Phosphorylated STAT1 and PKR and the total amounts of STAT1 and β-actin were determined. The asterisk indicates nonspecific bands.
Previous reports suggested that the ISDR of NS5A participates in conferring IFN sensitivity (10) and in an interaction with PKR (13). To determine the effect of the interaction of MyD88 with NS5A through the ISDR on the IFN signaling pathway, we examined the phosphorylation of STAT1 and PKR in response to treatment with murine IFN-α and infection with VSV. The expression of wild-type NS5A and the Δ280-300 mutant but not the Δ240-280 mutant reduced the phosphorylation of STAT1 in response to IFN-α treatment (Fig. 8C, top), suggesting that the ISDR/MyD88 binding region in NS5A is involved in the IFN signaling pathway. Although cells expressing wild-type NS5A reduced PKR phosphorylation, those expressing mutant NS5A (Δ240-280 or Δ280-300) did not inhibit PKR phosphorylation upon infection with VSV (Fig. 8C, bottom), which is consistent with the previous observation that the 66 ISDR-inclusive amino acid residues (amino acids 237 to 302) are required for interactions with PKR (13). These results suggest that the expression of HCV NS5A in macrophage cells counteracts the IFN signaling pathway through the repression of STAT1 and PKR due to the interaction with ISDR and its adjacent region.
DISCUSSION
The majority of HCV-infected individuals become chronic carriers; however, the mechanism of progression to chronicity remains unclear. Among HCV proteins, NS3 has been shown to be immunodominant, and T cells that are reactive to NS3 have been suggested to play a crucial role in viral clearance, while HCV core protein is immunosuppressive (8). Treatment of immature DCs with core or NS3 protein inhibited DC differentiation, and DCs transduced to express core or E1 protein exhibited poor allogeneic T-cell responses (43). The immunosuppressive potential of HCV proteins has been implicated as a mechanism of the functional subversion of T cells, natural killer (NK) cells, and DCs. The association of HCV core protein with the globular domain of the C1q receptor on T cells down-regulates T-cell proliferation and IL-2 production (25). Additionally, the HCV E2 protein displays a high affinity for the tetraspanin cell surface molecule human CD81, which is one of the candidates for an HCV entry receptor (40), and E2 cross-linking with cell surface human CD81 impairs the activation of NK cells (7, 52).
In the present study, we established macrophage cell lines stably expressing HCV proteins and examined the effects of viral proteins on TLR function. The expression of the NS5A protein specifically inhibits TLR-MyD88-induced signaling by associating with the death domain of MyD88 through the ISDR spanning amino acid residues 240 to 280 in macrophage cells. HCV NS5A is a phosphoprotein that appears to possess multiple and diverse functions in viral replication, IFN resistance, and pathogenesis (34). Mutation in the ISDR has been suggested to correlate with the responsiveness of patients chronically infected with HCV genotype 1b to IFN treatment (10). Furthermore, NS5A has been shown to rescue virus replication in IFN-treated cell cultures (41) and to inhibit the antiviral activity of IFN by binding to PKR through the ISDR and its adjacent region (amino acids 237 to 302) (13, 14). However, controversial observations that the ISDR sequence variation does not account for differences in IFN sensitivity in patients (9) and also in an HCV subgenomic RNA replicon system (15) have been made. Moreover, the expression of NS5A or the entire HCV polyprotein has been reported to counteract the antiviral effect of IFN in a PKR- and ISDR-independent manner (12). Therefore, the possibility remains that a molecule other than PKR may be involved in the NS5A-mediated inhibition of IFN (50). Restoration of the phosphorylation of STAT1 in cells expressing a deletion mutant lacking ISDR in response to IFN-α and that of PKR phosphorylation upon infection with VSV in cells expressing NS5A mutants lacking amino acid residues 240 to 280 (ISDR) or 280 to 300 may support the hypothesis that the ISDR and the adjacent region are involved in IFN sensitivity. Thus, the ISDR may participate not only in conferring IFN resistance but also in disrupting TLR-MyD88 signaling pathways in macrophage cells.
Several viral proteins have been shown to counteract TLRs and their downstream signaling cascade. The vaccinia virus A46R protein contains a Toll/IL-1 receptor domain that interacts with multiple Toll/IL-1 receptor-containing adaptor molecules, thereby inhibiting the activation of NF-κB and IRF3 (49). Measles virus and respiratory syncytial virus have been shown to inhibit the TLR7- and TLR9-dependent IFN-inducing pathways stimulated by R848 and CpG oligodeoxynucleotides in primary human pDCs (45). HCV NS3/4A has been shown to influence the functions of adaptor molecules mediating TLR-dependent and -independent signaling pathways, resulting in an impairment of the induction of IFN-β as well as the subsequent IFN-inducible genes (11). Recently, RIG-I and MDA5 have been identified as being cytoplasmic dsRNA detectors responding to viral RNAs and poly(I:C) in a TLR-independent manner and recruit IPS-1 as an adaptor molecule for signal transduction (24). The uncapped 5′-triphosphate RNA generated by viral polymerases was shown to be selectively recognized by RIG-I (18, 39). In this study, we could demonstrate that the invasion of VSV and poly(I:C) into RAW cell lines is preferentially recognized by RIG-1-IPS-1- and TLR3-TRIF-dependent signaling pathways, respectively, and that the expression of HCV NS3/4A protease selectively inhibits cytokine production upon infection with VSV through the cleavage of IPS-1. Therefore, it is feasible that the expression of NS5A and NS3/4A proteins in macrophage cells may disrupt TLR-dependent and -independent signaling pathways, respectively. However, the mechanism for the inhibition of the TLR signaling pathway in the macrophage cells by the expression of NS3 or NS4B remains unclear.
Although there have been reports suggesting a lack of DC dysfunction in both chimpanzees and humans chronically infected with HCV (26, 32), direct infection of DCs with HCV may be a plausible mechanism for the dysfunction of DCs in patients with chronic HCV infection (4, 21). Indeed, the HCV genome has been detected in DCs by PCR (4), and HCV was detected in a monocyte/macrophage subpopulation of peripheral blood mononuclear cells from patients with chronic HCV infection (5). Further experiments are needed to exclude the possibility of contamination of viral RNA in blood samples. Pseudotype VSV-bearing chimeric HCV E1 and E2 proteins have been shown to infect immature myeloid DCs isolated from healthy donors through interactions with lectins in a Ca-independent manner (20). Recently, the in vitro replication of the HCV JFH1 clone of genotype 2a isolated from an HCV-infected patient who developed fulminant hepatitis was reported (31, 54, 59). However, in vitro replication was limited in the combination of HCV clones derived from strain JFH1 and certain human hepatoma cell lines, and a robust cell culture of genotypes 1a and 1b, the most prevalent viruses in the world and resistant to IFN therapy, has not yet been successful except for a cell culture system for strain H77-S (genotype 1a) in which infectivity was significantly lower than that of the JFH1 clone (56). The establishment of a robust and reliable in vitro replication system for various HCV isolates is essential to determine the role of HCV infection in the modulation of TLR function in immunocompetent cells.
In conclusion, we have shown that the expression of the HCV nonstructural protein NS3, NS3/4A, NS4B, or NS5A impairs the activation of TLR signaling pathways in immunocompetent cells. Furthermore, the NS5A protein was shown to inhibit the TLR-MyD88 signaling pathway by a direct interaction with the death domain of MyD88 through the ISDR. These findings suggest new aspects of virus-cell interactions that may be explored to develop a greater understand of the mechanisms of escape of HCV from the host immune surveillance system and the establishment of persistent infection. However, it remains to be proven whether the results obtained using murine macrophage cell lines are applicable to immunocompetent cells in patients with HCV infection.
Acknowledgments
We gratefully thank H. Murase for her secretarial work.
This work was supported in part by grants-in-aid from the Ministry of Health, Labor, and Welfare; the Ministry of Education, Culture, Sports, Science, and Technology; the 21st Century Center of Excellence Program; and the Foundation for Biomedical Research and Innovation.
Footnotes
Published ahead of print on 13 June 2007.
REFERENCES
- 1.Aizaki, H., Y. Aoki, T. Harada, K. Ishii, T. Suzuki, S. Nagamori, G. Toda, Y. Matsuura, and T. Miyamura. 1998. Full-length complementary DNA of hepatitis C virus genome from an infectious blood sample. Hepatology 27:621-627. [DOI] [PubMed] [Google Scholar]
- 2.Akira, S., and K. Takeda. 2004. Toll-like receptor signalling. Nat. Rev. Immunol. 4:499-511. [DOI] [PubMed] [Google Scholar]
- 3.Applequist, S. E., R. P. Wallin, and H. G. Ljunggren. 2002. Variable expression of Toll-like receptor in murine innate and adaptive immune cell lines. Int. Immunol. 14:1065-1074. [DOI] [PubMed] [Google Scholar]
- 4.Bain, C., A. Fatmi, F. Zoulim, J. P. Zarski, C. Trepo, and G. Inchauspe. 2001. Impaired allostimulatory function of dendritic cells in chronic hepatitis C infection. Gastroenterology 120:512-524. [DOI] [PubMed] [Google Scholar]
- 5.Bouffard, P., P. H. Hayashi, R. Acevedo, N. Levy, and J. B. Zeldis. 1992. Hepatitis C virus is detected in a monocyte/macrophage subpopulation of peripheral blood mononuclear cells of infected patients. J. Infect. Dis. 166:1276-1280. [DOI] [PubMed] [Google Scholar]
- 6.Cerny, A., J. G. McHutchison, C. Pasquinelli, M. E. Brown, M. A. Brothers, B. Grabscheid, P. Fowler, M. Houghton, and F. V. Chisari. 1995. Cytotoxic T lymphocyte response to hepatitis C virus-derived peptides containing the HLA A2.1 binding motif. J. Clin. Investig. 95:521-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crotta, S., A. Stilla, A. Wack, A. D'Andrea, S. Nuti, U. D'Oro, M. Mosca, F. Filliponi, R. M. Brunetto, F. Bonino, S. Abrignani, and N. M. Valiante. 2002. Inhibition of natural killer cells through engagement of CD81 by the major hepatitis C virus envelope protein. J. Exp. Med. 195:35-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolganiuc, A., K. Kodys, A. Kopasz, C. Marshall, T. Do, L. Romics, Jr., P. Mandrekar, M. Zapp, and G. Szabo. 2003. Hepatitis C virus core and nonstructural protein 3 proteins induce pro- and anti-inflammatory cytokines and inhibit dendritic cell differentiation. J. Immunol. 170:5615-5624. [DOI] [PubMed] [Google Scholar]
- 9.Duverlie, G., H. Khorsi, S. Castelain, O. Jaillon, J. Izopet, F. Lunel, F. Eb, F. Penin, and C. Wychowski. 1998. Sequence analysis of the NS5A protein of European hepatitis C virus 1b isolates and relation to interferon sensitivity. J. Gen. Virol. 79:1373-1381. [DOI] [PubMed] [Google Scholar]
- 10.Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, N. Izumi, F. Marumo, and C. Sato. 1995. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J. Clin. Investig. 96:224-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 12.Francois, C., G. Duverlie, D. Rebouillat, H. Khorsi, S. Castelain, H. E. Blum, A. Gatignol, C. Wychowski, D. Moradpour, and E. F. Meurs. 2000. Expression of hepatitis C virus proteins interferes with the antiviral action of interferon independently of PKR-mediated control of protein synthesis. J. Virol. 74:5587-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gale, M., Jr., C. M. Blakely, B. Kwieciszewski, S. L. Tan, M. Dossett, N. M. Tang, M. J. Korth, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1998. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol. Cell. Biol. 18:5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gale, M. J., Jr., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230:217-227. [DOI] [PubMed] [Google Scholar]
- 15.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamamoto, I., Y. Nishimura, T. Okamoto, H. Aizaki, M. Liu, Y. Mori, T. Abe, T. Suzuki, M. M. Lai, T. Miyamura, K. Moriishi, and Y. Matsuura. 2005. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J. Virol. 79:13473-13482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honda, K., H. Yanai, T. Mizutani, H. Negishi, N. Shimada, N. Suzuki, Y. Ohba, A. Takaoka, W. C. Yeh, and T. Taniguchi. 2004. Role of a transductional-transcriptional processor complex involving MyD88 and IRF-7 in Toll-like receptor signaling. Proc. Natl. Acad. Sci. USA 101:15416-15421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornung, V., J. Ellegast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K. K. Conzelmann, M. Schlee, S. Endres, and G. Hartmann. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994-997. [DOI] [PubMed] [Google Scholar]
- 19.Jayakar, H. R., and M. A. Whitt. 2002. Identification of two additional translation products from the matrix (M) gene that contribute to vesicular stomatitis virus cytopathology. J. Virol. 76:8011-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaimori, A., T. Kanto, C. K. Limn, Y. Komoda, C. Oki, M. Inoue, H. Miyatake, I. Itose, M. Sakakibara, T. Yakushijin, T. Takehara, Y. Matsuura, and N. Hayashi. 2004. Pseudotype hepatitis C virus enters immature myeloid dendritic cells through the interaction with lectin. Virology 324:74-83. [DOI] [PubMed] [Google Scholar]
- 21.Kanto, T., N. Hayashi, T. Takehara, T. Tatsumi, N. Kuzushita, A. Ito, Y. Sasaki, A. Kasahara, and M. Hori. 1999. Impaired allostimulatory capacity of peripheral blood dendritic cells recovered from hepatitis C virus-infected individuals. J. Immunol. 162:5584-5591. [PubMed] [Google Scholar]
- 22.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131-137. [DOI] [PubMed] [Google Scholar]
- 23.Kawai, T., S. Sato, K. J. Ishii, C. Coban, H. Hemmi, M. Yamamoto, K. Terai, M. Matsuda, J. Inoue, S. Uematsu, O. Takeuchi, and S. Akira. 2004. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 5:1061-1068. [DOI] [PubMed] [Google Scholar]
- 24.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981-988. [DOI] [PubMed] [Google Scholar]
- 25.Kittlesen, D. J., K. A. Chianese-Bullock, Z. Q. Yao, T. J. Braciale, and Y. S. Hahn. 2000. Interaction between complement receptor gC1qR and hepatitis C virus core protein inhibits T-lymphocyte proliferation. J. Clin. Investig. 106:1239-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsson, M., E. Babcock, A. Grakoui, N. Shoukry, G. Lauer, C. Rice, C. Walker, and N. Bhardwaj. 2004. Lack of phenotypic and functional impairment in dendritic cells from chimpanzees chronically infected with hepatitis C virus. J. Virol. 78:6151-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lechner, F., D. K. Wong, P. R. Dunbar, R. Chapman, R. T. Chung, P. Dohrenwend, G. Robbins, R. Phillips, P. Klenerman, and B. D. Walker. 2000. Analysis of successful immune responses in persons infected with hepatitis C virus. J. Exp. Med. 191:1499-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, K., E. Foy, J. C. Ferreon, M. Nakamura, A. C. Ferreon, M. Ikeda, S. C. Ray, M. Gale, Jr., and S. M. Lemon. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. USA 102:2992-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, X. D., L. Sun, R. B. Seth, G. Pineda, and Z. J. Chen. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. USA 102:17717-17722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, R., J. Lacoste, P. Nakhaei, Q. Sun, L. Yang, S. Paz, P. Wilkinson, I. Julkunen, D. Vitour, E. Meurs, and J. Hiscott. 2006. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKɛ molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J. Virol. 80:6072-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 32.Longman, R. S., A. H. Talal, I. M. Jacobson, M. L. Albert, and C. M. Rice. 2004. Presence of functional dendritic cells in patients chronically infected with hepatitis C virus. Blood 103:1026-1029. [DOI] [PubMed] [Google Scholar]
- 33.Loo, Y. M., D. M. Owen, K. Li, A. K. Erickson, C. L. Johnson, P. M. Fish, D. S. Carney, T. Wang, H. Ishida, M. Yoneyama, T. Fujita, T. Saito, W. M. Lee, C. H. Hagedorn, D. T. Lau, S. A. Weinman, S. M. Lemon, and M. Gale, Jr. 2006. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 103:6001-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macdonald, A., and M. Harris. 2004. Hepatitis C virus NS5A: tales of a promiscuous protein. J. Gen. Virol. 85:2485-2502. [DOI] [PubMed] [Google Scholar]
- 35.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 36.Moriishi, K., and Y. Matsuura. 2003. Mechanisms of hepatitis C virus infection. Antivir. Chem. Chemother. 14:285-297. [DOI] [PubMed] [Google Scholar]
- 37.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 38.Okamoto, K., K. Moriishi, T. Miyamura, and Y. Matsuura. 2004. Intramembrane proteolysis and endoplasmic reticulum retention of hepatitis C virus core protein. J. Virol. 78:6370-6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pichlmair, A., O. Schulz, C. P. Tan, T. I. Naslund, P. Liljestrom, F. Weber, and C. Reis e Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997-1001. [DOI] [PubMed] [Google Scholar]
- 40.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 41.Polyak, S. J., D. M. Paschal, S. McArdle, M. J. Gale, Jr., D. Moradpour, and D. R. Gretch. 1999. Characterization of the effects of hepatitis C virus nonstructural 5A protein expression in human cell lines and on interferon-sensitive virus replication. Hepatology 29:1262-1271. [DOI] [PubMed] [Google Scholar]
- 42.Reis e Sousa, C. 2004. Toll-like receptors and dendritic cells: for whom the bug tolls. Semin. Immunol. 16:27-34. [DOI] [PubMed] [Google Scholar]
- 43.Sarobe, P., J. J. Lasarte, A. Zabaleta, L. Arribillaga, A. Arina, I. Melero, F. Borras-Cuesta, and J. Prieto. 2003. Hepatitis C virus structural proteins impair dendritic cell maturation and inhibit in vivo induction of cellular immune responses. J. Virol. 77:10862-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sasai, M., H. Oshiumi, M. Matsumoto, N. Inoue, F. Fujita, M. Nakanishi, and T. Seya. 2005. Cutting edge: NF-kappaB-activating kinase-associated protein 1 participates in TLR3/Toll-IL-1 homology domain-containing adapter molecule-1-mediated IFN regulatory factor 3 activation. J. Immunol. 174:27-30. [DOI] [PubMed] [Google Scholar]
- 45.Schlender, J., V. Hornung, S. Finke, M. Gunthner-Biller, S. Marozin, K. Brzozka, S. Moghim, S. Endres, G. Hartmann, and K. K. Conzelmann. 2005. Inhibition of Toll-like receptor 7- and 9-mediated alpha/beta interferon production in human plasmacytoid dendritic cells by respiratory syncytial virus and measles virus. J. Virol. 79:5507-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669-682. [DOI] [PubMed] [Google Scholar]
- 47.Shi, S. T., S. J. Polyak, H. Tu, D. R. Taylor, D. R. Gretch, and M. M. Lai. 2002. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology 292:198-210. [DOI] [PubMed] [Google Scholar]
- 48.Shoukry, N. H., A. Grakoui, M. Houghton, D. Y. Chien, J. Ghrayeb, K. A. Reimann, and C. M. Walker. 2003. Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection. J. Exp. Med. 197:1645-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stack, J., I. R. Haga, M. Schroder, N. W. Bartlett, G. Maloney, P. C. Reading, K. A. Fitzgerald, G. L. Smith, and A. G. Bowie. 2005. Vaccinia virus protein A46R targets multiple Toll-like-interleukin-1 receptor adaptors and contributes to virulence. J. Exp. Med. 201:1007-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taguchi, T., M. Nagano-Fujii, M. Akutsu, H. Kadoya, S. Ohgimoto, S. Ishido, and H. Hotta. 2004. Hepatitis C virus NS5A protein interacts with 2′,5′-oligoadenylate synthetase and inhibits antiviral activity of IFN in an IFN sensitivity-determining region-independent manner. J. Gen. Virol. 85:959-969. [DOI] [PubMed] [Google Scholar]
- 51.Thimme, R., D. Oldach, K. M. Chang, C. Steiger, S. C. Ray, and F. V. Chisari. 2001. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J. Exp. Med. 194:1395-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tseng, C. T., and G. R. Klimpel. 2002. Binding of the hepatitis C virus envelope protein E2 to CD81 inhibits natural killer cell functions. J. Exp. Med. 195:43-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uematsu, S., S. Sato, M. Yamamoto, T. Hirotani, H. Kato, F. Takeshita, M. Matsuda, C. Coban, K. J. Ishii, T. Kawai, O. Takeuchi, and S. Akira. 2005. Interleukin-1 receptor-associated kinase-1 plays an essential role for Toll-like receptor (TLR)7- and TLR9-mediated interferon-α induction. J. Exp. Med. 201:915-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727-740. [DOI] [PubMed] [Google Scholar]
- 56.Yi, M., R. A. Villanueva, D. L. Thomas, T. Wakita, and S. M. Lemon. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. USA 103:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y. M. Loo, M. Gale, Jr., S. Akira, S. Yonehara, A. Kato, and T. Fujita. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851-2858. [DOI] [PubMed] [Google Scholar]
- 58.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 59.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]