Abstract
Mutational analysis of the four conserved proline residues in human immunodeficiency virus type 1 (HIV-1) Vpr reveals that only Pro-35 is required for efficient replication of R5-tropic, but not of X4-tropic, viruses in human lymphoid tissue (HLT) cultivated ex vivo. While Vpr-mediated apoptosis and G2 cell cycle arrest, as well as the expression and subcellular localization of Vpr, were independent, the capacity for encapsidation of Vpr into budding virions was dependent on Pro-35. 1H nuclear magnetic resonance data suggest that mutation of Pro-35 causes a conformational change in the hydrophobic core of the molecule, whose integrity is required for the encapsidation of Vpr, and thus, Pro-35 supports the replication of R5-tropic HIV-1 in HLT.
Human immunodeficiency virus type 1 (HIV-1) Vpr induces G2 cell cycle arrest and apoptosis and facilitates the transport of the preintegration complex into the nuclei of nondividing cells, thereby mediating the integration of proviral DNA into the host genome. Although dispensable for growth of HIV-1 in cultured T cells, Vpr appears to play an important role in virus replication and pathogenesis in vivo (6, 12, 23). Vpr is packaged into virus particles via an interaction with the p6 domain of the Gag precursor Pr55 involving residues 32 to 46, which comprise the LXXLFG motif in p6 (1, 3, 7, 11, 13, 22). Mutational analysis of Vpr has shown that residues located between Ser-28 and His-40 are required for efficient binding to p6 (14, 15, 24, 27). Since Vpr is encapsidated into viral particles, it is present during early steps of infection, where it is essential for productive infection of nondividing cells such as terminally differentiated cells from the monocyte/macrophage lineage.
In order to investigate the biological function of the highly conserved proline residues in the N terminus of Vpr, single proline-to-alanine mutations were introduced at position 5, 10, 14, or 35 into an X4-tropic HIV-1NL4-3 molecular clone (2) and an otherwise isogenic R5-tropic derivative thereof (18). To assess the functional relevance of each proline residue, parallel cultures of human lymphoid tissue (HLT) cultivated ex vivo (8) were infected with virus stocks standardized for p24 content, and release of viral particles was determined by quantification of reverse transcriptase activity or p24 antigen (Fig. 1B to D). HLT was chosen, as it represents a relevant physiological system to study accessory proteins such as Vpr that exercise their functions particularly in primary cells (10, 19, 20).
FIG. 1.
Pro-35 enhances the replication of R5-tropic HIV-1 in HLT. (A) N-terminal structure and amino acid sequence of Vpr from HIV-1NL4-3. Proline residues in positions 5, 10, 14, and 35, as well as p6 binding regions, are indicated (4, 16). (B) Representative replication profiles of R5-tropic HIV-1 and Vpr mutants in HLT ex vivo. RT, reverse transcriptase. (C and D) For average virus production, tissue samples were inoculated with X4-tropic and R5-tropic viruses, respectively, and cumulative virus release by 18 to 21 tissue blocks over 15 days was measured. Bars show mean virus release relative to that for the wt virus. Error bars, SEMs.
A representative replication profile of R5-tropic HIV-1NL4-3 is depicted in Fig. 1B. Since the replication of HIV-1 in HLT generally exhibits donor-dependent differences (9), an average of replication profiles in tissues derived from several donors revealed that productive infection of both R5-tropic and X4-tropic HIV-1 variants resulted in the typical Vpr-dependent replication kinetics (19, 20): the inactivation of the vpr gene reduced the replication of X4-tropic HIV-1NL4-3 to 50% ± 8.8% (mean ± standard error of the mean [SEM]) (number of HLT donors [n] = 9; P < 0.0001), and the replication of R5-tropic viruses decreased to 37% ± 8.2% (n = 10; P < 0.0001) (Fig. 1B to D). Mutation of proline 5, 10, or 14 did not influence virus replication with either coreceptor (Fig. 1B to D). However, mutation of Pro-35 reduced the replication of R5-tropic HIV-1 variants to 62% ± 12.7% (n = 10; P = 0.009) compared with that of the wild-type (wt) virus or the other proline mutants (Fig. 1B and D). While the P35A mutation did not influence the replication of X4-tropic variants (Fig. 1C), Pro-35 supports Vpr-mediated replication of R5-tropic HIV-1.
Functional analyses showed that the proline mutations did not affect the de novo synthesis and subcellular localization of Vpr or biological activities such as the induction of G2 arrest and apoptosis (data not shown). To compare protein half-lives, Western blot kinetic analyses were performed in HeLa cells transfected with pCMV-FLAG-Vpr (28) encoding wt Vpr and mutants thereof. In order to completely block de novo synthesis of Vpr, cells were treated with a combination of the protein biosynthesis inhibitors puromycin, cycloheximide, and emetine 24 h posttransfection. Aliquots of cell cultures were removed during a 24-h treatment period, and equal amounts of cell lysates were analyzed by Western blotting. All Vpr proteins had comparable half-lives, except for the P35A mutant, which exhibited a slightly faster decay starting 6 h posttreatment (Fig. 2A).
FIG. 2.
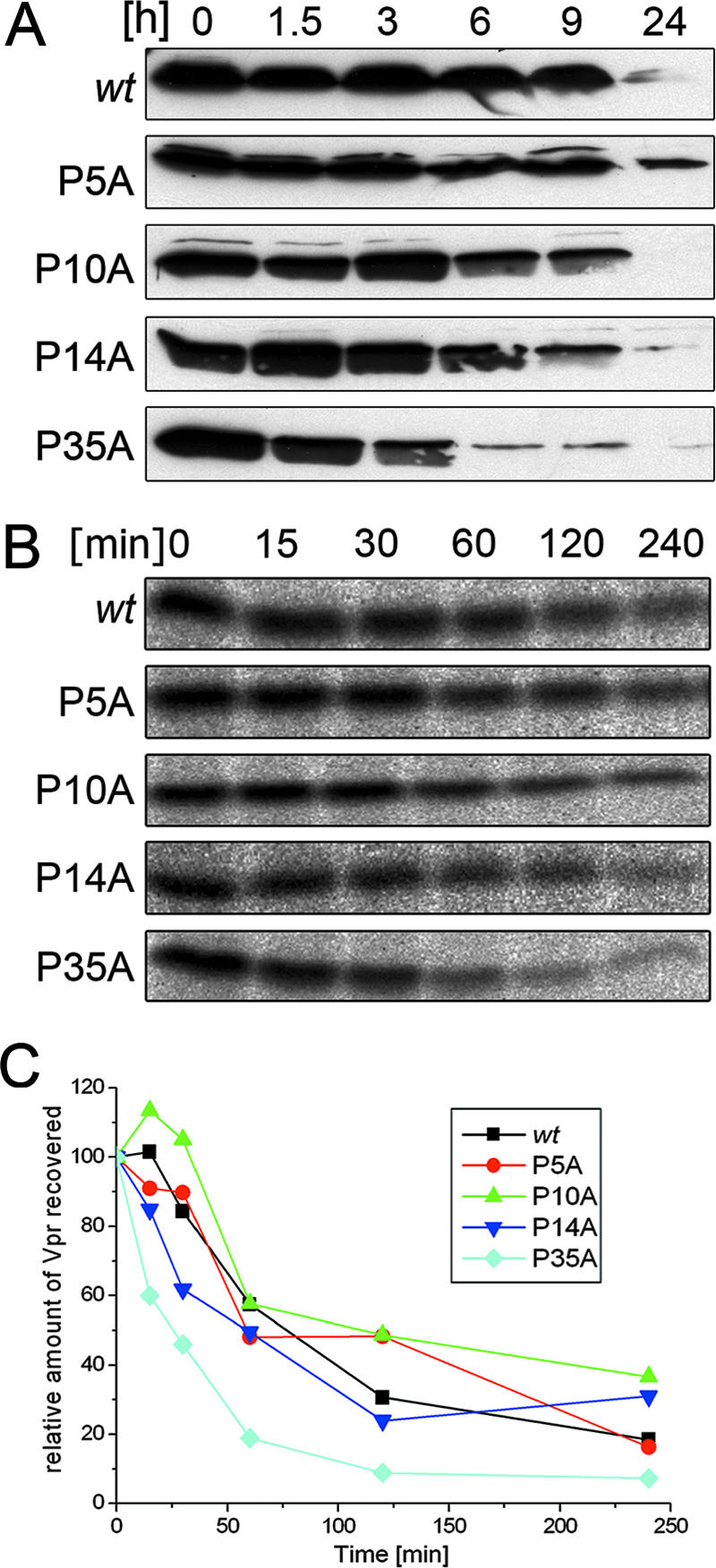
Effects of proline residues on the protein stability of Vpr. (A) In transfected HeLa cells, protein biosynthesis was blocked. Samples were taken at the times indicated, and cell lysates were subjected to Western blotting. (B) Transfected cells were pulse-labeled with [35S]methionine for 10 min and chased for the times indicated. After cell lysis, FLAG-Vpr was immunoprecipitated with anti-FLAG antibodies, separated by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and analyzed by fluorography. (C) The Vpr proteins for which results are shown in panel B were quantified with an image analyzer. The percentages of wt Vpr and proline mutants recovered relative to the amounts present at the end of the pulse (0 h) were plotted as a function of time.
For a more sensitive analysis of protein stability, pulse-chase metabolic radiolabeling experiments were performed using HeLa cells transfected with pCMV-FLAG-Vpr vectors as described previously (28). Following 10 min of pulse-labeling with [35S]methionine, cells were chased for as long as 4 h, and Vpr proteins were recovered from cell lysates by immunoprecipitation with FLAG-specific antibodies, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and analyzed by fluorography (Fig. 2B). The relative amounts of Vpr proteins recovered were plotted as a function of time (Fig. 2C). All mutants exhibited half-lives comparable to that of wt Vpr except for the P35A mutant, which had a ∼1-h shorter half-life. However, these differences in protein turnover did not affect the steady-state level of the P35A mutant (Fig. 3C).
FIG. 3.
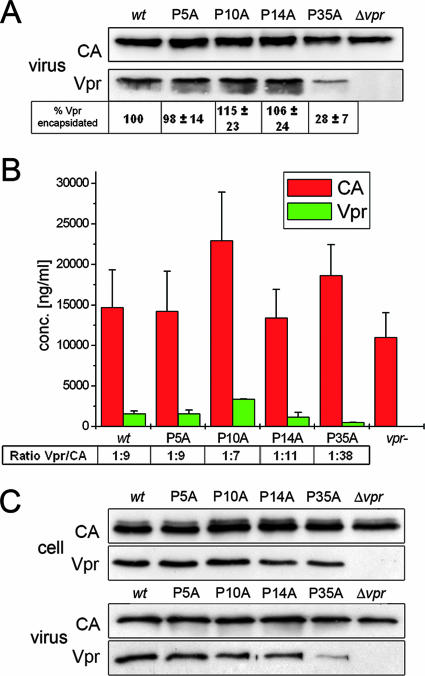
Effects of proline residues on Vpr incorporation into HIV-1 virions. (A) VLPs were produced in HeLa cells transfected with pNLenv1. At 48 h posttransfection, VLPs were pelleted and analyzed by Western blotting using antisera for Vpr and CA. Shown is a Western blot from one representative experiment. The relative amounts of Vpr incorporated ± SEMs from three independent experiments, as quantified by image analysis, are given below. (B) Absolute amounts of CA and Vpr in purified viral supernatants were determined by ELISA. 293T cells were transfected with HIV-1NL4-3 and its Vpr mutants, supernatants were purified, and virions were harvested by ultracentrifugation. The absolute quantities of Vpr and CA in virions were each determined by ELISA techniques. (C) Incorporation of FLAG-Vpr mutants into VLPs in cells cotransfected with pCMV-FLAG-Vpr and pNLΔenvΔvpr. The intracellular steady-state level and the amount of FLAG-Vpr incorporated into VLPs were determined by Western blotting using anti-FLAG and anti-CA specific antibodies.
In order to investigate a possible role of the proline residues in the encapsidation of Vpr, HeLa cells were transfected with the HIV-1NL4-3 subgenomic expression vector pNLenv1, in which env is deleted (21), encoding wt Vpr or its proline mutants. Virus-like particles (VLPs) were isolated from cell culture supernatants and standardized for CA, and the amounts of encapsidated Vpr were analyzed by Western blotting (Fig. 3A). While the P5A, P10A, and P14A mutants showed a wt phenotype, the level of incorporation of the P35A mutant into the virus was reduced to 28%. To further quantify the Vpr/CA ratio, the total amounts of Vpr and CA present in purified viruses were quantified by an enzyme-linked immunosorbent assay (ELISA) (Fig. 3B). The concentration of CA in virus lysates was determined by a p24 capture ELISA (Aalto, Dublin, Ireland). In parallel, the amount of Vpr determined by a solid-phase ELISA was calculated from a standard curve generated by using defined amounts of synthetic Vpr (sVpr). Consistent with results obtained from Western blotting, the encapsidation of the P5A, P10A, and P14A mutants was comparable to that of wt Vpr, yielding a Vpr/CA ratio between 1:7 and 1:11, which is in agreement with earlier reports for wt Vpr (17). However, encapsidation of the P35A mutant was again reduced to 24%, yielding a Vpr/CA ratio of 1:38.
In a further assay, we verified that the reduced encapsidation of the P35A mutant is not due to potential changes in the intracellular steady-state level. Since Vpr expression driven by the subgenomic HIV-1 vector pNLenv1 is too low for unambiguous quantification of Vpr in cell extracts, HeLa cells were cotransfected with a vpr- and env-deficient vector, pNLenvΔvpr, and the pCMV-FLAG-Vpr expression plasmids. As shown in Fig. 3C and in agreement with the results described above (Fig. 2 and data not shown), the intracellular steady-state levels of all proline mutants were comparable to that of wt Vpr and were not affected by the slight differences in protein half-life (Fig. 2). Nevertheless, encapsidation of the P35A mutant was again reduced to 30%, indicating a loss of Vpr binding to p6.
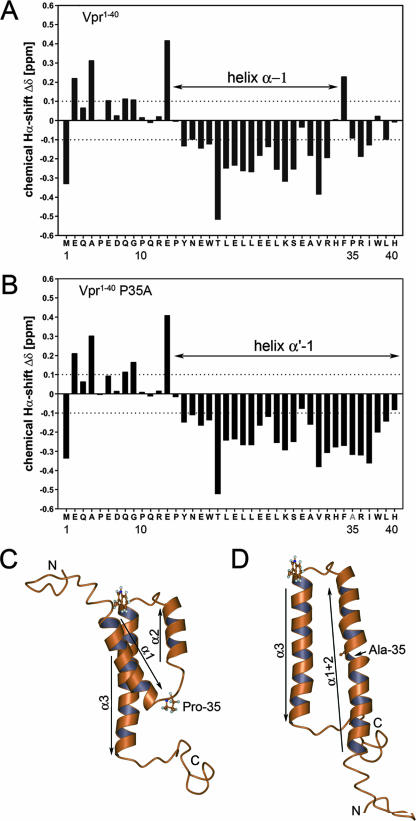
Previous nuclear magnetic resonance (NMR) structural studies of sVpr and fragments thereof have shown that Pro-35 terminates the first N-terminal helix and comprises the center of a turn-like structure leading to helix 2 (residues 38 to 50) (Fig. 4C) (4, 16, 25, 26). In order to investigate the consequences of the P35A mutation for the structure in this region, NMR data recorded in 50% trifluoroethanol were compared for the two related synthetic peptides sVpr1-40 and sVpr1-40 P35A (for experimental details, see the supplemental material). In the case of sVpr1-40 (Fig. 4A), the 1Hα chemical shift differences imply the presence of an α-helix comprising residues Tyr-15 to Arg-32 that is bounded N- and C-terminally by Pro-14 and Pro-35, respectively (4). The sVpr1-40 P35A mutant shows in its N-terminal and central regions 1Hα differences that are almost identical with those of the wt peptide. However, pronounced upfield shifts were identified for residues 32 to 39, providing strong evidence of a loss of the turn-like structure identified in sVpr1-40 (Fig. 4A and B). The formation of a C-terminal extension of helix 1 implies a fusion of helices 1 and 2 and a strong influence of Pro-35 on the structure of Vpr.
FIG. 4.
(A and B) 1H chemical shift differences (Δδ) of the α-protons between the experimentally determined values and those for residues in a random coil for sVpr1-40 (A) and sVpr1-40 P35A (B). (C) Experimental 3-dimensional structure of Vpr (available at http://www.pdb.org under PDB ID 1M8L and published in reference 16) showing the relative positions of the three helices and Pro-35. (D) Calculated conformation of the P35A mutant in which helix 1 (α1) has merged with helix 2 (α2) to give an antiparallel arrangement of helices. This was modeled by moving helix 1 and folding the interhelical residues, between helices 1 and 2, into a helical conformation to form a merged, extended structure, termed helix 1+2 (α1+2). The relative positions of the original helices 2 and 3 were unaltered in this conformation.
Generally, Vpr consists of three well-defined α-helices, located between residues 17 and 33 (helix 1), 38 and 50 (helix 2), and 56 and 77 (helix 3), in a defined tertiary structure (Fig. 4C) (16). This, taken together with the empirical predictions and our experimental data (Fig. 4), clearly suggests that the P35A mutation would increase the helical character in the region between the original two helices (helices 1 and 2) to give one long, extended helical region comprising residues 17 to 50 and termed helix 1+2 (Fig. 4D). This merging of the two helices will result in a rearrangement within the hydrophobic core of the molecule and thus in a significant global change of the structure of Vpr in the p6 binding region (Fig. 1 and 4).
Vpr has been reported to augment virus replication in terminally differentiated cells from the monocyte/macrophage lineage by facilitating the import of the viral preintegration complex across the nuclear membrane, thereby supporting the integration of HIV proviral DNA into the host genome (5). Here we show that a mutation of Pro-35 changes the structure of Vpr, which results in decreased encapsidation of Vpr and compromises the replication of R5-tropic HIV-1 in HLT.
Supplementary Material
Acknowledgments
This work was supported by a grant from the FORINGEN research network, funded by the State of Bavaria, Germany; by grant IE-S08T06 from the German Human Genome Research Project; and by grants SFB 466-A11 and SFB 643-A1, from the German Research Council, to U.S.
We thank Hans-Jürgen Hecht and Uwe Tessmer for helpful discussions and Prisca Kunert, Barbara Brecht, and Christel Kakoschke for excellent technical assistance.
Footnotes
Published ahead of print on 6 June 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Accola, M. A., A. A. Bukovsky, M. S. Jones, and H. G. Göttlinger. 1999. A conserved dileucine-containing motif in p6gag governs the particle association of Vpx and Vpr of simian immunodeficiency viruses SIVmac and SIVagm. J. Virol. 73:9992-9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachand, F., X. J. Yao, M. Hrimech, N. Rougeau, and E. A. Cohen. 1999. Incorporation of Vpr into human immunodeficiency virus type 1 requires a direct interaction with the p6 domain of the p55 gag precursor. J. Biol. Chem. 274:9083-9091. [DOI] [PubMed] [Google Scholar]
- 4.Bruns, K., T. Fossen, V. Wray, P. Henklein, U. Tessmer, and U. Schubert. 2003. Structural characterization of the HIV-1 Vpr N terminus: evidence of cis/trans-proline isomerism. J. Biol. Chem. 278:43188-43201. [DOI] [PubMed] [Google Scholar]
- 5.de Noronha, C. M., M. P. Sherman, H. W. Lin, M. V. Cavrois, R. D. Moir, R. D. Goldman, and W. C. Greene. 2001. Dynamic disruptions in nuclear envelope architecture and integrity induced by HIV-1 Vpr. Science 294:1105-1108. [DOI] [PubMed] [Google Scholar]
- 6.Fassati, A. 2006. HIV infection of non-dividing cells: a divisive problem. Retrovirology 3:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fossen, T., V. Wray, K. Bruns, J. Rachmat, P. Henklein, U. Tessmer, A. Maczurek, P. Klinger, and U. Schubert. 2005. Solution structure of the human immunodeficiency virus type 1 p6 protein. J. Biol. Chem. 280:42515-42527. [DOI] [PubMed] [Google Scholar]
- 8.Glushakova, S., B. Baibakov, L. B. Margolis, and J. Zimmerberg. 1995. Infection of human tonsil histocultures: a model for HIV pathogenesis. Nat. Med. 1:1320-1322. [DOI] [PubMed] [Google Scholar]
- 9.Glushakova, S., B. Baibakov, J. Zimmerberg, and L. B. Margolis. 1997. Experimental HIV infection of human lymphoid tissue: correlation of CD4+ T cell depletion and virus syncytium-inducing/non-syncytium-inducing phenotype in histocultures inoculated with laboratory strains and patient isolates of HIV type 1. AIDS Res. Hum. Retrovir. 13:461-471. [DOI] [PubMed] [Google Scholar]
- 10.Glushakova, S., Y. Yi, J. C. Grivel, A. Singh, D. Schols, E. De Clercq, R. G. Collman, and L. Margolis. 1999. Preferential coreceptor utilization and cytopathicity by dual-tropic HIV-1 in human lymphoid tissue ex vivo. J. Clin. Investig. 104:R7-R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenkins, Y., O. Pornillos, R. L. Rich, D. G. Myszka, W. I. Sundquist, and M. H. Malim. 2001. Biochemical analyses of the interactions between human immunodeficiency virus type 1 Vpr and p6gag. J. Virol. 75:10537-10542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Rouzic, E., and S. Benichou. 2005. The Vpr protein from HIV-1: distinct roles along the viral life cycle. Retrovirology 2:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu, Y. L., R. P. Bennett, J. W. Wills, R. Gorelick, and L. Ratner. 1995. A leucine triplet repeat sequence (LXX)4 in p6gag is important for Vpr incorporation into human immunodeficiency virus type 1 particles. J. Virol. 69:6873-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahalingam, S., S. A. Khan, M. A. Jabbar, C. E. Monken, R. G. Collman, and A. Srinivasan. 1995. Identification of residues in the N-terminal acidic domain of HIV-1 Vpr essential for virion incorporation. Virology 207:297-302. [DOI] [PubMed] [Google Scholar]
- 15.Mahalingam, S., S. A. Khan, R. Murali, M. A. Jabbar, C. E. Monken, R. G. Collman, and A. Srinivasan. 1995. Mutagenesis of the putative alpha-helical domain of the Vpr protein of human immunodeficiency virus type 1: effect on stability and virion incorporation. Proc. Natl. Acad. Sci. USA 92:3794-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morellet, N., S. Bouaziz, P. Petitjean, and B. P. Roques. 2003. NMR structure of the HIV-1 regulatory protein VPR. J. Mol. Biol. 327:215-227. [DOI] [PubMed] [Google Scholar]
- 17.Müller, B., U. Tessmer, U. Schubert, and H. G. Kräusslich. 2000. Human immunodeficiency virus type 1 Vpr protein is incorporated into the virion in significantly smaller amounts than Gag and is phosphorylated in infected cells. J. Virol. 74:9727-9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papkalla, A., J. Münch, C. Otto, and F. Kirchhoff. 2002. Nef enhances human immunodeficiency virus type 1 infectivity and replication independently of viral coreceptor tropism. J. Virol. 76:8455-8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajan, D., S. Wildum, E. Rücker, M. Schindler, and F. Kirchhoff. 2006. Effect of R77Q, R77A and R80A changes in Vpr on HIV-1 replication and CD4 T cell depletion in human lymphoid tissue ex vivo. AIDS 20:831-836. [DOI] [PubMed] [Google Scholar]
- 20.Rücker, E., J. C. Grivel, J. Münch, F. Kirchhoff, and L. Margolis. 2004. Vpr and Vpu are important for efficient human immunodeficiency virus type 1 replication and CD4+ T-cell depletion in human lymphoid tissue ex vivo. J. Virol. 78:12689-12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schubert, U., K. A. Clouse, and K. Strebel. 1995. Augmentation of virus secretion by the human immunodeficiency virus type 1 Vpu protein is cell type independent and occurs in cultured human primary macrophages and lymphocytes. J. Virol. 69:7699-7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selig, L., J. C. Pages, V. Tanchou, S. Preveral, C. Berlioz-Torrent, L. X. Liu, L. Erdtmann, J. Darlix, R. Benarous, and S. Benichou. 1999. Interaction with the p6 domain of the gag precursor mediates incorporation into virions of Vpr and Vpx proteins from primate lentiviruses. J. Virol. 73:592-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherman, M. P., C. M. De Noronha, S. A. Williams, and W. C. Greene. 2002. Insights into the biology of HIV-1 viral protein R. DNA Cell Biol. 21:679-688. [DOI] [PubMed] [Google Scholar]
- 24.Singh, S. P., B. Tomkowicz, D. Lai, M. Cartas, S. Mahalingam, V. S. Kalyanaraman, R. Murali, and A. Srinivasan. 2000. Functional role of residues corresponding to helical domain II (amino acids 35 to 46) of human immunodeficiency virus type 1 Vpr. J. Virol. 74:10650-10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wecker, K., N. Morellet, S. Bouaziz, and B. P. Roques. 2002. NMR structure of the HIV-1 regulatory protein Vpr in H2O/trifluoroethanol. Comparison with the Vpr N-terminal (1-51) and C-terminal (52-96) domains. Eur. J. Biochem. 269:3779-3788. [DOI] [PubMed] [Google Scholar]
- 26.Wecker, K., and B. P. Roques. 1999. NMR structure of the (1-51) N-terminal domain of the HIV-1 regulatory protein Vpr. Eur. J. Biochem. 266:359-369. [DOI] [PubMed] [Google Scholar]
- 27.Yao, X. J., R. A. Subbramanian, N. Rougeau, F. Boisvert, D. Bergeron, and E. A. Cohen. 1995. Mutagenic analysis of human immunodeficiency virus type 1 Vpr: role of a predicted N-terminal alpha-helical structure in Vpr nuclear localization and virion incorporation. J. Virol. 69:7032-7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zander, K., M. P. Sherman, U. Tessmer, K. Bruns, V. Wray, A. T. Prechtel, E. Schubert, P. Henklein, J. Luban, J. Neidleman, W. C. Greene, and U. Schubert. 2003. Cyclophilin A interacts with HIV-1 Vpr and is required for its functional expression. J. Biol. Chem. 278:43202-43213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.