Abstract
The human cytidine deaminase APOBEC3G (A3G) and other APOBEC3 proteins exhibit differential inhibitory activities against diverse endogenous retroelements and retroviruses, including Vif-deficient human immunodeficiency virus type 1. The potential inhibitory activity of human APOBEC proteins against long interspersed element 1 (LINE-1) has not been fully evaluated. Here, we demonstrate inhibition of LINE-1 by multiple human APOBEC3 cytidine deaminases, including previously unreported activity for A3DE and A3G. More ancient members of APOBEC, cytidine deaminases AID and APOBEC2, had no detectable activity against LINE-1. A3A, which did not form high-molecular-mass (HMM) complexes and interacted poorly with P bodies, was the most potent inhibitor of LINE-1. A3A specifically recognizes LINE-1 RNA but not the other cellular RNAs tested. However, in the presence of LINE-1, A3A became associated with HMM complexes containing LINE-1 RNA. The ability of A3A to recognize LINE-1 RNA required its catalytic domain and was important for its LINE-1 suppression. Although the mechanism of LINE-1 restriction did not seem to involve DNA editing, A3A inhibited the accumulation of nascent LINE-1 DNA, suggesting interference with LINE-1 reverse transcription and/or integration or intracellular movement of LINE-1 ribonucleoprotein. Thus, association with P bodies or cellular HMM complexes could not predict the potency of APOBEC3 anti-LINE-1 activities. The catalytic domain of APOBEC3 proteins may be important for proper folding and target factors such as RNA or protein interaction in addition to cytidine deamination.
A3G and other APOBEC3 proteins (22) belong to a family of proteins that also includes activation-induced cytidine deaminase (AID), apolipoprotein B-editing catalytic subunit 1 (APOBEC1), and APOBEC2 (1, 16, 19, 35, 39, 45). These proteins have cytidine deaminase activities that can modify RNA or DNA. A3G was the first APOBEC3 protein to be identified as a potent inhibitor of human immunodeficiency virus type 1 (HIV-1) in the absence of Vif (41). Subsequently, several other human APOBEC3 proteins, including APOBEC3A (A3A), APOBEC3B (A3B), APOBEC3C (A3C), APOBEC3DE (A3DE), and APOBEC3F (A3F), were identified as broad antiviral factors against HIV-1, simian immunodeficiency virus, murine leukemia virus, and various endogenous retroelements (3, 4, 11-14, 24, 26, 37, 40, 48, 49, 53) as well as hepatitis B virus (44).
Like AID, which edits single-stranded immunoglobulin gene DNA, APOBEC3 proteins prefer minus-strand retroviral DNAs as targets (1, 10, 16, 19, 29, 35, 39, 45, 51). In the absence of Vif-induced A3G degradation in virus-producing cells, virion-packaged A3G induces C-to-U mutations in minus-strand viral DNA during reverse transcription (18, 25, 30, 31, 43, 50, 52). Both cytidine deamination-dependent and -independent antiviral functions of APOBEC3 proteins have been reported (2, 8, 17, 20, 28, 33, 36, 44).
The potential inhibitory activity of certain human APOBEC3 proteins against LINE-1 has been reported. A3A and A3B are potent inhibitors of long interspersed element 1 (LINE-1), whereas A3G has no detectable activity (5, 7, 21, 34, 42, 46). Whether A3F has restriction activity against LINE-1 is controversial (5, 7, 21, 34, 42, 46), and the role of A3DE in LINE-1 restriction has not been evaluated.
Differential inhibition of LINE-1 by human APOBEC proteins.
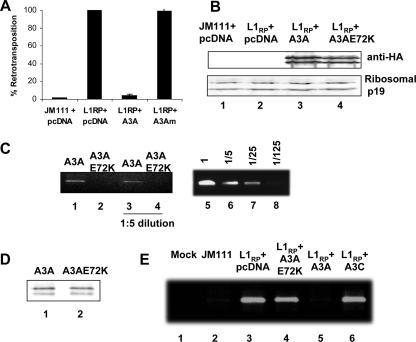
To further evaluate the effect of human APOBEC3 proteins on LINE-1 restriction, a cell culture-based retrotransposition assay was used (6). In this assay, a full-length LINE-1 element was tagged with a retrotransposition cassette containing a marker gene for enhanced green fluorescent protein (EGFP), interrupted by an intron in the opposite transcriptional orientation. The marker could become activated only after the full-length LINE-1 element was transcribed, the intron was removed by splicing, and the RNA was reverse transcribed and integrated into the host cell genome. 293T cells were cotransfected with a tagged LINE-1 construct (pL1RP-EGFP) and one of several expression vectors encoding APOBEC proteins or a control empty vector, and retrotransposition events were detected by examining GFP-positive cells 5 days after transfection. Retrotransposition events of pL1RP-EGFP were detected in 293T cells, with a 72-fold-higher level of EGFP-positive cells than in cells transfected with the negative control LINE-1 vector pL1RP(JM111)-EGFP (see Fig. 1A), which contains a LINE-1 retrotransposon with two missense mutations in open reading frame 1 that abolishes retrotransposition (38). LINE-1 retrotransposition was potently inhibited by A3A and A3B (Fig. 1A), consistent with other recent reports (5, 7, 21, 34, 42). The potential activity of A3DE in LINE-1 retrotransposition has not been reported. We observed that A3DE also inhibited LINE-1 retrotransposition effectively (Fig. 1A). Furthermore, although human APOBEC3 proteins could inhibit LINE-1 retrotransposition, the more ancient members of the APOBEC cytidine deaminase family, such as APOBEC2 and AID, had no activity against LINE-1 (Fig. 1A). All APOBEC proteins were expressed in transfected 293T cells (Fig. 1B). In contrast to other reports (5, 7, 21, 34, 42, 46), we also observed that A3G reduced LINE-1 retrotransposition in 293T cells (Fig. 1C) when A3G expression was comparable to that of A3A and A3F (Fig. 1D).
FIG. 1.
Effect of human APOBEC proteins on LINE-1 retrotransposition. (A) 293T cells were transfected with the negative control pLINE-1RPJM111-EGFP (JM111) or pLINE-1RP-EGFP in the absence (with pcDNA3.1+ empty vector) or presence of APOBEC expression vectors. Relative retrotransposition was measured by detecting EGFP expression in cells using fluorescence-activated cell sorting analysis 5 days after transfection and calculated by setting the value for pLINE-1RP-EGFP cotransfected with an empty vector at 100%. (B) Immunoblot showing similar expression levels of APOBEC proteins in 293T cells cotransfected with various plasmids expressing HA-tagged APOBEC proteins and pLINE-1RP-EGFP. APOBEC proteins were detected with anti-HA, and ribosomal p19 was detected as a loading control. The expression of AID-V5 from transfected 293T cells was detected with an anti-V5 antibody (data not shown). (A) 293T cells were transfected with the negative control pLINE-1RPJM111-EGFP (JM111) or pLINE-1RP-EGFP in the absence (with pcDNA3.1+ empty vector) or presence of APOBEC3 expression vectors. Relative retrotransposition was measured by detecting EGFP expression in cells using fluorescence-activated cell sorting analysis 5 days after transfection and calculated by setting the value for pLINE-1RP-EGFP cotransfected with an empty vector at 100%. (D) Immunoblot showing similar expression levels of APOBEC3 proteins in 293T cells cotransfected with various plasmids expressing HA-tagged APOBEC proteins and pLINE-1RP-EGFP. APOBEC proteins were detected with anti-HA, and ribosomal p19 was detected as a loading control.
Association with intracellular HMM complexes is not required for LINE-1 inhibition.
It was recently reported that A3G can interact with cellular mRNAs (9, 15, 23, 47) to form high-molecular-mass (HMM) complexes (8, 23, 27) and to associate with stress granules, staufen granules, or P bodies.
To explore whether other human APOBEC3 proteins are associated with HMM complexes, we purified lysates of APOBEC3-transfected 293T cells through velocity sucrose gradients, followed by immunoblotting. Like A3G, A3C and A3F formed HMM complexes in 293T cells (Fig. 2A). In sharp contrast to A3C, A3F, and A3G, A3A did not form HMM complexes and was found at the top of the sucrose gradient (Fig. 2A). When A3G was treated with 1% sodium dodecyl sulfate (A3G monomers), it also stayed at the top fractions of the sucrose gradient (data not shown). Thus, most of the A3A proteins in 293T cells apparently exist as monomers or small complexes. The original distribution of fractions seen in cells expressing only A3A was not affected by RNase treatment (Fig. 2B), suggesting that A3A did not interact with cellular RNAs (Fig. 2B). To characterize the interaction of APOBEC3 proteins with cellular mRNA, we transfected an expression vector for A3A-hemagglutinin (HA), A3C-HA, or A3G-HA or an empty control vector (pcDNA3.1) into 293T cells. The APOBEC3 proteins were then immunoprecipitated, and the coprecipitated RNA samples were analyzed by quantitative reverse transcription-PCR (qRT-PCR). A3C, A3F, and A3G were found to interact with β-actin (Fig. 2C) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Fig. 2D) compared to the negative control samples. However, we did not detect any association of A3A with the abundant cellular mRNA for β-actin (Fig. 2C) or GAPDH (Fig. 2D). Also, A3A showed no detectable interaction with polymerase (Pol) I-transcribed RNAs, such as 18S or 5.8S rRNAs, or with Pol III-transcribed RNAs, such as tRNAs, 5S rRNA, 7SL RNA, U6 snRNA, or Y RNAs (data not shown).
FIG. 2.
Formation of intracellular HMM complexes containing APOBEC3 proteins and selective interaction of APOBEC3 proteins with cellular mRNAs. (A) Velocity sucrose gradient fractionation of APOBEC3-containing complexes. 293T cells were transfected with APOBEC3 expression vectors. Three days later, the cells were lysed and subjected to 10-to-30% continuous sucrose gradient fractionation. Samples were collected and analyzed by immunoblotting with an anti-HA monoclonal antibody. (B) Velocity sucrose gradient fractionation of A3A was not affected by RNase treatment. Cell lysates expressing A3A-HA were treated in the absence or presence of RNase and then analyzed by sucrose gradient fractionation. (C and D) Coimmunoprecipitation of cellular mRNAs with APOBEC3 proteins. HA-tagged APOBEC3 proteins were expressed in transfected 293T cells. Cell lysates from transfected 293T cells were immunoprecipitated with anti-HA antibody conjugated to agarose beads. RNAs coprecipitated with various APOBEC3 proteins were analyzed by qRT-PCR using primers specific for beta-actin (C) or GAPDH (D). The nonspecific RNA (containing the control vector) binding to the assay system was set at 1.
Association with P bodies does not predict the potency of LINE-1 inhibition by APOBEC3 proteins.
To examine the colocalization of APOBEC3 proteins with P bodies, we transiently transfected 293T cells with various HA-tagged APOBEC3 expression vectors (A3A, A3C, A3DE, and A3G). Cells were fixed 48 h after transfection, stained with anti-HA or anti-DDX6 (RCK/p54), a protein known to localize to P bodies. We confirmed the localization of A3G to the cytoplasm, with a strong association with P bodies (15, 47), and report that A3DE has a similar localization (Fig. 3A). On the other hand, A3A and A3C appear to have a diffuse localization in both the cytoplasm and the nucleus, with occasional small focal points of staining that were colocalized with DDX6 (Fig. 3A). However, the apparent association of A3A and A3C with P bodies is by no means as strong as the colocalization of A3DE or A3G (Fig. 3A). It is possible that this weaker association can be explained by the fact that A3A and A3C are single-domain cytidine deaminases, while A3DE and A3G have double cytidine deamination domains (CDDs). The double-domain proteins may be able to interact more strongly with host RNA(s) that may direct them to P bodies.
FIG. 3.
Localization of APOBEC3 proteins and P bodies. (A) 293T cells were transfected with various APOBEC3 expression vectors. APOBEC3 proteins were stained with anti-HA (red). P bodies were visualized by anti-DDX6 (RCK/p54), a P-body marker (green). Some A3DE and A3G were strongly associated with P bodies, while A3A and A3C seemed to have a weaker association. Nuclei were counterstained by DAPI (4′,6′-diamidino-2-phenylindole). (B) Anti-HA antibodies staining A3A and the mutant A3AE72K. A3A appears to have a fairly homogeneous localization in both the cytoplasm and nucleus. The A3AE72K mutant shows aberrant localization, forming aggregate-like spots in the cytoplasm and nucleus. Nuclei were counterstained with DAPI.
A3A typically has a diffuse localization in both the cytoplasm and the nucleus (Fig. 3B). However, mutating an essential glutamate to a lysine in the catalytic site of A3A caused mislocalization of the mutant protein to aggregate-like spots in the cytoplasm and nucleus (Fig. 3B). While mutations of residues in the catalytic domain have been shown to inhibit cytidine deaminase activity, it is important to note that these mutations may also trigger structural changes in the protein, causing it to misfold and become mislocalized in the cell.
A3A interacts with HMM complexes containing LINE-1 RNA.
To study the potential interaction between A3A and LINE-1 RNA, we examined the intracellular complexes formed with A3A in the absence or presence of L1RP-EGFP (6). Although A3A-HA did not form HMM complexes and resided at the top of the sucrose gradient (Fig. 4A, top), we observed that in cells cotransfected with A3A-HA and L1RP-EGFP, A3A shifted into fractions of higher molecular mass (Fig. 4A, middle, lanes 6 to 9). It is of interest that mouse LINE-1 ribonucleoproteins (RNPs) were detected previously in fractions 6 to 9 of the same type of sucrose gradient (32). This shift appeared to be RNA dependent, since A3A-HA returned to the LMM fractions after the A3A/L1RP-EGFP cell lysates were treated with RNase (Fig. 4A, bottom). To confirm that the A3A-HA in HMM fractions is associated with L1RP mRNA, we pooled fractions 6 to 9, immunoprecipitated A3A-HA (Fig. 4B) from them, and then used RT-PCR to amplify L1RP-EGFP RNA from the precipitated A3A-HA sample (Fig. 4C, second and fourth lanes). The results indicated that A3A associated with L1RP-EGFP mRNA. L1RP-EGFP RNA was not detected when sucrose gradient samples (fractions 6 to 9) were prepared side by side using cell lysates containing L1RP-EGFP in the absence of A3A-HA (Fig. 4C, first and third lanes). Thus, A3A was associated with LINE-1 RNA in HMM complexes that presumably contain LINE-1 RNPs.
FIG. 4.
Association of A3A with LINE-1 RNA in HMM complexes. (A) Anti-HA immunoblot of sucrose velocity gradient fractions of lysed 293T cells cotransfected with HA-tagged A3A plus an empty vector (pcDNA3.1) (top), pLINE-1RP-EGFP (middle), or pLINE-1RP-EGFP with RNase treatment prior to sucrose velocity gradient centrifugation (bottom). A3A was shifted to fractions 6 to 9 in the presence of pLINE-1RP-EGFP. (B) Immunoprecipitation of A3A-HA from fractions 6 to 9 in the presence of pLINE-1RP-EGFP. (C) Coprecipitation of LINE-1 RNA with A3A-HA from fractions 6 to 9 in the presence of pLINE-1RP-EGFP. LINE-1 RNA was not detected in fractions 6 to 9 of the sucrose gradient samples containing LINE-1RP-EGFP in the absence of A3A-HA.
A mutation in the CDD of A3A blocks its interaction with LINE-1 RNA and abolishes its ability to inhibit LINE-1 retrotransposition.
A3A is a single-CDD enzyme with an HXEX28PCX4C catalytic site motif, in which the conserved histidine and cysteine residues function as a zinc-coordinating region and the glutamic acid residue serves as a proton donor during general acid catalysis. To determine whether the enzymatic function of A3A was necessary for LINE-1 inhibition, we used A3AE72K, a catalytically inactive A3A mutated at the essential glutamic acid residue (E72). In contrast to the almost complete inhibition that we observed with wild-type A3A, the mutant A3AE72K had almost no inhibitory effect on LINE-1 retrotransposition (Fig. 5A). Similar levels of expression of A3A and A3AE72K proteins were shown by immunoblotting with an anti-HA antibody (Fig. 5B), indicating that the differences observed in LINE-1 inhibition by A3A and A3AE72K were not due to expression differences.
FIG. 5.
An intact catalytic site in A3A is necessary for LINE-1 RNA binding and LINE-1 inhibition. (A) 293T cells were transfected with the negative control pLINE-1RP(JM111)-EGFP or pLINE-1RP-EGFP in the absence or the presence of A3A or A3Am. Retrotransposition was detected by fluorescence-activated cell sorting analysis of EGFP expression in 293T cells 5 days after transfection and was calculated by setting pLINE-1RP-EGFP cotransfected with an empty vector (pcDNA3.1) at 100%. (B) Anti-HA immunoblot showing similar levels of expression of A3A and A3Am in 293T cells cotransfected with pLINE-1RP-EGFP. (C) Coprecipitation of LINE-1 RNA with A3A-HA but not A3AE72K-HA. Ethidium bromide-stained agarose gel showing reverse-transcribed and PCR-amplified EGFP precipitated with A3A-HA but not with A3AE72K-HA. RT-PCR detection was within the detection sensitivity of the assay, as indicated by the dilution standard (lanes 5 to 8). (D) Immunoblot showing that both A3A-HA and A3Am-HA were immunoprecipitated. (E) A3A but not A3AE72K inhibits LINE-1 DNA accumulation. Ethidium bromide-stained agarose gel showing accumulation of LINE-1 DNA in 293T cells cotransfected with A3A or A3Am and pLINE-1RP-EGFP. LINE-1 DNA was amplified by nested PCR primers in EGFP.
Unlike A3A (Fig. 5C, lanes 1 and 3), A3AE72K was unable to interact with LINE-1 RNA (Fig. 5C, lanes 2 and 4), despite comparable immunoprecipitation of both proteins (Fig. 5D). These results suggest that the glutamic acid residue (E72) in A3A's CDD plays a critical role in inhibiting LINE-1 retrotransposition, even though we detected no G-to-A hypermutation in LINE-1 DNA in the presence of A3A (data not shown). The glutamic acid residue (E72) of A3A is necessary for interaction with LINE-1 RNA and inhibition of LINE-1 retrotransposition.
To determine whether the apparent lack of LINE-1 retrotransposition in cells cotransfected with A3A was due to an absence of reverse transcription and/or integration, we used PCR to amplify the EGFP marker included in the L1RP-EGFP plasmid. Amplifying EGFP through specific primers spanning the splice site, rather than LINE-1 DNA, allowed us to differentiate between plasmid-derived LINE-1 DNA and any endogenous LINE-1 DNA that might have been newly synthesized in the cells. Furthermore, the use of primers spanning the splice site ensured that reverse-transcribed, but not plasmid, DNA was amplified. We found that EGFP DNA could not be amplified in untransfected cells (Fig. 5E, lane 1), and only very low levels were detected in cells transfected with the LINE-1 mutant plasmid, JM111 (Fig. 5E, lane 2); as expected, EGFP could be amplified in cells cotransfected with L1RP-EGFP and the empty vector control (Fig. 5E, lane 3). In contrast, when we cotransfected cells with L1RP-EGFP and A3A, only background levels of EGFP were detected (Fig. 5E, lane 5). We further observed that A3AE72K, unlike A3A, was unable to reduce LINE-1 DNA levels (Fig. 5E, lane 4). Therefore, an intact catalytic site was necessary for the decrease in LINE-1 DNA observed in cells expressing A3A.
The present study indicated that all human APOBEC3 proteins tested could inhibit LINE-1 transposition, whereas the more ancient cytidine deaminases of APOBEC family members, including APOBEC2 and AID, lacked such activities. The lack of anti-LINE-1 activity for APOBEC2 has also been reported (7). The potency of LINE-1 suppression varied significantly among human APOBEC3 proteins and could not be explained simply by their association with P bodies or HMM intracellular complexes or by their intracellular localizations. A3G had weaker anti-LINE-1 activity than A3A or A3B. A3G interacted efficiently with P bodies (15, 47) compared to A3A (Fig. 3) or A3B (47). One could argue that association with P bodies may limit the amount of A3G available to inhibit LINE-1. However, A3A and A3C both interacted poorly with P bodies compared to A3G (Fig. 3), but A3A was the most potent inhibitor of LINE-1, while A3C was one of the weakest. The ability to form intracellular HMM complexes, or lack thereof, could not explain the differential LINE-1 restriction by APOBEC3 proteins, either. A3C, A3G, and A3F all formed intracellular HMM complexes, while their anti-LINE-1 activities differed significantly (Fig. 1).
The more potent LINE-1 inhibition by nucleus-localized A3A and A3B than by cytoplasmic A3F and A3G was previously attributed to the ability of A3A and A3B to recognize target-primed reverse transcription of LINE-1 in the nucleus (5, 7, 21, 34, 42, 46). However, both A3DE (largely cytoplasmic) and A3B (mainly nuclear) were potent inhibitors of LINE-1 (Fig. 1). Clearly, the determinants for various degrees of LINE-1 restriction by human APOBEC3 proteins and the mechanism(s) of LINE-1 restriction by various APOBEC3 proteins require further investigation.
We noted that A3A exists in a low-molecular-mass form, and its migration in a velocity sucrose gradient is not affected by RNase treatment. Consistent with this observation, we did not detect any interaction between A3A and Pol II-transcribed cellular mRNAs, such as GAPDH and β-actin (Fig. 2), Pol I-transcribed RNAs, such as 18S or 5.8S rRNAs, or Pol III-transcribed RNAs, such as tRNAs, 5S rRNA, 7SL RNA, U6 snRNA, or Y RNAs (data not shown). However, in the presence of LINE-1 RNA and proteins, A3A shifted to HMM complexes. Since mouse LINE-1 RNPs were detected previously in similar HMM complexes in the same type of velocity gradient fractions (32), these data suggest that A3A interacts with LINE-1 RNPs. Consistent with this argument, HMM A3A was associated with LINE-1 RNA, and A3A HMM complexes were destroyed by RNase treatment.
The APOBEC proteins are characterized as single- or double-domain cytidine deaminases on the basis of the presence of one or two copies of the conserved catalytic site amino acid motifs, with each motif comprising two cysteines and one histidine, which serve to coordinate a zinc ion, together with a glutamic acid residue, which acts as an acid-base catalyst. While A3A inhibits LINE-1 without any apparent DNA deamination (data not shown), the glutamic acid residue of the single catalytic site of A3A is critical for its activity against LINE-1 retrotransposition (Fig. 5). The critical glutamic acid residue of A3A is also required for the recognition of LINE-1 RNA. It is possible that the conserved H, C, and E residues in the catalytic site [(H/C)(A/V)E(X24-30)(PCX2-4C)] of APOBEC3 proteins have evolved to fill important roles such as RNA binding and/or protein-protein interaction in addition to cytidine deamination. It is also possible that critical residues in the catalytic site of APOBEC3 proteins are important for protein folding, as A3AE72K molecules were apparently mislocalized compared to the parental A3A (Fig. 3).
Although DNA editing does not seem to be involved in LINE-1 restriction, A3A inhibited the accumulation of nascent LINE-1 DNA. It has been observed that A3B can also inhibit the accumulation of LINE-1 DNA (42). Inhibition of HIV-1 reverse transcription and integration by A3G has also been reported (2, 8, 17, 20, 28, 31, 33). Similar to A3G-mediated restriction of Vif-deficient HIV-1, A3A may also interfere with LINE-1 reverse transcription/integration. Alternatively, the association of A3A or other APOBEC3 proteins with LINE-1 RNP could also impede the intracellular movement of LINE-1 RNP and therefore its retrotransposition. The exact step(s) of LINE-1 replication affected by APOBEC3 cytidine deaminases remains to be identified.
Acknowledgments
We thank A. Morrot, E. Ehrlich, and B. Liu for advice and technical assistance, M. Malim, N. Landau, Y. Zheng, and H. Kazazian for critical reagents, and D. McClellan for editorial assistance. MAGI-CCR5 cells and the NL4-3ΔVif construct were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
This work was supported by a grant from the NIH (AI062644) and funding from the National Science Foundation of China (NSFC-30425012) and Cheung Kong Scholars Program Foundation of the Chinese Ministry of Education to X.-F. Yu.
Footnotes
Published ahead of print on 20 June 2006.
REFERENCES
- 1.Bieniasz, P. D. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 5:1109-1115. [DOI] [PubMed] [Google Scholar]
- 2.Bishop, K. N., R. K. Holmes, and M. H. Malim. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 80:8450-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392-1396. [DOI] [PubMed] [Google Scholar]
- 4.Bogerd, H. P., H. L. Wiegand, B. P. Doehle, K. K. Lueders, and B. R. Cullen. 2006. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 34:89-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogerd, H. P., H. L. Wiegand, A. E. Hulme, J. L. Garcia-Perez, S. K. O'Shea, J. V. Moran, and B. R. Cullen. 2006. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. USA 103:8780-8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brouha, B., J. Schustak, R. M. Badge, S. Lutz-Prigge, A. H. Farley, J. V. Moran, and H. H. Kazazian, Jr. 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. USA 100:5280-5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, H., C. E. Lilley, Q. Yu, D. V. Lee, J. Chou, I. Narvaiza, N. R. Landau, and M. D. Weitzman. 2006. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 16:480-485. [DOI] [PubMed] [Google Scholar]
- 8.Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435:108-114. [DOI] [PubMed] [Google Scholar]
- 9.Chiu, Y. L., H. E. Witkowska, S. C. Hall, M. Santiago, V. B. Soros, C. Esnault, T. Heidmann, and W. C. Greene. 2006. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl. Acad. Sci. USA 103:15588-15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cullen, B. R. 2006. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 80:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang, Y., X. Wang, W. J. Esselman, and Y. H. Zheng. 2006. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 80:10522-10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doehle, B. P., A. Schafer, and B. R. Cullen. 2005. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology 339:281-288. [DOI] [PubMed] [Google Scholar]
- 13.Dutko, J. A., A. Schafer, A. E. Kenny, B. R. Cullen, and M. J. Curcio. 2005. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr. Biol. 15:661-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esnault, C., O. Heidmann, F. Delebecque, M. Dewannieux, D. Ribet, A. J. Hance, T. Heidmann, and O. Schwartz. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433:430-433. [DOI] [PubMed] [Google Scholar]
- 15.Gallois-Montbrun, S., B. Kramer, C. M. Swanson, H. Byers, S. Lynham, M. Ward, and M. H. Malim. 2007. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P-bodies and stress granules. J. Virol. 81:2165-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goff, S. P. 2004. Retrovirus restriction factors. Mol. Cell 16:849-859. [DOI] [PubMed] [Google Scholar]
- 17.Guo, F., S. Cen, M. Niu, J. Saadatmand, and L. Kleiman. 2006. Inhibition of formula-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 80:11710-11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 19.Harris, R. S., and M. T. Liddament. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4:868-877. [DOI] [PubMed] [Google Scholar]
- 20.Holmes, R. K., F. A. Koning, K. N. Bishop, and M. H. Malim. 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J. Biol. Chem. 282:2587-2595. [DOI] [PubMed] [Google Scholar]
- 21.Hulme, A. E., H. P. Bogerd, B. R. Cullen, and J. V. Moran. 2007. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene 390:199-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285-296. [DOI] [PubMed] [Google Scholar]
- 23.Kozak, S. L., M. Marin, K. M. Rose, C. Bystrom, and D. Kabat. 2006. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 281:29105-29119. [DOI] [PubMed] [Google Scholar]
- 24.Langlois, M. A., R. C. Beale, S. G. Conticello, and M. S. Neuberger. 2005. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 33:1913-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 26.Liddament, M. T., W. L. Brown, A. J. Schumacher, and R. S. Harris. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:1385-1391. [DOI] [PubMed] [Google Scholar]
- 27.Luo, K., B. Liu, Z. Xiao, Y. Yu, X. Yu, R. Gorelick, and X. F. Yu. 2004. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J. Virol. 78:11841-11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo, K., T. Wang, B. Liu, C. Tian, Z. Xiao, J. Kappes, and X. F. Yu. 2007. Cytidine deaminases APOBEC3G and APOBEC3F interact with HIV-1 integrase and inhibit proviral DNA formation. J. Virol. 81:7238-7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malim, M. H. 2006. Natural resistance to HIV infection: the Vif-APOBEC interaction. Crit. Rev. Biol. 329:871-875. [DOI] [PubMed] [Google Scholar]
- 30.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 31.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 32.Martin, S. L., and D. Branciforte. 1993. Synchronous expression of LINE-1 RNA and protein in mouse embryonal carcinoma cells. Mol. Cell. Biol. 13:5383-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mbisa, J. L., R. Barr, J. A. Thomas, N. Vandegraaff, I. J. Dorweiler, E. S. Svarovskaia, W. L. Brown, L. M. Mansky, R. J. Gorelick, R. S. Harris, A. Engelman, and V. K. Pathak. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 81:7099-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muckenfuss, H., M. Hamdorf, U. Held, M. Perkovic, J. Lower, K. Cichutek, E. Flory, G. G. Schumann, and C. Munk. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 281:22161-22172. [DOI] [PubMed] [Google Scholar]
- 35.Navarro, F., and N. R. Landau. 2004. Recent insights into HIV-1 Vif. Curr. Opin. Immunol. 16:477-482. [DOI] [PubMed] [Google Scholar]
- 36.Newman, E. N., R. K. Holmes, H. M. Craig, K. C. Klein, J. R. Lingappa, M. H. Malim, and A. M. Sheehy. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166-170. [DOI] [PubMed] [Google Scholar]
- 37.OhAinle, M., J. A. Kerns, H. S. Malik, and M. Emerman. 2006. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J. Virol. 80:3853-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ostertag, E. M., E. T. Prak, R. J. DeBerardinis, J. V. Moran, and H. H. Kazazian, Jr. 2000. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 28:1418-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rose, K. M., M. Marin, S. L. Kozak, and D. Kabat. 2004. The viral infectivity factor (Vif) of HIV-1 unveiled. Trends Mol. Med. 10:291-297. [DOI] [PubMed] [Google Scholar]
- 40.Schumacher, A. J., D. V. Nissley, and R. S. Harris. 2005. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc. Natl. Acad. Sci. USA 102:9854-9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 42.Stenglein, M. D., and R. S. Harris. 2006. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 281:16837-16841. [DOI] [PubMed] [Google Scholar]
- 43.Suspene, R., P. Sommer, M. Henry, S. Ferris, D. Guetard, S. Pochet, A. Chester, N. Navaratnam, S. Wain-Hobson, and J. P. Vartanian. 2004. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 32:2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turelli, P., B. Mangeat, S. Jost, S. Vianin, and D. Trono. 2004. Inhibition of hepatitis B virus replication by APOBEC3G. Science 303:1829. [DOI] [PubMed] [Google Scholar]
- 45.Turelli, P., and D. Trono. 2005. Editing at the crossroad of innate and adaptive immunity. Science 307:1061-1065. [DOI] [PubMed] [Google Scholar]
- 46.Turelli, P., S. Vianin, and D. Trono. 2004. The innate antiretroviral factor APOBEC3G does not affect human LINE-1 retrotransposition in a cell culture assay. J. Biol. Chem. 279:43371-43373. [DOI] [PubMed] [Google Scholar]
- 47.Wichroski, M. J., G. B. Robb, and T. M. Rana. 2006. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2:e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu, Q., D. Chen, R. Konig, R. Mariani, D. Unutmaz, and N. R. Landau. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 279:53379-53386. [DOI] [PubMed] [Google Scholar]
- 50.Yu, Q., R. Konig, S. Pillai, K. Chiles, M. Kearney, S. Palmer, D. Richman, J. M. Coffin, and N. R. Landau. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11:435-442. [DOI] [PubMed] [Google Scholar]
- 51.Yu, X. 2006. Innate cellular defenses of APOBEC3 cytidine deaminases and viral counter-defenses. Curr. Opin. HIV AIDS 1:187-193. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng, Y. H., D. Irwin, T. Kurosu, K. Tokunaga, T. Sata, and B. M. Peterlin. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78:6073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]