Abstract
SV40 large T antigen (T-ag) is a multifunctional protein that successively binds to 5′-GAGGC-3′ sequences in the viral origin of replication, melts the origin, unwinds DNA ahead of the replication fork, and interacts with host DNA replication factors to promote replication of the simian virus 40 genome. The transition of T-ag from a sequence-specific binding protein to a nonspecific helicase involves its assembly into a double hexamer whose formation is likely dictated by the propensity of T-ag to oligomerize and its relative affinities for the origin as well as for nonspecific double- and single-stranded DNA. In this study, we used a sensitive assay based on fluorescence anisotropy to measure the affinities of wild-type and mutant forms of the T-ag origin-binding domain (OBD), and of a larger fragment containing the N-terminal domain (N260), for different DNA substrates. We report that the N-terminal domain does not contribute to binding affinity but reduces the propensity of the OBD to self-associate. We found that the OBD binds with different affinities to its four sites in the origin and determined a consensus binding site by systematic mutagenesis of the 5′-GAGGC-3′ sequence and of the residue downstream of it, which also contributes to affinity. Interestingly, the OBD also binds to single-stranded DNA with an ∼10-fold higher affinity than to nonspecific duplex DNA and in a mutually exclusive manner. Finally, we provide evidence that the sequence specificity of full-length T-ag is lower than that of the OBD. These results provide a quantitative basis onto which to anchor our understanding of the interaction of T-ag with the origin and its assembly into a double hexamer.
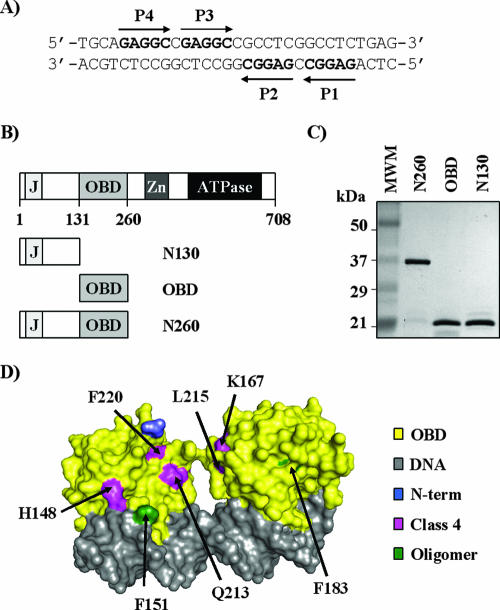
Eukaryotic DNA replication is a complex process that is initiated by the coordinated action of several factors, including the origin recognition complex, cdt1, cdc6, and the minichromosome maintenance complex, the likely cellular replicative helicase (21, 31, 40). Our understanding of the molecular events involved in the initiation of DNA replication has greatly benefited from studies performed with simpler model systems, like budding yeast or small-DNA tumor viruses, for which the origins of replication are of well-defined nucleotide sequences (47, 57). Small-DNA viruses also offer the advantage of encoding a single initiator protein that performs multiple functions during replication. A well-studied example is large T antigen (T-ag), the initiator protein from simian virus 40 (SV40). This multifunctional protein can successively recognize the viral origin of replication, melt it, interact with the host DNA replication factors polymerase α-primase, replication protein A, and topoisomerase I, and unwind DNA ahead of the replication fork (5). Whereas the smallest form of T-ag that has ATPase and minimal unwinding activity is the hexamer, processive unwinding and bidirectional replication require a double hexamer (1, 55, 66). T-ag, a member of helicase superfamily III (18), is a modular protein that contains three distinct domains implicated in viral DNA replication (Fig. 1B). The C-terminal half of T-ag (amino acids [aa] 261 to 708) contains the helicase domain and thus can assemble into hexamers and bind single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) nonspecifically (20, 65). The central part of the protein contains the origin-binding domain (OBD) (aa 131 to 260) which, as its name indicates, recognizes specific sequences in the origin (34, 52, 59, 67). Finally, the N-terminal region, which contains a J-domain (aa 1 to 82; reference 12), a nuclear localization signal (aa 126 to 132; references 23 and 24), and phosphorylation sites for cellular kinases, including one, Thr124, for cyclin/cdk (32, 33, 35, 38, 50), also contribute to viral DNA replication. Indeed, mutations that affect the J-domain or specific phosphorylation sites within the N-terminal domain abrogate replication in vivo and assembly of the protein into functional double hexamers in vitro (2, 9, 42, 44, 62, 64) (see below).
FIG. 1.
Sequence of site II and purified proteins used in this study. (A) Nucleotide sequence of site II located in the SV40 origin of replication. The positions of the four pentanucleotide binding sites, P1 to P4, are indicated. (B) Schematic representation of the 708-aa-long SV40 large T-ag and subfragments thereof used in this study. The following functional domains are shown: J-domain (J; aa 1 to 82), OBD (aa 131 to 260), zinc finger (Zn; aa 302 to 320), and ATPase/helicase domain (aa 418 to 616). (C) Coomassie-stained 15% SDS-PAGE gel of the purified T-ag fragments used in this study. Three-microgram portions of each protein, N260, OBD, and N130, were analyzed. (D) Structure of two OBD molecules bound independently to an oligonucleotide containing P1 and P3 (PDB, 2NTC) (37). Given that P1 and P3 are in opposite orientations, both faces of the OBD can be seen in this structure. Amino acids changed in class 4 mutants (pink) or located at a putative OBD oligomerization interface (green) (37) are indicated. The N-terminal residue of the OBD where the N-terminal domain would be connected is colored in blue.
The core of the SV40 origin is a 64-bp sequence that can be separated into three distinct regions (10, 41). In the middle, a fragment of approximately 30 bp, known as site II, contains four pentanucleotides (5′-GAGGC-3′) (pentanucleotide 1 [P1] to P4), arranged as two pairs of inverted repeats (Fig. 1A) (11, 13, 58). This fragment is flanked on one side by a sequence rich in adenine and thymine, the AT-rich region, and on the other by the early palindrome (EP), which encompasses the site of initial melting (5, 7, 41). Either flanking region is required in addition to site II for double hexamer assembly (46, 56). At the molecular level, the nucleotide sequences and domains of T-ag required for hexamer and double hexamer assembly are beginning to be unraveled, primarily from in vitro biochemical studies. Under the right conditions, formation of a single hexamer can occur on any one of the four pentanucleotides in the origin (22). Assembly of double hexamers occurs preferentially on P1 and P3 (22, 56) and is dependent on the integrity of the N-terminal domain (62, 64), the OBD (53, 64, 67) and C-terminal β-hairpin motif (residues 507 to 517) (16, 28, 46, 51). This motif is presumably needed to melt the region downstream of each T-ag binding site (TBS) in order to generate regions of ssDNA which promote the assembly of additional T-ag molecules into a complete double hexamer (28). The molecular details of how two specific binding sites, P1 and P3, nucleate the assembly of a double hexamer and specifically of how T-ag transits from a monomeric sequence-specific DNA binding protein to a nonspecific oligomeric helicase are still poorly understood.
Biochemical and genetic studies have provided evidence that specific binding of the OBD to its target pentanucleotide sites in the origin involves two structural elements, A1 and B2, that form part of the DNA binding surface (30, 54). Our previous studies further indicated that the OBD binds to the four pentanucleotides in site II independently, without any evidence of cooperativity (59). These findings have been corroborated by nuclear magnetic resonance (NMR) and recent X-ray studies of the OBD bound to DNA (4, 6, 37), which indicated that the OBD recognizes specific bases in the major groove of DNA and that OBD molecules bound on P1 and P3, or on all four pentanucleotides in site II, do not interact with each other. In contrast, the crystal structure of the OBD without DNA showed that the protein can assemble into a left-handed spiral that is wide enough to accommodate duplex DNA in a negatively charged central channel (36). These findings prompted Meinke et al. (36) to speculate that this spiral structure reflects how the OBDs are organized within a double hexamer.
As mentioned above, double hexamer assembly also relies on the integrity of the N-terminal domain. This is exemplified by the finding that a truncated T-ag lacking the first 82 aa oligomerizes into aberrant multimeric complexes either in solution or on DNA (62). Furthermore, double hexamer assembly is regulated by phosphorylation of the N-terminal domain, specifically of Thr124 by a cyclin/cdk (15). Indeed, a mutant T-ag in which Thr124 was changed to alanine was found to assemble into double hexamers with impaired cooperativity, and these complexes dissociated more rapidly from DNA (64). In addition, the T124A T-ag was shown to require longer sequences in the EP or AT region for its assembly on single assembly units (2, 46). Interestingly, specific amino acid substitutions in the OBD, known as class 4 mutations, have been identified that also interfere with double hexamer assembly but without affecting phosphorylation of the protein (64). These findings suggested that the N-terminal domain and the OBD play coordinated functions in double hexamer assembly. This prompted Kim et al. (26) to investigate if peptides containing Thr124 could bind DNA. Strikingly, peptides spanning aa 118 to 130 were found to bind nonspecifically to DNA and inhibit double hexamer assembly in vitro, but only when nonphosphorylated (26). These results provided the first evidence that the N-terminal domain may influence the affinity of the protein for DNA.
The transition of T-ag from a monomeric sequence-specific DNA binding protein to a nonspecific dodecameric helicase is likely dictated by a number of parameters, in particular its affinity for specific pentanucleotides in site II and for nonspecific ds- and ssDNA and its propensity to oligomerize. We believe that a quantitative analysis of these interactions will provide a useful framework to understand how a double hexameric helicase is assembled from individual monomeric components and how this process is regulated by phosphorylation. It was previously reported that the interaction of the OBD with its target site can be studied using a quantitative and sensitive DNA binding assay based on fluorescence anisotropy (59). Here we have extended these studies to measure the interaction of wild-type and mutant forms of the OBD (aa 131 to 260), and of a longer T-ag fragment also containing the N-terminal domain (aa 1 to 260), with different DNA substrates. We found that the N-terminal domain does not contribute directly to DNA binding but dampens the propensity of the OBD to form large oligomers in solution and on DNA. We also report the determination of a specific T-ag consensus binding sequence and that the OBD has a substantial affinity for ssDNA, which is 10-fold higher than its affinity for nonspecific dsDNA. These findings are discussed in light of the recent crystal structures of the OBD and models of double hexamer assembly.
MATERIALS AND METHODS
Expression plasmids.
Plasmids to express the SV40 T-ag N130, OBD, and N260 protein fragments fused to a six-histidine tag and glutathione S-transferase (GST) were constructed by inserting BamHI-EcoRI-digested PCR products encoding these fragments between the BamHI and EcoRI sites of plasmid AB-401. AB-401 is a modified version of pGEX-4T-1 (GE Healthcare, formerly Amersham Biosciences) in which a sequence encoding a His tag has been inserted downstream of the GST-coding region. Primers used for amplification were designed so as to encode EcoRI and BamHI restriction sites. The following pairs of primers were used: SV40 T-ag OBD (aa 131 to 260), 5′-GGCTGGATCCGAAGGTAGAAGACCCCAAGG-3′ and 5′-GGGGAATTCACTATTCTGGATTAAAATCATGCTCC-3′; SV40 T-ag N260 (aa 1 to 260), 5′-GGCTGGATCCGATGGATAAAGTTTTAAACAGAGAG-3′ and 5′-GGGGAATTCACTATTCTGGATTAAAATCATGCTCC-3′; and SV40 T-ag N130 (aa 1 to 130), 5′-GGCTGGATCCGATGGATAAAGTTTTAAACAGAGAG-3′ and 5′-GGGAATTCCT ATCATCTCTTCTTTTTTGGAGGAG-3′. All DNA constructs were verified by sequencing.
Protein expression and purification.
Expression vectors for GST-N130, GST-OBD, and GST-N260 were transformed into Escherichia coli BL21 (Novagen). Bacterial cultures were grown in Luria-Bertani medium supplemented with 2% ethanol at 25°C to an optical density of 0.5 to 0.8 (at 595 nm) and then induced for 4 h by the addition of 0.75 mM isopropyl-β-d-thiogalactopyranoside. Cells were harvested and frozen at −80°C for later purification. For each protein, a thawed pellet from a 2-liter culture (for OBD and N130 proteins) or a 4-liter culture (for N260 proteins) was resuspended in 160 ml or 320 ml, respectively, of lysis buffer (50 mM Tris-HCl [pH 7.6], 1 M NaCl, 5 mM EDTA, 5 mM dithiothreitol [DTT], 10% glycerol, 0.1% NP-40, antipain [10 μg/ml], leupeptin [2 μg/ml], pepstatin [1 μg/ml], aprotinin [2 μg/ml], 1 mM phenylmethylsulfonyl fluoride), and the bacterial cells were mechanically disrupted using a French press (40K cell at 20,000 lb/in2). The resulting cell lysate was cleared by centrifugation at 30,000 × g for 30 min and incubated with 2 ml or 4 ml of glutathione-Sepharose 4B beads (Amersham Pharmacia Biotech) for 3 h at 4°C. Beads were washed with 50 bed volumes of HS buffer (50 mM Tris-HCl [pH 8.0], 0.5 M NaCl, 5 mM EDTA, 5 mM DTT, 10% glycerol, 0.1% NP-40) followed by 50 bed volumes of LS buffer (same as HS buffer but containing 0.2 M NaCl). Fusion proteins were cleaved with thrombin to remove the GST moiety. For thrombin cleavage, bead-bound GST fusion proteins were resuspended in 1 bed volume of cleavage buffer (25 mM Tris-HCl [pH 8.0], 0.2 M NaCl, 1 mM EDTA, 5 mM DTT, 2.5 mM CaCl2, 10% glycerol, 0.1% NP-40) and incubated for 16 h at 4°C with 5 units/liter of a culture of biotinylated thrombin (Novagen). The supernatant was then recovered, and thrombin was removed by incubation with 100 μl of streptavidin agarose beads (Novagen) for 30 min at 4°C. The soluble cleaved OBD and N130 were then recovered after centrifugation of the supernatant at 14,000 × g for 15 min at 4°C and stored at −80°C. Although the N130 and OBD protein were essentially pure after this step, N260 required further purification by ion-exchange chromatography. Cleaved N260 protein was diluted in binding buffer (20 mM Tris-HCl [pH 8.0]) to a final NaCl concentration of 10 mM. The diluted protein was incubated with 500 mg of dry DEAE cellulose (Whatman International Ltd.) per 5 mg of cleaved protein for 3 h at 4°C. The resin was washed with 20 bed volumes of binding buffer and the protein eluted with binding buffer containing 75 mM NaCl. The eluted protein was then concentrated on a nickel column, specifically by incubation with 1 ml of Ni-nitrilotriacetic acid agarose (QIAGEN) per 5 mg of eluted protein for 3 h at 4°C and washed with 20 bed volumes of buffer composed of 20 mM Tris-HCl [pH 8.0] and 100 mM NaCl, and the protein was eluted in the same buffer containing 350 mM EDTA. Purified proteins (OBD, N260, and N130) were dialyzed against buffer containing 100 mM NaCl, 20 mM Tris-HCl [pH 8.0], 1 mM DTT, 0.01% NP-40, and 10% glycerol. All protein concentrations were determined by Bradford analysis.
Full-length T-ag was expressed in Sf9 cells with a baculovirus expression vector and subsequently purified by immunoaffinity techniques employing the polyomavirus antibody 419 monoclonal antibody as previously described (2). Purified T-ag was dialyzed against T-ag storage buffer (20 mM Tris-HCl [pH 8.0], 50 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1 mM phenylmethylsulfonyl fluoride, 0.2 μg of leupeptin per ml, 0.2 μg of antipain per ml, and 10% glycerol) and stored at −80°C until use.
DLS.
Dynamic light scattering (DLS) was performed using a DynaPro instrument (Protein Solutions, Wyatt Technology Corporation). Purified proteins (N130, OBD, and N260) were analyzed in 20 mM HEPES-NaOH (pH 7.4), 50 mM NaCl, 1 mM DTT, and 0.01% NP-40 at concentrations of 10 μM. GST and buffer alone were used as a positive and as a negative control, respectively (data not shown). Scattering data were analyzed using the Dynamics software (Protein Solutions). The hydrodynamic radius values reported in this study were obtained using the regularization algorithm in Dynamics, which performs a multiple exponential fit of the raw data and provides a size distribution analysis for samples containing two or more populations. In this study, results were an average of 20 readings of each sample.
Cross-linking assays.
Increasing concentrations (1.25, 2.5, 5, and 10 μM) of purified N130, OBD, and N260 were incubated alone or together (N130 and N260 or OBD) in 20 mM HEPES-NaOH (pH 7.4), 50 mM NaCl, 1 mM DTT, 0.01% NP-40, and 0 to 2.5% glycerol in a total volume of 80 μl for 1 h at room temperature. Samples were then cross-linked with 0.1% glutaraldehyde (Sigma) for 10 min. Subsequently, the protein samples were mixed with 20 μl of 5× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer and boiled for 5 min. Cross-linked proteins were resolved on 10% (N260), 12% (OBD), or 15% (N130) SDS-PAGE gels and detected by silver staining or Western blotting with an anti-His tag antibody (8036, H3; Santa Cruz) as indicated. In these experiments, GST was used as a positive control (data not shown).
Fluorescence anisotropy DNA binding assay.
Binding assays were performed by use of 15 nM fluorescein-labeled probe and the indicated concentrations of protein in 150-μl reaction mixtures with the following buffer: 20 mM HEPES-NaOH (pH 7.4), 50 mM NaCl, 0.01% NP-40, 1 mM DTT, and 0 to 2.5% glycerol. Fluorescence readings were taken after an hour of incubation at room temperature in OptiPlate-96F high-binding-affinity black 96-well plates (Perkin Elmer) by use of a Wallac 1420 multilabel high-throughput screening counter (Victor3V) equipped with a 485-nm/535-nm filter set. The background fluorescence from buffer was subtracted, and anisotropy values were defined as follows: P = (I‖ − I⊥)/(I‖ + I⊥) and A = (I‖ − I⊥)/(I‖ + 2I⊥), where I‖ and I⊥ are the fluorescence intensities recorded in orientations parallel and perpendicular to the excitation polarizer, respectively.
Fluorescent DNA probes and competitor oligonucleotides.
Fluorescein-labeled oligonucleotides were purchased from Invitrogen or Genset (Paris, France), with the fluorophore attached at the 5′ end by a six-carbon linker. Duplex DNA probes were prepared by annealing each fluorescein-labeled oligonucleotide to a complementary oligonucleotide in a ratio of 1:1.5 by heating at 95°C for 5 min in DNA binding assay buffer (20 mM HEPES-NaOH [pH 7.4], 50 mM NaCl, 0.01% NP-40, and 1 mM DTT) and then slowly cooling to room temperature. Competitor oligonucleotides were purchased from Invitrogen and annealed as described above except in a ratio of 1:1.
KD determination.
Equilibrium dissociation constant (KD) values were obtained from direct binding isotherms, with each data point performed in triplicate, and fitted by nonlinear least-squares regression with the GraphPad Prism program to the standard equation describing the following tight binding equilibrium, D + P ↔ DP (where D is DNA, P is the T-ag protein fragment, and DP is DNA-T-ag protein complex), in solution as follows: ΔA = ΔAmax {[DT + PT + KD] − [(DT + PT + KD)2 − (4DTPT)]1/2}/(2DT), where ΔA is the measured anisotropy gain (Abound − Afree) at given concentrations of protein (PT; T = total) and fluorescent probe (DT), and ΔAmax is the anisotropy gain when all of the probe is bound. No computational corrections for emission intensity were performed, since the quantum yield did not change significantly upon binding of the OBD, N130, and N260 proteins.
IC50 determinations and calculation of apparent Ki.
The 50% inhibitory concentration (IC50) values for various DNA competitors were measured in the fluorescence polarization (FP) assay. Competitors were titrated in binding reactions to reach 100% inhibition (each concentration point was performed in triplicate) in reactions performed with 250 μM of protein and 15 nM of fluorescent probe. IC50s were determined by a nonlinear least-squares regression fit of the inhibition curve (sigmoidal dose-response curve) by use of the GraphPad Prism program. Apparent inhibitor affinity constant (Ki) values were then calculated by resolving the standard equation that portrays the equilibrium PI ↔ I + P + D ↔ DP (where I is the inhibitor) by solving the following cubic equation: ax3 + bx2 + cx + d, where x is [P], a is 1, b is KD + Ki + [I]T − [P]T, c is KiKD + Ki[D]T + KD[I]T − Ki[P]T − KD[P]T, and d is −KiKD[P]T when [I] is IC50. For each competitor, this equation was resolved for a given KD (determined from direct binding isotherms), evaluated as previously described.
RESULTS
Purification and characterization of N130, OBD, and N260 by DLS.
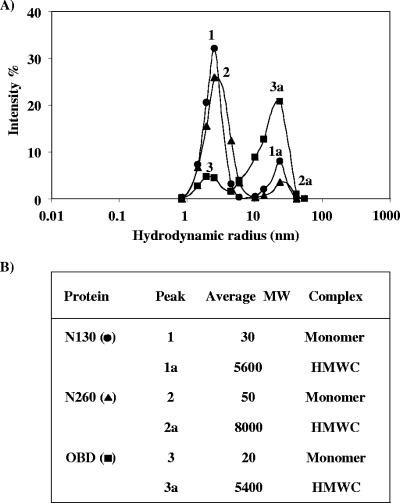
As a first step towards determining the contribution of the N-terminal domain of T-ag to the assembly of the protein at the viral origin, we expressed and purified from bacteria the N-terminal domain (N130; aa 1 to 130), the OBD (aa 131 to 260), or a protein fragment containing both the OBD and the N-terminal domain, N260 (aa 1 to 260). These proteins were produced as fusions with GST and a His tag, and the GST moiety was removed during purification by thrombin cleavage (Fig. 1C). As these proteins were produced in E. coli, they do not contain any posttranslational modification such as phosphorylation. Next, we investigated the oligomeric state of the purified OBD, N260, and N130 fragments by DLS. The observed profiles of N130 and N260, at a concentration of 10 μM, were characteristic of primarily monodisperse solutions with major peaks at hydrodynamic radii consistent with monomeric species (Fig. 2A and B). In contrast, the profile of the OBD, also at a concentration of 10 μM, showed a peak with a hydrodynamic radius most likely corresponding to much larger oligomers (high-molecular-weight complexes [HMWC]) (Fig. 2A and B). During the course of these experiments, we noted that the oligomeric state of the OBD is dependent on buffer composition; in particular, concentrations of glycerol above 5% reduce its multimerization (data not shown). However, these results suggested that the OBD, unlike N130 and N260, is capable of forming large oligomers under our assay conditions.
FIG. 2.
DLS analysis of N130, OBD, and N260. (A) The graph represents the scattering intensity (fraction of total scattering) as a function of the hydrodynamic radii obtained for the N130, OBD, and N260 proteins at a concentration of 10 μM. Each curve represents an average of 20 measurements. (B) Molecular weight (MW) and types of complexes estimated from each hydrodynamic radius.
Protein cross-linking assays.
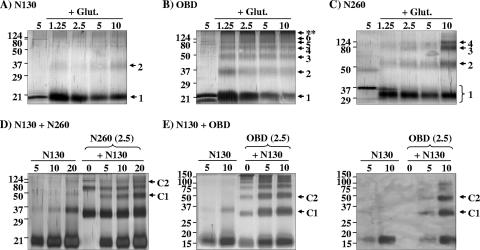
To further investigate the oligomeric state of the purified proteins, we performed a series of protein cross-linking assays. Increasing concentrations of N130, OBD, and N260 ranging from 1.25 to 10 μM were incubated in the presence or absence of 0.1% glutaraldehyde and analyzed by SDS-PAGE. GST, a known dimer, was used as a positive control (data not shown). As shown in Fig. 3A, N130 was primarily monomeric even at high concentrations. In contrast, the OBD was cross-linked primarily into dimers, trimers, and tetramers at low concentrations and into HMWC at higher ones (Fig. 3B). Similar results were obtained with an untagged OBD protein (data not shown). The analysis with N260 protein revealed that it could also assemble into dimers, trimers, and tetramers at high concentrations (Fig. 3C, lane 5) but never formed complexes as large as those generated with the OBD. These results were in agreement with those obtained by DLS. Collectively, they suggested that the N-terminal domain dampens the propensity of the OBD to form large complexes, perhaps, one may speculate, by obstructing an oligomerization interface on the OBD.
FIG. 3.
Glutaraldehyde cross-linking of N130, OBD, and N260. Cross-linking of N130 (A), OBD (B), and N260 (C) was performed using the indicated concentrations (in μM) of each protein. Samples were cross-linked with 0.1% glutaraldehyde (+ Glut.) for 10 min and analyzed by SDS-PAGE and silver staining. For each protein, a reaction performed in the absence of cross-linking agent was used as a control (left lane of each gel). Oligomeric forms of each protein are indicated by arrows on the right of the gels. HMWC formed by the OBD are indicated by asterisks (**). (D) Cross-linking of a mixture of N130 and N260. The three lanes at the left contain the products obtained by cross-linking of N130 at the indicated concentrations. The other lanes contain the products obtained by cross-linking N260, at a constant concentration of 2.5 μM, with increasing concentrations of N130 ranging from 0 to 20 μM. Complexes C1 and C2 correspond in size to heterodimers and heterotetramers, respectively, of N130 and N260. (E) Cross-linking of a mixture of N130 and OBD. Samples were analyzed by silver staining (left panel) and by Western blotting against the His-tagged N130 protein (right panel) with an anti-His tag antibody. The first two lanes contain the products obtained by cross-linking N130 alone at the indicated concentrations. The other lanes contain the products obtained by cross-linking the OBD, at a constant concentration of 2.5 μM, with increasing concentrations of N130 ranging from 0 to 10 μM. Complexes C1 and C2 correspond in size to heterodimers and heterotrimers, respectively, of N130 and OBD. Molecular weights in thousands are indicated to the left of the gels.
Interestingly, the cross-linking of N260 generated a species that migrated faster than the intact monomer (not cross-linked) and which was formed even at low protein concentrations (Fig. 3C; lanes 2 to 5 form the left of the gel). This faster-migrating species is possibly the result of intramolecular cross-linking events that would generate a more tightly packed protein, a phenomenon which has been reported for other proteins (14). Since no faster-migrating species were detected either with the OBD or with N130, it is possible that the one generated with N260 arises from intramolecular cross-linking between the OBD and the N-terminal domain. To investigate a possible interaction between the N-terminal domain and a region of the OBD, we asked whether N130 could be cross-linked to N260 or the OBD. This was found to be the case for N260, as determined by the appearance of new protein bands not detected when either protein was cross-linked alone. The presence of a novel band corresponding in size to a heterodimer of N130 and N260 was particularly striking (Fig. 3D, lanes 5 to 7). The same protocol could not be used to determine if N130 can be cross-linked to the OBD, since both proteins are of comparable molecular weights. To circumvent this problem, we used an untagged OBD and followed its cross-linking to increasing concentrations of His-tagged N130 by Western blotting with an anti-His tag antibody. These experiments reveled the formation of hetero-oligomers, in particular dimers and trimers, between the OBD and N130 (Fig. 3E right, lanes 4 and 5). Interestingly, the formation of these hetero-oligomers was accompanied by a concomitant decrease in the assembly of the OBD into HMWC, suggesting that N130 reduces in trans oligomerization of the OBD (Fig. 3E left, lanes 3 to 5). These results provided further evidence that the N-terminal domain can associate with the OBD.
DNA binding properties of purified N130, OBD, and N260.
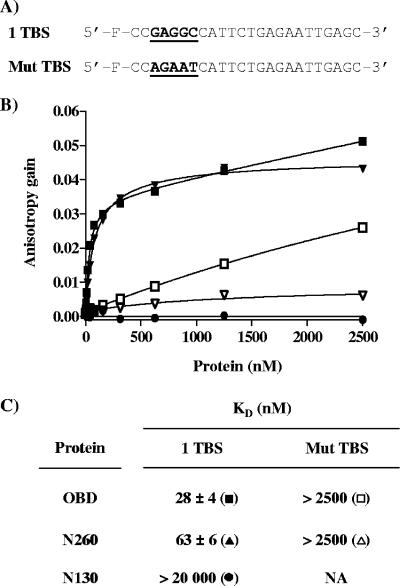
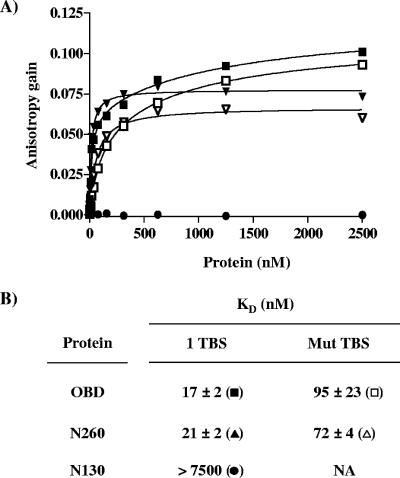
The DNA binding properties of these purified proteins were then characterized using an assay based on FP (17, 29, 59). In comparison to other common methods to monitor DNA binding, such as electrophoretic mobility shift assays, FP assays offer the advantages of being performed in solution and at equilibrium and hence are ideally suited for affinity measurements (17, 29). FP DNA binding assays were used to determine the affinity of the three proteins for a duplex oligonucleotide probe containing one pentanucleotide (TBS; 5′-GAGGC-3′). As a control for specificity, a similar probe in which the TBS was inactivated by five mutations (5′-AGAAT-3′) was used (Fig. 4A). Throughout this study, we will when necessary refer to the TBS probe as “specific DNA” and to the mutant probe as “nonspecific DNA.” Titration of the purified OBD and N260 proteins resulted in a dose-dependent increase in polarization (expressed as an anisotropy gain) with the TBS probe (Fig. 4B), indicative of their binding to DNA. In contrast, titration of purified N130 had no effect on polarization, demonstrating that this protein does not bind significantly to DNA, even at concentrations as high as 20 μM (Fig. 4B and data not shown). From a series of binding isotherms similar to those shown in Fig. 4B, KD values of 28 ± 4 nM and 63 ± 6 nM were obtained for the binding of N260 and OBD proteins to the TBS-containing probe (Fig. 4C), respectively. Binding of these two proteins to a control probe lacking a TBS was much weaker, confirming that both bind DNA in a sequence-specific manner. Interestingly, a significant increase in anisotropy was reproducibly observed at high concentrations of the OBD (>500 nM) (Fig. 4B), an observation which we made with several nonspecific probes of differing sequences (data not shown). This effect is likely due to nonspecific binding of the OBD to DNA and/or its ability to oligomerize at high concentrations (>500 nM), as determined above by DLS and cross-linking studies. This property of the OBD is also likely the reason why its binding to the TBS-containing probe did not reach a plateau at high protein concentrations. To rule out a possible effect of the His tag, we performed similar experiments with an untagged OBD and confirmed that it has affinities similar to those of the tagged protein for the one TBS probe (KD values of 24 ± 9 nM) and for nonspecific DNA (data not shown). In contrast to the OBD, N260 showed little binding to the control probe, and its binding to the TBS-containing probe was clearly saturable. Collectively, these results indicate that the N260 and OBD proteins bind to a single TBS with comparable affinities, which diverge by less than threefold. However, their binding to nonspecific DNA appears to be more dissimilar. Namely, the significant gain in anisotropy observed at high concentrations of the OBD suggests that more molecules of the OBD are bound on the nonspecific DNA probe than for N260.
FIG. 4.
Binding of the N130, OBD, and N260 proteins to DNA measured by fluorescence anisotropy. (A) Probes used in this study. Nucleotide sequences (underlined and in boldface) of the probe containing a single TBS (1 TBS) and of the control nonspecific probe containing a mutated TBS (Mut TBS). (B) Binding isotherms were performed either with the TBS probe (filled symbols) or with the control probe (open symbols) and increasing concentrations of the OBD (squares), N260 (triangles), and N130 (circles) proteins. For each isotherm, 15 nM of duplex DNA probe was incubated with increasing concentrations of N130, OBD, or N260 protein in triplicate. Error bars are not visible on the graph, as they are smaller than the symbols. (C) KD values for the N130, OBD, and N260 proteins for the specific and nonspecific probe were calculated from the data by nonlinear regression and fitting to a one-binding-site equilibrium. The values and standard deviations reported were calculated from over 20 independent binding isotherms, with each data point performed in triplicate.
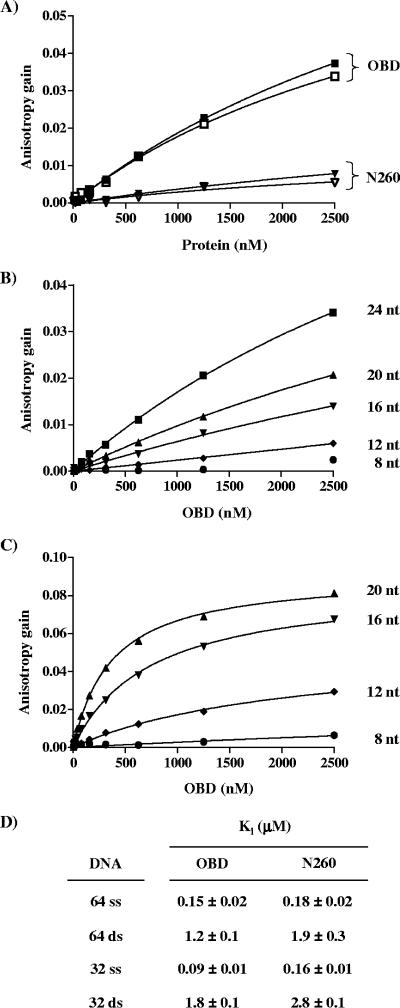
We then investigated the impact of the length of the probe on the binding of the N260 and OBD proteins to a single TBS. These studies showed that both proteins could not bind to a probe containing 8 bp (Table 1) but could do so on a 12-bp probe. However, maximum binding affinity was reached only with probes greater than 16 bp (Table 1). These observations are in agreement with the crystal structure reported by Bochkareva et al. (4) as well as Meinke et al. (37), which demonstrated that the OBD spans approximately 10 bp when bound to dsDNA. Moreover, the fact that the same length of DNA is needed for binding of the OBD and the N260 protein supports the idea that the N-terminal domain of the protein does not significantly interact with DNA.
TABLE 1.
Effect of probe length and ionic strength on the DNA binding affinities of the OBD and N260 proteinsa
| Probe or buffer constituent | Probe length or ionic strength |
KD (nM) for:
|
|
|---|---|---|---|
| OBD | N260 | ||
| 5′-F-CCGAGGCC-3′ | 8 bp | >2,500 | >2,500 |
| 5′-F-CCGAGGCCATTC-3′ | 12 bp | 41 ± 3 | 121 ± 5 |
| 5′-F-CCGAGGCCATTCTGAG-3′ | 16 bp | 26 ± 2 | 60 ± 3 |
| 5′-F-CCGAGGCCATTCTGAGAATT-3′ | 20 bp | 21 ± 2 | 52 ± 3 |
| 5′-F-CCGAGGCCATTCTGAGAATTGAGC-3′ | 24 bp | 29 ± 2 | 61 ± 3 |
| NaCl | 50 nM | 22 ± 2 | 46 ± 2 |
| 100 nM | 269 ± 39 | 828 ± 99 | |
| 150 nM | >2,500 | >2,500 | |
| 200 nM | >2,500 | >2,500 | |
The KD values for the OBD and N260 proteins for the indicated probes (with TBS underlined and in boldface) at 50 mM NaCl (top) and for a 24-bp probe at the indicated concentrations of salt (NaCl; bottom) were calculated from binding isotherms performed as described for Fig. 4. Standard deviations were calculated from two independent binding isotherms, with each data point performed in triplicate.
Influence of pH and ionic strength on the binding of the OBD and N260 to DNA.
The binding of polyomavirus T-ag to origin DNA has been shown to increase at lower pH (43). We used our FP assay to determine if acidity also increases the affinities of the SV40 T-ag OBD and N260 proteins for specific as well as nonspecific DNA. Assays were performed as before, except that the final pH of the binding reactions was 6.8 rather than 7.4. This lower pH increased binding of the OBD and N260 to a single TBS-containing probe only slightly, by approximately twofold (Fig. 5A and B). In contrast, binding to a nonspecific DNA probe (lacking a TBS) was drastically increased by 25- to 40-fold. We also investigated the effect of increasing the ionic strength on the affinities of the proteins for specific DNA. As expected, increasing the NaCl concentration reduced the affinities of the OBD and N260 proteins for the TBS-containing probe (Table 1). Specifically, 100 mM NaCl reduced DNA binding affinity 10- to 20-fold, and a further increase to 150 mM almost completely abolished DNA binding of both proteins to the probe (Table 1). These effects of pH and salt are consistent with the recent crystallographic data indicating that the interaction of the OBD with DNA involves specific base contacts through hydrogen bonds and salt bridges as well as extensive nonspecific contacts with phosphate groups of the DNA. Unless mentioned otherwise, all of the binding reactions shown in this work were performed as described for Fig. 4, i.e., using 50 mM NaCl at a pH of 7.4.
FIG. 5.
Effect of pH and ionic strength on the binding of the OBD and N260 proteins to DNA. (A) Effect of pH. Binding isotherms were performed as described for Fig. 4 but at the lower pH of 6.8. Binding isotherms were performed either with the TBS probe (filled symbols) or with the control probe (open symbols) and increasing concentrations of the OBD (squares), N260 (triangles), and N130 (circles) proteins. (B) KD values for the OBD and N260 for each probe at pH 6.8. Standard deviations were calculated from two independent binding isotherms, with each data point done in triplicate.
Binding affinities of the OBD and N260 for individual pentanucleotides in site II.
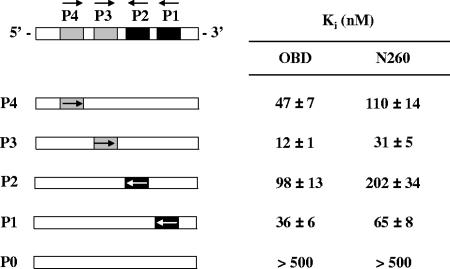
Site II of the SV40 origin contains four pentanucleotides (P1 to P4) needed for the assembly of the protein into a double hexamer capable of supporting viral genome replication (Fig. 1A) (13). To determine if the N-terminal domain of T-ag affects binding of the OBD to any one of these pentanucleotides, we measured and compared the affinities of the OBD and N260 proteins for each of these four sites. Affinity measurements were made with the FP assay described above but using unmodified duplex DNAs carrying one of the four pentanucleotides as competitors. Because we wanted to determine the affinity for each binding site in the context of the origin, we used competitor versions of the 31-bp site II in which three of the four pentanucleotides were inactivated by mutations (Fig. 6). A version of site II with all TBS mutated served as a negative control. For each competitor DNA, increasing amounts were used to inhibit the binding of the OBD and N260 to a TBS-containing probe in order to determine an IC50, from which an apparent Ki was derived (see Materials and Methods) (Fig. 6). Three observations were made. First, we confirmed that the binding affinities of the OBD and N260 proteins for P3 measured in these competition experiments (Ki values) were comparable, within twofold, to those obtained in direct binding experiments to fluorescent probes (KD values; Fig. 4), as expected if the fluorescein moiety on the probe does not impair binding. Second, we observed that the affinities of N260 for each of the four pentanucleotides were consistently two- to threefold lower than those measured for the OBD (Fig. 6), consistent with the data from Fig. 4C. Third, we reproducibly found that both N260 and the OBD bind with low affinities to P2 and to a lesser extent to P4, compared to what was seen for P1 and P3, despite the fact that all four sites have identical sequences. This last finding provides a rational explanation for why double hexamers have been found to preferentially assemble on sites 1 and 3 rather than 2 and 4 (22).
FIG. 6.
Affinities of the OBD and N260 proteins for each individual pentanucleotide in site II. Schematic representations of site II and competitor DNAs are shown on the left. All competitors contain only one of four pentanucleotides (P1 to P4; 5′-GAGGC-3′). P0 is a competitor in which all four pentanucleotides were mutated to 5′-AGAAT-3′. Arrows represent the direction of the 5′-GAGGC-3′ sequence. Apparent Ki values measured for each competitor DNA are indicated on the right. Ki and standard deviations were calculated from the IC50 of each competitor DNA obtained in triplicate in assays performed at 250 nM of protein and 15 nM of one TBS probe.
Determination of a consensus T-ag binding sequence.
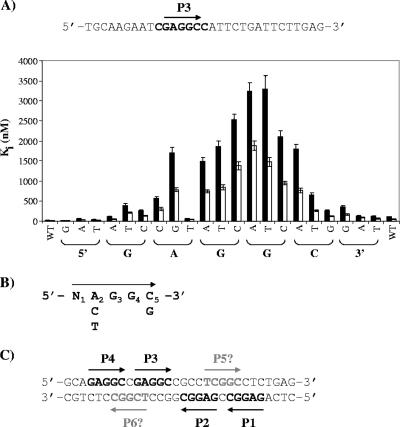
The observation that P1 and P3 are bound with affinities differing from those for P2 and P4, despite having the same sequence, prompted us to investigate if the nucleotides immediately flanking each 5′-GAGGC-3′ site could also contribute to affinity (Fig. 7A). To test this, we performed a systematic mutagenesis of the two nucleotides immediately upstream and downstream of P3, respectively. In addition, we also mutated each nucleotide of the 5′-GAGGC-3′ sequence in order to determine the contribution of each to affinity and generate a consensus T-ag binding sequence, a study which has never been done systematically and quantitatively. We performed this systematic analysis on P3 in the context of a site II oligonucleotide in which only P3 had not been inactivated (Fig. 7A). The different mutated duplex DNAs were used in competition experiments, and their Ki values against the OBD and N260 were determined (Fig. 7A). This study revealed that (i) the nucleotide immediately 5′ of P3 (C) and the first position of the pentanucleotide (G) can be changed to any of the three other nucleotides without a drastic effect on binding affinity, (ii) the second position (A) can be changed to a cytosine or thymine but not to a guanine, (iii) the third and fourth positions (GG) cannot be substituted by any of the other three nucleotides without a drastic loss of affinity, (iv) the fifth position (C) cannot accommodate an adenine or a thymine, and (v) the nucleotide immediately 3′ of the pentanucleotide can be changed to an adenine or a thymine but a guanine at this position reduces binding. Based on this last finding, we infer that the presence of a guanine immediately 3′ of P2, rather than a cytosine for all three other pentanucleotides, is likely the reason why this site is bound with the lowest affinity (Fig. 6). Collectively, these results indicate that the preferred binding site for the OBD is 5′-GAGGC-3′, followed by A, C, and T. However, significant binding can be observed on the following consensus sequence: 5′-N(ACT)GG(CG)-3′ (Fig. 7B). Interestingly, inspection of the sequence of site II revealed the presence of two additional TBS in addition to the four already known pentanucleotides (Fig. 7C). Determining whether these two potential TBSs are of any biological significance will necessitate further experimentation.
FIG. 7.
Systematic mutagenesis of P3. (A) Nucleotide sequence of the oligonucleotide containing P3 (indicated by an arrow). Nucleotides shown in bold were systematically changed for all three other nucleotides. The apparent Ki values associated with each change are shown in the bar graph. Apparent Ki and standard deviations were calculated from the IC50 of each competitor DNA obtained in triplicate in assays performed at 250 nM of N260 (filled bars) or OBD (open bars) and 15 nM of one TBS probe. (B) Consensus binding site determined from the data in panel A. (C) Location of two putative novel TBSs (P5 and P6) in site II suggested by the consensus sequence obtained as described above.
Binding affinities of the OBD and N260 for pairs of pentanucleotides.
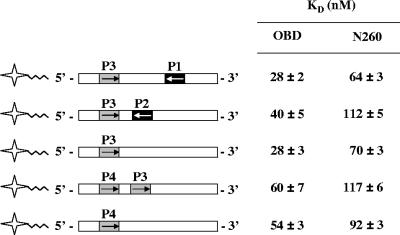
We previously reported that the OBD does not bind to a palindromic combination of two pentanucleotides, either P1 and P3 or P2 and P4, with increased affinity (59). These results led us to suggest that monomers of the OBD bind to these sites independently rather than cooperatively, a suggestion that was recently supported by the two crystal structures of the OBD bound to DNA (4, 37). To verify if the same was true for N260 and extend these studies to other combination of sites, we measured the affinities of N260 and the OBD for the three different types of combinations of pentanucleotides that can be found within site II. These combinations correspond to inverted repeats with a spacing of 7 bp (such as the combination of sites P3 and P1), inverted repeats with a spacing of 1 bp (such as sites P3 and P2), and direct repeats with a spacing of 1 bp (such as sites P4 and P3) (Fig. 8). As can be seen in Fig. 8, the presence of P1 or P2 did not enhance the affinity (KD) of the OBD or N260 for P3. Similarly, binding to P4 was not significantly changed by the presence of P3 downstream of it. These results strongly suggest that N260, like the OBD, binds independently to each of the four pentanucleotides within site II.
FIG. 8.
Affinities of the OBD and N260 proteins for pairs of pentanucleotides. Schematic representations of the 24-bp probes used in this analysis are shown on the left. KD values for the OBD and N260 were obtained from the titration of the given probe with each of the proteins and are reported on the right. Binding reactions were performed as described for Fig. 4. Standard deviations were calculated from two independent binding isotherms, with each data point done in triplicate.
Binding of the OBD and N260 to ssDNA.
It was shown recently by NMR that the OBD can bind to poly(dT)25 ssDNA (45). We used FP to measure the affinity of the OBD and N260 proteins for different ssDNA substrates and assess the role of the N-terminal domain on this interaction. We first measured the affinities of the proteins for two 24-nucleotide-long ssDNA probes, one containing and the other lacking a 5′-GAGGC-3′ sequence (these probes correspond to the fluorescein-labeled strands shown in Fig. 4A). Titration of increasing concentrations of proteins resulted in a dose-dependent gain in anisotropy that was much larger for the OBD than for N260 and that was independent of the presence of the 5′-GAGGC-3′ sequence. These results indicated that the OBD does bind to ssDNA in a nonspecific manner (Fig. 9A). Furthermore, the larger gain in anisotropy observed for the OBD suggests that at the highest concentrations tested, more OBD proteins are bound to the probe than N260. When N130 was incubated with the same probe, no binding was detected (data not shown), indicating that the N-terminal domain of T-ag does not bind to ssDNA.
FIG. 9.
Binding affinity of wild-type OBD and N260 for ssDNA. (A) Binding isotherms obtained with ssDNA probes either containing (filled symbols) or lacking (open symbols) a 5′-GAGGC-3′ sequence and with the indicated protein. (B and C) Binding isotherms generated from the binding of the OBD to ssDNA probes of the indicated length (in nucleotides [nt]) at pH 7.4 (B) or pH 6.8 (C). (D) Apparent Ki values for single-stranded and double-stranded nonspecific DNA competitors of the indicated lengths against the OBD and N260 proteins. Ki and standard deviations were calculated from the IC50 of each competitor DNA obtained in triplicate with assays performed with 250 nM of protein and 15 nM of one TBS probe.
We then characterized further the binding of the OBD and N260 to ssDNA. We found that the magnitude of the anisotropy gain was a function of the probe length, as expected if the OBD and N260 bind to ssDNA in a nonspecific manner (Fig. 9B and data not shown). We also found that binding to ssDNA of increasing length was increased at lower pH, similar to what we observed for duplex DNA (Fig. 5B and data not shown). Even at this lower pH, binding to a probe of 8 or 12 nucleotides was weaker than on longer probes of 16 and 20 nucleotides. The affinities of the OBD for the 16- and 20-nucleotide-long probes at pH 6.8 were measured to be 741 and 370 nM, respectively. These results with ssDNA probes were reminiscent of those obtained with nonspecific duplex DNA (compare Fig. 5 and 9C), suggesting that the OBD might use similar surfaces to interact with ss- and dsDNA. If so, we would expect that binding to both types of nucleic acids might be mutually exclusive. We tested this possibility by investigating if binding of N260 and the OBD could be competed by ssDNA. As can be seen in Fig. 9D, binding to a duplex DNA probe carrying a single TBS could be efficiently inhibited by ssDNA. These results indicate that N260 and the OBD bind to ss- and dsDNA in a mutually exclusive manner. Remarkably, both proteins showed an affinity for ssDNA 10-fold greater than that for nonspecific dsDNA. These results have important implications when the mechanism by which T-ag transitions from a sequence-specific DNA binding protein to a nonspecific helicase is considered (see Discussion).
DNA binding affinities of mutant OBD and N260 proteins.
Several amino acid substitutions in T-ag have been identified that specifically affect its binding to the origin or its phosphorylation-dependent assembly into functional double hexamers. The availability of a sensitive FP assay that monitors binding at equilibrium prompted us to reexamine the effect of some of these substitutions on the binding of the OBD and N260 proteins to a single TBS. We focused our attention on substitutions that do not lie directly in the DNA binding surface but rather on those in other regions of the OBD or in the N-terminal domain and for which the mechanism of action is not completely understood. The locations of these amino acid substitutions in the structure of the OBD (37) are shown in Fig. 1D, and their affinities for a single TBS-containing probe are described below and reported in Table 2.
TABLE 2.
DNA binding affinities of mutant OBD and N260 proteinsa
| Amino acid substitution |
KD for one TBS (nM) for:
|
KD ratio | |
|---|---|---|---|
| OBD | N260 | ||
| None (wild type) | 28 ± 4 | 63 ± 6 | 2.7 |
| Phosphorylation site substitutions | |||
| S120D/S123D | NA | 83 ± 13 | NA |
| T124A | NA | 78 ± 9 | NA |
| T124D | NA | 59 ± 10 | NA |
| T124E | NA | 54 ± 12 | NA |
| Class 4 substitutions | |||
| H148N | 38 ± 10 | 105 ± 4 | 2.7 |
| K167R | 30 ± 5 | 92 ± 16 | 2.7 |
| Q213H | 28 ± 2 | 59 ± 6 | 2.2 |
| L215V | 24 ± 3 | 63 ± 11 | 2.6 |
| F220Y | 25 ± 6 | 61 ± 4 | 2.4 |
| Oligomerization substitutions | |||
| F151A | 23 ± 6 | 56 ± 17 | 2.4 |
| F151Y | 25 ± 5 | 65 ± 10 | 2.7 |
| F183A | 44 ± 10 | 43 ± 1 | 1.0 |
| F151A/F183A | 28 ± 4 | 46 ± 2 | 1.6 |
KD values and standard deviations were obtained from at least three independent binding isotherms as described in Fig. 4 and with each data point done in triplicate. For each amino acid substitution, the ratio of the KD value of N260 over that of the corresponding OBD is indicated on the right.
Substitutions of serines 120 and 123.
Previous studies have found that dephosphorylation of Ser120 and Ser123 by protein phosphatase 2A increases the affinity of T-ag for the origin and stimulates its assembly into double hexamers (3, 27, 39, 49, 50, 60, 61). Accordingly, phosphorylation of S120 and S123 by casein kinase I was found to inhibit viral DNA replication (8). Since our FP assays are performed with recombinant proteins made in E. coli and hence not phosphorylated, we investigated if the double amino acid substitutions of S120 and S123 for aspartic acids (S120D/S123D) as phosphomimetics would have an effect on binding affinity. Under our assay conditions, the S120D/S123D substitutions reduced only slightly the affinity of N260 for the one TBS probe (Table 2), indicating that negative charges at positions 120 and 123 are not sufficient to reduce the interaction of the OBD with its target site.
Substitutions of threonine 124 and class 4 substitutions.
Phosphorylation of Thr124 by a cyclin/cdk was previously shown to be essential for viral DNA replication (15, 32, 33, 35, 38, 48) and to affect double hexamer assembly in vitro (2, 46, 64) (see the introduction). Similarly, specific amino acid substitutions in the OBD, known as class 4 mutations, have been identified that interfere with double hexamer assembly but without affecting phosphorylation of the protein (64). An interpretation of these findings is that the N-terminal domain and the OBD cooperate during the double hexamer assembly in a phosphorylation-dependent manner. We produced and purified N260 mutant proteins carrying substitutions of T124 for either an alanine (T124A) or for aspartic or glutamic acid as phosphomimetics (T124D and T124E). We also produced N260 and OBD mutant proteins carrying one of five different class 4 substitutions: H148N, K167R, Q213H, L215V, and F220Y (described in reference 67). In the FP assay, all of the T124 substitutions had little effect on the affinity of N260 and the OBD for specific DNA (Table 2). Similar results were obtained for class 4 substitutions. The two substitutions with the greatest effects were H148N and K167R, which reproducibly decreased binding but by less than twofold (Table 2). Importantly, we also observed very little effect of T124 and class 4 substitutions on the binding of the OBD or N260 to pairs of binding sites, as was done for the wild-type proteins as shown in Fig. 8 (data not shown). From these studies, we conclude that T124 and class 4 mutations do not affect binding to the origin but rather affect a later step in double hexamer assembly. It is also noteworthy that a mutant T-ag carrying the T124E substitution was shown to be replication defective by an unknown mechanism (50). Our data suggest that this defect is not related to origin binding.
Substitutions in a putative OBD-OBD interface.
A recent crystal structure of the OBD has suggested that it can assemble into a left-handed spiral through two complementary binding surfaces (36). To determine the importance of these interaction surfaces for DNA binding, we produced mutant proteins carrying substitutions predicted to disrupt spiral formation. Specifically, phenylalanines 151 and 183 (F151, F183) of the OBD and N260 proteins were each mutated to alanine or tyrosine to maintain the aromatic character of the residue. We also characterized the effect of combining both substitutions, F151A/F183A. In the FP assay, these substitutions reduced by less than half the affinities of N260 or the OBD for the TBS probe. Throughout this study, we found that N260 binds to DNA with an affinity two- to threefold lower than that of the OBD and that nearly all of substitutions failed to alter this difference (Table 2). The only exception we observed was for F183A, which, either alone or in combination with F151A, appears to reduce this affinity difference between N260 and the OBD. Furthermore, the KD values suggest that it does so primarily by increasing the affinity of N260 for DNA, albeit very slightly. For all three substitutions tested, we also observed very little effect on the binding of the OBD or N260 to pairs of binding sites (data not shown). The fact that these substitutions in a putative OBD-OBD interface have little effect in our assays is consistent with the idea that spiral formation occurs at a step subsequent to origin binding.
DNA binding affinities of full-length T-ag.
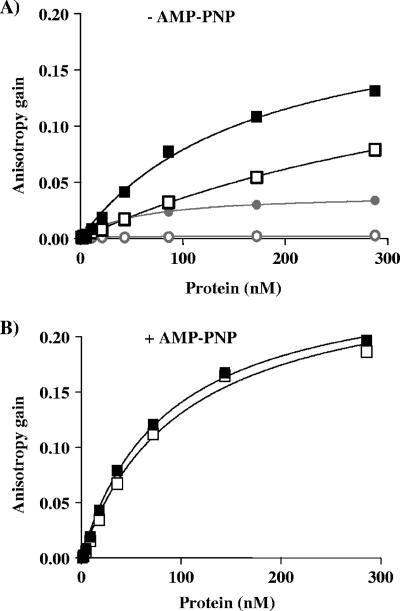
We purposely performed all of the studies presented above with fragments of T-ag lacking the C-terminal helicase domain in order to investigate only the DNA binding characteristics of the OBD and avoid any possible confounding effect from the nonspecific interaction of the helicase domain with DNA (20, 65). Nevertheless, we still wished to compare the binding specificity of N260 to that of the complete T-ag protein. We therefore characterized the binding of full-length T-ag, produced in insect cells by use of a baculovirus system, in our FP assay. Specifically, we measured its binding to a 24-bp probe containing a single TBS and to a control probe lacking a specific binding site. These studies were performed in the presence or absence of 5′-adenylyl-β, γ-imidodiphosphate (AMP-PNP), which promotes oligomerization of T-ag. In the absence of AMP-PNP, increasing concentrations of T-ag generated a dose-dependent gain in anisotropy (Fig. 10A) that was larger than the one generated by N260. This large anisotropy gain is likely due to T-ag being a bigger protein than N260. Interestingly, T-ag also bound substantially to the nonspecific probe much more than N260. It is entirely possible that this nonspecific binding occurs through the helicase domain of the protein. Regardless, these findings indicate that the specificity of full-length T-ag, as defined by its ability to discriminate between specific and nonspecific DNA, is less than that of N260. KD values for the binding of T-ag to the specific and nonspecific probes were estimated to be 170 ± 22 nM and 494 ± 38 nM, respectively. These values contrast with those obtained with N260 under the same conditions, which were 63 ± 6 nM and >2,500 nM, respectively (Fig. 4C). The addition of AMP-PNP increased the binding of T-ag to both specific and nonspecific DNA (Fig. 10B), to the point of becoming too tight to be accurately measured under these assay conditions. AMP-PNP also gave rise to a greater gain in anisotropy, as would be expected if multimers (for example, hexamers) of T-ag are binding to the probe. Regardless of the precise stoichiometry, the important point from the above-described studies is that full-length T-ag has a substantial affinity for nonspecific DNA, in contrast to N260. This raises the question of how full-length T-ag finds the origin in vivo in the presence of a vast excess of competitor genomic DNA. Perhaps there exist mechanisms to reduce the nonspecific binding of the protein in vivo (see Discussion).
FIG. 10.
Binding of full-length T-ag to DNA measured by fluorescence anisotropy. Binding isotherms of full-length T-ag (squares) by use of a 24-bp probe containing (filled symbols) or lacking (open symbols) a single TBS and performed in the absence (A) or presence (B) of 4 mM AMP-PNP and 7 mM MgCl2. In panel A, binding isotherms generated with the N260 protein (circles) under the same conditions are shown in gray for comparison. Binding isotherms were performed in duplicate.
DISCUSSION
In this study, we have measured the affinity of wild-type and mutant versions of the T-ag OBD and of a larger N-terminal fragment, N260, for different DNA substrates by fluorescence anisotropy as well as determined the oligomeric states of these proteins in solution by DLS and cross-linking assays. These biophysical studies have provided a quantitative basis onto which to anchor our understanding of the interaction of T-ag with the origin and suggested a role for the N-terminal domain of the protein in regulating the oligomerization of the OBD.
Effect of the N-terminal domain on the OBD.
Earlier studies suggested that the N-terminal region of T-ag modulates the interaction and/or assembly of the protein at the origin (see the introduction). Our characterization of purified N130 indicated that this T-ag fragment is primarily monomeric in solution and does not bind with any significant affinity to ds- or ssDNA. In addition, we found that N260 had affinities comparable to those of the OBD for all DNA substrates tested, which consistently differ by less than threefold. Hence, we conclude that the N-terminal domain has only a modest effect on the affinity of the OBD for specific and nonspecific ss- and dsDNA and thus that its role in double hexamer assembly must occur at a step subsequent to origin binding and/or be dependent on the C-terminal helicase domain, which is missing from our protein fragments (see below).
Interestingly, preparations of N260 were more monodisperse than those of the OBD in DLS experiments, suggesting that the N-terminal domain reduces the multimerization of the OBD under our assay conditions. This was also observed in glutaraldehyde cross-linking assays, which showed that the OBD, unlike N260, could be cross-linked into HMWC. We believe that the tendency of the OBD to self-associate under our assay conditions accounts for its ability to generate larger anisotropy gains than N260 upon binding to nonspecific ds- and ssDNA in FP assays. We cannot rule out that the N-terminal domain stabilizes the OBD and prevents its aggregation in solution. However, we note that at low concentrations (<5 μM), the OBD is mostly monodisperse and monomeric, as assessed by DLS and cross-linking, respectively (data not shown), arguing against it being aggregated at the concentrations used in our assays. A recent crystal structure has suggested that the OBD can oligomerize into a left-handed spiral (36). Whether the multimerization of the OBD that we observe in solution or on DNA is related to spiral formation is unknown. If it were, it would raise the possibility that the N-terminal domain regulates spiral formation by masking an oligomerization interface on the OBD. Along these lines, we have found that glutaraldehyde cross-linking of N260 generates a faster-migrating species on SDS-PAGE gels, which we interpreted as a more compact form of the protein. This phenomenon was not observed with the OBD and N130, suggesting that it was due to intermolecular cross-linking of the N-terminal domain against the OBD. In support of this notion, we found that N130 could be cross-linked to either N260 or the OBD. These results indicate that the N-terminal domain can interact in trans with another T-ag molecule, albeit at high concentrations, as would be the case for two T-ag molecules bound next to each other on DNA. Collectively, our findings support the notion that the N-terminal domain of T-ag regulates oligomerization of the OBD, in agreement with the finding by Weisshart et al. (62) that truncation of the first 82 aa from full-length T-ag generates a protein that forms aberrant multimers in solution and on DNA.
Interaction of the OBD and N260 with their target pentanucleotide binding site.
Our systematic and quantitative analysis of the importance of each nucleotide of the 5′-GAGGC-3′ sequence led to the determination of the following consensus TBS: 5′-N(ACT)GG(CG)-3′. This consensus highlights the importance of the GGC triplet for high-affinity binding, in agreement with two recent crystal structures of the OBD bound to DNA showing that 70% of the protein-DNA contacts occur through these three nucleotides (4, 37). Our analysis also indicated that the nature of the first residues has little effect on affinity. This is in agreement with previous in vivo studies which showed that mutation of this residue from a G to an A reduced viral DNA replication by 5- to 10-fold, whereas mutations at positions 3, 4, and 5 completely abolished it (11). These findings are in good agreement with our in vitro data showing that changing position 1 from a G to an A reduces binding by approximately 5-fold, whereas mutations at positions 3, 4, and 5 lower affinity by 50- to 200-fold. These data also suggest that a relatively small reduction in DNA binding affinity (fivefold) is sufficient to impair DNA replication in vivo. Our binding analysis also showed that position 2 can be mutated without a drastic loss in affinity, although not to a guanine, whose amide group may sterically exclude a water molecule found in the structure of Bochkareva et al. (4) from hydrogen bonding with both the OBD and DNA. Our analysis also suggested that T-ag could interact, albeit more weakly, with two noncanonical sequences, both 5′-TCGGC-3′, present in the origin (Fig. 7C). Bochkareva et al. (4) also provided evidence for a noncanonical binding site in the origin, in their case 5′-ATGGC-3′. Whether these noncanonical sequences are of any functional significance will require further experimentation. However, we note that mutation of these two sites independently to 5′-TCAGC-3′ (where underlining indicates the mutation) was previously shown to reduce viral DNA replication in vivo by 40%, whereas an identical substitution in P4 (to 5′-GAAGC-3′) completely abolished it (11). These results point to these noncanonical sites being of an importance lesser than that of P1 to P4. We also observed that the affinity of the OBD for its target 5′-GAGGC-3′ sequence is influenced to some extent by the nucleotide located immediately downstream of it. As a result, the OBD has a higher affinity for P1 and P3 than for P2 and P4 in site II, a finding that may explain in part why double hexamers assemble preferentially on P1 and P3. Mutagenesis of the residue downstream of P3 indicated that a G, as found in P2, is deleterious to binding. Satisfyingly, the structure of Meinke et al. (37) revealed that stabilizing van der Waals interactions occur between the OBD and the nucleotide complementary to that located 3′ of the 5′-GAGGC-3′ sequence. Finally, we found that the OBD requires additional sequences on each side of the TBS for high-affinity binding and that its interaction with specific and nonspecific DNA is dramatically increased at lower pH and reduced at higher ionic strength. These results are consistent with both crystal structures (4, 37), which showed that the protein spans not only the 5′-GAGGC-3′ sequence but also the upstream two and the downstream three nucleotides and that the interaction of the OBD with DNA is stabilized through a number of hydrogen bonds with the phosphate backbone (4, 37). Thus, our analysis has provided experimental evidence for many of the OBD-DNA interactions observed in the crystal structures.
N260 and the OBD bind independently to pairs of pentanucleotide binding sites.
We previously reported that the OBD binds to palindromic pairs of TBS, either P1/P3 or P2/P4, independently, suggesting that OBD molecules bound at these sites do not interact significantly with each other (59). This was recently confirmed by crystal structures of the OBD bound to P1/P3 or to all four TBSs (4, 37). In this study, we have obtained similar results for N260 and for other pairs of binding sites, namely, P2/P3 and P3/P4. We also obtained similar results with mutant proteins carrying amino acid substitutions known or suspected to impair protein-protein interactions important for double hexamer assembly, including changes at Ser120, Ser123, and Thr124, as well as substitutions in the OBD found in class 4 mutants or predicted to disrupt spiral assembly. Since none of them had any deleterious consequence, we surmise that they affect later steps of double hexamer assembly. These results are consistent with those of others indicating that Thr124 and class 4 substitutions, in the context of full-length T-ag, do not affect origin binding and hexamer assembly but rather specifically affect the formation of a double hexamer (2, 64).
Binding of N260 and the OBD to ssDNA.
Remarkably, we found that N260 and the OBD bound to ssDNA in a sequence-independent manner with a substantial affinity of 100 to 200 nM (Fig. 9D), approximately 10-fold more tightly than to nonspecific dsDNA (∼1 to 2 μM; Fig. 9D) and only 5-fold or less more weakly than to any of the four pentanucleotides in site II (Fig. 6). We also found that binding to ssDNA was mutually exclusive with binding to a specific TBS, in agreement with recent NMR data indicating that many of the same A1 and B2 residues are used for binding ss- and dsDNA (45). Thus, we imagine that the sequence-specific contacts made between the OBD and each pentanucleotide during initiation are eventually lost at the expense of binding to ssDNA regions generated upon double hexamer assembly and melting of the origin.
Sequence specificity of full-length T-ag.
Interestingly, we determined that full-length T-ag produced in insect cells with a baculovirus expression system, which allows phosphorylation of Thr124 (19), binds significantly to nonspecific dsDNA with an estimated affinity of 494 ± 38 nM, only threefold lower than that for a specific pentanucleotide (170 ± 22 nM). This lack of sequence specificity was even more pronounced in the presence of AMP-PNP and is likely due to the strong nonspecific binding of the C-terminal helicase domain to DNA (20, 25, 65). These results raise the question of how T-ag finds the origin in vivo and suggests that there must be mechanisms to favor its specific binding to the origin. Specificity could be enhanced by preventing the nonspecific interaction of the T-ag helicase domain with DNA and/or favoring its interaction with specific sequences flanking site II, namely, the AT-rich region and the EP, which are missing from our fluorescent probe. It is tempting to speculate that phosphorylation of the N-terminal domain could be involved in regulating the interaction of the helicase domain with DNA. This notion is supported by the findings that the N-terminal domain can be cross-linked to the helicase domain (63) and that mutant T124A T-ag molecules assemble into double hexamers that are less stable (64) and require longer sequences in the EP or AT region for their assembly on single assembly units (46).
Acknowledgments
We thank Daniel Simmons (University of Delaware) for the gift of class 4 mutant DNA, Albert M. Berghuis and Magdalena Korczynska (McGill University) for the use of the DLS instrument and advice on data analysis, and Steve Titolo for advice on FP data analysis. We also thank Andrew Bohm and Gretchen Meinke (Tufts University School of Medicine) for communicating results prior to publication.
J.A. is a senior scholar from the Fonds de la Recherche en Santé du Québec (FRSQ). This work was supported by grants from the Canadian Institutes for Health Research to J.A. and the National Institutes of Health to P.A.B. (9R01GM55397).
Footnotes
Published ahead of print on 27 June 2007.
REFERENCES
- 1.Alexandrov, A. I., M. R. Botchan, and N. R. Cozzarelli. 2002. Characterization of simian virus 40 T-antigen double hexamers bound to a replication fork. The active form of the helicase. J. Biol. Chem. 277:44886-44897. [DOI] [PubMed] [Google Scholar]
- 2.Barbaro, B. A., K. R. Sreekumar, D. R. Winters, A. E. Prack, and P. A. Bullock. 2000. Phosphorylation of simian virus 40 T antigen on Thr 124 selectively promotes double-hexamer formation on subfragments of the viral core origin. J. Virol. 74:8601-8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baur, C. P., K. Klausing, M. Scheffner, H. Stahl, and R. Knippers. 1988. Protein-DNA interactions at the simian virus 40 origin of replication. Biochim. Biophys. Acta 951:388-395. [DOI] [PubMed] [Google Scholar]
- 4.Bochkareva, E., D. Martynowski, A. Seitova, and A. Bochkarev. 2006. Structure of the origin-binding domain of simian virus 40 large T antigen bound to DNA. EMBO J. 25:5961-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borowiec, J. A., F. B. Dean, P. A. Bullock, and J. Hurwitz. 1990. Binding and unwinding—how T antigen engages the SV40 origin of DNA replication. Cell 60:181-184. [DOI] [PubMed] [Google Scholar]
- 6.Bradshaw, E. M., D. G. Sanford, X. Luo, J. L. Sudmeier, Z. A. Gurard-Levin, P. A. Bullock, and W. W. Bachovchin. 2004. T antigen origin-binding domain of simian virus 40: determinants of specific DNA binding. Biochemistry 43:6928-6936. [DOI] [PubMed] [Google Scholar]
- 7.Bullock, P. A., W. S. Joo, K. R. Sreekumar, and C. Mello. 1997. Initiation of SV40 DNA replication in vitro: analysis of the role played by sequences flanking the core origin on initial synthesis events. Virology 227:460-473. [DOI] [PubMed] [Google Scholar]
- 8.Cegielska, A., I. Moarefi, E. Fanning, and D. M. Virshup. 1994. T-antigen kinase inhibits simian virus 40 DNA replication by phosphorylation of intact T antigen on serines 120 and 123. J. Virol. 68:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, R., K. Peden, J. M. Pipas, D. Nathans, and R. Tjian. 1983. Biochemical activities of T-antigen proteins encoded by simian virus 40 A gene deletion mutants. Mol. Cell. Biol. 3:220-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dean, F. B., J. A. Borowiec, Y. Ishimi, S. Deb, P. Tegtmeyer, and J. Hurwitz. 1987. Simian virus 40 large tumor antigen requires three core replication origin domains for DNA unwinding and replication in vitro. Proc. Natl. Acad. Sci. USA 84:8267-8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deb, S., S. Tsui, A. Koff, A. L. DeLucia, R. Parsons, and P. Tegtmeyer. 1987. The T-antigen-binding domain of the simian virus 40 core origin of replication. J. Virol. 61:2143-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeCaprio, J. A. 1999. The role of the J domain of SV40 large T in cellular transformation. Biologicals 27:23-28. [DOI] [PubMed] [Google Scholar]
- 13.DeLucia, A. L., B. A. Lewton, R. Tjian, and P. Tegtmeyer. 1983. Topography of simian virus 40 A protein-DNA complexes: arrangement of pentanucleotide interaction sites at the origin of replication. J. Virol. 46:143-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enami, I., M. Kamo, H. Ohta, S. Takahashi, T. Miura, M. Kusayanagi, S. Tanabe, A. Kamei, A. Motoki, M. Hirano, T. Tomo, and K. Satoh. 1998. Intramolecular cross-linking of the extrinsic 33-kDa protein leads to loss of oxygen evolution but not its ability of binding to photosystem II and stabilization of the manganese cluster. J. Biol. Chem. 273:4629-4634. [DOI] [PubMed] [Google Scholar]
- 15.Fanning, E., and R. Knippers. 1992. Structure and function of simian virus 40 large tumor antigen. Annu. Rev. Biochem. 61:55-85. [DOI] [PubMed] [Google Scholar]
- 16.Gai, D., R. Zhao, D. Li, C. V. Finkielstein, and X. S. Chen. 2004. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 119:47-60. [DOI] [PubMed] [Google Scholar]
- 17.Heyduk, T., Y. Ma, H. Tang, and R. H. Ebright. 1996. Fluorescence anisotropy: rapid, quantitative assay for protein-DNA and protein-protein interaction. Methods Enzymol. 274:492-503. [DOI] [PubMed] [Google Scholar]
- 18.Hickman, A. B., and F. Dyda. 2005. Binding and unwinding: SF3 viral helicases. Curr. Opin. Struct. Biol. 15:77-85. [DOI] [PubMed] [Google Scholar]
- 19.Hoss, A., I. Moarefi, K. H. Scheidtmann, L. J. Cisek, J. L. Corden, I. Dornreiter, A. K. Arthur, and E. Fanning. 1990. Altered phosphorylation pattern of simian virus 40 T antigen expressed in insect cells by using a baculovirus vector. J. Virol. 64:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiao, J., and D. T. Simmons. 2003. Nonspecific double-stranded DNA binding activity of simian virus 40 large T antigen is involved in melting and unwinding of the origin. J. Virol. 77:12720-12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, A., and M. O'Donnell. 2005. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 74:283-315. [DOI] [PubMed] [Google Scholar]
- 22.Joo, W. S., H. Y. Kim, J. D. Purviance, K. R. Sreekumar, and P. A. Bullock. 1998. Assembly of T-antigen double hexamers on the simian virus 40 core origin requires only a subset of the available binding sites. Mol. Cell. Biol. 18:2677-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalderon, D., W. D. Richardson, A. F. Markham, and A. E. Smith. 1984. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311:33-38. [DOI] [PubMed] [Google Scholar]
- 24.Kalderon, D., B. L. Roberts, W. D. Richardson, and A. E. Smith. 1984. A short amino acid sequence able to specify nuclear location. Cell 39:499-509. [DOI] [PubMed] [Google Scholar]
- 25.Kim, H. Y., B. A. Barbaro, W. S. Joo, A. E. Prack, K. R. Sreekumar, and P. A. Bullock. 1999. Sequence requirements for the assembly of simian virus 40 T antigen and the T-antigen origin binding domain on the viral core origin of replication. J. Virol. 73:7543-7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, R. J., S. Moine, D. K. Reese, and P. A. Bullock. 2002. Peptides containing cyclin/Cdk-nuclear localization signal motifs derived from viral initiator proteins bind to DNA when unphosphorylated. J. Virol. 76:11785-11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klausing, K., K. H. Scheidtmann, E. A. Baumann, and R. Knippers. 1988. Effects of in vitro dephosphorylation on DNA-binding and DNA helicase activities of simian virus 40 large tumor antigen. J. Virol. 62:1258-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar, A., G. Meinke, D. K. Reese, S. Moine, P. J. Phelan, A. Fradet-Turcotte, J. Archambault, A. Bohm, and P. A. Bullock. 2007. Model for T-antigen-dependent melting of the simian virus 40 core origin based on studies of the interaction of the beta-hairpin with DNA. J. Virol. 81:4808-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundblad, J. R., M. Laurance, and R. H. Goodman. 1996. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol. Endocrinol. 10:607-612. [DOI] [PubMed] [Google Scholar]
- 30.Luo, X., D. G. Sanford, P. A. Bullock, and W. W. Bachovchin. 1996. Solution structure of the origin DNA-binding domain of SV40 T-antigen. Nat. Struct. Biol. 3:1034-1039. [DOI] [PubMed] [Google Scholar]
- 31.Masai, H., Z. You, and K. Arai. 2005. Control of DNA replication: regulation and activation of eukaryotic replicative helicase, MCM. IUBMB Life 57:323-335. [DOI] [PubMed] [Google Scholar]
- 32.McVey, D., L. Brizuela, I. Mohr, D. R. Marshak, Y. Gluzman, and D. Beach. 1989. Phosphorylation of large tumour antigen by cdc2 stimulates SV40 DNA replication. Nature 341:503-507. [DOI] [PubMed] [Google Scholar]
- 33.McVey, D., S. Ray, Y. Gluzman, L. Berger, A. G. Wildeman, D. R. Marshak, and P. Tegtmeyer. 1993. cdc2 phosphorylation of threonine 124 activates the origin-unwinding functions of simian virus 40 T antigen. J. Virol. 67:5206-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McVey, D., M. Strauss, and Y. Gluzman. 1989. Properties of the DNA-binding domain of the simian virus 40 large T antigen. Mol. Cell. Biol. 9:5525-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McVey, D., B. Woelker, and P. Tegtmeyer. 1996. Mechanisms of simian virus 40 T-antigen activation by phosphorylation of threonine 124. J. Virol. 70:3887-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meinke, G., P. A. Bullock, and A. Bohm. 2006. Crystal structure of the simian virus 40 large T-antigen origin-binding domain. J. Virol. 80:4304-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meinke, G., P. Phelan, S. Moine, E. Bochkareva, A. Bochkarev, P. A. Bullock, and A. Bohm. 2007. The crystal structure of the SV40 T-antigen origin binding domain in complex with DNA. PLoS Biol. 5:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moarefi, I. F., D. Small, I. Gilbert, M. Hopfner, S. K. Randall, C. Schneider, A. A. Russo, U. Ramsperger, A. K. Arthur, H. Stahl, et al. 1993. Mutation of the cyclin-dependent kinase phosphorylation site in simian virus 40 (SV40) large T antigen specifically blocks SV40 origin DNA unwinding. J. Virol. 67:4992-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohr, I. J., B. Stillman, and Y. Gluzman. 1987. Regulation of SV40 DNA replication by phosphorylation of T antigen. EMBO J. 6:153-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishitani, H., and Z. Lygerou. 2002. Control of DNA replication licensing in a cell cycle. Genes Cells 7:523-534. [DOI] [PubMed] [Google Scholar]
- 41.Parsons, R., M. E. Anderson, and P. Tegtmeyer. 1990. Three domains in the simian virus 40 core origin orchestrate the binding, melting, and DNA helicase activities of T antigen. J. Virol. 64:509-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peden, K. W., and J. M. Pipas. 1992. Simian virus 40 mutants with amino-acid substitutions near the amino terminus of large T antigen. Virus Genes 6:107-118. [DOI] [PubMed] [Google Scholar]
- 43.Peng, Y. C., and N. H. Acheson. 1998. Polyomavirus large T antigen binds cooperatively to its multiple binding sites in the viral origin of DNA replication. J. Virol. 72:7330-7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pipas, J. M., K. W. Peden, and D. Nathans. 1983. Mutational analysis of simian virus 40 T antigen: isolation and characterization of mutants with deletions in the T-antigen gene. Mol. Cell. Biol. 3:203-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reese, D. K., G. Meinke, A. Kumar, S. Moine, K. Chen, J. L. Sudmeier, W. Bachovchin, A. Bohm, and P. A. Bullock. 2006. Analyses of the interaction between the origin binding domain from simian virus 40 T antigen and single-stranded DNA provide insights into DNA unwinding and initiation of DNA replication. J. Virol. 80:12248-12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reese, D. K., K. R. Sreekumar, and P. A. Bullock. 2004. Interactions required for binding of simian virus 40 T antigen to the viral origin and molecular modeling of initial assembly events. J. Virol. 78:2921-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson, N. P., and S. D. Bell. 2005. Origins of DNA replication in the three domains of life. FEBS J. 272:3757-3766. [DOI] [PubMed] [Google Scholar]
- 48.Scheidtmann, K. H., M. Buck, J. Schneider, D. Kalderon, E. Fanning, and A. E. Smith. 1991. Biochemical characterization of phosphorylation site mutants of simian virus 40 large T antigen: evidence for interaction between amino- and carboxy-terminal domains. J. Virol. 65:1479-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheidtmann, K. H., M. Hardung, B. Echle, and G. Walter. 1984. DNA-binding activity of simian virus 40 large T antigen correlates with a distinct phosphorylation state. J. Virol. 50:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider, J., and E. Fanning. 1988. Mutations in the phosphorylation sites of simian virus 40 (SV40) T antigen alter its origin DNA-binding specificity for sites I or II and affect SV40 DNA replication activity. J. Virol. 62:1598-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen, J., D. Gai, A. Patrick, W. B. Greenleaf, and X. S. Chen. 2005. The roles of the residues on the channel beta-hairpin and loop structures of simian virus 40 hexameric helicase. Proc. Natl. Acad. Sci. USA 102:11248-11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simmons, D. T., G. Loeber, and P. Tegtmeyer. 1990. Four major sequence elements of simian virus 40 large T antigen coordinate its specific and nonspecific DNA binding. J. Virol. 64:1973-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simmons, D. T., R. Upson, K. Wun-Kim, and W. Young. 1993. Biochemical analysis of mutants with changes in the origin-binding domain of simian virus 40 tumor antigen. J. Virol. 67:4227-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simmons, D. T., K. Wun-Kim, and W. Young. 1990. Identification of simian virus 40 T-antigen residues important for specific and nonspecific binding to DNA and for helicase activity. J. Virol. 64:4858-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smelkova, N. V., and J. A. Borowiec. 1997. Dimerization of simian virus 40 T-antigen hexamers activates T-antigen DNA helicase activity. J. Virol. 71:8766-8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sreekumar, K. R., A. E. Prack, D. R. Winters, B. A. Barbaro, and P. A. Bullock. 2000. The simian virus 40 core origin contains two separate sequence modules that support T-antigen double-hexamer assembly. J. Virol. 74:8589-8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stenlund, A. 2003. Initiation of DNA replication: lessons from viral initiator proteins. Nat. Rev. Mol. Cell Biol. 4:777-785. [DOI] [PubMed] [Google Scholar]
- 58.Tegtmeyer, P., B. A. Lewton, A. L. DeLucia, V. G. Wilson, and K. Ryder. 1983. Topography of simian virus 40 A protein-DNA complexes: arrangement of protein bound to the origin of replication. J. Virol. 46:151-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Titolo, S., E. Welchner, P. W. White, and J. Archambault. 2003. Characterization of the DNA-binding properties of the origin-binding domain of simian virus 40 large T antigen by fluorescence anisotropy. J. Virol. 77:5512-5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Virshup, D. M., M. G. Kauffman, and T. J. Kelly. 1989. Activation of SV40 DNA replication in vitro by cellular protein phosphatase 2A. EMBO J. 8:3891-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Virshup, D. M., A. A. Russo, and T. J. Kelly. 1992. Mechanism of activation of simian virus 40 DNA replication by protein phosphatase 2A. Mol. Cell. Biol. 12:4883-4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weisshart, K., M. K. Bradley, B. M. Weiner, C. Schneider, I. Moarefi, E. Fanning, and A. K. Arthur. 1996. An N-terminal deletion mutant of simian virus 40 (SV40) large T antigen oligomerizes incorrectly on SV40 DNA but retains the ability to bind to DNA polymerase alpha and replicate SV40 DNA in vitro. J. Virol. 70:3509-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weisshart, K., S. Friedl, P. Taneja, H. P. Nasheuer, B. Schlott, F. Grosse, and E. Fanning. 2004. Partial proteolysis of simian virus 40 T antigen reveals intramolecular contacts between domains and conformation changes upon hexamer assembly. J. Biol. Chem. 279:38943-38951. [DOI] [PubMed] [Google Scholar]
- 64.Weisshart, K., P. Taneja, A. Jenne, U. Herbig, D. T. Simmons, and E. Fanning. 1999. Two regions of simian virus 40 T antigen determine cooperativity of double-hexamer assembly on the viral origin of DNA replication and promote hexamer interactions during bidirectional origin DNA unwinding. J. Virol. 73:2201-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu, C., D. Edgil, and D. T. Simmons. 1998. The origin DNA-binding and single-stranded DNA-binding domains of simian virus 40 large T antigen are distinct. J. Virol. 72:10256-10259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wun-Kim, K., and D. T. Simmons. 1990. Mapping of helicase and helicase substrate-binding domains on simian virus 40 large T antigen. J. Virol. 64:2014-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wun-Kim, K., R. Upson, W. Young, T. Melendy, B. Stillman, and D. T. Simmons. 1993. The DNA-binding domain of simian virus 40 tumor antigen has multiple functions. J. Virol. 67:7608-7611. [DOI] [PMC free article] [PubMed] [Google Scholar]