Abstract
The UL3.5 and UL48 genes, which are conserved in most alphaherpesvirus genomes, are important for maturation of pseudorabies virus (PrV) particles in the cytoplasm of infected cells (W. Fuchs, B. G. Klupp, H. J. Rziha, and T. C. Mettenleiter, J. Virol. 70:3517-3527, 1996; W. Fuchs, H. Granzow, B. G. Klupp, M. Kopp and T. C. Mettenleiter, J. Virol. 76:6729-6742, 2002). In bovine herpesvirus 1 (BoHV-1), the homologous gene products pUL3.5 and pUL48 have been demonstrated to interact physically (N. Lam and G. Letchworth, J. Virol. 74:2876-2884, 2000). Moreover, BoHV-1 pUL3.5 partially complemented a pUL3.5 defect in PrV (W. Fuchs, H. Granzow, and T. C. Mettenleiter, J. Virol. 71:8886-8892, 1997). By using coimmunoprecipitation and yeast two-hybrid studies, we observed a similar interaction between pUL3.5 and pUL48 of PrV, as well as a heterologous interaction between the PrV and BoHV-1 gene products. The relevant domain could be confined to the first 43 amino acids of PrV pUL3.5. Unlike its BoHV-1 homologue, PrV pUL3.5 is processed by proteolytic cleavage, and only an abundant 14-kDa fragment consisting of amino acids 1 to ≥116 could be detected by peptide mass fingerprint analysis of purified wild-type PrV particles, which also contain the pUL48 tegument component. To determine the biological relevance of the protein-protein interaction, pUL3.5-, pUL48-, and double-negative PrV mutants were analyzed in parallel. All deletion mutants were replication competent but exhibited significantly reduced plaque sizes and virus titers in cultured rabbit kidney cells compared to wild-type and rescued viruses, which correlated with a delayed neuroinvasion in intranasally infected mice. Remarkably, the defects of the double-negative mutant were similar to those of pUL48-negative virus. Electron microscopy of cells infected with either deletion mutant revealed the retention of naked nucleocapsids in the cytoplasm and the absence of mature virus particles. In summary, our studies for the first time demonstrate the relevance of the pUL3.5-pUL48 interaction for secondary envelopment of an alphaherpesvirus, give a molecular basis for the observed trans-complementation between the PrV and BHV-1 pUL3.5 homologs, yield conclusive evidence for the incorporation of a proteolytically processed pUL3.5 into PrV virions, and demonstrate the importance of both proteins for neuroinvasion and neurovirulence of PrV.
Pseudorabies virus (PrV), also designated suid herpesvirus 1, represents a member of the genus Varicellovirus within the Alphaherpesvirinae subfamily of the Herpesviridae (9). PrV causes Aujeszky's disease in pigs and also leads to fatal neurological disorders in many other mammalian species, excluding higher primates and humans (55). Although PrV has been eradicated in several industrialized countries, it remains a problem in most parts of the world. Moreover, PrV has been developed into a helpful tool in neurophysiology as well as a valuable model for investigations of alphaherpesvirus gene functions for replication in tissue culture and for pathogenesis in laboratory animals (17, 56-58).
Most of the 72 genes identified within the 143-kbp genome of PrV (43) possess homologs in other alphaherpesviruses and partly also in beta- and gammaherpesviruses. This apparent conservation of gene and protein structure suggests a functional relationship. In particular, the genes required for viral DNA replication, capsid formation, and DNA encapsidation are highly conserved throughout the Herpesviridae (63). Furthermore, all hitherto-characterized members of the virus family possess two nonstructural proteins homologous to the UL31 and UL34 gene products of herpes simplex virus type 1 (HSV-1), which are involved in nuclear egress (54, 58).
In contrast, many structural components of tegument and envelope, which are acquired predominantly during virion morphogenesis in the cytoplasm, exhibit a higher degree of variability. Whereas several tegument proteins like those encoded by HSV-1 open reading frames (ORFs) UL7, UL11, UL16, UL21, UL36, UL37, and UL51 are conserved in all herpesvirus subfamilies, other major tegument components, encoded by UL41, UL46, UL47, UL48, and UL49, are apparently restricted to alphaherpesviruses (54, 56). Nevertheless, as shown by the generation of respective deletion mutants of HSV-1 and PrV, the presence of not only the conserved inner tegument components pUL36 and pUL37 but also the nonconserved pUL48 is crucial for final envelopment in the trans-Golgi region (11, 12, 23, 26, 42, 62, 70). Furthermore, the UL48 proteins of several alphaherpesviruses function as potent activators of immediate-early gene expression (1, 3, 23, 33, 61). However, whereas pUL48 has been shown to be required for efficient productive replication of HSV-1, PrV, and equid herpesvirus 1 (EHV-1), it proved to be dispensable for replication of varicella-zoster virus (VZV) and Marek's disease virus (MDV), and no UL48 homologue has been found in the genome of another avian alphaherpesvirus, psittacid herpesvirus 1 (PsHV-1) (5, 15, 23, 62, 68, 69, 70).
Besides pUL36, pUL37, and pUL48, several other PrV proteins are involved in secondary envelopment and virus egress. Virion formation of deletion mutants lacking the tegument proteins encoded by UL7, UL11, UL16, UL21, UL47, or UL51 was less efficient or delayed compared to that of wild-type PrV (27, 45, 46, 48, 49), whereas deletion of the interacting alphaherpesvirus-specific membrane proteins encoded by UL20 and UL53 (glycoprotein K) affected the release of enveloped particles from Golgi membrane-derived vesicles into the extracellular space (13, 20, 39). Drastic egress defects associated with huge aggregations of unenveloped capsids and associated tegument proteins as well as membrane dilations in the cytoplasm of infected cells have been observed after the simultaneous deletion of several individually nonessential proteins, including the conserved glycoprotein gM (pUL10) and membrane-associated pUL11, or gM and the alphaherpesvirus-specific glycoproteins gI (pUS7) and gE (pUS8) (2, 50).
An even less-conserved gene, UL3.5, has also been demonstrated to play a very important role during final PrV envelopment in the trans-Golgi region (19). UL3.5 has originally been considered a unique gene of PrV (10), but careful reinvestigations identified homologous ORFs at corresponding genome positions of many other alphaherpesviruses, including bovine herpesvirus 1 (BoHV-1), EHV-1, VZV, avian infectious laryngotracheitis virus, and MDV (8, 18, 34, 36, 67). In contrast, HSV genomes do not contain a corresponding ORF (14, 54). Moreover, unlike deletion of PrV UL3.5, deletion of the homologous ORF57 of VZV did not significantly affect virus replication in cultured cells (6). Other alphaherpesvirus UL3.5 deletion mutants have not been described up to now. However, replication of BoHV-1 was severely impaired in cells expressing a C-terminally truncated BoHV-1 pUL3.5 (52), and UL3.5-negative PrV could be partly rescued by insertion of the UL3.5 gene of BoHV-1 (21). These findings indicated that functions of pUL3.5 in PrV and BoHV-1 might be related. Furthermore, for BoHV-1, a physical interaction between pUL3.5 and pUL48 has been demonstrated (51); this interaction, if also present in PrV, would explain the similar phenotypes of UL3.5- and UL48-negative virus mutants (19, 23). However, a direct interaction between the two PrV proteins seemed implausible, since in our previous studies only pUL48, but not pUL3.5, could be detected in purified virions (19, 23), whereas both proteins have been identified as abundant components of mature BoHV-1 virions (65). However, an apparently pUL3.5-related polypeptide of 14 kDa was recently detected in purified PrV virions whose origin remained obscure (59).
To investigate this finding in more depth, proteome analyses of highly purified PrV particles were repeated and refined, including analysis of a virus mutant expressing a functional UL3.5 gene product which had been tagged with enhanced green fluorescent protein (EGFP). Yeast two-hybrid and coimmunoprecipitation studies were performed to unravel possible interactions between PrV pUL3.5 and pUL48. Furthermore, isogenic UL3.5-, UL48-, and double-negative virus recombinants were generated and analyzed in vitro and in vivo.
MATERIALS AND METHODS
Viruses and cells.
All mutants used in this study were derived from the laboratory PrV strain Kaplan (PrV-Ka) (35). The genome of PrV-Ka has been cloned as bacterial artificial chromosome (BAC) pPrV-ΔgB (49), which permits convenient mutagenesis in Escherichia coli. Two previously described deletion mutants, PrV-ΔUL48 (23) and PrV-ΔUL43G (47), were used in our investigations. All viruses were propagated in porcine kidney (PSEK) cells for production of high-titer virus stocks and for purification of virions, or in rabbit kidney (RK13) cells for determination of in vitro replication properties. Cells were grown at 37°C in minimum essential medium (MEM) supplemented with 10% fetal calf serum (Invitrogen). After infection, the medium was replaced by MEM which contained only 5% fetal calf serum and, for plaque assays, 6 g/liter methyl cellulose (Sigma).
Plasmid construction.
For mutagenesis of UL3.5, the described plasmid pUC-S2.4 (28), which contains a genomic 2,390-bp SalI fragment of PrV-Ka, was used (Fig. 1B). To generate the deletion plasmid pUC-ΔUL3.5KF, UL3.5 codons 7 to 199 were removed from plasmid pUC-S2.4 by PshAI/NotI double digestion and replaced by a 1,258-bp BstBI fragment of pKD13 (7), which contains a kanamycin resistance gene flanked by Flp recombinase recognition target sites (FRT). To permit in-frame fusion of the gene encoding EGFP to the 5′ end of UL3.5, an AflIII site was removed from the vector part of pUC-S2.4 by PciI digestion followed by Klenow treatment and religation, and a second AflIII site was removed by the subsequent deletion of a 677-bp XcmI/HindIII fragment (Fig. 1B, HindIII site in vector polylinker). Plasmid pUC-UL3.5G was obtained by insertion of a 770-bp HindIII/BsrGI fragment of pEGFP-N1 (Clontech) in the remaining single AflIII site located 15 bp upstream of the UL3.5 start codon. For yeast two-hybrid studies, UL3.5 was recloned from pUC-S2.4 within a 1,079-bp AflIII fragment and fused in-frame to the 3′ end of the LexA ORF of plasmid pLexA (Clontech), which had been digested with BamHI (Fig. 1C). In pLex-UL3.5NN, -UL3.5AN, and -UL3.5ES, the insert of pLex-UL3.5 was truncated by digestions with NotI, ApaI and NotI, or EcoRI and SbfI, followed by religation (Fig. 1C).
FIG. 1.
Maps of virus mutants and plasmids. (A) The PrV genome, consisting of long (UL) and short (US) unique regions and inverted repeat sequences (IR, TR), is depicted (B) Enlarged sections show the viral ORFs (pointed rectangles) within the analyzed genome regions. Recombinant viruses were generated by mutagenesis in E. coli of an infectious full-length clone of the PrV-Ka genome, which led to insertion of resistance genes (KanR/TetR) or FRT. In PrV-UL3.5G, the EGFP ORF was fused in-frame to UL3.5, which also affected the 3′ end (shaded) of the overlapping UL3 gene. (C) Plasmids used for yeast two-hybrid studies contained UL48 of PrV, and complete or truncated UL3.5 genes of PrV or BoHV-1 fused to the ORFs of a transcription-activating protein (B42) or a promoter-specific DNA-binding protein (LexA), respectively. Relevant restriction sites and retained amino acid sequences of the UL3.5 and UL48 proteins are shown in panels B and C. (D) Amino acid sequence of PrV pUL3.5. Peptides detected by MALDI-TOF analyses of the virion proteins are underlined, and identified features of different parts of the protein are indicated by shaded rectangles.
Plasmid pUC-B1K containing a genomic 3,605-bp BamHI/KpnI fragment of PrV-Ka (23) was used for deletion of UL48 (Fig. 1B). To obtain pUC-ΔUL48K, a 924-bp NcoI/NheI fragment containing UL48 codons 58 to 365 was substituted by the kanamycin resistance gene (nucleotides 1800 to 2769) of pACYC177 (New England BioLabs), which had been amplified by PCR using custom-made primers and Pfx DNA polymerase (Invitrogen). For yeast two-hybrid experiments, pB42-UL48 (Fig. 1C) was generated by recloning of the UL48 ORF as a 1,270-bp EcoRI/XhoI fragment from expression plasmid pcDNA-UL48 (23) into the similarly digested shuttle vector pB42AD.
The UL3.5 gene of BoHV-1 strain Schönböken was isolated in a 523-bp NcoI/XhoI subfragment of pSP-BHKSX (21) and inserted into pLexA, which had been digested with EcoRI and XhoI (Fig. 1C). Noncompatible fragment ends were routinely blunt-ended prior to ligation using Klenow polymerase.
Isolation of virus mutants.
For deletion of UL3.5, the insert fragment of pUC-ΔUL3.5KF was amplified by PCR with the vector-specific M13/pUC (−47) and M13/pUC reverse (−48) primers (New England BioLabs). The PCR product was used for Red recombinase-mediated mutagenesis of pPrV-ΔgB in E. coli as described previously (7, 49). The kanamycin resistance gene was then removed by mutagenesis with Flp recombinase encoded by plasmid pCP20 (4), which led to retention of a single FRT site (Fig. 1B). To generate a UL3.5 and UL48 double-deletion mutant, a kanamycin resistance gene was stably inserted into the resulting BAC by using Red recombinase and a PCR product obtained after amplification of the mutated UL48 gene from pUC-ΔUL48K with primers PUL48-F and PUL48-R (23). Finally, the mini F-plasmid vector sequences were removed by cotransfection (29) of RK13 cells with the mutagenized BAC DNAs and with plasmid pUC-B1BclI, which led to restoration of the essential gB gene of PrV (49). From the virus progenies, single plaque isolates of PrV-ΔUL3.5F and PrV-ΔUL3.5/48 (Fig. 1B) were further propagated. Revertant PrV-UL3.5R and mutant PrV-UL3.5G, which expresses a functional EGFP-tagged UL3.5 protein (Fig. 1B), were obtained after cotransfection of RK13 cells with genomic DNA of PrV-ΔUL3.5F and pUC-S2.4 or pUC-UL3.5G, respectively. Isolation of the mutants was facilitated by their increased plaque sizes and, in the latter case, by autofluorescence of infected cells. All generated PrV recombinants were characterized by restriction analyses of viral DNA and Southern blot hybridization, as well as by PCR amplification and sequencing of the mutated genome regions (results not shown).
In vitro replication studies.
For determination of one-step growth kinetics, confluent monolayers of RK13 cells were infected with 2 PFU per cell of wild-type PrV-Ka or mutants PrV-ΔUL48, PrV-ΔUL3.5F, PrV-ΔUL3.5/48, PrV-UL3.5R, and PrV-UL3.5G. After adsorption for 1 h on ice, prewarmed medium was added, and incubation was continued at 37°C. One hour after the temperature shift, nonpenetrated virus was inactivated by low-pH treatment, and after 3, 6, 9, 12, 24, 48, and 72 h, cells were scraped into the medium and lysed by freezing (−70°C) and thawing (37°C) (49). Progeny virus titers were determined by plaque assays using RK13 cells which were fixed at 48 h postinfection (p.i.) for 30 min with a 1:1 mixture of methanol and acetone at −20°C. Plaques were visualized by indirect immunofluorescence with a gC-specific monoclonal antibody (40) and Alexa Fluor 488-conjugated anti-mouse secondary antibodies (Invitrogen) at dilutions of 1:100 and 1:1,000, respectively. The mean titers for four parallel kinetics studies per virus were calculated. Furthermore, the diameters of 30 plaques per virus were measured, and average plaque sizes as well as standard deviations were plotted.
Western blot and radioimmunoprecipitation.
For Western blot analyses, RK13 cells were harvested 16 h after infection with PrV-Ka, PrV-ΔUL48, PrV-ΔUL3.5F, PrV-ΔUL3.5/48, PrV-UL3.5R, or PrV-UL3.5G at a multiplicity of infection (MOI) of 5. Samples of infected and noninfected cells as well as of PrV virions were prepared as described previously (13), separated by discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and electrotransferred to nitrocellulose membranes (Mini-Protean II and Transblot SD cell; Bio-Rad). Blots were blocked with 5% low-fat milk in Tris-buffered saline (TBS-T; 150 mM NaCl, 10 mM Tris-HCl [pH 8.0], 0.25% Tween 20), and incubated for 1 h with monospecific rabbit antisera against pUL3 (44), pUL3.5 (19), pUL31 (22), pUL48 (23), and pUL49 (2) or EGFP (kindly provided by C. Höhle and G. M. Keil, Insel Riems, Germany) at dilutions of 1:20,000 to 1:100,000 in TBS-T. Bound antibody was detected with peroxidase-conjugated anti-rabbit antibodies (Dianova) and visualized by chemiluminescence (Super Signal, Pierce) recorded on X-ray film.
For immunolabeling, RK13 cells were infected with PrV-Ka or PrV-UL3.5G at an MOI of 5 and incubated for 2 h in MEM without methionine and cysteine prior to the addition of 3.7 MBq/ml [35S]methionine and [35S]cysteine (Tran35S-Label; MP Biochemicals). Sixteen hours after infection, the cells were lysed, and lysates were precipitated with monospecific rabbit antisera against gH (40), pUL48, or EGFP as described previously (53). Samples were separated by SDS-PAGE, and labeled proteins were detected in dried gels by radioluminography (FLA-3000; Fuji).
Proteome analyses of PrV particles.
Virions were purified and separated by one-dimensional SDS-PAGE as described previously (59). Protein bands were isolated from Coomassie brilliant blue-stained gels and, after in-gel tryptic digestion (64), mass spectra of the obtained peptides were registered with a Bruker Ultraflex instrument. Proteins were identified using flexAnalysis 2.0 software (Bruker) with an in-house database of the PrV proteome, which had been deduced from the published genome sequence (43).
Yeast two-hybrid studies.
A Matchmaker LexA two-hybrid system (Clontech) was used for investigation of interactions between pUL48 and pUL3.5 of PrV and BoHV-1. Yeast cells containing the reporter plasmid p8op-lacZ were simultaneously transformed with pB42-UL48 and one of the pLex-UL3.5 constructs shown in Fig. 1C. Furthermore, cells transformed with the empty vectors (pLexA, pB42AD) and pLexA-Pos were used as negative or positive controls, respectively. Yeast clones were selected on plates lacking amino acids which required plasmid-encoded genes for synthesis, and the presence of the plasmids was verified by PCR amplification and DNA sequencing of the insert fragments. According to the manufacturers’ protocols, liquid cultures of two clones per plasmid combination were assayed for reporter gene expression using o-nitrophenyl-β-d-galactopyranoside (ONPG) as substrate, and β-galactosidase activities were calculated (60).
Electron microscopy.
RK13 cells were infected with 1 PFU per cell of PrV-ΔUL3.5F, PrV-ΔUL3.5/48, or PrV-UL3.5R. After 1 h on ice and an additional hour at 37°C, the inoculum was replaced by fresh medium, and incubation was continued for 13 h at 37°C. Fixation and embedding were performed as described previously (41), and counterstained ultrathin sections were analyzed with an electron microscope (Tecnai 12; Philips).
Animal experiments.
The relevance of pUL48 and pUL3.5 for virulence and neuronal spread of PrV was investigated by infection of 10 7-week-old CD1 mice, each with PrV-ΔUL3.5F, PrV-ΔUL48, PrV-UL3.5/48, or PrV-Ka. After bilateral intranasal instillation of 5 μl of virus suspensions containing 106 PFU per ml, the animals were observed three times a day for clinical signs. Mean survival times were determined, and standard deviations were calculated. Single mice were necropsied every 24 h after inoculation, and viral spread in the trigeminal circuit was analyzed by immunohistochemistry of paraffin sections of the brains by using an antiserum against the major capsid protein (pUL19) of PrV as described previously (37).
RESULTS
Protein expression of virus mutants.
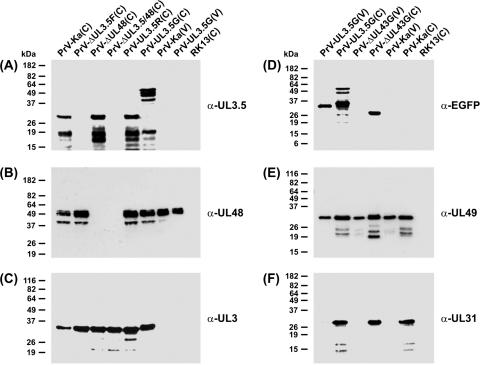
The previously described UL3.5 mutants of PrV contain large foreign gene insertions encoding either gB of BoHV-1 (19) or β-galactosidase of E. coli (21). To exclude any possible effects of these heterologous sequences on other PrV genes or viral DNA replication, a novel gene deletion mutant was generated by BAC mutagenesis in bacteria. The resulting recombinant PrV-ΔUL3.5F lacks the major part (codons 7 to 199) of the UL3.5 ORF of PrV-Ka, which is translated into a 224-amino-acid protein (Fig. 1B). Since the deleted sequence was replaced by only 36 bp of foreign DNA, representing an FRT site, an impairment of mRNA stability or expression rate of the coterminally transcribed upstream genes UL1, UL2, and UL3 (10) was unlikely. This could be confirmed by Western blot analyses of infected cell lysates, which revealed that pUL3 is expressed at similar levels by wild-type PrV-Ka and PrV-ΔUL3.5F (Fig. 2C). In contrast, neither the described 30-kDa pUL3.5 (19) nor smaller products of this protein were detectable in cells infected with PrV-ΔUL3.5F (Fig. 2A).
FIG. 2.
Western blot analyses. RK13 cells were harvested 16 h after infection with the indicated viruses (MOI = 5). Lysates of infected and noninfected cells (C) and of sucrose gradient-purified virions (V) were separated by SDS-PAGE. Blots were incubated with monospecific rabbit antisera against the C-terminal part of the UL3.5 gene product (A), the viral tegument components encoded by UL48 (B) and UL49 (E), the nonstructural UL3 (C) and UL31 (F) proteins, or EGFP (D). Molecular mass markers are indicated at the left.
Three additional mutants were derived from PrV-ΔUL3.5F: double-deletion mutant PrV-ΔUL3.5/48, rescuant PrV-UL3.5R, and PrV-UL3.5G (Fig. 1B). As expected, the double mutant did also not express any detectable pUL3.5, whereas pUL3.5 of the rescuant and of the previously described mutant PrV-ΔUL48 (23) were undistinguishable from that of wild-type PrV (Fig. 2A). PrV-UL3.5G expresses an EGFP-UL3.5 fusion gene under control of the PrV UL3.5 promoter, which consists of codons 1 to 238 of EGFP, five originally nonexpressed codons, and the complete UL3.5 ORF of 225 codons (Fig. 1B). The deduced 467-amino-acid protein has a predicted molecular mass of 51 kDa. This roughly fits the apparent mass of ca. 55 kDa of the largest protein found by Western blot analyses with pUL3.5-specific (Fig. 2A) or EGFP-specific (Fig. 2D) antisera. However, like the native pUL3.5, the EGFP-UL3.5 fusion protein appears to be highly unstable, since numerous smaller protein fragments were detectable.
The insertion of PrV-UL3.5G also affects the 3′ end of the overlapping UL3 gene, since it replaces its last 5 codons with 21 codons from the pEGFP-N1 polylinker region (Fig. 1B). Thus, the size and molecular mass of the deduced protein increased from 237 to 253 amino acids and from 25.6 to 27.6 kDa, respectively. Consequently, the apparent mass of pUL3 of PrV-UL3.5G was slightly increased compared to the respective gene products of all other investigated viruses (Fig. 2C).
In PrV-ΔUL48, the UL48 ORF had been replaced by a tetracycline resistance gene (23), whereas PrV-ΔUL3.5/48 contains a kanamycin resistance gene at the corresponding genome position (Fig. 1B). However, both mutants possess identical deletions of a viral NcoI/NheI DNA fragment comprising codons 58 to 365 of UL48. In Western blot analyses, a UL48-specific antiserum did not react with lysates of cells infected with PrV-ΔUL48 or PrV-ΔUL3.5/48 (Fig. 2B), whereas all other investigated viruses expressed multiple forms of pUL48 with apparent masses of ca. 53 to 57 kDa, as has previously been shown for wild-type PrV (23).
Virion localization of PrV pUL3.5.
In the past, pUL3.5 of PrV has been considered a nonstructural protein (19), since repeated Western blot analyses with the available monospecific antiserum did not detect any UL3.5 gene products in purified particles of PrV-Ka or PrV-UL3.5G (Fig. 2A). Whereas pUL3 and pUL31 were also absent from virion preparations (Fig. 2C and F), as had been described previously (22, 44), tegument components like pUL48 and pUL49 were efficiently incorporated (Fig. 2B and E). However, an EGFP-specific specific antiserum detected a protein of ca. 35 kDa in PrV-UL3.5G virions (Fig. 2D). Although this polypeptide did not react with the UL3.5-specific antiserum (Fig. 2A), it appeared to be the most abundant form of the fusion protein also in infected cells (Fig. 2D). The different reaction patterns of the two antisera can be explained by the pronounced instability of the full-length pUL3.5 (see above) and by the fact that the anti-UL3.5 serum had been prepared against the C terminus (amino acids 137 to 223) of the protein (Fig. 1D) (19), whereas EGFP had been fused to its N terminus (Fig. 1C). To clarify which part of the EGFP-UL3.5 fusion protein targets its virion incorporation, a PrV recombinant expressing native EGFP was included in the present study. The 27-kDa EGFP was detectable only in cells infected with PrV-ΔUL43G (47), not in purified virus particles (Fig. 2D). Thus, virion incorporation is dependent on the presence of an N-terminal fragment of the UL3.5 protein, which was not detectable by our monospecific antiserum.
This finding was confirmed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry of one-dimensionally separated proteins from PrV particles, which identified an abundant 14-kDa virion component of PrV-Ka as the N-terminal part of the UL3.5 gene product. In contrast, only traces of the 30-kDa full-length protein were detectable. Peptides representing pUL3.5 amino acids 7 to 116 were identified after tryptic digestion of the 14-kDa fragment as well as of the 35-kDa EGFP-UL3.5 fusion protein from PrV-UL3.5G (Fig. 1D). However, the precise C terminus of the EGFP-UL3.5 protein present in virions and the responsible processing mechanism remain to be determined.
Physical interactions between pUL3.5 and pUL48.
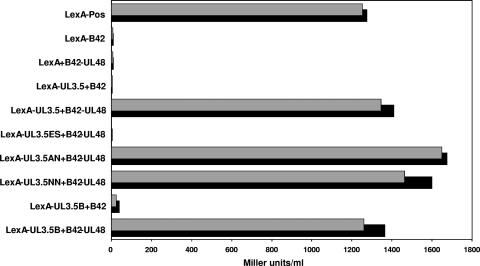
Since a direct interaction between pUL3.5 and pUL48 of BoHV-1 has been demonstrated (51), yeast two-hybrid studies were performed to test whether this also applies to the respective PrV homologs. For this purpose, an expression plasmid was constructed in which the complete UL3.5 ORF of PrV had been fused in-frame to the gene of the sequence-specific DNA-binding LexA protein (pLex-UL3.5) (Fig. 1C). A second plasmid contained the entire UL48 ORF fused to the coding sequence of a transcription-activating protein (pB42-UL48; Fig. 1C). Cotransfection of yeast cells with these two plasmids led to pronounced expression of a lacZ reporter gene under control of an inducible promoter containing LexA-binding elements. Using ONPG as a substrate, β-galactosidase activities of 1,200 to 1,400 Miller units per ml in liquid cultures could be determined (Fig. 3). This level of activity was similar to that obtained with a positive control plasmid provided by the manufacturer (Fig. 3, pLexA-pos). However, if the UL3.5 or UL48 plasmid was replaced by the empty vector pLexA or pB42AD, respectively, lacZ expression was almost completely abolished (Fig. 3). These findings strongly indicated that a physical interaction between the UL3.5 and UL48 parts of the fusion proteins was responsible for reporter gene activation.
FIG. 3.
Two-hybrid interactions of pUL3.5 and pUL48. Yeast cells were cotransformed with plasmids expressing DNA-binding LexA and transcription-activating B42 fusion proteins (see Fig. 1C) and an inducible LacZ reporter plasmid to detect specific protein interactions. The empty vectors expressing B42 and LexA were used as controls, and the β-galactosidase activity of two clones per plasmid combination was assayed using ONPG as a substrate (Miller units/ml).
To investigate the interaction in more detail, three truncated pLexA-UL3.5 plasmids containing UL3.5 codons 1 to 139 (pLex-UL3.5NN), 1 to 43 (pLex-UL3.5AN), or 20 to 224 (pLex-UL3.5ES) were generated (Fig. 1C). Coexpression with pB42-UL48 revealed that the N terminus of the UL3.5 protein (amino acids 1 to 20) is essential for transactivation, whereas amino acids 44 to 223 are dispensable (Fig. 3). Remarkably, the N-terminal part is highly conserved between the UL3.5 proteins of different alphaherpesviruses (18); therefore, it was conceivable that the homologues of other virus species might also interact with the UL48 gene product of PrV. This could be confirmed for the 126 codon UL3.5 ORF of BoHV-1, which was also cloned and expressed as LexA fusion protein (pLexA-UL3.5B; Fig. 1C) and which induced lacZ expression via interaction with PrV pUL48 as efficiently as the autologous constructs (Fig. 3).
We also attempted to localize the interacting domain of pUL48. However, all tested 5′- or 3′-terminal truncations of the insert of pB42-UL48 completely abolished transactivation (results not shown). Thus, for interaction with pUL3.5, a discontinuous sequence motif and/or a proper tertiary structure of pUL48 might be required. It was also impossible to confirm the yeast two-hybrid interactions by reversal studies using a LexA-UL48 fusion protein, since this construct induced reporter gene expression on its own. Most likely, this was a consequence of the known transcription-activating properties of the UL48 gene products of different herpesviruses, including that of PrV (23).
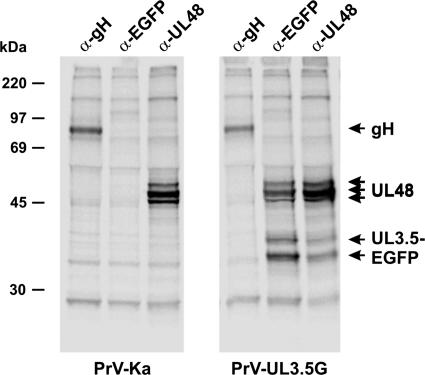
To provide further evidence for the interaction between pUL3.5 and pUL48 of PrV, immunoprecipitation experiments with [35S]methionine- and [35S]cysteine-labeled lysates of infected cells were performed. Since our UL3.5-specific antiserum is not suitable for these studies, the virus mutant expressing the EGFP-UL3.5 fusion protein (PrV-UL3.5G) and an EGFP-specific serum were used. This serum precipitated two proteins of ca. 35 and 37 kDa, which likely represent truncated EGFP-UL3.5 fusion proteins, but also three proteins of 53 to 57 kDa, which correspond to different forms of pUL48 (Fig. 4), as shown by precipitation by an anti-UL48 serum but not by a gH-specific control antiserum (Fig. 4). The 53- to 57-kDa UL48 proteins were also precipitated from lysates of cells infected with wild-type PrV-Ka, but the 35- and 37-kDa proteins were not coprecipitated by the anti-UL48 serum (Fig. 4), which confirmed that they indeed represent EGFP-UL3.5 fusion proteins. Remarkably, there was no clear evidence for coprecipitation of the full-length wild-type pUL3.5. However, since the anti-EGFP serum did not specifically precipitate any proteins from PrV-Ka (Fig. 4), the coprecipitation of pUL48 from PrV-UL3.5G-infected cells was apparently caused by its interaction with the EGFP-UL3.5 fusion protein. Immunofluorescence analyses of RK13 cells infected with PrV-UL3.5G revealed a cytoplasmic colocalization of the EGFP-UL3.5 and pUL48, which was not seen in cells infected with PrV mutants expressing native EGFP (data not shown). Thus, taken together, our studies provide strong evidence for the existence of a pUL3.5-pUL48 complex in PrV-infected cells, which is also incorporated into virions.
FIG. 4.
Coimmunoprecipitation of pUL3.5-EGFP and pUL48. RK13 cells were infected with PrV-Ka or PrV-UL3.5G at a MOI of 5 and incubated at 37°C for 14 h in the presence of [35S]methionine and [35S]cysteine. Labeled proteins were precipitated with monospecific antisera against gH, EGFP, or pUL48 and separated by SDS-PAGE. Molecular masses of marker proteins are indicated.
In vitro growth properties of UL3.5- and UL48-negative PrV mutants.
PrV mutants lacking either pUL3.5 or pUL48 have already been described (19, 21, 23). However, these mutants originating from distinct parental viruses had never been investigated in parallel under identical conditions. For the present study, the growth properties of isogenic mutants derived from the BAC-cloned PrV-strain Ka were determined in rabbit kidney cells. Furthermore, an UL3.5 and UL48 double-deletion mutant was analyzed for the first time.
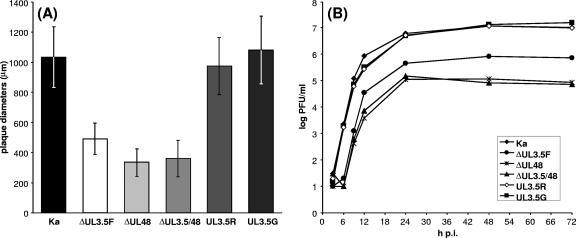
In agreement with the results of earlier investigations, plaque diameters measured 2 days after infection of RK13 cells with UL3.5- or UL48-negative PrV were significantly reduced, by 50 or 65%, respectively, compared to the plaque diameters for wild-type PrV-Ka (Fig. 5A). Rescuant PrV-UL3.5R as well as PrV-UL3.5G exhibited wild-type-like plaque sizes, demonstrating that the observed defect of the deletion mutant was indeed caused by the absence of the UL3.5 gene and that the EGFP-UL3.5 fusion protein can functionally substitute for the native pUL3.5 (Fig. 5A). This was also confirmed by one-step growth studies, in which maximum titers of PrV-ΔUL3.5F and PrV-ΔUL48 were reduced ca. 15-fold and 100-fold, respectively, whereas the two rescuants reached wild-type-like titers (Fig. 5B). A rescuant of PrV-ΔUL48 was not tested in the present study, since it had already been shown that the defects of this mutant can be fully complemented by pUL48 (23).
FIG. 5.
In vitro growth properties of PrV mutants. (A) For determination of plaque sizes, infected RK13 cells were incubated for 48 h under semisolid medium. The average diameters of 30 plaques and standard deviations were calculated. (B) For determination of growth kinetics, RK13 cells were infected at a MOI of 2 and scraped into the medium after 3, 6, 9, 12, 24, 48, and 72 h at 37°C. The cells were lysed by freezing and thawing, and progeny virus titers were determined by plaque assays. The mean results of two experiments are shown.
PrV-ΔUL3.5/48, which was derived from PrV-ΔUL3.5F by the additional deletion of UL48, exhibited replication defects similar to those of PrV-ΔUL48 (Fig. 5A and B). Thus, the functions of pUL3.5 and pUL48 during replication of PrV are obviously not independent of each other, since the presence or absence of pUL3.5 is no longer relevant after the deletion of pUL48. However, the functions may not be completely identical, since the defects of pUL3.5-negative PrV are less pronounced than those of pUL48-negative mutants.
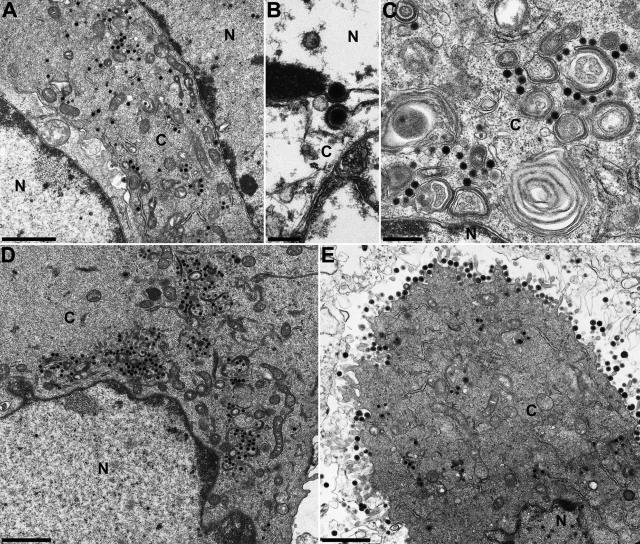
Electron microscopy of RK13 cells which were fixed during the late replication phase of PrV-ΔUL3.5/48 (Fig. 6A to C) or PrV-ΔUL3.5F (Fig. 6D) revealed very similar phenotypes. Nucleocapsid formation in the nucleus (Fig. 6A and D) and nuclear egress by sequential envelopment and deenvelopment at the inner and outer lamellae of the nuclear membrane (Fig. 6B) were not detectably affected. In contrast, secondary envelopment of cytoplasmic nucleocapsids was almost completely blocked (Fig. 6A, C, and D). Remarkably, the infected cells exhibited no tight aggregations of capsids or electron-dense tegument material, as had been described for several other PrV mutants possessing egress defects (56). Unlike PrV-ΔUL48 (23), neither the double mutant nor PrV-ΔUL3.5F showed an enhanced release of tegument-containing but capsidless light (L) particles from the cells (Fig. 6A and D). As expected from the restoration of plaque formation and virus replication, the release of mature particles and all preceding steps of virion morphogenesis of the rescue mutant PrV-UL3.5R (Fig. 6E) appeared similar to those observed for cells infected with wild-type PrV-Ka (30). Taken together, these results indicate that pUL3.5 and pUL48 of PrV are relevant for secondary envelopment of virus particles in the cytoplasm of infected cells, and, coinciding with the physical interaction of the proteins, their functions are interdependent.
FIG. 6.
Electron microscopy. RK13 cells were fixed 14 h after infection with PrV-ΔUL3.5/48 (A to C), PrV-ΔUL3.5F (D), or PrV-UL3.5R (E) at a MOI of 1 and analyzed. Whereas nucleocapsids were still formed and released from the host cell nucleus in the absence of UL3.5, or UL3.5 and UL48 (A to D), secondary envelopment of cytoplasmic nucleocapsids and release of mature virus particles were detectable only in cells infected by the UL3.5 rescuant (E). Bars represent 1.5 μm (A, D, and E), 500 nm (C), or 200 nm (B). N, nucleus; C, cytoplasm.
Relevance of pUL3.5 and pUL48 of PrV for in vivo neurovirulence.
Previous studies revealed that the deletion of the UL48 gene severely affected the kinetics of neuroinvasion of PrV after intranasal infection of mice (37). To investigate the in vivo relevance of pUL3.5, similar animal experiments including PrV-ΔUL3.5F and the double-deletion mutant PrV-ΔUL3.5/48 were performed. In these studies, the first clinical symptoms like depression, anorexia, cowering in a hunched position, and scratching could be observed 3 to 4 days after infection with either of the deletion mutants, which was approximately 48 h later than in mice infected with wild-type PrV-Ka (Table 1). Correspondingly, the mean survival times of animals infected with mutants lacking UL3.5, UL48, or both were within the same range and were prolonged ca. 2.5-fold compared to those of wild-type-infected mice (Table 1). Correlating with the less-pronounced in vitro growth defects of PrV-ΔUL3.5F, the animals died earlier than those infected with either of the two other deletion mutants. Although significantly delayed in the absence of any part of the pUL3.5-pUL48 complex, neurovirulence was not completely abolished, since the infection was still fatal in all animals investigated.
TABLE 1.
Virulence in mice
| Virus | Mean time to death (SD)a | Clinical symptoms at indicated day p.i.b
|
Immunohistochemistry (day p.i.)c
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | Nasal cavity | First-order neurons | Second-order neurons | Third-order neurons | ||
| PrV-Ka | 49.1 (1.58) | − | +++ | † | 1 | 1 | 2 | ND | |||
| PrV-ΔUL48 | 125.7 (3.26) | − | − | − | + | +++ | † | 1 | 3 | 4 | 5 |
| PrV-ΔUL3.5F | 117.7 (9.93) | − | − | + | +++ | † | 1 | 3 | 4 | ND | |
| PrV-ΔUL3.5/48 | 127.7 (6.42) | − | − | − | + | +++ | † | 1 | 4 | 4 | ND |
Average times to death (in hours) after intranasal infection with 104 PFU of the indicated virus were calculated for 10 animals for each virus.
Clinical symptoms were scored as follows: −, clinically inconspicuous; +, slight depression, hunched position, and ruffled hair coat; ++, apathy, anorexia, moderate dyspnea, and slight facial pruritus; +++, severe attacks of excitation, self-mutilation, skin erosions, and heavy dyspnea; †, animals moribund or dead.
Time in days p.i. of first UL19 antigen detection at the subsequent levels of the trigeminal pathway: nasal cavity (respiratory mucosal epithelium), first-order neurons (trigeminal ganglion), second-order neurons (spinal trigeminal nucleus, Sp5), third-order neurons (ectorhinal cortex). ND, not detected.
In skull sections of mice infected with either virus, the major capsid protein of PrV (pUL19) could be detected by immunohistochemistry in epithelial cells of the nasal mucosa from day 1 p.i. (Table 1). At this time, PrV-Ka was also found in first-order neurons of the trigeminal ganglia, and at day 2 p.i., it reached second-order neurons of the spinal trigeminal nuclei. In contrast, the UL3.5 and UL48 deletion mutants were not detectable in first-order neurons before 3 or 4 days p.i. (Table 1). However, all deletion mutants were able to infect second-order neurons, and PrV-ΔUL48 could be additionally detected in third-order neurons of the ectorhinal cortex. This finding clearly demonstrates that pUL3.5 and pUL48, although important for neuroinvasion, are not required for transneuronal spread of PrV.
DISCUSSION
The salient findings of this study are the following. (i) As previously observed for the homologous BoHV-1 proteins (51), pUL3.5 and pUL48 of PrV physically interact directly. (ii) pUL3.5 of BoHV-1 forms a complex with pUL48 of PrV, which constitutes the molecular basis for the observed functional complementation (21). (iii) PrV pUL3.5 is proteolytically processed, and only the processed 14-kDa N-terminal portion is incorporated into virions, explaining our previous failure to detect pUL3.5 in virions by probing with an antiserum directed against the C terminus (19). (iv) The interaction between pUL3.5 and pUL48 is important for virion formation, since deletion mutants for either gene and a double-deletion mutant exhibit similar phenotypes. (v) The interaction between both proteins is also critical for neuroinvasion and neurovirulence.
Despite the important roles of pUL3.5 and pUL48 during efficient envelopment of PrV nucleocapsids in the trans-Golgi region, these proteins are not conserved throughout the Herpesviridae but are restricted to members of the subfamily Alphaherpesvirinae (9). Moreover, no UL3.5 homologues have been found in HSV genomes (14, 54), and a recently sequenced avian alphaherpesvirus, PsHV-1, possesses none of the two genes (68). In contrast, in the genomes of most other alphaherpesviruses of the genera Varicellovirus (e.g., VZV, BoHV-1, EHV-1), Mardivirus (MDV-2), and Iltovirus (avian infectious laryngotracheitis virus), UL3.5 and UL48 homologues have been identified (8, 10, 18, 34, 36, 67). However, these proteins apparently differ with respect to their importance for viral replication. pUL3.5 and pUL48 are dispensable for replication of VZV in cell culture (5, 6), as is pUL48 of MDV (15). Remarkably, in vitro these two viruses do not form infectious extracellular particles at detectable levels but, rather, spread only directly from cell to cell. Thus, the secondary envelopment of VZV capsids as observed in the cytoplasm of cultured cells (32) is obviously mediated, at least partly, by other viral proteins. This is in stark contrast to PrV and BoHV-1, which form extracellular infectious virions at high levels. Although it is unclear whether expression of a pUL3.5-like protein in VZV or a pUL48 homolog in MDV would enhance the formation of extracellular virions, our studies clearly show that these proteins and their interaction are crucial for efficient virion morphogenesis of BoHV-1 and PrV.
In contrast, HSV-1 also readily forms extracellular virions without a pUL3.5 homolog. Thus, pUL3.5 function must have been adopted by another protein of HSV-1, perhaps by pUL48 itself. Like PrV pUL48, the HSV-1 homolog not only exhibits transcription-activating functions but also is relevant for virion morphogenesis (1, 3, 23, 62, 70). For closer cognates of PrV, like BoHV-1 and EHV-1, the functional relevance of the UL3.5 gene has not yet been investigated. A UL48-negative EHV-1 mutant has been described and shown to be defective in viral egress, like the corresponding PrV and HSV-1 mutants (69). To assess the role of pUL3.5 in these viruses, construction and characterization of corresponding deletion mutants is required, as has been done for PrV (19; this study).
The UL48 gene products of several alphaherpesviruses, including PrV, have been identified as abundant tegument proteins (23, 63). pUL3.5 was also detected in mature BoHV-1 virions (65); full-length pUL3.5 had not been found in PrV virions (19), although a smaller product was recently detected (59). We now demonstrate that the 14-kDa pUL3.5 incorporated into PrV virions (59) results from proteolytic processing of the primary 30-kDa translation product. The virion component comprises the N-terminal part of the UL3.5 protein (amino acids 1 to ≥116) as identified by peptide mass fingerprint analyses of purified PrV particles. This fragment could not be detected in our previous studies by an antiserum raised against the C-terminal part of the protein (amino acids 158 to 224) (Fig. 1D) (19). Proteolytic cleavage of the primary translation product is suggested, since RNA analyses (10) provided no evidence for the existence of a spliced UL3.5 transcript, which would permit translation of a C-terminally truncated polypeptide. However, the deduced amino acid sequence of PrV pUL3.5 contains no clear indication of a putative cleavage site; therefore, the precise terminus of the virion protein and the type of protease involved in processing remain to be identified. The UL3.5 gene product of BoHV-1 is obviously not processed in a similar manner, but its overall length of 126 amino acids, resulting in an apparent mass of 13 kDa (36, 65), fits the size of the protein fragment incorporated into PrV virions.
The hitherto-identified UL3.5 genes of other alphaherpesviruses are significantly smaller than the UL3.5 ORF of PrV, and detectable amino acid sequence homologies are limited to the N-terminal 57 residues (18). However, our yeast two-hybrid studies revealed that the conserved amino acids 1 to 43 of PrV pUL3.5 are sufficient for interaction with pUL48, whereas the interaction was completely abolished after deletion of the N-terminal 19 residues. In similar truncation analyses, amino acids 1 to 86 but not amino acids 21 to 126 of the BoHV-1 UL3.5 protein interacted with pUL48 in pull-down experiments (51). Thus, the conserved N-terminal domains of other alphaherpesvirus UL3.5 proteins might function in similar specific protein interactions. Interestingly, our yeast two-hybrid studies further demonstrated a strong interaction between pUL3.5 of BoHV-1 and pUL48 of PrV. This interaction explains the previously described heterologous cis complementation of UL3.5-negative PrV by the corresponding gene of BoHV-1 (21). To date, it is unclear whether the unique C-terminal part of PrV pUL3.5, which is removed during processing, possesses any biological relevance. In earlier studies, a PrV mutant expressing a truncated UL3.5 gene product consisting of the first 158 amino acids exhibited no detectable in vitro replication defects compared to the parental virus expressing the complete protein (19). However, in vivo virulence of this mutant has not been investigated.
To determine more precisely the biological role of the interaction between the N-terminal part of pUL3.5 with pUL48, a PrV-mutant lacking UL3.5 codons 7 to 199 was investigated in parallel with UL48-negative virus (23) and a double mutant carrying deletions of both genes. Electron microscopy of rabbit kidney cells infected with either mutant revealed similar defects during virus morphogenesis. As in earlier studies with single deletion mutants (19, 23), capsid formation, DNA-encapsidation, and nuclear egress were not detectably affected. In contrast, the secondary envelopment of nucleocapsids in the cytoplasm was blocked in the absence of UL48, UL3.5, or both. An inhibition of final envelopment was also observed with several other gene deletion mutants of PrV. However, semicrystalline aggregations of cytoplasmic capsids like those observed in the absence of pUL37 (42) were not detectable with UL3.5 or UL48 mutants. The deletion of UL3.5 and/or UL48 also failed to induce electron-dense accumulations of the pUL46, pUL47, and pUL49 tegument components in the cytoplasm, or pronounced dilations of Golgi membranes, which had been observed after deletion of gM in combination with gE and gI (2), or gM and pUL11 (50). Whereas from cells infected with these double and triple mutants no virions or virus-like particles were secreted at detectable levels, UL48-negative PrV has been shown to release a high amount of enveloped, tegument-containing but capsidless particles (23). Such L particles were not produced at similar levels by UL3.5-negative, or UL3.5 and UL48 double-negative PrV, which was the only marked difference detected by electron microscopic analyses between these deletion mutants. Nevertheless, our results indicate closely related functions of pUL3.5 and pUL48 during an egress step which occurs after primary tegumentation of cytoplasmic nucleocapsids but that is independent of targeting of at least several other tegument components to Golgi-derived vesicles, which has been suggested to be mediated by interactions between, e.g., the UL49 tegument protein and envelope glycoproteins gE and gM (24, 56).
Although almost no mature virus particles were detectable in or around noncomplementing cells infected with UL3.5, UL48, or double-negative PrV mutants, plaque assays and one-step growth analyses revealed that none of the deletions caused a complete inhibition of direct cell-to-cell spread or formation of infectious progeny virus. It is still unclear whether this apparent discrepancy can be explained by the insensitivity of electron microscopy or by the infectivity of subviral structures like nucleocapsids, primary enveloped virions, or even naked virus DNA. Irrespective of the precise reason, UL3.5 and UL48 of PrV are, strictly speaking, nonessential either singly or in combination.
In rabbit kidney cells, plaque diameters of UL3.5-negative PrV were reduced by ca. 50%, and maximum virus titers were ca. 15-fold lower than those of wild-type PrV or rescue mutants. Previous studies performed with African green monkey (Vero) cells indicated much more pronounced replication defects of other UL3.5 deletion mutants of PrV (19, 21). It remains to be investigated whether these differences are exclusively cell type dependent or are caused partly by the large foreign gene insertions at the UL3.5 gene locus of the previous mutants, which might have affected transcription efficiency or mRNA stability of the coterminally transcribed upstream genes UL1, UL2, and UL3 (10). Remarkably, UL1, which encodes gL, is known to be essential for formation of infectious virions and for cell-to-cell spread (38). However, since the mutant used in the present study contains only an FRT site consisting of 36 bp of foreign DNA, and since expression of upstream genes proved to be unaffected, the observed replication defects could be attributed exclusively to the deletion of UL3.5.
Plaque diameters of UL48-negative PrV in RK13 cells were reduced by ca. 65%, and titers were 100-fold lower than those of wild-type or rescued viruses. Thus, deletion of UL48 caused more severe in vitro replication defects than deletion of UL3.5. This could indicate that the functions of pUL48 and pUL3.5 during viral egress are overlapping but distinct, despite the similar ultrastructural patterns of the deletion mutants. On the other hand, PrV pUL48 is at least bifunctional (23), having transcription-activating properties in newly infected cells. For this function, the presence of pUL3.5 is dispensable. Thus, in the absence of pUL48, pUL3.5 is no longer relevant, as shown by almost identical in vitro replication deficiencies of the double-deletion mutant and UL48-negative PrV.
The observed in vitro growth defects correlate with significantly delayed neuroinvasion in vivo, leading to prolonged survival times after intranasal infection of mice. Again, UL3.5-negative PrV was slightly less impaired than the UL48 and double-deletion mutants. Therefore, it appears unlikely that the two proteins function during independent steps of virion morphogenesis and neuronal spread or that they possess redundant functions, since in both cases the defects caused by the two mutations should be additive or synergistic, like those observed for several other PrV mutants with multiple deletions of tegument and/or envelope proteins involved in viral egress (2, 25, 50).
Thus, most likely, pUL3.5 and pUL48 execute functions during the same process, which requires their physical interaction. Fractionation of PrV-infected cells indicated a tight association of pUL3.5 with cytoplasmic membranes, although the nature of this interaction remained unclear (19). For HSV-1 pUL48, physical and functional interactions with other tegument proteins encoded by UL41, UL46, UL47, and UL49 have been reported (16, 66, 71). Similar interactions of the PrV homolog, together with its binding to the membrane-associated pUL3.5, might be a prerequisite for proper assembly of virion components at the future budding site in the trans-Golgi region. In HSV-1, another membrane-associated protein might execute the role of the missing UL3.5 gene product. In this context, the proposed interactions of HSV-1 pUL48 with envelope glycoproteins gB, gD, and gH should be considered (31, 72).
In summary, we show here that pUL3.5 and pUL48 of PrV interact physically and demonstrate for the first time the importance of this interaction for virion formation of an alphaherpesvirus. Heterologous complex formation between the BoHV-1 pUL3.5 and PrV pUL48 explains the observed trans-complementation of UL3.5-deficient PrV by the BoHV-1 homolog. In contrast to other pUL3.5 homologs, the PrV protein is proteolytically processed prior to incorporation into virions, retaining only the N-terminal portion which is responsible for complex formation. Thus, the pUL3.5-pUL48 complex apparently plays a prominent role during cytoplasmic virion morphogenesis of these alphaherpesviruses.
Acknowledgments
This study was supported by the the Deutsche Forschungsgemeinschaft (Me 854/8-1).
The EGFP-specific antiserum was kindly provided by C. Höhle and G. M. Keil. The technical help of C. Ehrlich, M. Jörn, P. Meyer, K. Müller, D. Werner, and E. Zorn is greatly appreciated.
Footnotes
Published ahead of print on 20 June 2007.
REFERENCES
- 1.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 180:1-19. [DOI] [PubMed] [Google Scholar]
- 4.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, J. I., and K. Seidel. 1994. Varicella-zoster virus (VZV) open reading frame 10 protein, the homolog of the essential herpes simplex virus protein VP16, is dispensable for VZV replication in vitro. J. Virol. 68:7850-7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox, E., S. Reddy, I. Iofin, and J. I. Cohen. 1998. Varicella-zoster virus ORF57, unlike its pseudorabies virus UL3.5 homolog, is dispensable for viral replication in cell culture. Virology 250:205-209. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 9.Davison, A. J., R. Eberle, G. S. Hayward, D. J. McGeoch, A. C. Minson, P. E. Pellet, B. Roizman, M. Studdert, and E. Thiry. 2005. Family Herpesviridae, p. 193-212. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. Academic Press, San Diego, CA.
- 10.Dean, H. J., and A. K. Cheung. 1993. A 3′ coterminal gene cluster in pseudorabies virus contains herpes simplex virus UL1, UL2, and UL3 gene homologs and a unique UL3.5 open reading frame. J. Virol. 67:5955-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai, P., G. L. Sexton, J. M. McCaffery, and S. Person. 2001. A null mutation in the gene encoding the herpes simplex virus type 1 UL37 polypeptide abrogates virus maturation. J. Virol. 75:10259-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desai, P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dietz, P., B. G. Klupp, W. Fuchs, B. Köllner, E. Weiland, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein K requires the UL20 gene product for processing. J. Virol. 74:5083-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorange, F., B. K. Tischer, J. F. Vautherot, and N. Osterrieder. 2002. Characterization of Marek's disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J. Virol. 76:1959-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enquist, L. W. 2002. Exploiting circuit-specific spread of pseudorabies virus in the central nervous system: insights to pathogenesis and circuit tracers. J. Infect. Dis. 186:S209-S214. [DOI] [PubMed] [Google Scholar]
- 18.Fuchs, W., and T. C. Mettenleiter. 1996. DNA sequence and transcriptional analysis of the UL1 to UL5 gene cluster of infectious laryngotracheitis virus. J. Gen. Virol. 77:2221-2229. [DOI] [PubMed] [Google Scholar]
- 19.Fuchs, W., B. G. Klupp, H. Granzow, H. J. Rziha, and T. C. Mettenleiter. 1996. Identification and characterization of the pseudorabies virus UL3.5 protein, which is involved in virus egress. J. Virol. 70:3517-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 1997. The UL20 gene product of pseudorabies virus functions in virus egress. J. Virol. 71:5639-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuchs, W., H. Granzow, and T. C. Mettenleiter. 1997. Functional complementation of UL3.5-negative pseudorabies virus by the bovine herpesvirus 1 UL3.5 homolog. J. Virol. 71:8886-8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuchs, W., B. G. Klupp, H. Granzow, A. Mundt, C. Hengartner, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuchs, W., H. Granzow, and T. C. Mettenleiter. 2003. A pseudorabies virus recombinant simultaneously lacking the major tegument proteins encoded by the UL46, UL47, UL48, and UL49 genes is viable in cultured cells. J. Virol. 77:12891-12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 78:11879-11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuchs, W., H. Granzow, R. Klopfleisch, B. G. Klupp, D. Rosenkranz, and T. C. Mettenleiter. 2005. The UL7 gene of pseudorabies virus encodes a nonessential structural protein which is involved in virion formation and egress. J. Virol. 79:11291-11299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuchs, W., H. Granzow, R. Klopfleisch, B. G. Klupp, and T. C. Mettenleiter. 2006. The UL4 gene of pseudorabies virus encodes a minor infected-cell protein that is dispensable for virus replication. J. Gen. Virol. 87:2517-2525. [DOI] [PubMed] [Google Scholar]
- 29.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 30.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gross, S. T., C. A. Harley, and D. W. Wilson. 2003. The cytoplasmic tail of herpes simplex virus glycoprotein H binds to the tegument protein VP16 in vitro and in vivo. Virology 317:1-12. [DOI] [PubMed] [Google Scholar]
- 32.Hambleton, S., M. D. Gershon, and A. A. Gershon. 2004. The role of the trans-Golgi network in varicella zoster virus biology. Cell. Mol. Life Sci. 61:3047-3056. [DOI] [PubMed] [Google Scholar]
- 33.Helferich, D., J. Veits, T. C. Mettenleiter, and W. Fuchs. 2007. Identification of transcripts and protein products of the UL31, UL37, UL46, UL47, UL48, UL49 and US4 gene homologues of avian infectious laryngotracheitis virus. J. Gen. Virol. 88:719-731. [DOI] [PubMed] [Google Scholar]
- 34.Izumiya, Y., H. K. Jang, M. Ono, and T. Mikami. 2001. A complete genomic DNA sequence of Marek's disease virus type 2, strain HPRS24. Curr. Top. Microbiol. Immunol. 255:191-221. [DOI] [PubMed] [Google Scholar]
- 35.Kaplan, A. S., and A. Vatter. 1959. A comparison of herpes simplex and pseudorabies virus. Virology 7:394-407. [DOI] [PubMed] [Google Scholar]
- 36.Khattar, S. K., S. van Drunen Littel-van den Hurk, L. A. Babiuk, and S. K. Tikoo. 1995. Identification and transcriptional analysis of a 3′-coterminal gene cluster containing UL1, UL2, UL3, and UL3.5 open reading frames of bovine herpesvirus-1. Virology 213:28-37. [DOI] [PubMed] [Google Scholar]
- 37.Klopfleisch, R., J. P. Teifke, W. Fuchs, M. Kopp, B. G. Klupp, and T. C. Mettenleiter. 2004. Influence of tegument proteins of pseudorabies virus on neuroinvasion and transneuronal spread in the nervous system of adult mice after intranasal inoculation. J. Virol. 78:2956-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klupp, B. G., J. Baumeister, A. Karger, N. Visser, and T. C. Mettenleiter. 1994. Identification and characterization of a novel structural glycoprotein in pseudorabies virus, gL. J. Virol. 68:3868-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klupp, B. G., J. Baumeister, P. Dietz, H. Granzow, and T. C. Mettenleiter. 1998. Pseudorabies virus glycoprotein gK is a virion structural component involved in virus release but is not required for entry. J. Virol. 72:1949-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klupp, B. G., and T. C. Mettenleiter. 1999. Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J. Virol. 73:3014-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klupp, B. G., C. J. Hengartner, T. C. Mettenleiter, and L. W. Enquist. 2004. Complete, annotated sequence of the pseudorabies virus genome. J. Virol. 78:424-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klupp, B. G., H. Granzow, W. Fuchs, E. Mundt, and T. C. Mettenleiter. 2004. Pseudorabies virus UL3 gene codes for a nuclear protein which is dispensable for viral replication. J. Virol. 78:464-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klupp, B. G., S. Böttcher, H. Granzow, M. Kopp, and T. C. Mettenleiter. 2005. Complex formation between the UL16 and UL21 tegument proteins of pseudorabies virus. J. Virol. 79:1510-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klupp, B. G., H. Granzow, R. Klopfleisch, W. Fuchs, M. Kopp, M. Lenk, and T. C. Mettenleiter. 2005. Functional analysis of the pseudorabies virus UL51 protein. J. Virol. 79:3831-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klupp, B. G., J. Altenschmidt, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2005c. Identification and characterization of the pseudorabies virus UL43 protein. Virology. 334:224-233. [DOI] [PubMed] [Google Scholar]
- 48.Kopp, M., B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2002. Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm. J. Virol. 76:8820-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, E. Mundt, A. Karger, and T. C. Mettenleiter. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 77:5339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, and T. C. Mettenleiter. 2004. Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. J. Virol. 78:3024-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lam, N., and G. J. Letchworth. 2000. Bovine herpesvirus 1 UL3.5 interacts with bovine herpesvirus 1 α-transinducing factor. J. Virol. 74:2876-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lam, N., and G. Letchworth. 2004. A derivative of bovine herpesvirus 1 (BoHV-1) UL3.5 lacking the last forty amino acids inhibits replication of BoHV-1. Arch. Virol. 149:2295-2306. [DOI] [PubMed] [Google Scholar]
- 53.Lukàcs, N., H. J. Thiel, T. C. Mettenleiter, and H. J. Rziha. 1985. Demonstration of three major species of pseudorabies virus glycoproteins and identification of a disulfide-linked glycoprotein complex. J. Virol. 53:166-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 55.Mettenleiter, T. C. 2000. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis—state of the art, June 1999. Vet. Res. 31:99-115. [DOI] [PubMed] [Google Scholar]
- 56.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mettenleiter, T. C. 2003. Pathogenesis of neurotropic herpesviruses: role of viral glycoproteins in neuroinvasion and transneuronal spread. Virus Res. 92:197-206. [DOI] [PubMed] [Google Scholar]
- 58.Mettenleiter, T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106:167-180. [DOI] [PubMed] [Google Scholar]
- 59.Michael, K., B. G. Klupp, T. C. Mettenleiter, and A. Karger. 2006. Composition of pseudorabies virus particles lacking tegument protein US3, UL47, or UL49 or envelope glycoprotein E. J. Virol. 80:1332-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 61.Misra, V., A. C. Bratanich, D. Carpenter, and P. O'Hare. 1994. Protein and DNA elements involved in transactivation of the promoter of the bovine herpesvirus (BHV) 1 IE-1 transcription unit by the BHV α gene trans-inducing factor. J. Virol. 68:4898-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 64.Rosenfeld, J., J. Capdevielle, J. C. Guillemot, and P. Ferrara. 1992. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203:173-179. [DOI] [PubMed] [Google Scholar]
- 65.Schikora, B., Z. Lu, G. F. Kutish, D. Rock, G. Magyar, and G. J. Letchworth. 1998. The bovine herpesvirus type 1 UL3.5 open reading frame encodes a virion structural protein. Virology 240:76-82. [DOI] [PubMed] [Google Scholar]
- 66.Smibert, C. A., B. Popova, P. Xiao, J. P. Capone, and J. R. Smiley. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 68:2339-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Telford, E. A., M. S. Watson, K. McBride, and A. J. Davison. 1992. The DNA sequence of equine herpesvirus-1. Virology 189:304-316. [DOI] [PubMed] [Google Scholar]
- 68.Thureen, D. R., and C. L. Keeler Jr. 2006. Psittacid herpesvirus 1 and infectious laryngotracheitis virus: comparative genome sequence analysis of two avian alphaherpesviruses. J. Virol. 80:7863-7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.von Einem, J., D. Schumacher, D. J. O'Callaghan, and N. Osterrieder. 2006. The alpha-TIF (VP16) homologue (ETIF) of equine herpesvirus 1 is essential for secondary envelopment and virus egress. J. Virol. 80:2609-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weinheimer, S. P., B. A. Boyd, S. K. Durham, J. L. Resnick, and D. R. O'Boyle. 1992. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J. Virol. 66:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, Y., D. A. Sirko, and J. L. McKnight. 1991. Role of herpes simplex virus type 1 UL46 and UL47 in αTIF-mediated transcriptional induction: characterization of three viral deletion mutants. J. Virol. 65:829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu, Q., and R. J. Courtney. 1994. Chemical cross-linking of virion envelope and tegument proteins of herpes simplex virus type 1. Virology 204:590-599. [DOI] [PubMed] [Google Scholar]