Abstract
To test the importance of the hydrophobic residues within the putative Epstein-Barr virus (EBV) glycoprotein B (gB) fusion loops in membrane fusion, WY112-113 and WLIW193-196 were mutated into alanine, glutamic acid, or the analogous residues from herpes simplex virus type 1 (HSV-1) gB (HR and RVEA). All gB variants exhibited cell surface expression, demonstrating that the substitutions did not perturb gB trafficking. None of six gB variants was, however, capable of mediating fusion with either epithelial or B cells. These data demonstrate that the bulky and hydrophobic EBV loop residues, which differ from the more hydrophilic HSV-1 residues and appear more compatible with membrane insertion, are essential for EBV gB-dependent fusion.
Envelope glycoprotein B (gB) and glycoproteins H and L (gH/gL) form the core fusion machinery of all herpesviruses (32). The mechanism by which the three glycoproteins function to orchestrate membrane fusion is not fully understood. In varicella-zoster virus, cytomegalovirus, and human herpesvirus 8 (HHV-8), the action of gB or the gH/gL complex alone can result in fusion, although at a lower level than when all three glycoproteins are present (6, 17, 25). A truncated variant of Epstein-Barr virus (EBV) gB mediates fusion with epithelial cells at levels up to 60% of what is observed when gB, gH, and gL are transfected together (23, 25). In the case of herpes simplex virus type 1 (HSV-1), the gH/gL complex seems to be responsible for the formation of a hemifusion intermediate, whereas gB is required to resolve the intermediate and complete fusion (33). The involvement of multiple proteins distinguishes herpesviruses from most other viruses where membrane merger is typically mediated by one fusion protein (16).
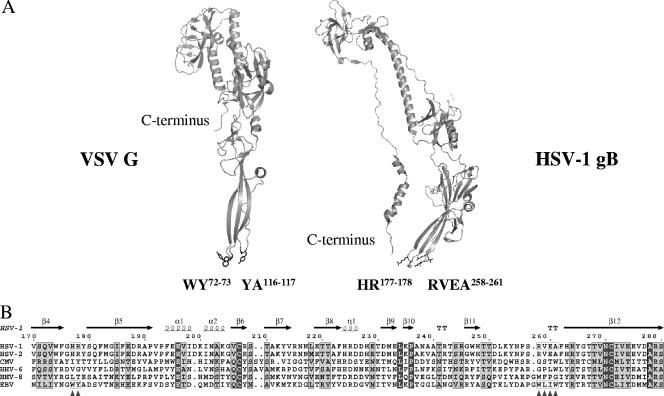
Glycoprotein B is highly conserved throughout the herpesvirus family. HSV-1 gB exhibits 86% sequence identity with HSV-2 gB and 29% with EBV gB, while EBV and HHV-8 gB share 40% sequence identity. Although HSV-1 gB does not share any similarity with the fusion protein (G) of vesicular stomatitis virus (VSV) at the protein sequence level, the structural homology between the two proteins is notable (Fig. 1A) (11, 29). The only available structure of HSV-1 gB (11) was proposed to represent a postfusion conformation based on the similarity with the postfusion form of G.
FIG. 1.
(A) Structures of the ectodomains of HSV-1 gB and G protein of VSV in postfusion conformations. Structural homology is notable between HSV-1 gB and VSV G protein, despite the lack of similarity at the protein sequence level. For clarity reasons, only monomers are shown. Residues forming a bipartite fusion peptide in VSV G protein (fusion loop 1, WY72-73; fusion loop 2, YA116-117) are labeled, and their side chains are shown as sticks. Fusion loops in the VSV G protein adopt a hairpin conformation that is typical for the internal fusion peptides of class II fusion proteins and that is compatible with membrane penetration. Residues located in the structurally homologous loops in HSV-1 gB are marked, and their side chains are shown as sticks (fusion loop 1, HR177-178; fusion loop 2, RVEA258-261). The conformations of HSV-1 gB loops do not resemble a hairpin fold and seem to be suboptimal for membrane insertion. The corresponding residues in EBV gB (fusion loop 1, WY112-113; fusion loop 2, WLIW193-196) were mutated in this study to evaluate their importance for the ability of EBV gB to mediate fusion. Both HSV-1 gB and VSV G ectodomains used for crystallization were truncated at the C terminus just before the stem regions. C termini are marked in both structures to indicate the putative location of the stem regions. Protein Data Bank files used for this figure are 2gum and 2cmx. The figure was generated using PyMOL (4). (B) Sequence alignment of gB fragments containing putative fusion loops. In contrast to the highly conserved fusion peptides identified for G protein and class I (5) and class II fusion proteins (1), the fusion loops of different gB proteins are not well conserved. Protein sequences are shown for representative herpesviruses known to infect humans: HSV-1 and HSV-2, cytomegalovirus (CMV) HHV-6, HHV-8, and EBV. Secondary structure elements, extracted from the HSV-1 gB ectodomain X-ray structure (Protein Data Bank file 2gum), are shown on top. Numbering is shown for unprocessed HSV-1 gB. Locations of the residues proposed to form fusion loops in gB are marked with triangles at the bottom of the alignment. Amino acids in the putative fusion loops in HSV-1 and EBV gB are boxed. Amino acids are shaded according to their conservation. The Risler matrix (28) was used to calculate similarity scores. Residues showing strict conservation are shown in dark gray, and residues with a similarity score of 0.7 and higher are shown in light gray. Alignment was generated using the ESPript program (8). Swiss-Prot entry numbers for the sequences shown are (from top to bottom) P10211, P06763, P06473, P36319, P03188, and P88906.
Fusion peptides of class I and II fusion proteins are rich in hydrophobic and aromatic residues and directly insert into the membrane after the conformational change is triggered. The residues critical for the ability of VSV G protein to cause fusion fall within two internal regions and give rise to a bipartite fusion peptide made of WY72-73 and YA116-117 (7, 35, 37). The conformation of the two fusion loops resembles the typical hairpin fold adopted by fusion peptides of class II fusion proteins (16). Regions structurally homologous to the fusion peptide of G were proposed to form putative fusion loops in HSV-1 gB (11). Most of the corresponding residues in HSV-1 gB, however, are not hydrophobic (HR177-178 and RVEA258-261), and the putative fusion loops appear in the crystal structure to be in a conformation suboptimal for membrane penetration. Rather, aromatic residues adjacent to the tips of the loops were proposed to be brought to interact with membranes through a conformational change (11). The residues forming the analogous loops in EBV gB, WY112-113 and WLIW193-196, were identified based on the alignment of gB protein sequences shown in Fig. 1B. The EBV gB fusion loops have a greater resemblance to the fusion peptides of class I and II fusion proteins and are more compatible with membrane insertion.
To investigate the importance of the aromatic and hydrophobic EBV residues WY112-113 and WLIW193-196 for the fusion activity of EBV gB, a series of mutants was constructed. Mutations were introduced by using a PCR overlap extension method (12). The plasmid encoding wild-type gB in the Stratagene pSG5 vector was used as a template (9). The bulky and hydrophobic residues were replaced with three types of amino acids, differing in hydrophobicity, size, and charge. The residues introduced into each of the loops were the analogous residues from HSV-1 gB (HR and RVEA), smaller but still hydrophobic alanine residues, and negatively charged glutamic acid residues (Table 1).
TABLE 1.
Design of gB variants
| Putative FLa | gB variant designation | EBV wild-type gB residues | Mutated residuesb |
|---|---|---|---|
| FL1 | FL1-1 | WY112-113 | AA |
| FL1-2 | HR* | ||
| FL1-3 | EE | ||
| FL2 | FL2-1 | WLIW193-196 | RVEA* |
| FL2-2 | AAAA | ||
| FL2-3 | EEEE |
FL, fusion loop.
The asterisk indicates substitutions corresponding to the residues found in the analogous regions in HSV-1 gB.
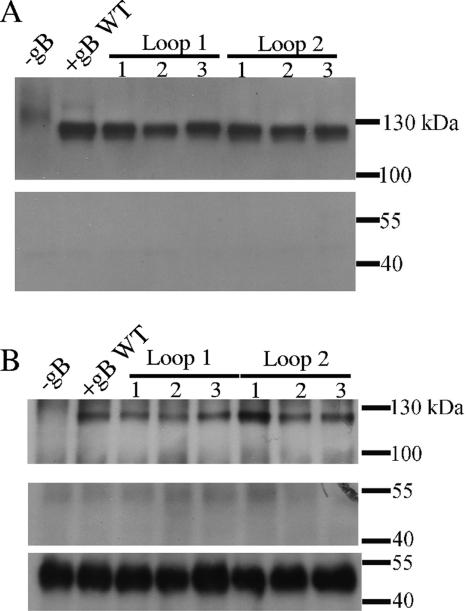
In contrast to the highly surface-expressed gB of HSV-1 and other herpesviruses, EBV gB is primarily retained in nuclear and endoplasmic reticulum membranes, with low expression levels at the cell surface. Biotinylation of the surface proteins from Chinese hamster ovary (CHO)-K1-transfected cells (3, 24), followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (20) and Western blotting (2, 18), was performed to investigate the localization of the wild-type and gB variants and also to estimate their surface expression level compared to the wild-type protein. This approach was established (3) and successfully used for detection of surface-expressed EBV gH/gL complexes (24). The membrane-impermeable biotinylation agent, sulfosuccinimidyl-6-(biotinamido) hexanoate (Pierce), was used to ensure labeling of only surface proteins (3). Actin served as a negative control for labeling of intracellular proteins.
The mixture of biotinylated proteins was bound to UltraLink immobilized neutravidin protein (Pierce), eluted, separated by SDS-PAGE, and then blotted with antibodies recognizing gB or actin (Fig. 2A). In parallel experiments, gB was immunoprecipitated with protein G-Sepharose beads (GE Healthcare) loaded with the polyclonal anti-gB antibody, and subjected to SDS-PAGE, and then the biotinylated gB was detected with avidin conjugated to horseradish peroxidase (Fig. 2B). Polyclonal anti-gB antibody was made by genetic immunization of rabbits with EBV gB expression vectors (Aldevron, North Dakota). Anti-actin antibody (Sigma) bound to protein G beads was used to pull down actin from these samples as well.
FIG. 2.
Surface expression of EBV gB variants. Surface proteins were biotinylated, and neutravidin beads were used to precipitate all biotinylated proteins (A); alternatively, protein G beads loaded with anti-gB antibodies (upper panel) or anti-actin antibodies (middle and lower panels) were used to precipitate gB or actin, respectively (B). In the upper part of panel A, biotinylated gB was detected by Western blotting using anti-gB antibody, while biotinylated actin could not be detected when anti-actin antibody was used instead (lower panel). In panel B avidin conjugated to horseradish peroxidase was used for detection of biotinylated proteins in the upper and middle blots. Biotinylated gB was detected (upper panel), and there was no biotinylated actin in the samples (middle panel). Actin was detected when anti-actin antibody was used for detection, confirming the presence of actin. Samples are labeled as shown in Table 1. WT, wild type.
The six gB variants exhibited similar surface expression to wild-type gB, as shown in Fig. 2. A band corresponding to biotinylated gB, with an observed molecular mass of 120 kDa, was detected in all samples, both when biotin label incorporated in gB was used to pull down the gB (Fig. 2A) and to detect it (Fig. 2B). Moreover, the surface expression levels of gB mutants were comparable to the expression level of the wild-type gB, indicating that the substitution of putative fusion loop residues did not affect the production and trafficking of the proteins to the cell surface. Biotinylated actin was not detected in any of the samples, demonstrating that the cells stayed intact during labeling. To demonstrate that actin, however, was present in cells, actin was precipitated with protein G beads and anti-actin antibody and then subjected to Western blotting with the anti-actin antibody. A strong intensity band corresponding to actin was observed at 42 kDa (Fig. 2B, bottom panel).
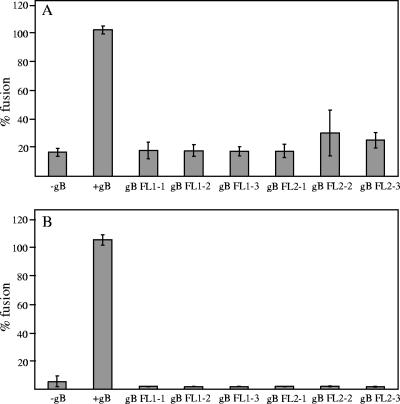
A virus-free cell-based fusion assay (18, 22) was employed to evaluate the ability of gB variants to mediate fusion with two target cell types that EBV infects in vivo: B cells (Daudi B lymphocytes; American Type Culture Collection) and epithelial cells (human embryonic kidney 293 cells; American Type Culture Collection). The cells were maintained in culture as described previously (18, 24, 31). The effector CHO-K1 cells were transfected (18, 22) with the combination of plasmids encoding the glycoproteins required for fusion (gB, gH, and gL for epithelial cell fusion and gB, gH, gL, and gp42 for B-cell fusion) and the luciferase gene under the control of T7 polymerase. The target cells were stably transfected with T7 polymerase, and the luciferase gene was activated only in the event of fusion of the effector and the target cells. The amount of expressed luciferase was measured using a chemiluminescent substrate, and it allowed for quantification of the fusion activity of the variants. Luciferase activity was measured for triplicate aliquots of cell lysates transferred to a 96-well plate. For each experiment, each of the triplicate measurements was expressed as a percentage of the average value calculated for the positive control sample. The results shown in Fig. 3 represent an average of data collected in three independent experiments (total of nine data points). Error bars correspond to standard deviations of the normalized values.
FIG. 3.
Effect of the amino acid substitutions in EBV gB putative fusion loops on epithelial cell fusion (A) and B-cell fusion (B). CHO-K1 cells were transiently transfected with plasmids encoding gB (wild-type or mutant protein), gH/gL, and T7-driven luciferase for epithelial cells fusion or gB (wild-type or mutant protein), gH/gL, gp42, and T7-driven luciferase for B-cell fusion. The CHO-K1 cells were overlaid with the same number of 293T epithelial cells or Daudi B cells expressing T7 polymerase. Fusion was allowed to proceed for 24 h, and luciferase activity was measured to quantify the level of fusion. The first sample lacked gB (gB−) and served as a background control. The second sample (+gB) refers to the positive control (transfected with wild-type gB, gH/gL, and T7-driven luciferase for epithelial cell fusion and gp42 for B-cell fusion). gB mutant proteins are marked according to the type of putative fusion loop (FL) substitution shown in Table 1. Luciferase activity measured for the positive control is set to 100%, and the rest of the measurements are expressed as a percentage of the positive control.
The ability of gB variants to mediate fusion with epithelial cells is shown in Fig. 3A. The level of fusion when the wild-type gB, gH, and gL were transfected was set to 100%, and the variants exhibited 16 to 34% of the wild-type gB fusion activity, which is not significantly different from the background levels of fusion observed in the absence of wild-type gB. This demonstrated that the presence of hydrophobic residues in the putative fusion loops of EBV gB was essential for the function of gB and its ability to cause fusion with epithelial cells. All six gB variants were also entirely defective in their ability to cause fusion with B cells, consistent with what was observed in epithelial cell fusion (Fig. 3B). Higher levels of fusion with the adherent 293T epithelial cells, in contrast to the nonadherent B cells, were observed, possibly because of the involvement of cell-cell contact in epithelial cell fusion (13, 30). These data are consistent with the interpretation that the putative hydrophobic fusion loops in EBV gB are necessary for its membrane fusion activity.
Although gB is highly conserved in herpesviruses, functional complementation between gB proteins from different herpesviruses has not been found (21, 26). This lack of functional complementation is further highlighted by the inability of the EBV gB to retain its function when the homologous HSV-1 gB residues are introduced and points to other virus-specific differences that are important for membrane fusion to occur. Even introduction of hydrophobic, albeit smaller, alanines decreases fusion close to the background level. The aromatic and hydrophobic amino acids (W, Y, I, and L) forming putative fusion loops in EBV gB strongly resemble the residues typically present in the fusion peptides of class I and class II fusion proteins. Moreover, tryptophan and tyrosine side chains are often found at the interface between charged phospholipids and hydrophobic fatty acid chains of lipid membranes (36). The abundance of these amino acids in the EBV gB putative fusion loops suggests that the residues provided by gB alone might cause sufficient lipid mixing that would result in membrane fusion. This would be consistent with the enhanced inherent fusogenicity of EBV gB and its ability to cause fusion in a gH/gL-independent manner (23).
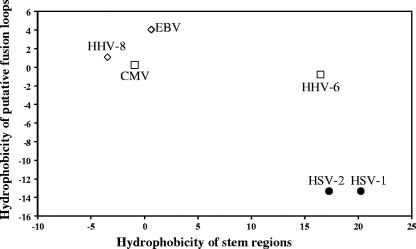
The membrane-proximal regions of the VSV G protein, called stems, have been shown to be important for the fusion mechanism and viral infectivity (14, 15). When expressed alone, these fragments, rich in hydrophobic residues, potentiate the activity of unrelated fusion proteins and can cause hemifusion in the absence of the G ectodomain. The importance of the analogous regions in herpesvirus gBs for fusion is not clear, although swapping these domains between gB proteins of different herpesviruses results in chimeras that do not functionally complement each other (21), reminiscent of our results with the HSV-1 fusion loops present in the EBV gB protein. By analogy with the G protein of VSV, it seems possible that some of the energy required for lipid mixing that leads to fusion could be provided by the gB stem regions. These stem regions might be of particular importance for those gB proteins that lack hydrophobic residues in the fusion loops. Indeed, gB proteins of HSV-1 and HSV-2 have more hydrophobic stem regions than the EBV and HHV-8 gB proteins, whose fusion loops are rich in hydrophobic amino acids compatible with membrane insertion. The comparison is shown in Fig. 4 and is based on the calculation of the average hydrophobicity of the mentioned regions, performed using the Kyte and Doolittle hydrophobicity scale (19, 34).
FIG. 4.
Negative correlation of hydrophobicities of gB putative fusion loops and stem regions. Total hydrophobicities were calculated using the Kyle and Doolittle scale. Protein sequences and abbreviations used here are the same as shown in Fig. 1B. Residues found in the putative fusion loops and used in calculations are labeled with filled triangles at the bottom in Fig. 1B. The region spanning 50 residues located immediately upstream of the transmembrane anchor (residues 774 to 795) in HSV-1 gB (27) contains two hydrophobic helical regions which, we hypothesized, might serve as stem regions in HSV-1 gB. The borders of homologous segments in gB proteins of other herpesviruses were determined based on the gB sequence alignment (data not shown). The hydrophobicity values calculated for putative fusion loops are plotted as a function of hydrophobicity values obtained for the stem regions. Due to the higher hydrophobicity of stem regions and lower hydrophobicity of the fusion loops, gB of α-herpesviruses HSV-1 and HSV-2 cluster at the opposite side of the plot from the gB of γ-herpesviruses EBV and HHV-8, which have more hydrophobic fusion loops and fewer hydrophobic stem regions.
In a recently reported mutagenesis study of the HSV-1 gB (10), the aromatic and hydrophobic amino acids investigated for their importance for fusion (W174, Y179, V259, A261, and F262) included residues at the edges of the two loop regions, i.e., W174, Y179, and F262 (Fig. 1B). The latter two residues are less hydrophobic in EBV gB (Y109, A114, and T197), and it is possible that these edge hydrophobic amino acids in HSV-1 gB somehow locally compensate for the greater hydrophilicity of the HSV-1 fusion loops or that the HSV-1 loops interact with membranes in a manner or orientation that differs from that in EBV gB. However, it is undoubtedly the case that herpesvirus gB proteins share a generally similar mechanism for membrane fusion despite these apparent differences in their fusion loop regions.
Acknowledgments
We thank Austin N. Kirschner for excellent advice and technical assistance with the fusion assays and A. Silva Lowrey and Jessica Reimer for sharing their reagents.
This research was supported by Public Health Service grants CA93444 (R.L. and T.S.J.) and CA117794 (R.L. and T.S.J.) from the National Cancer Institute.
Footnotes
Published ahead of print on 6 June 2007.
REFERENCES
- 1.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, and F. X. Heinz. 2001. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J. Virol. 75:4268-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burnette, W. N. 1981. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112:195-203. [DOI] [PubMed] [Google Scholar]
- 3.Daniels, G. M., and S. G. Amara. 1998. Selective labeling of neurotransmitter transporters at the cell surface. Methods Enzymol. 296:307-318. [DOI] [PubMed] [Google Scholar]
- 4.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
- 5.Dutch, R. E., T. S. Jardetzky, and R. A. Lamb. 2000. Virus membrane fusion proteins: biological machines that undergo a metamorphosis. Biosci. Rep. 20:597-612. [DOI] [PubMed] [Google Scholar]
- 6.Duus, K. M., C. Hatfield, and C. Grose. 1995. Cell surface expression and fusion by the varicella-zoster virus gH:gL glycoprotein complex: analysis by laser scanning confocal microscopy. Virology 210:429-440. [DOI] [PubMed] [Google Scholar]
- 7.Fredericksen, B. L., and M. A. Whitt. 1995. Vesicular stomatitis virus glycoprotein mutations that affect membrane fusion activity and abolish virus infectivity. J. Virol. 69:1435-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gouet, P., E. Courcelle, D. I. Stuart, and F. Metoz. 1999. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15:305-308. [DOI] [PubMed] [Google Scholar]
- 9.Haan, K. M., S. K. Lee, and R. Longnecker. 2001. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology 290:106-114. [DOI] [PubMed] [Google Scholar]
- 10.Hannah, B. P., E. E. Heldwein, F. C. Bender, G. H. Cohen, and R. J. Eisenberg. 2007. Mutational evidence of internal fusion loops in herpes simplex virus glycoprotein B. J. Virol. 81:4858-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heldwein, E. E., H. Lou, F. C. Bender, G. H. Cohen, R. J. Eisenberg, and S. C. Harrison. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217-220. [DOI] [PubMed] [Google Scholar]
- 12.Horton, R. M., S. N. Ho, J. K. Pullen, H. D. Hunt, Z. Cai, and L. R. Pease. 1993. Gene splicing by overlap extension. Methods Enzymol. 217:270-279. [DOI] [PubMed] [Google Scholar]
- 13.Imai, S., J. Nishikawa, and K. Takada. 1998. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J. Virol. 72:4371-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeetendra, E., K. Ghosh, D. Odell, J. Li, H. P. Ghosh, and M. A. Whitt. 2003. The membrane-proximal region of vesicular stomatitis virus glycoprotein G ectodomain is critical for fusion and virus infectivity. J. Virol. 77:12807-12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeetendra, E., C. S. Robison, L. M. Albritton, and M. A. Whitt. 2002. The membrane-proximal domain of vesicular stomatitis virus G protein functions as a membrane fusion potentiator and can induce hemifusion. J. Virol. 76:12300-12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kielian, M., and F. A. Rey. 2006. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 4:67-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinzler, E. R., and T. Compton. 2005. Characterization of human cytomegalovirus glycoprotein-induced cell-cell fusion. J. Virol. 79:7827-7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirschner, A. N., J. Omerovic, B. Popov, R. Longnecker, and T. S. Jardetzky. 2006. Soluble Epstein-Barr virus glycoproteins gH, gL, and gp42 form a 1:1:1 stable complex that acts like soluble gp42 in B-cell fusion but not in epithelial cell fusion. J. Virol. 80:9444-9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 21.Lee, S. K., T. Compton, and R. Longnecker. 1997. Failure to complement infectivity of EBV and HSV-1 glycoprotein B (gB) deletion mutants with gBs from different human herpesvirus subfamilies. Virology 237:170-181. [DOI] [PubMed] [Google Scholar]
- 22.McShane, M., and R. Longnecker. 2004. Analysis of fusion using a virus-free cell fusion assay, p. 187-196. In Paul M. Lieberman (ed.), DNA viruses: methods and protocols. Methods in molecular biology, vol. 292. Springer-Verlag, New York, NY. [DOI] [PubMed] [Google Scholar]
- 23.McShane, M. P., and R. Longnecker. 2004. Cell-surface expression of a mutated Epstein-Barr virus glycoprotein B allows fusion independent of other viral proteins. Proc. Natl. Acad. Sci. USA 101:17474-17479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Omerovic, J., L. Lev, and R. Longnecker. 2005. The amino terminus of Epstein-Barr virus glycoprotein gH is important for fusion with epithelial and B cells. J. Virol. 79:12408-12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pertel, P. E. 2002. Human herpesvirus 8 glycoprotein B (gB), gH, and gL can mediate cell fusion. J. Virol. 76:4390-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pertel, P. E., P. G. Spear, and R. Longnecker. 1998. Human herpesvirus-8 glycoprotein B interacts with Epstein-Barr virus (EBV) glycoprotein 110 but fails to complement the infectivity of EBV mutants. Virology 251:402-413. [DOI] [PubMed] [Google Scholar]
- 27.Rasile, L., K. Ghosh, K. Raviprakash, and H. P. Ghosh. 1993. Effects of deletions in the carboxy-terminal hydrophobic region of herpes simplex virus glycoprotein gB on intracellular transport and membrane anchoring. J. Virol. 67:4856-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Risler, J. L., M. O. Delorme, H. Delacroix, and A. Henaut. 1988. Amino acid substitutions in structurally related proteins: a pattern recognition approach. Determination of a new and efficient scoring matrix. J. Mol. Biol. 204:1019-1029. [DOI] [PubMed] [Google Scholar]
- 29.Roche, S., S. Bressanelli, F. A. Rey, and Y. Gaudin. 2006. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 313:187-191. [DOI] [PubMed] [Google Scholar]
- 30.Shannon-Lowe, C. D., B. Neuhierl, G. Baldwin, A. B. Rickinson, and H. J. Delecluse. 2006. Resting B cells as a transfer vehicle for Epstein-Barr virus infection of epithelial cells. Proc. Natl. Acad. Sci. USA 103:7065-7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silva, A. L., J. Omerovic, T. S. Jardetzky, and R. Longnecker. 2004. Mutational analyses of Epstein-Barr virus glycoprotein 42 reveal functional domains not involved in receptor binding but required for membrane fusion. J. Virol. 78:5946-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian, R. P., and R. J. Geraghty. 2007. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc. Natl. Acad. Sci. USA 104:2903-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tossi, A., L. Sandri, and A. Giangaspero. 2002. New consensus hydrophobicity scale extended to non-proteinogenic amino acids, p. 416-417. In E. Benedetti and C. Pedone (ed.), Peptides 2002: proceedings of the 27th European Peptide Symposium. Edizioni Ziino, Naples, Italy.
- 35.Whitt, M. A., P. Zagouras, B. Crise, and J. K. Rose. 1990. A fusion-defective mutant of the vesicular stomatitis virus glycoprotein. J. Virol. 64:4907-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wimley, W. C., and S. H. White. 1992. Partitioning of tryptophan side-chain analogs between water and cyclohexane. Biochemistry 31:12813-12818. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, L., and H. P. Ghosh. 1994. Characterization of the putative fusogenic domain in vesicular stomatitis virus glycoprotein G. J. Virol. 68:2186-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]